Abstract
Infectious salmon anemia virus (ISAV) has emerged as a virus of great concern to the aquaculture industry since it can lead to highly contagious and lethal infections in farm-raised salmon populations. While little is known about the transcription/replication cycle of ISAV, initial evidence suggests that it follows molecular mechanisms similar to those found in other orthomyxoviruses, which include the highly pathogenic influenza A (inf A) virus. During the life cycle of orthomyxoviruses, a panhandle structure is formed by the pairing of the conserved 5′ and 3′ ends of each genomic RNA. This structural motif serves both as a promoter of the viral RNA (vRNA)-dependent RNA polymerase and as a regulatory element in the transcription/replication cycle. As a first step toward characterizing the structure of the ISAV panhandle, here we have determined the secondary structures of the vRNA and the cRNA panhandles on the basis of solution nuclear magnetic resonance (NMR) and thermal melting data. The vRNA panhandle is distinguished by three noncanonical U·G pairs and one U·U pair in two stem helices that are linked by a highly stacked internal loop. For the cRNA panhandle, a contiguous stem helix with a protonated C·A pair near the terminus and tandem downstream U·U pairs was found. The observed noncanonical base pairs and base stacking features of the ISAV RNA panhandle motif provide the first insight into structural features that may govern recognition by the viral RNA polymerase.
INTRODUCTION
Infectious salmon anemia virus (ISAV) has emerged as a great threat to aquaculture of Atlantic salmon (Salmo salar) and, more recently, Coho salmon (Oncorhynchus kisutch). ISAV infection leads to disease with mortality rates of up to 90% in farm-raised salmon (53). First identified in Norway in 1984, the virus has since spread throughout fish farms in the European Union, North America, and Chile (4, 19, 35, 54). ISAV has been classified as a member of the family Orthomyxoviridae due to similarities in its genetic, morphological, and biochemical properties to those of the influenza and thogoto viruses (14, 26, 44). However, ISAV is a distant cousin of influenza virus and has been categorized as the only species in the genus Isavirus (26).
Orthomyxoviruses, including ISAV, are generally composed of eight genomic segments of single-stranded RNA with negative polarity, although influenza C and Dhori viruses have seven segments, and the thogoto virus has six segments (26). The mRNAs derived from these genomic RNA segments encode proteins that include the RNA-dependent RNA polymerase proteins (PB1, PB2, PA) and one nucleoprotein (NP) (7, 10, 23, 51, 55). While not much is known about the transcription/replication cycle of ISAV, it has been proposed to utilize mechanisms (Fig. 1) similar to those of influenza virus (8, 58), which includes the highly pathogenic influenza A (inf A) virus.
Fig. 1.
Schematic for synthesis of mRNA and replication of the viral RNA of all eight segments (consensus sequence shown) from ISAV (18, 28). The mRNA is defined by a 5′ catabolite gene activator protein (CAP) structure that is derived from cellular mRNA and a 3′ poly(A) tail. In contrast, the cRNA from which copies of the genomic vRNA are made does not have these modifications.
During the inf A viral life cycle, the viral RNA (vRNA) replication occurs through a highly regulated synthesis of both mRNA and cRNA, the latter of which is used to produce full-length genomic copies, through a complex interplay between the RNA, RNA polymerase, and NP (42, 45, 48). Highly conserved sequences at the 5′ and 3′ ends of the noncoding regions of inf A virus (12 to 13 nucleotides [nt]) and ISAV (7 to 8 nt) base pair to form a structure known as a panhandle motif that is important for regulation of both transcription and replication (7, 18, 28, 42, 52). While the exact regulatory mechanism underlying the RNA-polymerase interactions is not fully understood, formation of the inf A virus panhandle has been demonstrated in vivo (24) and determined to be essential for the initiation of transcription (17). Subsequent mutational analyses of inf A virus vRNA and cRNA promoter sequences using a chloramphenicol acetyltransferase (CAT) reporter assay showed that disruption of panhandle formation greatly reduces the efficacy of or altogether halts transcription (9, 15, 16, 46). These studies also revealed that nine terminal base pairs of the RNA panhandles melt upon interaction with the viral polymerase and subsequently form hairpin structures at both the 5′ and 3′ termini, a rearrangement referred to as the “corkscrew” structural model. CAT activity-based assays also demonstrated that the inf A virus panhandles act as modular motifs since they promote viral transcription, replication, and packaging of a foreign gene in the absence of the coding sequence within a viral genome segment (37). Such features may be general characteristics of panhandle motifs for orthomyxoviruses, including ISAV.
Unlike the inf A virus panhandle motif, for which many biochemical and structural studies have been reported, to date there are very limited data on the ISAV panhandle. An important study from Sandvik et al., in which the nucleotide sequences of the termini of two ISAV vRNA genomic segments were determined and secondary structures for these sequences were predicted using nearest-neighbor thermodynamic rules, established ISAV to be an orthomyxovirus (56). The secondary structure prediction was performed for the 25 terminal nucleotides on genomic segments 6 (originally misidentified as segment 7 [28, 41]) and 8 using MulFold, an earlier version of Mfold (59), and predicted a temperature-dependent switch in RNA secondary structures between 15°C, the temperature of the North Atlantic and the optimal temperature for ISAV replication, and 37°C. In this respect, it should be noted that ISAV replication does not occur above 25°C (14). For genomic segment 8 of the vRNA, 5′ and 3′ ends were predicted to form a panhandle structure at 15°C, whereas at 37°C, no panhandle structure was predicted. In comparison, genomic segment 6 was predicted to adopt similar panhandle folds at the two temperatures; however, there were some differences in the size of the internal loop, suggesting a more subtle temperature dependence of the structure for this RNA. Given the importance of the panhandle motif for inf A virus replication, the structure prediction results for the ISAV RNAs suggest that the ability of ISAV to replicate efficiently may be contingent upon formation of a specific, temperature-dependent RNA structure(s) in the promoter elements in the genomic segments. To test the predicted temperature dependency of the RNA panhandle structures, high-resolution nuclear magnetic resonance (NMR) spectroscopy has been utilized to determine the secondary structures of both the vRNA and cRNA segment 8 terminal regions. Fluorescence and UV thermal melts were then carried out to assess the thermodynamic stability of the RNAs and confirm secondary structure assignments. The results of the NMR and thermal melting analyses provide the first insight into the unique structural features that may govern recognition of the ISAV panhandle RNA motif by the viral RNA polymerase.
MATERIALS AND METHODS
Sample preparation.
Unlabeled, [5-2H]uridine-labeled, 13C/15N doubly labeled G/C, 13C/15N doubly labeled A/U, and 13C/15N doubly labeled C/U/G RNA hairpin constructs of the vRNA panhandle (49-mer) and cRNA panhandle (33-mer) were synthesized by in vitro transcription using T7 RNA polymerase and hemiduplex DNA templates (Integrated DNA Technologies, Inc.), as previously described (43). RNAs substituted with 2-aminopurine (2-AP) were purchased from Dharmacon, Inc. (Thermo Fisher), and deprotected using standard procedures. Four RNAs corresponding to the 5′ and 3′ strands of the putative vRNA and cRNA panhandles (all 23-mers) were designed such that each resulting duplex panhandle construct has an A-U terminal pair and purchased from Integrated DNA Technologies, Inc. All unlabeled and 13C/15N uniformly labeled nucleotide triphosphates were purchased from Sigma-Aldrich. [5-2H]UMP was synthesized from UMP (Sigma-Aldrich) according to literature methods (21, 47) and was then converted enzymatically to [5-2H]UTP (3). RNAs were purified using preparative denaturing polyacrylamide gel electrophoresis (PAGE), electroeluted, and exchanged into NMR buffer (80 mM sodium chloride, 10 mM sodium phosphate, pH 6.0 or 7.0) using Amicon Ultracel-3 or -10 centrifugal filters (Millipore, Inc.) for cRNA panhandle and vRNA panhandle constructs, respectively. The RNAs were folded by heating the samples to 95°C for 3 min and allowing the samples to cool slowly to ambient temperature in a water bath. NMR samples typically contained 1 mM RNA, 80 mM sodium chloride, 10 mM sodium phosphate, pH 6.0 or 7.0 (uncorrected), and 97% H2O–3% 2H2O or 99.96% 2H2O (Cambridge Isotope Laboratories).
NMR spectroscopy.
Data were collected on a either a Bruker Avance II 700-MHz or a Bruker Avance II 800-MHz spectrometer, both equipped with triple resonance, inverse cryoprobes. All spectra were processed using the NMRPipe program (11) on a Linux workstation and analyzed using the Sparky program on a Windows workstation. With a few exceptions, all pulse sequences were implemented from the Biomolecular NMR pulse program catalogue on Topspin software (version 2.0; Bruker).
Exchangeable 1H assignments were made on the 800-MHz spectrometer at 10°C in 97% H2O–3% 2H2O on unlabeled RNAs using a two-dimensional (2D) nuclear Overhauser effect spectroscopy (NOESY)-Watergate spectra with mixing times (τmix) of 50 ms and 150 ms, a sweep width of 17,500 Hz in both dimensions, 4,096 by 800 complex points, and 32 scans per increment. The proton carrier frequency was set to 4.70 ppm. A 2D 1H-15N jump-return heteronuclear multiple-quantum coherence spectrum was acquired on the 13C/15N doubly labeled A/U sample and the 13C/15N doubly labeled G/C sample. For the vRNA panhandle hairpin construct (49-mer), 2D 15N filtered/edited NOESY-Watergate (τmix = 50 ms) data were also collected on the 13C/15N doubly labeled A/U sample.
For assignment of nonexchangeable protons, the following 2D experiments were recorded in 99.96% 2H2O at 20°C on unlabeled RNAs and [5-2H]uridine RNAs: double-quantum coherence (DQF)-correlation spectroscopy (COSY), NOESY (τmix = 50 ms, 100 ms, 250 ms), and 1H, 1H total correlation spectroscopy (1H, 1H-TOCSY; τmix = 60 ms). For the 13C/15N-labeled G/C and A/U samples, data from the following 2D and three-dimensional (3D) experiments were collected: 2D 13C edited/filtered NOESY (τmix = 50 ms, 250 ms), 2D 13C edited/filtered 1H, 1H-TOCSY (τmix = 60 ms), 2D 1H-13C constant time (CT) heteronuclear single-quantum coherence (HSQC), 3D multiple-quantum coherence-HCNCH-transverse relaxation optimized spectroscopy, 3D 1H-13C NOESY-CT-HSQC (τmix = 200 ms), 2D HCCH-COSY (where underlining indicates the correlated nuclei), and 2D HCCH-COSY. The 2D NOESY data were acquired at 700 MHz with a sweep width of 7,000 Hz in both dimensions, 4,096 by 800 complex points, and 32 scans per increment. The 2D 1H-13C CT-HSQC data were collected with a sweep width of 5,280 Hz in F1 and 1,750 Hz in F2, 512 by 200 complex points, and 32 scans per increment. For both experiments, the proton carrier frequency was set to 4.71 ppm. Using these experiments, nearly complete resonance assignments of H8, H6, H5, H2, H1′, and H2′ protons were obtained for both NMR vRNA panhandle and NMR cRNA panhandle hairpin constructs derived from segment 8.
UV-detected thermal melts.
UV-detected thermal melting was measured using a Lambda 25 UV-visible spectrometer (Perkin-Elmer) equipped with Peltier thermostattable multicell holder and a temperature probe. The melts were typically performed with 1.0 μM RNA in 80 mM sodium chloride and 10 mM sodium cacodylate at pH 7.0. All samples were slow cooled as described above, transferred to quartz cuvettes, and equilibrated at 5°C in the spectrometer for 15 min. Absorbance at 260 nm was monitored as a function of temperature at 0.2°C intervals from 5°C to 90°C using the TempLab (version 2.0) software package (Perkin-Elmer). The resulting melting curves were smoothed over five points, and then first derivatives were calculated with respect to temperature and inspected for melting transitions.
Fluorescence-detected thermal melts.
2-AP fluorescence-detected melting experiments were measured using a FluoroMax-3 spectrofluorometer (Horiba Scientific) equipped with a programmable water bath and 3-mm-path-length quartz cuvettes. The melts were typically performed with 2.0 μM RNA in 80 mM sodium chloride and 10 mM sodium cacodylate at pH 7.0. All samples were slow cooled as described above. Each sample was transferred to a cuvette and then equilibrated in the fluorometer at 15°C for 10 min. An emission spectrum (excitation λ = 310 nm, emission λ = 330 to 450 nm) was recorded for each sample prior to melting to check the fluorescence intensity using standard right-angle emission collection in 1-nm increments with a 0.5-s integration time and 5-nm excitation and emission slit widths (band pass). Fluorescence spectra were corrected by subtracting a blank buffer spectrum. Samples were melted from 15°C to 85°C (excitation λ = 310 nm, emission λ = 371 nm) with a heating rate of approximately 1°C/min in 1-nm increments with a 5-s integration time and 5-nm excitation and emission slit widths (band pass) and a sample equilibration time of 1 min. For all melts, the sample volume was checked prior to melting. After the melt was finished and the sample was cooled to room temperature, the volume was always 150 μl (±2%), so no corrections were applied to the fluorescence data. Each melting curve was smoothed over five points and then normalized by dividing by the fluorescence at the highest temperature. First derivatives were calculated with respect to temperature and inspected for melting transitions.
RESULTS
Design and synthesis of model RNAs for biophysical studies.
The vRNA panhandle construct was designed on the basis of the secondary structure predictions reported by Sandvik et al., which predicted different RNA secondary structures at 15°C and 37°C for segment 8 (56). Initially, two RNAs corresponding to the 5′ and 3′ strands of the putative vRNA panhandle (Fig. 2 A) were designed so that the duplex panhandle construct, formed by annealing the strands, would have the native sequence with an A-U terminal pair (referred to as native duplex vRNA). For the NMR vRNA panhandle hairpin construct (referred to as NMR vRNA), the RNA was designed and prepared as a single RNA strand that contained the 5′ and 3′ sequences connected by a polypyrimidine heptaloop that was intended to simulate the remainder of the genomic segment. We did not want this linker to affect the structural stability of the upper stem, so we avoided using any structured loops, such as a UUCG tetraloop. In addition, the terminal base pair was changed from the native A-U pair to a G-C pair due to the requirements for efficient in vitro transcription of the RNA using T7 RNA polymerase; RNAs synthesized using T7 RNA polymerase tend to be produced more efficiently when they begin with 5′-GG or -GC (43), so given that milligram quantities of RNA are required for NMR experiments, we could not have our transcripts start with an A. We were initially concerned that making nonnative sequence changes to the panhandle construct termini could make it difficult to assess the biological relevance of our results. However, it has been demonstrated for inf A virus that its RNA polymerase has a high tolerance for sequence variability at the terminal position (16), so the identity of the base pair at the terminus is not crucial for function in inf A virus and, by extension, ISAV, since ISAV is known to follow similar molecular mechanisms to inf A virus (58).
Fig. 2.
ISAV panhandle sequences from segment 8. (A and B) Native and mutant vRNA panhandle constructs used for NMR analysis (A) and thermal melting analysis (B). AP, positions where adenosine was substituted with 2-aminopurine. (C) cRNA panhandle constructs used for NMR analysis. Nonnative bases are in lowercase. Canonical base pairing is indicated by solid lines. In panel B, base changes made relative to the NMR vRNA construct are in bold.
Several 2-AP-substituted and mutant vRNA panhandle hairpin constructs—NMR-ap4, NMR-ap32, C49U-ap4, C49U-ap32, C49U, U3C, U17C/U20C, and U15A—were also designed to probe differentially the UV-detected and 2-AP-detected melting transitions of the upper and lower stems (Fig. 2B).
For the cRNA panhandle construct, two RNAs corresponding to the 5′ and 3′ strands of the putative cRNA panhandle (Fig. 2C) were designed so that the duplex panhandle construct, formed by annealing the strands, would have the native sequence with an A-U terminal pair and the poly(A) complement (referred to as native duplex cRNA). For the NMR cRNA panhandle hairpin construct (referred to as NMR cRNA), the RNA was designed and prepared as a single RNA strand that contained the 5′ and 3′ sequences connected by a polypyrimidine pentaloop that was intended to simulate the remainder of the genomic segment in a manner similar to the NMR vRNA design. As will be discussed in more detail later, we did not include the complementary upper stem in the final NMR cRNA construct.
To obtain assignment of 1H, 13C, and 15N resonances by NMR spectroscopy, unlabeled, 13C/15N A/U-labeled, and 13C/15N G/C-labeled vRNA and cRNA samples were synthesized. A 13C/15N C/U/G NMR vRNA hairpin construct was also prepared to facilitate analysis of the internal loop. In addition, due to the uridine-rich nature of the NMR vRNA panhandle, a [5-2H]uridine-NMR vRNA construct was prepared to eliminate the strong H6-H5 cross-peak signals observed in NOESY and DQF-COSY NMR experiments (21, 47).
Secondary structure analysis of vRNA panhandle.
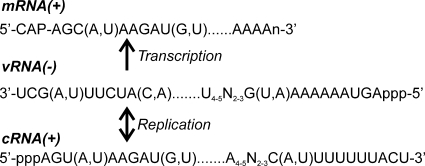
Initial analysis of the exchangeable protons of the native duplex vRNA and NMR vRNA panhandle constructs in 97% H2O–3% 2H2O at pH 7.0 revealed a remarkable similarity in the one-dimensional (1D) imino 1H spectra (Fig. 3 A). Both unlabeled RNAs were analyzed using a standard set of NOESY NMR experiments that correlate protons through space within 5 Å of each other. For each base pair in the RNA structure, an imino proton resonance is expected to be observed, while no observable signals are expected for imino protons from unpaired bases in loop regions due to enhanced solvent accessibility and exchange broadening. From the NOESY experiments of both vRNA panhandle constructs, two separate networks of interresidue correlations of imino-to-imino proton nuclear Overhauser effects (NOEs), the so-called NOE walk, confirmed the stacking of base pairs in the lower and the upper helical stems separated by an internal loop. A representative 2D NOESY spectrum for the NMR vRNA panhandle is given in Fig. 3B. From these experiments, it could also be determined that the stem helices of both vRNA panhandle constructs contained a number of non-Watson-Crick base pairs: two U·G pairs (U17·G33 and U20·G30) and one U·U pair (U15·U35) in the upper stem and one U-G pair (U3·G47) in the lower stem. These base pairs were further confirmed by correlations observed in 15N-filtered/edited 1H NOESY and 2D 1H-15N HSQC spectra using 13C/15N A/U- and G/C-labeled NMR vRNA panhandles (data not shown). Another unique characteristic of the NMR vRNA panhandle was the observation of solvent accessibility of the U14 imino proton, as evidenced by the near absence of a peak in the 1D 1H spectrum. The assignment of U14 H3 was confirmed by A36 H2 NOEs (data not shown). This observation, which suggests some degree of flexibility and/or instability at this site, was not expected since U14 is predicted to base pair with A36 and is located within a helical region next to a stabilizing G·C base pair. From these analyses, it was determined that the native duplex and NMR vRNA panhandle constructs have similar structures. Any observed chemical shift deviations can be attributed to small local changes in the chemical environment of the resonances resulting from modifying the terminus from an A-U pair to a G-C pair. To facilitate the production of isotopically labeled RNA with high yield, further NMR analysis was undertaken with the NMR vRNA panhandle construct substituted with the G-C terminal pair.
Fig. 3.
NMR characterization of the ISAV vRNA panhandle from segment 8. (A) Imino proton region of 1D Watergate at 10°C. Upper spectrum, 23-mer native duplex vRNA with the native A-U terminal pair; lower spectrum, final NMR vRNA hairpin construct (49-mer) with a G-C terminal pair. Sequential imino proton assignments of the lower stem are given in green, and the upper stem assignments are in red and were determined by the 2D NOESY in panel B. (B) Imino proton region of a 2D NOESY-Watergate spectrum (τmix = 50 ms) at 10°C in 3% 2H2O for the NMR vRNA panhandle construct. The imino assignments are listed on the 1D projection. (C) Aromatic-anomeric region of a 2D NOESY spectrum (τmix = 250 ms) at 20°C in 99.96% 2H2O. A representative sequential NOE walk, H6/H8-H1′, from G29 to C49 is shown as a red trace. The NOE walk from G1 to C21 is not shown for clarity. Adenosine H2-H1′ NOE correlations are shown in cyan. Adenosine H2-H1′ NOEs in the internal loop are highlighted in green. (D) Summary of vRNA secondary structure determined from the NMR data. As discussed in the text, both native A-U and nonnative G-C terminal pairs were used in NMR constructs. Solid lines and dots, canonical and noncanonical base pairing, respectively. Nonnative bases are in lowercase. The arrows represent observed interstrand H2-H1′ NOEs within the internal loop.
Analyses of nonexchangeable resonances were performed in 99.96% 2H2O, in which all exchangeable protons exchange rapidly with deuterons and hence are not observed in the NMR spectra. The correlation of aromatic to anomeric proton signals in the NOESY spectrum provides a second NOE walk that confirmed the helical stacking assigned from the imino-imino correlations. Additionally, observation of strong intra- and cross-strand anomeric to H2 NOEs and strong H2′ to aromatic NOEs indicated that the helical regions of the NMR vRNA panhandle adopt standard A-form geometries. The NOE walk for residues G29 to C49 is given in Fig. 3C. A similar NOE walk can be traced for residues G1 to C21. The connecting polypyrimidine heptaloop had weaker interresidue cross peaks due to its greater flexibility, as expected. In addition to the expected H2 to H1′ interresidue NOE correlations observed for both stem regions, similar correlations were observed for the adenosines within the internal loop (Fig. 3C and D, arrows). The presence of these cross peaks indicated that the average conformation of the internal loop orients each strand such that the bases are in a stacked conformation that does not deviate significantly from A-form geometry. Of particular note was the absence of H2 to H1′ NOEs to both G11 and U42; indeed, at 20°C both residues resided within an intermediate-exchange regime on the NMR time scale. At 30°C, the A10 H2-G11 H1′ cross peak was still absent, despite a subtle shift of other G11 NOESY cross peaks toward the fast-exchange regime (data not shown). However, additional analyses of NOESY data at 30°C indicated a destacking of the internal loop, as was especially evident by the shift of A40 and A41 H2 resonances toward the intermediate-exchange regime (data not shown). This observation was not surprising, considering that 30°C is within 10°C of the melting temperature of the lower stem (see following section). Due to the evidence that the NMR vRNA panhandle structure started to melt at 30°C, all further NMR analyses were performed at 20°C. Taken together, the exchangeable and nonexchangeable NOE data were used to determine the secondary structure of the NMR vRNA panhandle at 20°C (Fig. 3D).
Thermodynamic analysis of vRNA panhandle constructs.
UV and 2-AP fluorescence melting experiments were performed on the native and mutated vRNA panhandle constructs to determine the stability of the panhandle structure and dissect the melting transitions (TM) associated with the upper and lower stems of RNA structure (Fig. 2; Table 1). For the thermodynamic comparative analysis, three different constructs which contained native A-U (native hairpin), G·U (C49U), and G-C (NMR vRNA) terminal pairs were initially examined. From nearest-neighbor rules, G·U and A-U terminal pairs receive the same 0.45-kcal/mol penalty as a G-C pair (39), so the native hairpin vRNA panhandle construct was made to confirm that its stability was indeed comparable to that of the C49U vRNA panhandle. Constructs comprised of only the isolated lower or upper stems (ls-vRNA panhandle and us-vRNA panhandle, respectively) were also synthesized to obtain independent melting data for each stem. For design of the ls-vRNA panhandle, A10 was linked to A40 to create an octaloop, and the G-C terminal pair was maintained from the full NMR construct. The us-vRNA panhandle was comprised of G11 through C39, which includes the seven-nucleotide genome-mimicking loop. In addition, the U·G and U·U pairs were changed to C-G pairs and to an A-U pair, respectively, to yield the following samples: U3C, U17C/U20C, and U15A (Fig. 2B). Both U17C/U20C and U15A constructs were designed to stabilize the upper stem, while the U3C construct was crafted to afford greater stability to the lower stem. In all experiments discussed below, the melting transitions were determined to be concentration independent, confirming that the results report on a unimolecular process.
Table 1.
UV- and fluorescence-detected melting transitions of vRNA panhandle constructs
| vRNA panhandle construct | Method of detection | Transition temp (°C) |
||
|---|---|---|---|---|
| Lower stem | Singlea | Upper stem | ||
| Native hairpin | UV | 37.9 | 49.1 | |
| NMR vRNA | UV | 48.1 | ||
| Lower stem | UV | 39.9 | ||
| Upper stem | UV | 49.4 | ||
| C49U | UV | 33–40b | 48.2 | |
| U15A | UV | 47.0 | 60.2 | |
| U3C | UV | 53.7 | ||
| U17C/U20C | UV | 45.7 | 69.9 | |
| NMR-ap4 | Fluorescencec | 43.7 | ||
| NMR-ap32 | Fluorescencec | 47.6 | ||
| C49U-ap4 | Fluorescencec | 35.1 | ||
| C49U-ap32 | Fluorescencec | 46.7 | ||
The melting transitions of the upper and lower stems overlapped.
A lower shoulder was observed. See Fig. 4.
The major transition was assigned to melting of the stem.
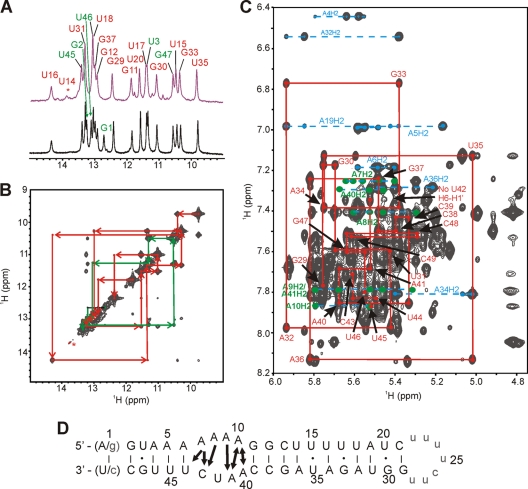
For the NMR vRNA panhandle, a single melting transition was observed at 48.1°C, which suggested that the panhandle structure formed by this construct melted cooperatively. However, upon analysis of the C49U vRNA panhandle construct, two melting transitions were observed, one at 48.2°C, similar to the NMR vRNA panhandle, and a shoulder between 33 and 40°C (Fig. 4 A and B; Table 1). The native hairpin vRNA panhandle construct gave similar results to the C49U vRNA panhandle, as expected, although the lower transition was resolved (TM1, 37.9°C; TM2, 49.1°C). Thermal melts of the us-vRNA panhandle and ls-vRNA panhandle constructs revealed that these RNA duplexes melted with single transitions at 49.4°C and 39.9°C, respectively. Taken together, these data suggest that the higher- and lower-temperature melting transitions for the native hairpin and C49U vRNA panhandle constructs correspond to the melting of the upper and lower stems, respectively, in the panhandle structure. The apparent single cooperative melting transition observed for the NMR vRNA panhandle construct therefore appeared to depend, at least in part, on the introduction of a nonnative terminal G-C base pair, which increased the stability of the lower stem by decreasing the fraying at the terminus. Since the NMR data showed that the internal loop is partially destacked at 30°C (data not shown), the apparent single melting transition for the NMR construct could be the result of base pair unfolding that originates from the internal loop into both stems. In contrast, for the C49U and native hairpin vRNA panhandle constructs, the decreased stability of the lower stem due to increased fraying of the G·U and A·U terminal pairs, respectively, resulted in separate melting transitions for each stem in the panhandle.
Fig. 4.
UV and fluorescence melting data for vRNA panhandle constructs. (A and B) UV-detected melts (A) and first-derivative plots (B) of native hairpin, NMR vRNA, and C49U vRNA panhandles. Arrow, the shoulder observed for the melt of the lower stem in the C49U vRNA panhandle. (C and D) Fluorescence-detected melts (C) and first-derivative plots (D) for the NMR-ap4 and-ap32 vRNA panhandle constructs. (E and F) Fluorescence-detected melts (E) and first-derivative plots (F) for the C49U-ap4 and-ap32 vRNA panhandle constructs. Melting temperatures derived from the melting transitions are given in Table 1.
These results were further corroborated by experiments with additional mutants of the NMR vRNA panhandle. For the U15A and U17C/U20C mutants, two melting transitions were observed at 47.0/60.2°C and 45.7/69.9°C, respectively (Table 1). Since both mutants were designed to stabilize the upper stem, the second, higher-temperature transitions were assigned to this stem. As expected, the U17C/U20C mutant had the most stable upper stem, since mutating two U·G pairs to two C-G pairs, as is the case for the U17C/U20C construct, affords greater thermodynamic stability than the mutation of a single U·U pair to an A-U pair, as is the case for the U15A construct. The lower-temperature transition, which is very similar in both mutants, was therefore assigned to the lower stem. Melts were also performed for the U3C mutant and yielded a single melting transition at 53.7°C. In this case, the U3C mutant, designed to confer greater structural stability to the lower stem, increased the overall stability of the RNA, as is evident by the 5.6°C increase in the single cooperative melting transition compared with the NMR vRNA construct. This is in contrast to the observation that further increasing the stability of the upper stem does not confer greater stability on the lower stem, as demonstrated by the melting data of the U17C/U20C and U15A constructs.
To further investigate the local thermal melting of the upper and lower stems of the vRNA panhandle, fluorescence-detected melting experiments were performed using NMR and C49U vRNA panhandles substituted with 2-AP at positions 4 and 32 in the sequence (referred to as NMR-ap4, NMR-ap32, C49U-ap4, and C49U-ap32) to monitor independently the local melting of each stem (Fig. 4C to F). 2-AP is known to pair with thymine in DNA and uracil in RNA in Watson-Crick geometry without a loss of helical stability compared to the native deoxyribo(A-T) or ribo(A-U) base pairs (25). For the NMR vRNA panhandle, the major melting transitions occurred at 43.7°C and 47.6°C for the ap4 and ap32 constructs, respectively, suggesting that the two stems melt at slightly different temperatures (Table 1). Comparing the fluorescent data to the UV melting data, these results suggest that the UV melting curve for the unlabeled NMR vRNA panhandle construct may not be a simple two-state cooperative melting curve but rather that the two melting transitions, as detected via fluorescence melts, could not be resolved in the UV detection experiments (compare Fig. 4B and D). For the C49U vRNA panhandle construct, the G-U terminal pair resulted in a large destabilization (8.6°C, comparing NMR-ap4 to C49U-ap4) of the lower-temperature melting transition but only a slight destabilization (0.9°C, comparing NMR-ap32 to C49U-ap32) of the higher-temperature melting transition, consistent with the UV melt for the C49U vRNA panhandle in which the lower- and upper-stem melting transitions were resolved. In all cases, additional minor melting transitions were detected, likely related to the local structural change in the 2-AP–U base pair before helix unfolding. For example, the melt of C49U-ap4 exhibited three peaks in the derivative plot at 28°C, 32°C, and 35.1°C (Fig. 4F). Melts performed in the presence of MgCl2 yielded the same peaks at 28°C and 32°C, but the third transition was stabilized to 41°C (data not shown). Since Mg2+ is known to stabilize helices, the transition at 35.1°C that was shifted in the presence of Mg2+ could be tentatively assigned to the melting of the lower stem.
Secondary structure analysis of cRNA panhandle.
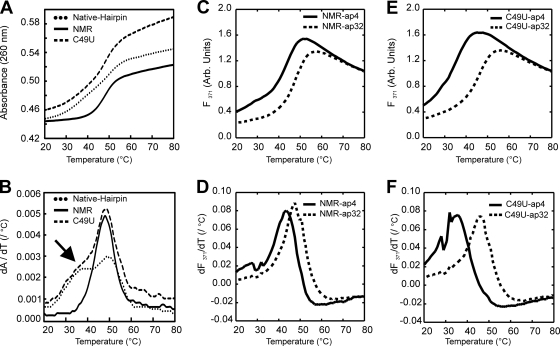
As for the vRNA panhandle constructs, examination of the 1D imino spectra of both the native duplex and NMR cRNA panhandle hairpin constructs (Fig. 2C) revealed a remarkable spectral similarity (Fig. 5). Indeed, the minor peaks, indicated by an asterisk at approximately 12.0 ppm in Fig. 5A, were the only hint of additional base-pairing interactions in the upper stem region in the native duplex cRNA panhandle (Fig. 5A, upper spectrum). Additionally, the imino peak for U14 was absent, suggesting that this proton is exposed to solvent and adjacent to a highly dynamic region. All other chemical shift deviations (e.g., G2 and G21) were attributed to small local changes in the chemical environmental from the slight difference in sequence. The general broadening of all resonances in the native duplex cRNA panhandle duplex results from a slower correlation time; i.e., larger molecules tumble more slowly in solution, leading to broader peaks. Since these data confirmed that the upper stem is indeed more dynamic, we decided to use the smaller construct, referred to as NMR cRNA panhandle hairpin, for full NMR characterization.
Fig. 5.
NMR characterization of ISAV cRNA panhandle. (A) Imino proton region of 1D Watergate at 10°C and pH 6.0. Upper spectrum, 23-mer native duplex cRNA with the native A-U terminal pair and the upper stem; lower spectrum, final NMR cRNA construct (33-mer) with a G-C terminal pair. Imino proton assignments were determined by the 2D NOESY in panel B. Green asterisk, minor peaks from insignificant base-pairing interactions in the upper stem of the native duplex cRNA. See the text for details. (B) Imino proton region of a 2D NOESY-Watergate spectrum (τmix = 150 ms) at 10°C in 3% 2H2O. Sequential imino proton assignments are given in red. (C) Aromatic-anomeric region of a 2D NOESY spectrum (τmix = 250 ms) at 20°C in 99.96% 2H2O. An NOE walk, H6/H8-H1′, from G1 to U14 is shown as a red trace. The NOE walk from U15 to C33 is not shown for clarity. Adenosine H2-H1′ correlations are shown in cyan.
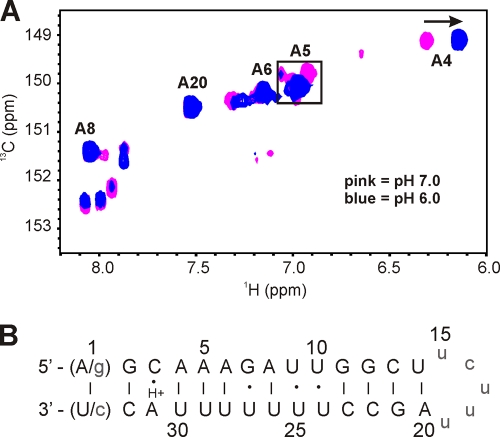
Attempts to characterize the NMR cRNA panhandle using NMR methods similar to those applied to the NMR vRNA panhandle at pH 7.0 failed due to severe spectral heterogeneity, suggesting the presence of multiple RNA conformers for the NMR cRNA panhandle under these conditions in the NMR sample. Analysis of the NMR cRNA panhandle construct via denaturing polyacrylamide gel electrophoresis revealed only a single band, ruling out the possibility of RNA degradation (data not shown). Further inspection of the NMR cRNA panhandle sequence suggested a possible protonation site on the A31 base which could stabilize a C3-A31 wobble pair. After reducing the pH to 6.0, the NMR data indicated a nearly homogeneous sample that is consistent both with the formation of a single RNA fold and with pKa values of 6.0 to 6.5 previously reported for A-C pairs (5, 33). Our pH titration corroborated these literature values, although we could not accurately determine a pKa for the protonation of the A31 due to multiple conformations above pH 6.5 (data not shown). Analysis of the H2-C2 region of the 1H-13C constant-time HSQC at pH 6.0 illustrates the effect of dropping the pH from 7.0 to 6.0 (Fig. 6 A). While there are only six adenosines in the NMR cRNA panhandle construct, there are 18 H2-C2 correlations observed at pH 7.0. At pH 6.0, the number of observed peaks is halved, and there are clearly five intense H2-C2 peaks. The H2-C2 correlation of A4 exhibited one intense peak that was significantly shifted upfield from the rest of the C2 resonances. Four peaks at about 6.95 ppm coalesced into one intense peak at pH 6.0. While the assignment of all the peaks could not be determined at pH 7.0, the coalesced peak was attributed to A5. The H2-C2 cross peaks for A6, A8, and A20 were unchanged from pH 7.0 to pH 6.0. However, while the A31 H2-C2 cross peak could not be located in the 13C HSQC at pH 6.0, assignment of the H2 resonance was determined from NOE cross peaks to the H1′ protons of C32 and A4 in the NOESY spectrum. The presence of the A31 H2-A4 H1′ NOE correlation was another strong indication that the C3-A31 wobble pair is well stacked within the helix. The additional unassigned H2-C2 cross peaks in the 13C HSQC represent other minor conformational states of the deprotonated RNA. These unassigned resonances were either weak or nonexistent in the NOESY spectra, allowing facile assignment of the major RNA species.
Fig. 6.
(A) Overlay of adenosine H2-C2 region of a 1H-13C constant-time HSQC spectrum of a 13C/15N A/U-labeled NMR cRNA panhandle at 20°C. The proton carrier frequency was set to 4.71 ppm. Listed assignments were made at pH 6.0. The boxed region indicates the coalescing of 4 peaks into one peak from pH 7.0 to 6.0. An arrow indicates the upfield shift of A4 after the pH is dropped to pH 6.0. (B) Summary of cRNA secondary structure as determined from the NMR data. As discussed in the text, both native A-U and nonnative G-C terminal pairs were used in the NMR constructs. Solid lines and dots, canonical and noncanonical base pairings, respectively; H+, the protonated A31. Nonnative bases are in lowercase.
Analysis of the NMR cRNA panhandle at pH 6.0 revealed one network of interresidue imino-to-imino NOE correlations indicating a contiguous helical structure and ruling out the presence of an internal loop. The imino NOE data also revealed one G·U pair and tandem U·U noncanonical pairs in the NMR cRNA panhandle (Fig. 5B). Additional analysis of NOESY data in 2H2O further supported the expected A-form helical geometry. A portion of the aromatic-anomeric NOE walk in the 1H NOESY spectrum is shown in Fig. 5C for residues G1 to U14. A similar NOE walk can be traced for the pentapyrimidine loop U15-U19 and for A20-C33. For A31H8 and C3H6 of the C-A wobble pair, both aromatic protons were observed to have one peak, further suggesting the presence of one major RNA species. On the basis of these observations, the secondary structure of the NMR cRNA panhandle construct is given in Fig. 6B.
DISCUSSION
In the present study, we sought to characterize ISAV vRNA and cRNA panhandle structures and gain insight into ISAV biology by comparing these RNA structures to those determined for a related and better-studied virus, namely, the inf A virus. For inf A virus, the exact regulatory switch between transcription and replication remains elusive but likely involves a complex interplay between the RNAs (9, 17), nucleoprotein (NP) (27, 40), viral polymerase (12, 31, 48), and, potentially, virus-encoded short RNAs (svRNAs) (50). Early studies to define the minimal molecular components for transcription and replication using conventional or modified chloramphenicol acetyltransferase (CAT) reporter assays revealed that the inf A virus promoter functions independently of the rest of the particular genomic segment (9, 15, 16, 36, 37, 46). In the case of ISAV, critical elements similar to those of inf A virus have been identified in the ISAV transcription/replication cycle, particularly the formation of the panhandle motif in the promoter regions of the genomic segments (28, 56) and the importance of the NP (20). Biochemical and biophysical investigation of the transcription/replication cycle of ISAV is still an emerging field, and this report represents the first detailed description of the vRNA and cRNA panhandle RNA secondary structures of an ISAV genomic segment.
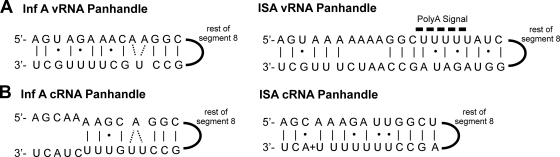
While both inf A virus and ISAV share the conserved 3′-UCGUUU-5′ sequence motif at the 3′ termini of their vRNA panhandles, our results indicate that there is very little resemblance between the secondary structures other than the general formation of a panhandle motif. For inf A virus, a previous NMR investigation of the terminal region of genomic segment 8, which did not include the poly(A) signal, revealed that the vRNA panhandle contained an (AA)·U motif (2), in which a uridine is base paired to two adenosines, which has been shown to be essential for the endonucleolytic activity of the viral polymerase (30) and for packaging of the vRNA panhandle (57) (Fig. 7 A). In addition, the inf A virus vRNA panhandle was found to contain a 46° bend in its stem 4 bp from the terminus that may influence the regulation of transcription and replication (2). In the case of the ISAV segment 8 vRNA panhandle studied here, an (AA)·U motif was not observed. However, there is an internal loop which is highly structured, as is evident from the cross-strand H2-H1′ NOEs. Since the internal loop is the linker region between the lower stem and the poly(A) signal-containing upper stem, which was included due to the predicted pairing in all eight genome segments (18, 28), it may be an important structural signal in protein-RNA interactions. In addition to the internal loop, the three U·G pairs and the one U·U pair could potentially serve as recognition elements for the viral polymerase since these noncanonical pairs are known protein binding motifs in other RNAs (22, 34).
Fig. 7.
Schematic representations of the NMR-determined secondary structures for the segment 8 vRNA panhandle motifs from influenza A/PR/8/34 virus (2) and ISAV (A) and segment 8 cRNA panhandle motifs from influenza A/PR/8/34 virus (49) and ISAV (B). Solid lines and dots, canonical and noncanonical base pairings, respectively; solid semicircle, the rest of genomic segment 8.
While few RNA polymerase binding studies have been reported for ISAV, it has been shown that preformation of the inf A virus vRNA panhandle facilitates recognition of the RNA by the inf A virus RNA polymerase, as opposed to a sequentially assembled ribonucleoprotein complex, and increases the polymerase's endonuclease activity (32). As discussed earlier, nine terminal base pairs of the inf A virus vRNA panhandles melt upon interaction with the viral polymerase and subsequently form hairpin structures at both the 5′ and 3′ termini, a rearrangement referred to as the corkscrew secondary structure model (9, 16, 46). This phenomenon has also been reported for the thogoto viruses (29). Formation of such a corkscrew structure in ISAV seems unlikely on the basis of Mfold structural predictions; however, the difference in stability that we observed for the upper and lower stems suggests an alternative mechanism for forming the complex, in that the propensity of the lower stem to melt and/or rearrange upon polymerase binding may allow enhanced recognition by the polymerase and may substitute for the lack of an RNA corkscrew. Our NMR and thermal melting data also indicate that the ISAV vRNA panhandle starts to melt at ∼30°C, which, in turn, may result in loss of structural features necessary for recognition and binding of the RNA by the ISAV polymerase. This hypothesis is consistent with the observation that ISAV functions optimally at 15°C and ceases to replicate above 25°C (14) and is further corroborated by the fact that inf A virus replicates at higher temperatures and contains vRNA panhandles that are 5 to 6 bp longer than those in ISAV, thereby making them more stable above 30°C.
Investigation of the ISAV segment 8 cRNA panhandle suggested the formation of a protonated C·A wobble pair that was not present in the corresponding inf A virus cRNA panhandle (49), although an A·C pair was reported for the inf A virus vRNA panhandle (2, 6) (Fig. 7B). This feature suggests a possible mechanism in which binding of the viral proteins to the ISAV cRNA panhandle involves a pH-dependent RNA structural switch. While the pKa value for the N1 of a free adenosine is ∼3.5, the pKa can shift significantly to physiological pH, depending upon the local structural context (1, 33). While such a pH switch has not been documented in vivo for the ISAV cRNA panhandle, a protonation event due to a locally induced shift in the pKa of A31 and formation of a C·A base pair could facilitate stabilization of the cRNA panhandle and protein recognition. Coupled with the tandem U·U pairs and the highly dynamic nature of the upper stem, the different secondary structures of the vRNA and cRNA panhandles could be responsible for any differences in the interactions between these RNAs and the viral polymerase. These interactions could be key regulatory components in the transcription/replication cycle of ISAV. Indeed, the inf A virus polymerase has differential modes of binding to the vRNA and cRNA panhandles, so it is possible that this is the case in the ISAV system (13, 38).
In conclusion, our characterization of the secondary structures of the ISAV vRNA and cRNA panhandles represents a key first step toward determining high-resolution, three-dimensional solution structures for both of these RNAs. The observation of noncanonical secondary structural elements within these panhandle structures provides the first indication of how specific viral protein recognition of the panhandles may occur and gives new structural clues about the mechanisms governing the ISAV transcription/replication cycle.
ACKNOWLEDGMENTS
We acknowledge, in part, the support of the Hollings Marine Laboratory NMR facility, and A.L.S. was supported by the National Research Council Research Associates Program Fellowship.
Certain commercial equipment, instruments, and materials are identified in this paper in order to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the material or equipment identified is necessarily the best available for the purpose.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Acharya S., et al. 2004. Significant pKa perturbation of nucleobases is an intrinsic property of the sequence context in DNA and RNA. J. Am. Chem. Soc. 126:8674–8681 [DOI] [PubMed] [Google Scholar]
- 2. Bae S. H., et al. 2001. Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 98:10602–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Batey R. T., Battiste J. L., Williamson J. R. 1995. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 261:300–322 [DOI] [PubMed] [Google Scholar]
- 4. Bouchard D. A., Brockway K., Giray C., Keleher W., Merrill P. L. 2001. First report of infectious salmon anemia (ISA) in the United States. Bull. Eur. Assoc. Fish Pathol. 21:86–88 [Google Scholar]
- 5. Cai Z., Tinoco I., Jr 1996. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35:6026–6036 [DOI] [PubMed] [Google Scholar]
- 6. Cheong H. K., Cheong C., Lee Y. S., Seong B. L., Choi B. S. 1999. Structure of influenza virus panhandle RNA studied by NMR spectroscopy and molecular modeling. Nucleic Acids Res. 27:1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheung T. K., Poon L. L. 2007. Biology of influenza A virus. Ann. N. Y. Acad. Sci. 1102:1–25 [DOI] [PubMed] [Google Scholar]
- 8. Cottet L., Rivas-Aravena A., Cortez-San Martin M., Sandino A. M., Spencer E. 2011. Infectious salmon anemia virus—genetics and pathogenesis. Virus Res. 155:10–19 [DOI] [PubMed] [Google Scholar]
- 9. Crow M., Deng T., Addley M., Brownlee G. G. 2004. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 78:6263–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Das K., Aramini J. M., Ma L. C., Krug R. M., Arnold E. 2010. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 17:530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delaglio F., et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 12. Deng T., Sharps J. L., Brownlee G. G. 2006. Role of the influenza virus heterotrimeric RNA polymerase complex in the initiation of replication. J. Gen. Virol. 87:3373–3377 [DOI] [PubMed] [Google Scholar]
- 13. Deng T., Vreede F. T., Brownlee G. G. 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 80:2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falk K., Namork E., Rimstad E., Mjaaland S., Dannevig B. H. 1997. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.). J. Virol. 71:9016–9023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flick R., Hobom G. 1999. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J. Gen. Virol. 80:2565–2572 [DOI] [PubMed] [Google Scholar]
- 16. Flick R., Neumann G., Hoffmann E., Neumeier E., Hobom G. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 2:1046–1057 [PMC free article] [PubMed] [Google Scholar]
- 17. Fodor E., Pritlove D. C., Brownlee G. G. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fourrier M., Heuser S., Munro E., Snow M. 2011. Characterization and comparison of the full 3′ and 5′ untranslated genomic regions of diverse isolates of infectious salmon anaemia virus by using a rapid and universal method. J. Virol. Methods 174:136–143 [DOI] [PubMed] [Google Scholar]
- 19. Godoy M. G., et al. 2008. First detection, isolation and molecular characterization of infectious salmon anaemia virus associated with clinical disease in farmed Atlantic salmon (Salmo salar) in Chile. BMC Vet. Res. 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goic B., et al. 2008. The nucleoprotein and the viral RNA of infectious salmon anemia virus (ISAV) are localized in the nucleolus of infected cells. Virology 379:55–63 [DOI] [PubMed] [Google Scholar]
- 21. Hayatsu H., Wataya Y., Kai K., Iida S. 1970. Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry 9:2858–2865 [DOI] [PubMed] [Google Scholar]
- 22. Hermann T., Westhof E. 1999. Non-Watson-Crick base pairs in RNA-protein recognition. Chem. Biol. 6:R335–R343 [DOI] [PubMed] [Google Scholar]
- 23. Hoffmann E., Neumann G., Hobom G., Webster R. G., Kawaoka Y. 2000. “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology 267:310–317 [DOI] [PubMed] [Google Scholar]
- 24. Hsu M. T., Parvin J. D., Gupta S., Krystal M., Palese P. 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl. Acad. Sci. U. S. A. 84:8140–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jean J. M., Hall K. B. 2001. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc. Natl. Acad. Sci. U. S. A. 98:37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kawaoka Y., et al. 2005. Infectious salmon anaemia virus, p. 681–693 In Fauquet C. M., Mayo M. A., Desselberger J. Maniloff, U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy Viruses. Elsevier Academic Press, New York, NY [Google Scholar]
- 27. Klumpp K., Ruigrok R. W., Baudin F. 1997. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 16:1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kulshreshtha V., et al. 2010. Identification of the 3′ and 5′ terminal sequences of the 8 RNA genome segments of European and North American genotypes of infectious salmon anaemia virus (an Orthomyxovirus) and evidence for quasispecies based on the non-coding sequences of transcripts. Virol. J. 7:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leahy M. B., Dessens J. T., Nuttall P. A. 1997. Striking conformational similarities between the transcription promoters of thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J. Virol. 71:8352–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leahy M. B., Zecchin G., Brownlee G. G. 2002. Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters. J. Virol. 76:2019–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee M. T., et al. 2002. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee M. T., Klumpp K., Digard P., Tiley L. 2003. Activation of influenza virus RNA polymerase by the 5′ and 3′ terminal duplex of genomic RNA. Nucleic Acids Res. 31:1624–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Legault P., Pardi A. 1994. In situ probing of adenine protonation in RNA by 13C NMR. J. Am. Chem. Soc. 116:8390–8391 [Google Scholar]
- 34. Lietzke S. E., Barnes C. L., Berglund J. A., Kundrot C. E. 1996. The structure of an RNA dodecamer shows how tandem U-U base pairs increase the range of stable RNA structures and the diversity of recognition sites. Structure 4:917–930 [DOI] [PubMed] [Google Scholar]
- 35. Lovely J. E., et al. 1999. First identification of infectious salmon anaemia virus in North America with haemorrhagic kidney syndrome. Dis. Aquat. Organ. 35:145–148 [DOI] [PubMed] [Google Scholar]
- 36. Luo G. X., Luytjes W., Enami M., Palese P. 1991. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J. Virol. 65:2861–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luytjes W., Krystal M., Enami M., Parvin J. D., Palese P. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 59:1107–1113 [DOI] [PubMed] [Google Scholar]
- 38. Maier H. J., Kashiwagi T., Hara K., Brownlee G. G. 2008. Differential role of the influenza A virus polymerase PA subunit for vRNA and cRNA promoter binding. Virology 370:194–204 [DOI] [PubMed] [Google Scholar]
- 39. Mathews D. H., Sabina J., Zuker M., Turner D. H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911–940 [DOI] [PubMed] [Google Scholar]
- 40. Medcalf L., Poole E., Elton D., Digard P. 1999. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J. Virol. 73:7349–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Merour E., et al. 2011. Completion of the full-length genome sequence of the infectious salmon anemia virus, an aquatic orthomyxovirus-like, and characterization of mAbs. J. Gen. Virol. 92:528–533 [DOI] [PubMed] [Google Scholar]
- 42. Mikulasova A., Vareckova E., Fodor E. 2000. Transcription and replication of the influenza A virus genome. Acta Virol. 44:273–282 [PubMed] [Google Scholar]
- 43. Milligan J. F., Uhlenbeck O. C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180:51–62 [DOI] [PubMed] [Google Scholar]
- 44. Mjaaland S., Rimstad E., Falk K., Dannevig B. H. 1997. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J. Virol. 71:7681–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Neumann G., Brownlee G. G., Fodor E., Kawaoka Y. 2004. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 283:121–143 [DOI] [PubMed] [Google Scholar]
- 46. Neumann G., Hobom G. 1995. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76:1709–1717 [DOI] [PubMed] [Google Scholar]
- 47. Nikonowicz E. P. 2001. Preparation and use of 2H-labeled RNA oligonucleotides in nuclear magnetic resonance studies. Methods Enzymol. 338:320–341 [DOI] [PubMed] [Google Scholar]
- 48. Olson A. C., Rosenblum E., Kuchta R. D. 2010. Regulation of influenza RNA polymerase activity and the switch between replication and transcription by the concentrations of the vRNA 5′ end, the cap source, and the polymerase. Biochemistry 49:10208–10215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park C. J., Bae S. H., Lee M. K., Varani G., Choi B. S. 2003. Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase. Nucleic Acids Res. 31:2824–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perez J. T., et al. 2010. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U. S. A. 107:11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Resa-Infante P., Jorba N., Coloma R., Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rimstad E., Mjaaland S. 2002. Infectious salmon anaemia virus. APMIS 110:273–282 [DOI] [PubMed] [Google Scholar]
- 53. Ritchie R. J., et al. 2009. Comparative virulence of infectious salmon anaemia virus isolates in Atlantic salmon, Salmo salar L. J. Fish Dis. 32:157–171 [DOI] [PubMed] [Google Scholar]
- 54. Rodger H. D., Richards R. H. 1998. Haemorrhagic smolt syndrome: a severe anaemic condition in farmed salmon in Scotland. Vet. Rec. 142:538–541 [DOI] [PubMed] [Google Scholar]
- 55. Ruigrok R. W., Crepin T., Hart D. J., Cusack S. 2010. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 20:104–113 [DOI] [PubMed] [Google Scholar]
- 56. Sandvik T., Rimstad E., Mjaaland S. 2000. The viral RNA 3′- and 5′-end structure and mRNA transcription of infectious salmon anaemia virus resemble those of influenza viruses. Arch. Virol. 145:1659–1669 [DOI] [PubMed] [Google Scholar]
- 57. Tchatalbachev S., Flick R., Hobom G. 2001. The packaging signal of influenza viral RNA molecules. RNA 7:979–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Toennessen R., Lauscher A., Rimstad E. 2009. Comparative aspects of infectious salmon anemia virus, an orthomyxovirus of fish, to influenza viruses. Indian J. Microbiol. 49:308–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]