Abstract
Apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) proteins are encapsidated by assembling HIV-1 virions and edit viral cDNA in the next round of infection. Using alpha interferon (IFN-α)-treated monocyte-derived macrophages, we show that infrequent editing of HIV-1 reverse transcripts can also be mediated by APOBEC3 proteins supplied by the targets of infection. Based on the local sequence contexts of these mutations and the established characteristics of APOBEC3 protein expression in myeloid cells, we speculate that APOBEC3A may be responsible for a substantial proportion of this activity.
TEXT
The apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) proteins are cell-encoded cytidine deaminases that postsynthetically edit single-stranded polynucleotides by deaminating cytidine to uridine (C to U). The APOBEC3 subfamily of proteins target extrachromosomal DNA and can potently suppress retrovirus and hepadnavirus infections, as well as endogenous element retrotransposition, and can also help clear experimentally introduced plasmid DNA (2, 5, 7, 11, 18, 30). These processes are associated with excessive DNA editing or hypermutation, with a concomitant loss of genetic integrity, and, in some cases, with impeded DNA synthesis. In the context of human immunodeficiency virus type 1 (HIV-1) infection, the major substrate for editing is nascent minus-strand cDNA formed in newly infected cells; the mutations then register as guanosine-to-adenosine (G-to-A) changes in plus-strand viral cDNA, and causative APOBEC3 proteins are recruited to virions assembled during the preceding round of virus replication. Thus, prevailing dogma indicates that cDNA from incoming virus infections is edited by passenger APOBEC3 proteins and not by proteins residing in target cells. Wild-type HIV-1 strains largely avert the incorporation of APOBEC3 proteins through the action of the Vif protein. However, infrequent, or sublethal, editing has also been shown to occur under certain circumstances, and it has been suggested that this can contribute to viral sequence diversification (13, 29).
Some recent observations prompted us to reconsider the absoluteness of the model that APOBEC3 proteins affecting HIV-1 infection are derived from virus-producing cells. First, purified DNA that is transfected into cells expressing APOBEC3 proteins, most notably APOBEC3A, can be hypermutated (30). Second, alpha interferon (IFN-α) treatment of myeloid cells imparts a potent antiretroviral state (6, 9, 10, 20) that correlates with a substantial (e.g., 1,000-fold) induction in APOBEC3A expression (14, 22, 27, 28). Third, enzymatic analysis of cell lysates suggests that APOBEC3A is the predominant DNA cytidine deaminase in IFN-α-treated monocytes (32). Fourth, small interfering RNA (siRNA)-mediated silencing of APOBEC3A in monocytes, cells that are refractory to HIV-1 infection, confers a degree of susceptibility to infection (26).
We therefore asked whether APOBEC3 proteins present in the cellular targets of HIV-1 infection have the capacity to edit reverse transcripts generated by incoming wild-type HIV-1 infection. Because we were interested in APOBEC3 protein function during infection of natural cell targets of HIV-1, we focused our efforts on cell populations isolated from peripheral blood mononuclear cells (PBMCs) and particularly on monocyte-derived macrophages (MDMs). CD14-positive cells were purified from the PBMCs of four healthy donors using MicroBeads (Miltenyi Biotec), differentiated into macrophages by plastic adherence, and cultured for 4 days in 25-mm wells (∼8 × 105 cells per well) with medium containing 100 ng/ml granulocyte-macrophage colony-stimulating factor (R&D Systems) (10). Cultures were treated (or not) with 2,000 U/ml IFN-α (PBL Interferon Source) for 24 h prior to infection. Cultures were challenged with v3SF162/IIIB/567A, a vif-expressing, CCR5-tropic derivative of HIV-1HXB3 (10) that carries a G-to-A mutation at position 567 of the U5 region of the 5′ long terminal repeat (5′-LTR), which enables cDNA fragments that include the 3′-LTR to be distinguished from contaminating plasmid DNA (3). Stocks were generated by transfection of 293T cells, filtration of the culture supernatant, treatment with 20 U/ml RQ1 DNase (Promega) in 10 mM MgCl2 for 3 h at 37°C, and purification by ultracentrifugation through a 20% sucrose cushion, and infections were initiated with virus corresponding to 390 ng p24Gag by spin infection for 2 h at 4°C. The cultures were maintained in the presence or absence of IFN-α, and cell lysates were prepared at 3, 6, 24, 48, and 72 h for DNA extraction.
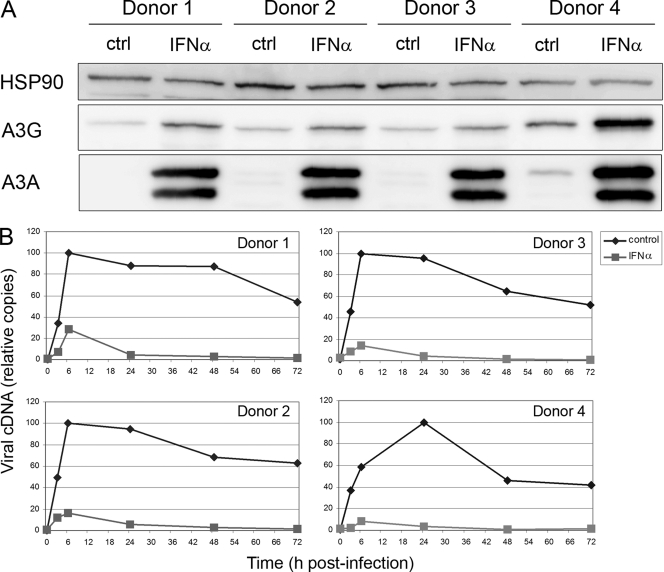
Immunoblot analysis of samples harvested at the time of viral challenge with an APOBEC3A/G-specific serum (23) confirmed that IFN-α induced a marked increase in APOBEC3A levels in the MDMs of all donors (though a low level of expression was evident in the untreated cells of donor 4) and a modest enhancement of APOBEC3G expression (Fig. 1A) (14). We next used quantitative real-time PCR (qRT-PCR) to monitor the accumulation of HIV-1 strong-stop reverse transcripts, an early product of reverse transcription (Fig. 1B). As demonstrated previously (10), IFN-α induces an impressive antiviral state in MDMs, with the levels of cDNA at 6 h being 10 to 30% of those in untreated cultures and <4% by 72 h.
Fig. 1.
IFN-α induces APOBEC3A expression in monocyte-derived macrophages and promotes resistance to HIV-1 infection. (A) Immunoblot analysis of APOBEC3A and APOBEC3G. Whole-cell lysates of MDMs that had been treated (or not) with IFN-α for 24 h were analyzed using a rabbit polyclonal serum that recognizes APOBEC3A and APOBEC3G or a rabbit anti-HSP90 serum (loading control) (Santa Cruz Biotechnologies), an anti-rabbit secondary antibody conjugated to horseradish peroxidase, and enhanced chemiluminescence (GE Healthcare). (B) qRT-PCR analysis of HIV-1 reverse transcripts. Untreated (diamonds) and IFN-α-treated (squares) cells were challenged with v3SF162/IIIB/567A, and the relative levels of strong-stop DNA at subsequent time points (set to a maximum value of 100) were determined using qRT-PCR (10).
To scrutinize the nascent cDNAs for APOBEC3 protein-mediated editing, as well as other sequence changes, we examined DNAs isolated at 3 or 24 h postinfection using PCR-mediated single-molecule amplification of a 604-bp nef/3′-LTR fragment. In brief, DNA samples were diluted to the point where ≤33% of derivative aliquots were predicted to yield a virus-specific product. Nested PCR was then performed using Platinum Taq high-fidelity DNA polymerase (Invitrogen) and two HIV-1-specific primer pairs: in the first round, EKf_Nef_HIV1_IIIB (5′-CAGGTACCTTTAAGACCAATGACT) and EKr_U5_HIV1_IIIB (5′-TGCTAGAGATTTTCCACACTGACT), and in the second round, oKG82 (5′-AGGCAGCTGTAGATCTTAGCCACTT) and oKG81-C (5′-GGTCTGAGGGATCTCTAGTTA). The resulting products were identified by agarose gel electrophoresis and sequenced (forward and reverse directions).
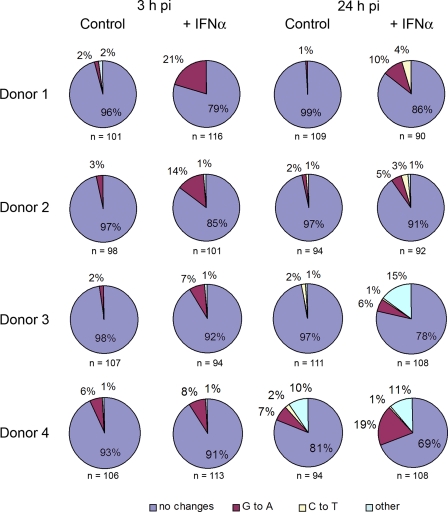
Approximately 100 cDNAs were analyzed from each sample once individual residual plasmid sequences that did not harbor the aforementioned G-to-A change in U5 had been discarded (zero to four out of any set). Table 1 provides the numbers of all possible mutations for all the samples, as well as the numbers of mutated and total cDNAs per sample. Because the majority of cDNAs from all samples contained either no changes or one mutation, each sequence was assigned according to the identity of that mutation as it appeared in the plus strand (specifically, cytidine deamination-driven G-to-A or C-to-thymidine [T] changes or others). For the low number of sequences containing more than one mutation, we scored them as G-to-A or C-to-T mutants if at least one such mutation was present. The percentages of cDNAs containing particular mutations are displayed in pie charts, together with the total number of sequences reported for each sample (Fig. 2).
Table 1.
Mutational analysis of HIV-1 cDNA during macrophage infection in the presence or absence of IFN-αa
| Donor | Time p.i. (h) | IFN-α | No. of occurrences of mutation: |
No. of sequences with at least 1 mutation | No. of sequences with at least 1 G-to-A mutation | Total no. of sequences | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G to A | C to T | A to G | T to C | C to A | A to T | A to C | G to T | T to A | C to G | ||||||
| 1 | 3 | − | 2 | ND | 2 | ND | ND | ND | ND | ND | ND | ND | 4 | 2 | 101 |
| + | 35 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 24 | 24 | 116 | ||
| 24 | − | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | 1 | 109 | |
| + | 13 | 8 | ND | ND | ND | ND | ND | ND | ND | 1 | 13 | 9 | 90 | ||
| 2 | 3 | − | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 3 | 3 | 98 |
| + | 17 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 15 | 14 | 101 | ||
| 24 | − | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | 3 | 2 | 94 | |
| + | 5 | 3 | 1 | ND | ND | ND | ND | ND | ND | ND | 9 | 5 | 92 | ||
| 3 | 3 | − | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | 2 | 107 |
| + | 7 | ND | ND | ND | 1 | ND | ND | ND | ND | ND | 8 | 7 | 94 | ||
| 24 | − | ND | 2 | 1 | ND | ND | ND | ND | 1 | ND | ND | 3 | ND | 111 | |
| + | 6 | 2 | 11 | 7 | ND | ND | ND | ND | 1 | ND | 23 | 6 | 108 | ||
| 4 | 3 | − | 6 | ND | ND | ND | 1 | ND | ND | ND | ND | ND | 7 | 6 | 106 |
| + | 13 | 1 | ND | 1 | ND | ND | ND | ND | ND | ND | 10 | 9 | 113 | ||
| 24 | − | 7 | 2 | 8 | 4 | ND | ND | 1 | ND | ND | ND | 18 | 7 | 94 | |
| + | 25 | 3 | 6 | 5 | 1 | 2 | ND | ND | ND | ND | 34 | 21 | 108 | ||
| Total | 144 | 23 | 29 | 17 | 3 | 2 | 1 | 1 | 1 | 1 | |||||
p.i., postinfection; ND, none detected.
Fig. 2.
Effects of IFN-α on HIV-1 sequence variation during macrophage infection. HIV-1 cDNAs isolated at 3 or 24 h postinfection (pi) (Fig. 1) were subjected to single-molecule amplification. The total number of sequences for each sample is indicated, together with the percentage of sequences containing no changes (mauve) or G-to-A (maroon), C-to-T (yellow), or other (blue) mutations. Based on the low mutational loads seen in the 3-h samples of donors 1, 2, and 3 in the absence of IFN-α, we infer that the cumulative error rate for RNA polymerase II, reverse transcriptase, and Platinum Taq polymerase was ∼5 × 10−5.
The extents of mutagenesis were varied between donors, as were the effects of sampling time and IFN-α treatment. Nevertheless, there were clear increases in the frequencies of hallmark APOBEC3 protein-induced G-to-A plus-strand mutations in all donors in response to IFN-α (maroon sectors in Fig. 2); specifically, combining the data from all donors, there were 13 sequences containing at least one G-to-A change in the absence of IFN-α and 54 in its presence at 3 h, compared to 396 and 367 unmutated sequences, respectively (P value = 1.4 × 10−7 according to Fisher's exact test). At 24 h, there were 10 sequences with G-to-A mutations in the absence of IFN-α and 41 in its presence, compared to 383 and 319 unmutated sequences (P = 8.1 × 10−7). These edits were well distributed across the amplified region, though three particular G residues were mutated in at least 10 of the cDNAs carrying G-to-A changes: 9309GAATG (mutated in 17 cDNAs out of 118), 9434GGACT (11/118), and 9465GTGGC (15/118) (the edited nucleotide is underlined, with its position in the pIIIB provirus indicated).
The cells from donor 4 behaved slightly differently in that cultures that had not received IFN-α also supported recognizable levels of G-to-A mutations. We suspect that this was due to the higher basal expression of APOBEC3A in these cells (Fig. 1A). There were also hints of increased levels of C-to-T mutations at 24 h in the cells of all donors, potentially due to low levels of plus-strand editing. Importantly, these patterns of infrequent mutagenesis are profoundly different from the lethal G-to-A hypermutation that characterizes infections of vif-deficient viruses produced in the presence of APOBEC3 proteins (2, 7, 11, 18) or the APOBEC3A-mediated hypermutation of transfected plasmid DNA (30).
Different APOBEC3 proteins possess different local sequence preferences for editing substrates: APOBEC3G has a strong bias for 5′-CC (5′-GG on the plus strand), whereas the remaining APOBEC3 proteins each favor 5′-TC (5′-GA) but can edit any 5′-NC dinucleotide (2, 7, 11, 18). Table 2 shows a compilation of the dinucleotide contexts for G-to-A changes found in all samples and demonstrates that 5′-GA (∼50%) is the preferred target, with 5′-GG (∼25%) as the second most common. This strongly implies that APOBEC3G is not the APOBEC3 protein responsible for the bulk of the observed G-to-A editing. Which of the other APOBEC3 proteins contribute, and it may be more than one, cannot be stated with certainty, as our efforts to deplete selective APOBEC3 proteins in IFN-α-treated MDMs using siRNA have not been successful. However, based on the link between this editing phenotype and IFN-α addition, the far greater induction of APOBEC3A levels and activity seen in IFN-α-treated myeloid cells than of all the other APOBEC3 proteins (14, 28, 32), and concordance with the substrate sequence preferences described for APOBEC3A (30, 33), our findings are most consistent with APOBEC3A being responsible for a substantial fraction of the G-to-A editing of HIV-1 seen in IFN-α-stimulated MDMs.
Table 2.
Local sequence preferences for HIV-1 G-to-A mutations during macrophage infection
| Dinucleotide contextb | No. of sequencesa |
Sum | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor 1 |
Donor 2 |
Donor 3 |
Donor 4 |
||||||||||||||
| 3 h |
24 h |
3 h |
24 h |
3 h |
24 h |
3 h |
24 h |
||||||||||
| − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | ||
| GA | ND | 19 | ND | 6 | 1 | 7 | ND | 2 | 1 | 3 | ND | 5 | 5 | 6 | 7 | 15 | 77 |
| GC | 2 | 3 | ND | 1 | ND | ND | 1 | ND | ND | 1 | ND | 1 | 1 | 2 | ND | 2 | 14 |
| GG | ND | 7 | 1 | 4 | 2 | 7 | 1 | 1 | 1 | 3 | ND | ND | ND | 3 | ND | 4 | 34 |
| GT | ND | 6 | ND | 2 | ND | 3 | ND | 2 | ND | ND | ND | ND | ND | 2 | ND | 4 | 19 |
ND, none detected. − and + indicate that IFN was not or was added to the culture, respectively.
The mutated nucleotide is underlined.
In addition to G-to-A and C-to-T mutations, a number of A-to-G and T-to-C transitions were also seen 24 h postinfection in two donors (Table 1). There are some possible explanations for these transitions, though our view is that each is implausible. First, the fidelity of reverse transcription may have been altered by host cell factors in some infections. This is unlikely because the observed G-to-A changes (the most frequent mutation caused by reverse transcriptase [1]) are largely attributable to APOBEC3 protein-mediated editing on the basis of their sequence context (Table 2), meaning that an unprecedented skewing of reverse transcriptase-mediated errors toward A to G and T to C would be required. Second, mutations on the plus strand that register as A-to-G mutations may be mediated by A-to-inosine deamination of viral RNA, a reaction that is mediated by the adenosine deaminase that acts on RNA (ADAR) enzymes, one of which, ADAR1, is IFN inducible (24). Potentially arguing against this is the late appearance of A-to-G changes, when substrate viral RNA levels would be lower, and the lack of evident clustering of mutations to regions of the RNA secondary structure (data not shown) (34). Further work is required to understand the origin of the A-to-G and T-to-C mutations, though it is noteworthy that these transition mutations are the two most common RNA edits seen in human mRNA (17).
In this paper, we report that APOBEC3 proteins expressed in the cellular targets of infection can mediate the G-to-A editing of HIV-1 cDNA. To our knowledge, this is the first account of such effects being imparted by a naturally expressed APOBEC3 protein in primary cells, rather than in a cell line, as has been reported previously for APOBEC3C (4). Whether (or not) the editing shown here is an important effector of the IFN-α-induced antiviral effect that is seen in macrophages (Fig. 1B) cannot be ascertained at present, since we have been unable to use siRNA to eliminate expression of APOBEC3 proteins in this system. Given the low mutational frequency noted in these experiments, our inclination is that it may well not be and, instead, is more likely to provide an additional mechanism to augment other sources of HIV-1 sequence diversification (13, 19, 21, 29). It is formally possible that excessively edited reverse transcripts may be rapidly degraded such that they would evade detection and characterization by the approaches described here; however, we consider DNA turnover of this nature to be unlikely, as attempts to manipulate DNA repair pathways and thereby alter the fate of uridine-containing HIV-1 cDNA have not yielded positive results (12, 15).
In sum, APOBEC3 proteins, and most likely APOBEC3A, can therefore be added to the growing constellation of host factors that are recruited to postentry HIV-1 replication complexes (8, 16, 25, 31, 35). Future work is required to determine the mechanism(s) for engagement as well as the full scope of the functional consequences, though it seems likely that these may be more significant in, and/or specific to, myeloid cells than CD4 T cells since the addition of IFN-α results in much higher levels of APOBEC3A expression in these cells, and treatment of CD4 T cells with IFN-α promotes little, if any, editing of incoming HIV-1 cDNA (data not shown).
Acknowledgments
We are grateful to Reiner Schulz for insightful discussion.
This work was supported by the United Kingdom Medical Research Council, the National Institutes of Health (grant AI070072), the European Community's Seventh Framework Programme (grant FP7/2007-2013]) under grant agreement no. PIEF-GA-2009-237501 (to C.G.), and the Department of Health via a National Institute for Health Research comprehensive Biomedical Research Centre award to Guy's and St. Thomas' NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust. F.A.K. is a fellow of the European Molecular Biology Organization.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Abram M. E., Ferris A. L., Shao W., Alvord W. G., Hughes S. H. 2010. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 84:9864–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albin J. S., Harris R. S. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert. Rev. Mol. Med. 12:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 4. Bourara K., Liegler T. J., Grant R. M. 2007. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS Pathog. 3:1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bulliard Y., et al. 2011. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 85:1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheney K. M., McKnight A. 2010. Interferon-α mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS One 5:e13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiu Y. L., Greene W. C. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317–353 [DOI] [PubMed] [Google Scholar]
- 8. Doitsh G., et al. 2010. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 143:789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gendelman H. E., et al. 1990. Regulation of HIV replication in infected monocytes by IFN-alpha. Mechanisms for viral restriction. J. Immunol. 145:2669–2676 [PubMed] [Google Scholar]
- 10. Goujon C., Malim M. H. 2010. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J. Virol. 84:9254–9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holmes R. K., Malim M. H., Bishop K. N. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32:118–128 [DOI] [PubMed] [Google Scholar]
- 12. Kaiser S. M., Emerman M. 2006. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 80:875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim E. Y., et al. 2010. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J. Virol. 84:10402–10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koning F. A., et al. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Langlois M. A., Neuberger M. S. 2008. Human APOBEC3G can restrict retroviral infection in avian cells and acts independently of both UNG and SMUG1. J. Virol. 82:4660–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee K., et al. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li M., et al. 2011. Widespread RNA and DNA sequence differences in the human transcriptome. Science 333:53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malim M. H. 2009. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malim M. H., Emerman M. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469–472 [DOI] [PubMed] [Google Scholar]
- 20. Meylan P. R., Guatelli J. C., Munis J. R., Richman D. D., Kornbluth R. S. 1993. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology 193:138–148 [DOI] [PubMed] [Google Scholar]
- 21. Moya A., Holmes E. C., Gonzalez-Candelas F. 2004. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Narvaiza I., et al. 2009. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 5:e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Newman E. N., et al. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 24. Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79:321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ocwieja K. E., et al. 2011. HIV integration targeting: a pathway involving transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 7:e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng G., et al. 2007. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood 110:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng G., Lei K. J., Jin W., Greenwell-Wild T., Wahl S. M. 2006. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 203:41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Refsland E. W., et al. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sadler H. A., Stenglein M. D., Harris R. S., Mansky L. M. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 84:7396–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17:222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Swanson C. M., Malim M. H. 2008. SnapShot: HIV-1 proteins. Cell 133:742. [DOI] [PubMed] [Google Scholar]
- 32. Thielen B. K., et al. 2010. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J. Biol. Chem. 285:27753–27766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vartanian J. P., Guetard D., Henry M., Wain-Hobson S. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320:230–233 [DOI] [PubMed] [Google Scholar]
- 34. Watts J. M., et al. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yan N., Cherepanov P., Daigle J. E., Engelman A., Lieberman J. 2009. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]