Abstract
Hepatitis C virus (HCV) is a major cause of chronic liver diseases. A high risk of chronicity is the major concern of HCV infection, since chronic HCV infection often leads to liver cirrhosis and hepatocellular carcinoma. Infection with the HCV genotype 1 in particular is considered a clinical risk factor for the development of hepatocellular carcinoma, although the molecular mechanisms of the pathogenesis are largely unknown. Autophagy is involved in the degradation of cellular organelles and the elimination of invasive microorganisms. In addition, disruption of autophagy often leads to several protein deposition diseases. Although recent reports suggest that HCV exploits the autophagy pathway for viral propagation, the biological significance of the autophagy to the life cycle of HCV is still uncertain. Here, we show that replication of HCV RNA induces autophagy to inhibit cell death. Cells harboring an HCV replicon RNA of genotype 1b strain Con1 but not of genotype 2a strain JFH1 exhibited an incomplete acidification of the autolysosome due to a lysosomal defect, leading to the enhanced secretion of immature cathepsin B. The suppression of autophagy in the Con1 HCV replicon cells induced severe cytoplasmic vacuolation and cell death. These results suggest that HCV harnesses autophagy to circumvent the harmful vacuole formation and to maintain a persistent infection. These findings reveal a unique survival strategy of HCV and provide new insights into the genotype-specific pathogenicity of HCV.
INTRODUCTION
Hepatitis C virus (HCV) is a major causative agent of blood-borne hepatitis and currently infects at least 180 million people worldwide (58). The majority of individuals infected with HCV develop chronic hepatitis, which eventually leads to liver cirrhosis and hepatocellular carcinoma (25, 48). In addition, HCV infection is known to induce extrahepatic diseases such as type 2 diabetes and malignant lymphoma (20). It is believed that the frequency of development of these diseases varies among viral genotypes (14, 51). However, the precise mechanism of the genotype-dependent outcome of HCV-related diseases has not yet been elucidated. Despite HCV's status as a major public health problem, the current therapy with pegylated interferon and ribavirin is effective in only around 50% of patients with genotype 1, which is the most common genotype worldwide, and no effective vaccines for HCV are available (35, 52). Although recently approved protease inhibitors for HCV exhibited a potent antiviral efficacy in patients with genotype 1 (36, 43), the emergence of drug-resistant mutants is a growing problem (16). Therefore, it is important to clarify the life cycle and pathogenesis of HCV for the development of more potent remedies for chronic hepatitis C.
HCV belongs to the genus Hepacivirus of the family Flaviviridae and possesses a single positive-stranded RNA genome with a nucleotide length of 9.6 kb, which encodes a single polyprotein consisting of approximately 3,000 amino acids (40). The precursor polyprotein is processed by host and viral proteases into structural and nonstructural (NS) proteins (34). Not only viral proteins but also several host factors are required for efficient replication of the HCV genome, where NS5A is known to recruit various host proteins and to form replication complexes with other NS proteins (39). In the HCV-propagating cell, host intracellular membranes are reconstructed for the viral niche known as the membranous web, where it is thought that progeny viral RNA and proteins are concentrated for efficient replication and are protected from defensive degradation, as are the host protease and nucleases (38).
Autophagy is a bulk degradation process, wherein portions of cytoplasm and organelles are enclosed by a unique membrane structure called an autophagosome, which subsequently fuses with the lysosome for degradation (37, 60). Autophagy occurs not only in order to recycle amino acids during starvation but also to clear away deteriorated proteins or organelles irrespective of nutritional stress. In fact, the deficiency of autophagy leads to the accumulation of disordered proteins that can ultimately cause a diverse range of diseases, including neurodegeneration and liver injury (12, 29, 30), and often to type 2 diabetes and malignant lymphoma (9, 32).
Recently, it has been shown that autophagy is provoked upon replication of several RNA viruses and is closely related to their propagation and/or pathogenesis. Coxsackievirus B3 utilizes autophagic membrane as a site of genome replication, whereas influenza virus attenuates apoptosis through the induction of autophagy (10, 59). Moreover, several groups have reported that HCV induces autophagy for infection or replication (5, 49); however, the role(s) of autophagy in the propagation of HCV is still controversial and the involvement of autophagy in the pathogenesis of HCV has not yet been clarified. In this study, we examined the biological significance of the autophagy observed in cells in which the HCV genome replicates.
MATERIALS AND METHODS
Plasmids.
The plasmids pmStrawberry-C1, pmStrawberry-Atg4BC74A, pmRFP-GFP-LC3, pEGFP-LC3, and pEGFP-Atg16L were described previously (7, 8, 24). The plasmids pFGR-JFH1 and pSGR-JFH1 were kind gifts from T. Wakita.
Cell culture.
All cell lines were cultured at 37°C under a humidified atmosphere with 5% CO2. Huh7 cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), nonessential amino acids, 100 U/ml penicillin, and 100 mg/ml streptomycin. For the starvation, the cells were cultivated with Earle's balanced salt solution (EBSS) (Sigma) for 6 h. HCV replicon cells were established as described previously (53). The plasmid pairs pFK-I389 neo/NS3-3′/NK5.1 and pFK-I389 neo/FGR/NK5.1 and pFGR-JFH1 and pSGR-JFH1 were linearized with ScaI or XbaI. The plasmids pFGR-JFH1 and pSGR-JFH1 were treated with mung bean exonuclease. The linearized DNA was transcribed in vitro by using the MEGAscript T7 kit (Applied Biosystems) according to the manufacturer's protocol. The transcribed RNA was electroporated into cells under conditions of 270 V and 960 mF using a Gene Pulser (Bio-Rad). All HCV replicon cells were maintained in DMEM containing 10% FBS, nonessential amino acids, and 1 mg/ml G418 (Nacalai).
Reagents and antibodies.
Concanamycin A and bafilomycin A1 were purchased from Sigma and Fluka, respectively. E64D and pepstatin A were from Peptide Institute Inc. Rabbit anti-HCV NS5A polyclonal antibody was described previously (45). Mouse monoclonal anti-JEV NS3 antibody was prepared by immunization using the recombinant protein spanning amino acid residues 171 to 619 of JEV NS3. Rabbit polyclonal anti-LC3 (PM036), mouse monoclonal anti-RFP (8D6), and anti-62/SQSTM1 (5F2) antibodies were purchased from Medical & Biological Laboratories. Rabbit polyclonal anti-cathepsin B (FL-339) and mouse monoclonal anti-LAMP1 (H4A3) antibodies were from Santa Cruz Biotechnology. Mouse monoclonal anti-HCV NS5A (HCM-131-5), rabbit polyclonal anti-β-actin, and mouse monoclonal anti-Golgin97 (CDF4) antibodies were from Austral Biologicals, Sigma, and Invitrogen, respectively. Mouse monoclonal and rabbit polyclonal anti-cathepsin B antibodies were from Calbiochem. Mouse monoclonal anti-p62/SQSTM1 (5F2) and anti-ATP6V0D1 (ab56441) antibodies were from Abcam. Rabbit polyclonal anti-Atg4B antibody was from Sigma. Mouse anti-double-stranded RNA (dsRNA) IgG2a (J2 and K1) antibodies were from Biocenter Ltd. (Szirak, Hungary).
Transfection, infection, and immunoblotting.
Transfection and infection were carried out as described previously (53). Each lysosome-enriched fraction was isolated by using the Lysosome Enrichment Kit for Tissue and Cultured Cells (Pierce) according to the manufacturer's protocol. Samples were subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were transferred to polyvinylidene difluoride membranes (Millipore) and were reacted with the appropriate antibodies. The immune complexes were visualized with Super Signal West Femto substrate (Pierce) and detected by an LAS-3000 image analyzer system (Fujifilm). The protein bands of LC3 and β-actin were quantified by Multi Gauge software (Fujifilm), and the values of LC3 were normalized to those of β-actin.
Fluorescence microscopy.
Cells were cultured on glass slides and then fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 30 min. After being washed twice with PBS, the cells were permeabilized at room temperature for 20 min with PBS containing 0.25% saponin and then blocked with PBS containing 0.2% gelatin (gelatin-PBS) for 60 min at room temperature. The cells were incubated with gelatin-PBS containing appropriate antibodies at 37°C for 60 min and washed three times with PBS containing 1% Tween 20 (PBST). The resulting cells were incubated with gelatin-PBS containing corresponding fluorescent-conjugated secondary antibodies at 37°C for 60 min and then washed three times with PBST. The stained cells were covered with Vectashield mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories Inc.) and observed with a FluoView FV1000 laser scanning confocal microscope (Olympus). Time-lapse video microscopy was performed at 37°C with a DeltaVision microscope system (Applied Precision Inc.) equipped with a ΔTC3 culture dish system (Bioptechs) for temperature control.
Quantification of pro-cathepsin B.
Each cell line was seeded on 12-well type I collagen-coated dishes (IWAKI) and cultured for 48 h. The supernatant and the cells were harvested and subjected to quantification of pro-cathepsin B by using Quantikine human pro-cathepsin B immunoassay (R&D Systems) according to the manufacturer's protocol.
Statistical analysis.
Estimated values were represented as the means ± standard deviations. The significance of differences in the means was determined by Student's t test.
RESULTS
Autophagy is induced in the HCV replicating cell in a strain-dependent manner.
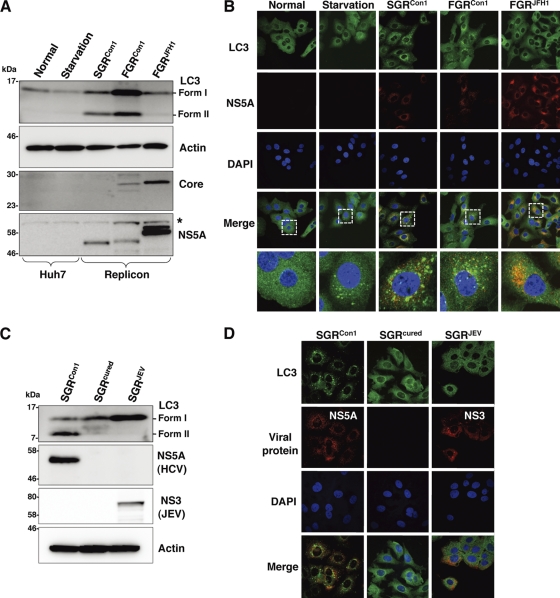
To determine whether autophagy is induced during the replication of HCV, we investigated the phosphoethanolamine (PE) conjugation of LC3 in HCV replicon cells in which HCV RNA was autonomously replicating. As shown in Fig. 1A, the amounts of PE-conjugated LC-3 (LC3-II), a conventional marker for an autophagosomal membrane, in Huh7 cells were slightly increased by starvation, in conjunction with a reduction of the unmodified LC-3 (LC3-I). In contrast, the amount of LC3-II was significantly increased in the subgenomic and full genomic HCV replicon cells of the genotype lb strain Con1 (SGRCon1 and FGRCon1), whereas a small amount of LC3-II was detected in the full genomic replicon cells of the genotype 2a strain JFH1 (FGRJFH1). We also examined the subcellular localization of LC3 by using confocal microscopy. Although LC3 was diffusely detected in the cytoplasm of naïve Huh7 cells, small foci of the accumulated LC3 appeared after starvation (Fig. 1B), whereas many LC3 foci that were larger in size than those in the starved cells appeared in the cytoplasm, particularly near the nucleus, in both SGRCon1 and FGRCon1 cells. However, a low level of LC3 focus formation comparable to that in the starved cells was observed in the FGRJFH1 cells. Most of the LC3 foci were not colocalized with NS5A, an HCV protein of the viral replication complex, in the HCV replicon cells, as reported previously (49). Elimination of HCV RNA from the SGRCon1 cells by treatment with alpha interferon (SGRcured) abrogated the lipidation and accumulation of LC3 (Fig. 1C and D). Interestingly, overexpression of the HCV polyprotein of genotype 1b by an expression plasmid induced no autophagy (data not shown), suggesting that replication of viral RNA is required for induction of autophagy. Furthermore, neither lipidation nor accumulation of LC3 was observed in SGRJEV cells harboring subgenomic replicon RNA cells of Japanese encephalitis virus (JEV), which is also a member of the family Flaviviridae (Fig. 1C and D). These results suggest that replication of HCV but not that of JEV induces autophagy.
Fig. 1.
Induction of autophagy in the HCV replicon cells. (A) The starved Huh7 cells and HCV replicon cells harboring a sub- or full genomic RNA of strain Con1 or strain JFH1 were subjected to immunoblotting using the appropriate antibodies. The asterisk indicates a nonspecific band. (B) Subcellular localizations of LC3 and NS5A were determined by confocal microscopy. The replicon cells and the starved Huh7 cells were stained with DAPI and then reacted with rabbit polyclonal anti-LC3 and mouse monoclonal anti-NS5A antibodies, respectively, followed by Alexa Fluor 488- and 594-conjugated secondary antibodies, respectively. The boxed areas in the merged images are magnified. (C) SGRCon1 cells were treated with alpha interferon for 1 week to remove the HCV replicon RNA. The resulting cells were designated SGRcured cells. The SGRCon1, SGRcured, and SGRJEV cells were lysed and subjected to immunoblotting using the appropriate antibodies. (D) Subcellular localization of LC3 and JEV NS3 and HCV NS5A was determined by confocal microscopy after staining with DAPI, followed by staining with rabbit polyclonal anti-LC3 and anti-JEV NS3 antibodies and mouse monoclonal anti-NS5A antibodies and then with the appropriate secondary antibodies. The data shown are representative of three independent experiments.
The autophagy flux is impaired in the replicon cells of HCV strain Con1 after a step of autophagosome formation.
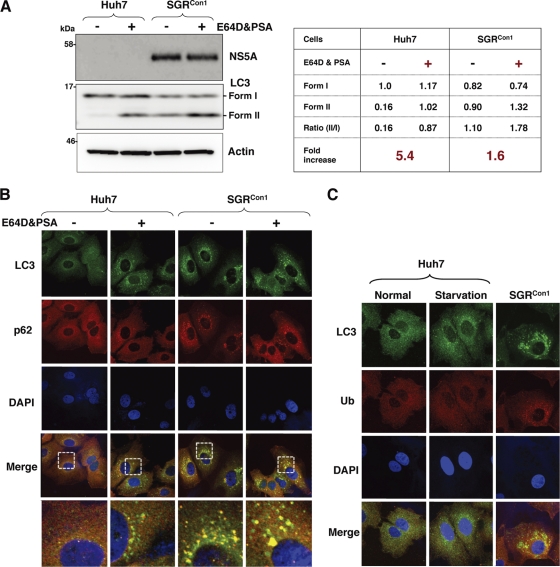
To further examine the autophagy induced in the HCV replicon cells in more detail, Huh7 and SGRCon1 cells were treated with pepstatin A and E64D, inhibitors of aspartic protease and cysteine protease, respectively. In this assay, treatment of intact cells capable of inducing autophagy with the inhibitors increases the amount of LC3-II, whereas no increase is observed in cells impaired in the autophagic degradation. The amount of LC3-II was significantly increased in the naïve Huh7 cells by treatment with the inhibitors, whereas only a slight increase was observed in the SGRCon1 cells (5.4-fold versus 1.6-fold) (Fig. 2A), suggesting that autophagy is suppressed in the HCV replicon cells. Furthermore, cytoplasmic accumulation of LC3 was significantly increased in the naïve Huh7 cells by treatment with the inhibitors, in contrast to the only slight increase induced by treatment in the SGRCon1 cells (Fig. 2B). In SGRCon1 cells, the LC3 foci were colocalized with the polyubiquitin-binding protein p62/SQSTM1, a specific substrate for autophagy (18), suggesting that most of the autophagosomes were distributed in the cytoplasm of the SGRCon1 cells (Fig. 2B and C). Next, to examine the autophagy flux in the SGRCon1 cells, we monitored the green fluorescent protein (GFP)-conjugated LC3 dynamics in living cells by using time-lapse imaging techniques (see movies in the supplemental material). A large number of small GFP-LC3 foci were detected in the starved Huh7 cell, moved quickly, and finally disappeared within 30 min. Although small foci of GFP-LC3 exhibited characteristics similar to those in the starved cells, some large foci exhibited confined movement and maintained constant fluorescence for at least 3 h in the SGRCon1 cells. The GFP-LC3 foci in the SGRJFH1 cells showed characteristics similar to those in the starved cells. These results support the notion that autophagy flux is suppressed in the SGRCon1 cells at some step after autophagosome formation.
Fig. 2.
Autophagy flux is impaired in the HCV replicon cells. Autophagy flux assay using lysosomal protease inhibitors. (A) Huh7 and SGRCon1 cells were treated with 20 μM E64D and pepstatin A (PSA) for 6 h, and the cell lysates were subjected to immunoblotting. The density of the protein band was estimated by Multi Gauge version 2.2 (Fujifilm). (B) After nuclear staining with DAPI, the intracellular localizations of LC3 and p62 in each cell were determined by staining with rabbit polyclonal anti-LC3 and mouse monoclonal anti-62 antibodies, respectively, followed by staining with Alexa Fluor 488- and 594-conjugated secondary antibodies, respectively. The resulting cells were observed by confocal microscopy. (C) Colocalization of accumulated LC3 with ubiquitinated proteins (Ub) in SGRCon1 cells. Nontreated and starved Huh7 cells and SGRCon1 cells were fixed and stained with DAPI and rabbit anti-LC3 and anti-ubiquitin (6C1.17) (BD) polyclonal antibodies, respectively, and then with the appropriate secondary antibodies. Subcellular localizations of LC3 and Ub were determined by confocal microscopy. The data shown are representative of three independent experiments.
Impairment of autolysosomal acidification causes incomplete autophagy in the replicon cell of strain Con1.
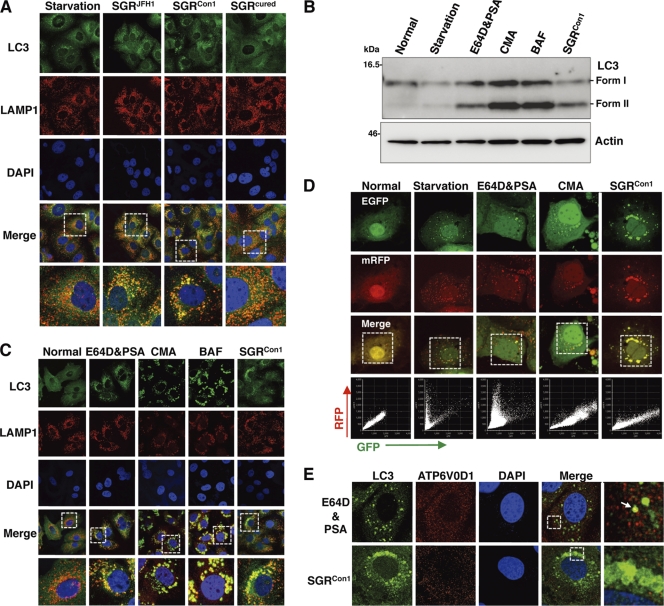
Recent studies have shown that some viruses inhibit the autophagy pathway by blocking the autolysosome formation (10, 42). Therefore, we determined the autolysosome formation in the HCV replicon cells through the fusion of autophagosome with lysosome. Colocalization of small foci of LC3 with LAMP1, a lysosome marker, was observed in the starved Huh7 cells, SGRCon1 cells, and SGRJFH1 cells but not in the SGRcured cells (Fig. 3A), suggesting that autolysosomes are formed in the HCV replicon cells of both Con1 and JFH1 strains. The autolysosome is acidified by the vacuolar-type H+ ATPase (V-ATPase) and degrades substrates by the lysosomal acidic hydrolases in the vesicle (2). Next, to determine the possibility of a deficiency in the acidification of the autolysosome on the autophagic dysfunction in the Con1 replicon cells, Huh7 cells were treated with the protease inhibitors E64D and pepstatin A (PSA) or with each of the V-ATPase inhibitors concanamycin A (CMA) and bafilomycin A1 (BAF). The amount of LC3-II was significantly increased in Huh7 cells treated with the inhibitors just as in the SGRCon1 cells (Fig. 3B). Furthermore, the large foci of LC3 colocalized with LAMP1 appeared in the cells treated with the V-ATPase inhibitors, as seen in SGRCon1 cells (Fig. 3C). These results suggest that stacked autophagosome flux caused by the inhibition of lysosomal degradation or acidification exhibits characteristics similar to those observed in the Con1 replicon cells.
Fig. 3.
Inhibition of autophagy maturation in HCV replicon cells. (A) After nuclear staining with DAPI, starved Huh7 cells, replicon cells, and SGRcured cells were stained with rabbit polyclonal anti-LC3 and mouse monoclonal anti-LAMP1 antibodies followed by Alexa Fluor 488- and 594-conjugated secondary antibodies, respectively, and examined by confocal microscopy. The boxed regions in the merged images are magnified. (B and C) Huh7 cells were treated with 20 μM protease inhibitors (E64D and PSA) or a 20 nM concentration of a V-ATPase inhibitor (CMA or BAF) for 6 h. (B) Cell lysates were subjected to immunoblotting using antibodies against LC3 and β-actin. (C) Intracellular localization of LAMP1 and LC3 was determined by confocal microscopy after staining with DAPI and appropriate antibodies. The boxed areas in the merged images are magnified. (D) Tandem fluorescence-tagged LC3 assay. The expression plasmid encoding mRFP-GFP-tandem-tagged LC3 was transfected into naïve and starved Huh7 cells or into the SGRCon1 cells treated with the indicated inhibitors at 36 h posttransfection. The resulting cells were fixed at 42 h posttransfection, and the relative GFP and RFP signals were determined by confocal microscopy. The fluorescent values in the boxes of the merged images were determined and shown as dot plots in the bottom column of the grid, in which the x and y axes indicate the signals of GFP and RFP, respectively. (E) Huh7 cells treated with E64D and PSA and the SGRCon1 cells were stained with DAPI and then with rabbit polyclonal anti-LC3 and mouse monoclonal anti-ATP6V0D1 antibodies followed by Alexa Fluor 488- and 594-conjugated secondary antibodies, respectively. The boxed regions in the merged images are magnified. A white arrow indicates colocalization of LC3 and ATP6V0D1. The data shown are representative of three independent experiments.
Since the fluorescence of GFP but not that of monomeric red fluorescent protein (mRFP) disappears under the acidic environment, expression of mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) is capable of being used to monitor the acidic status of the autolysosome (24). Both GFP and mRFP fluorescent signals were unfused, some of them accumulated as small foci in Huh7 cells after starvation or by treatment with the protease inhibitors, and half of the foci of mRFP were not colocalized with those of GFP (Fig. 3D), indicating that half of the foci are in an acidic state due to maturation into an autolysosome after fusion with a lysosome. On the other hand, the large foci of GFP and mRFP were completely colocalized in Huh7 cells treated with CMA or in the SGRCon1 cells. These results suggest that the large foci of LC3 in the SGRCon1 cells are not under acidic conditions. Recently, it was shown that the lack of lysosomal acidification in human genetic disorders due to dysfunction in assembly/sorting of V-ATPase induces incomplete autophagy similar to that observed in SGRCon1 cells (31, 45). Therefore, to explore the reason for the lack of acidification of the autolysosome in the SGRCon1 cells, we examined the subcellular localization of ATP6V0D1, a subunit of the integral membrane V0 complex of V-ATPase. Colocalization of ATP6V0D1 with large foci of LC3 was observed in Huh7 cells treated with the protease inhibitors but not in SGRCon1 cells (Fig. 3E), suggesting that dislocation of V- ATPase may participate in the impairment of the autolysosomal acidification in the SGRCon1 cells.
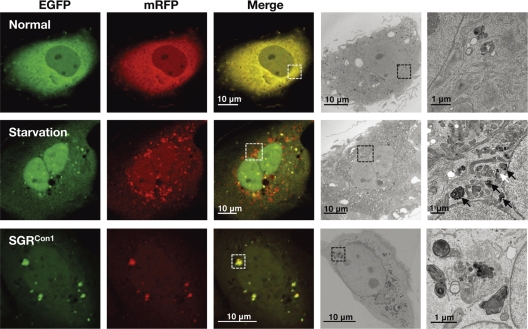
We further examined the morphological characteristics of the LC3-positive compartments by using correlative fluorescence microscopy-electron microscopy (FM-EM) (Fig. 4). The starved Huh7 cells exhibited a small double-membrane vesicle (white arrow) and high-density single-membrane structures (black arrows) in close proximity to the correlative position of the GFP- and mRFP-positive LC3 compartments, which are considered to be the autophagosome and lysosome/autolysosome, respectively. In contrast, many high-density membranous structures were detected in the correlative position of the large GFP- and mRFP-positive LC3 compartment in the SGRCon1 cells, which is well consistent with the observation in the time-lapse imaging in which small foci of LC3 headed toward and assembled with the large LC3-positive compartment (see movies in the supplemental material). These results suggest that the formation of large aggregates with aberrant inner structures in the SGRCon1 cells may impair maturation of the autolysosome through the interference of further fusion with functional lysosomes for the degradation.
Fig. 4.
Correlative fluorescence microscopy-electron microscopy (FM-EM) analysis. The expression plasmid encoding mRFP-GFP-tandem-tagged LC3 was transfected into naïve and starved Huh7 cells or into the SGRCon1 cells as described in the legend to Fig. 3D, and the mRFP-GFP-tandem-tagged LC3 signals were observed at 36 h posttransfection. The boxed regions in the merged images are magnified. The data shown are representative of three independent experiments.
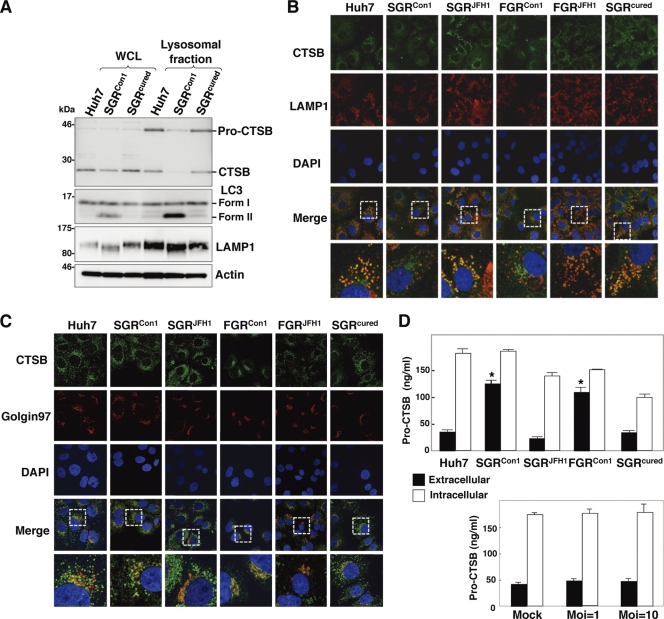
The secretion of immature cathepsin B is enhanced in the replicon cell of strain Con1.
Lysosomal acidification is required for the cleavage of cathepsins for activation, and cathepsin B (CTSB) is processed under acidic conditions (13). Although a marginal decrease of CTSB was detected in the whole lysates of the SGRCon1 cells, a significant reduction in the expression of both unprocessed (pro-CTSB) and matured CTSB was observed in the lysosomal fractions of the SGRCon1 cells compared with those of the naïve Huh7 and the SGRcured cells (Fig. 5A). LAMP1 was concentrated at a similar level in the lysosomal fractions of the cells, whereas LC-II was detected in the fractions of the SGRCon1 cells but not in those of Huh7 and the SGRcured cells, suggesting that autophagosomes and/or autolysosomes in the SGRCon1 cells are fractionated in the lysosomal fraction. Colocalization of CTSB with LAMP1 was observed in the naïve Huh7 cells, in the SGRcured cells, and in the replicon cells harboring a sub- or a full genomic RNA of strain JFH1 (SGRJFH1 and FGRJFH1, respectively) but not in those of strain Con1 (SGRCon1 and FGRCon1) (Fig. 5B). On the other hand, CTSB was colocalized with Golgin97, a marker for the Golgi apparatus, in the SGRCon1 and FGRCon1 cells but not in other cells (Fig. 5C). Since previous reports suggested that the alkalization in the lysosome triggers secretion of the unprocessed lysosomal enzymes (19, 41), we next determined the secretion of pro-CTSB in the replicon cells. Secretion of the pro-CTSB was significantly enhanced in the replicon cells of strain Con1 but not in those of strain JFH1 and naïve and cured cells (Fig. 5D, top). Furthermore, secretion of pro-CTSB was not observed in the cured cells infected with HCVcc, an infectious HCV strain derived from strain JFH1 (Fig. 5D, bottom). Collectively, these results suggest that the dysfunction of lysosomal acidification contributes to the impairment of autophagy in the HCV replicon cells of strain Con1.
Fig. 5.
Enhanced secretion of pro-CTSB in the HCV replicon cells. (A) The whole-cell lysate (WCL) and lysosomal fraction prepared from Huh7, SGRCon1, and SGRcured cells were subjected to immunoblotting. (B and C) Huh7 cells, HCV replicon cells, and SGRcured cells were stained with DAPI, rabbit polyclonal anti-CTSB antibody, and mouse anti-LAMP1 (B) or anti-Golgin97 (C) antibody. The boxed areas in the merged images are magnified. (D) Expression of pro-cathepsin B in the culture supernatants (black bars) and cell lysates (white bars) of the Huh7, SGRCon1, SGRJFH1, FGRCon1, and SGRcured cells and the SGRcured cells infected with HCVcc at a multiplicity of infection (Moi) of 1 or 10 and incubated for 72 h was determined by enzyme-linked immunosorbent assay (ELISA). The error bars indicate standard deviations. The asterisks indicate significant differences (P < 0.01) versus the control value. The data shown are representative of three independent experiments.
Autophagy induced in cells replicating HCV is required for cell survival.
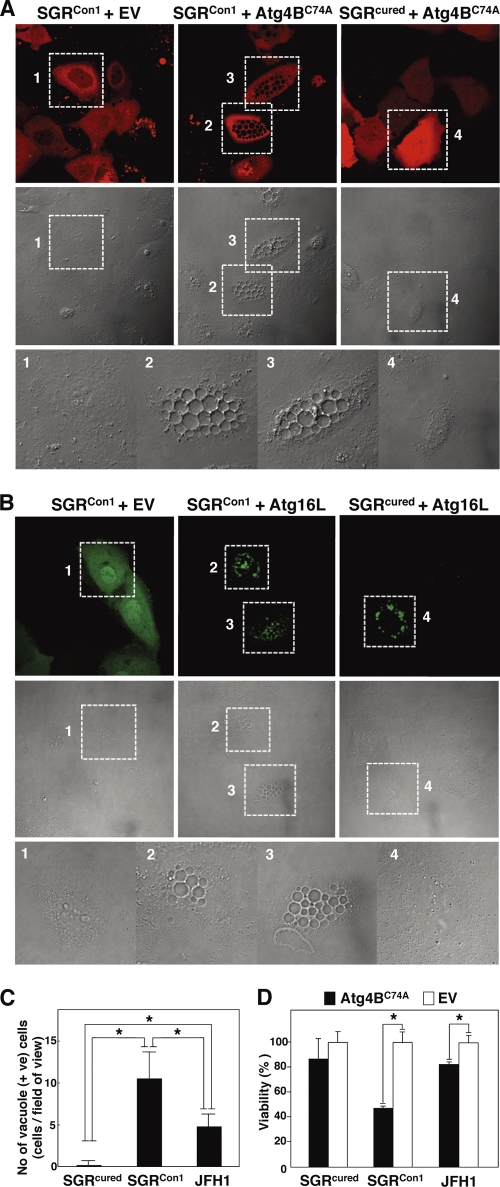
Finally, we examined the pathological significance of autophagy during HCV replication. Atg4B is known as an LC3-processing protease, and overexpression of its protease-inactive mutant (Atg4BC74A) results in inhibition of the autophagosome formation (7). To our surprise, severe cytoplasmic vacuolation was observed in the SGRCon1 cells expressing Atg4BC74A (Fig. 6A). These vacuolations were also observed in the SGRCon1 cells by the expression of Atg16L (Fig. 6B), a molecule that is an essential component of the autophagy complex and that, if expressed in excess amounts, can disrupt the autophagosome formation (8). Expression of Atg4BC74A induced a higher level of vacuole formation in the Con1 replicon cells than in cells infected with JFH1 virus but not in the cured cells (Fig. 6C). Along with these vacuolations, cell viability was significantly decreased by the expression of Atg4BC74A in SGRCon1 cells and slightly in JFH1 virus-infected cells (Fig. 6D). These results suggest that autophagy induced by the RNA replication of HCV is required for host cell survival.
Fig. 6.
Inhibition of autophagosome formation induces severe cytoplasmic vacuolations leading to cell death in the HCV replicon cells. (A) SGRCon1 and SGRcured cells transfected with pStrawberry-Atg4BC74A or empty vector pStrawberry (EV) were fixed at 48 h posttransfection and examined by fluorescence microscopy. The boxed areas in the phase-contrast images are magnified. (B) SGRCon1 and SGRcured cells transfected with pEGFP-Atg16L or EV were examined by fluorescence microscopy at 48 h posttransfection. The boxed areas in the phase-contrast images are magnified. (C) SGRcured, SGRCon1, and SGRcured cells infected with JFH1 virus were transfected with pStrawberry-Atg4BC74A, and the number of vacuole-positive cells in each of nine fields of view was counted at 48 h posttransfection and taken as the number in one field. (D) SGRcured, SGRCon1, and SGRcured cells infected with JFH1 virus were transfected with pStrawberry-Atg4BC74A (black bars) or EV (white bars), and cell viability was determined at 6 days posttransfection by using CellTiter-Glo (Promega) according to the manufacturer's protocol. The asterisks indicate significant differences (P < 0.05) versus the control value. The data shown are representative of three independent experiments.
DISCUSSION
In the present study, we demonstrated that two genotypes of HCV induce autophagy, whereas intact autophagy flux is required for the host cell to survive. The cell death characterized by cytoplasmic vacuolation that was induced in the HCV replicon cells by the inhibition of the autophagosome formation is similar to type III programmed cell death, which is distinguishable from apoptosis and autophagic cell death (4). Type III programmed cell death has been observed in the neurodegenerative diseases caused by the deposit of cytotoxic protein aggregates (15).
We previously reported that HCV hijacks chaperone complexes, which regulates quality control of proteins into the membranous web for circumventing unfolded protein response during efficient genome replication (53); in other words, the replication of HCV exacerbates the generation of proteins associated with cytotoxicity. In the experiments using a chimpanzee model, HCV of genotype 1 was successfully used to reproduce acute and chronic hepatitis similar to that in the human patients (3, 57), and transgenic mice expressing viral proteins of HCV of genotype 1b have been shown to develop Sjögren syndrome, insulin resistance, hepatic steatosis, and hepatocellular carcinoma (27, 28). In contrast, HCVcc, based on the genotype 2a strain JFH1 isolated from a patient with fulminant hepatitis C (33, 56), was unable to establish chronic infection in chimpanzees (56) or to induce cell damage and inflammation in chimeric mice xenotransplanted with human hepatocytes (17). These results imply that the onset of HCV pathogenesis could be dependent not only upon an amount but also on a property of deposited proteins, and they might explain the aggravated vacuolations under the inhibition of autophagosome formation in strain Con1 compared to that in strain JFH1. Interestingly, the overexpression of Atg4BC74A or Atg16L causes eccentric cell death in the Con1 replicon cells in which autophagy flux is already disturbed. Thus, we speculated that the quarantine of undefined abnormalities endowed with high cytotoxicity by the engulfing of the autophagic membrane might be sufficient for the amelioration of HCV-induced degeneration. The autophagosomal dysfunction observed in the Con1 replicon cells may suggest that a replicant of strain Con1 was more sensitive to the lysosomal vacuolation than that of strain JFH1. Because a limitation of our study was that we were unable to use infectious HCV of other strains, it is still unclear whether the autophagic degradation can be impaired only in the replicon of HCV strain Con1 or genotype 1.
We also demonstrated that HCV replication of strain Con1 but not that of strain JFH1 facilitates the secretion of pro-CTSB. It has been well established that the secretion of pro-CTSB is enhanced in several types of tumors (26, 50). The secretion of CTSB, like the secretion of matrix metalloproteases, is a marker of the progression of the proteolytic degradation of the extracellular matrix, which plays an important part in cancer invasion and metastasis. Since infection with HCV of genotype 1 is clinically considered a risk factor for the development of hepatocellular carcinoma (14, 51), the enhanced secretion of pro-CTSB by the replication of genotype 1 strains might synergistically promote infiltration of hepatocellular carcinoma.
As shown elsewhere (see movies in the supplemental material), although most degradations of the autophagosome were impaired due to a dislocalization of a V-ATPase subunit, some autophagic degradation was achieved in the SGRCon1 cells similar to that in the starved Huh7 cells. Moreover, the stagnated autophagy flux was rescued by the treatment of alpha interferon accompanied by elimination of HCV (Fig. 1C and D). Interestingly, we observed neither a significant impairment of lysosomal degradation nor the intracellular activity of cathepsins in the replicon cells of HCV strain Con1 (data not shown). Therefore, there might be a specific dysfunction within the autolysosome during the replication of HCV strain Con1. Detailed studies are needed to elucidate how HCV strain Con1 disturbs the sorting of V-ATPase.
A close relationship between autophagy and the immune system has been gradually unveiled (47). Autophagy assists not only in the direct elimination of pathogens by hydrolytic degradation but also in antigen processing in antigen-presenting cells such as macrophage and dendritic cells (DC) for presentation by major histocompatibility complex (MHC) I and II (11). Moreover, autophagy plays important roles in T lymphocyte homeostasis (44). As such, in some instances, interruptions of autophagy can allow microorganisms to escape from the host immune system. Indeed, the immune response against herpes simplex virus was suppressed by blocking the autophagy (6). With regard to HCV, functionally impaired DC dysfunctions marked by poor DC maturation, impaired antigen presentation, and attenuated cytokine production have been reported in tissue culture models and chronic hepatitis C patients (1, 22, 46). In addition, reduction of cell surface expression of MHC-I in HCV genotype 1b replicon cells has been reported (55). We confirmed that levels of cell surface expression of MHC-I in the replicon cells of genotype 1b, but not of genotype 2a, were reduced in comparison with those in the cured cells (data not shown). Hence it might be feasible to speculate that the replication of HCV RNA of genotype 1 induces an incomplete autophagy for attenuating antigen presentation to establish persistent infection. In contrast, autophagy is known to serve as a negative regulator of innate immunity (21, 54). A recent report demonstrated that autophagy induced by infection with strain JFH1 or dengue virus attenuates innate immunity to promote viral replication (23), indicating that an HCV genotype 2a strain may facilitate autophagy to evade innate immunity.
In this study, we demonstrated that HCV utilizes autophagy to circumvent the cell death induced by vacuole formation for its survival. This unique strategy of HCV propagation may provide new clues to the virus-host interaction and, ultimately, to the pathogenesis of infection by various genotypes of HCV.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Murase and M. Tomiyama for their secretarial work. We also thank R. Bartenschlager and T. Wakita for providing cell lines and plasmids.
This work was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare (Research on Hepatitis), the Ministry of Education, Culture, Sports, Science, and Technology, and the Osaka University Global Center of Excellence Program.
Footnotes
Published ahead of print on 12 October 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Auffermann-Gretzinger S., Keeffe E. B., Levy S. 2001. Impaired dendritic cell maturation in patients with chronic, but not resolved, hepatitis C virus infection. Blood 97:3171–3176 [DOI] [PubMed] [Google Scholar]
- 2. Beyenbach K. W., Wieczorek H. 2006. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209:577–589 [DOI] [PubMed] [Google Scholar]
- 3. Bradley D. W. 2000. Studies of non-A, non-B hepatitis and characterization of the hepatitis C virus in chimpanzees. Curr. Top. Microbiol. Immunol. 242:1–23 [DOI] [PubMed] [Google Scholar]
- 4. Clarke P. G. 1990. Developmental cell death: morphological diversity and multiple mechanisms. Anat. Embryol. (Berl.) 181:195–213 [DOI] [PubMed] [Google Scholar]
- 5. Dreux M., Gastaminza P., Wieland S. F., Chisari F. V. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. English L., et al. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10:480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fujita N., et al. 2008. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19:4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujita N., et al. 2008. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19:2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujitani Y., Ebato C., Uchida T., Kawamori R., Watada H. 2009. β-cell autophagy: a novel mechanism regulating β-cell function and mass: lessons from β-cell-specific Atg7-deficient mice. Islets 1:151–153 [DOI] [PubMed] [Google Scholar]
- 10. Gannage M., et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gannage M., Munz C. 2009. Autophagy in MHC class II presentation of endogenous antigens. Curr. Top. Microbiol. Immunol. 335:123–140 [DOI] [PubMed] [Google Scholar]
- 12. Hara T., et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889 [DOI] [PubMed] [Google Scholar]
- 13. Hasilik A. 1992. The early and late processing of lysosomal enzymes: proteolysis and compartmentation. Experientia 48:130–151 [DOI] [PubMed] [Google Scholar]
- 14. Hatzakis A., et al. 1996. Hepatitis C virus 1b is the dominant genotype in HCV-related carcinogenesis: a case-control study. Int. J. Cancer 68:51–53 [DOI] [PubMed] [Google Scholar]
- 15. Hirabayashi M., et al. 2001. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 8:977–984 [DOI] [PubMed] [Google Scholar]
- 16. Hiraga N., et al. 2011. Rapid emergence of telaprevir resistant hepatitis C virus strain from wildtype clone in vivo. Hepatology (Baltimore, Md.) 54:781–788 [DOI] [PubMed] [Google Scholar]
- 17. Hiraga N., et al. 2007. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis C virus and its susceptibility to interferon. FEBS Lett. 581:1983–1987 [DOI] [PubMed] [Google Scholar]
- 18. Ichimura Y., Kominami E., Tanaka K., Komatsu M. 2008. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 4:1063–1066 [DOI] [PubMed] [Google Scholar]
- 19. Isidoro C., et al. 1995. Altered intracellular processing and enhanced secretion of procathepsin D in a highly deviated rat hepatoma. Int. J. Cancer 60:61–64 [DOI] [PubMed] [Google Scholar]
- 20. Jacobson I. M., Cacoub P., Dal Maso L., Harrison S. A., Younossi Z. M. 2010. Manifestations of chronic hepatitis C virus infection beyond the liver. Clin. Gastroenterol. Hepatol. 8:1017–1029 [DOI] [PubMed] [Google Scholar]
- 21. Jounai N., et al. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 104:14050–14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanto T., et al. 1999. Impaired allostimulatory capacity of peripheral blood dendritic cells recovered from hepatitis C virus-infected individuals. J. Immunol. 162:5584–5591 [PubMed] [Google Scholar]
- 23. Ke P. Y., Chen S. S. 2011. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Invest. 121:37–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kimura S., Fujita N., Noda T., Yoshimori T. 2009. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 452:1–12 [DOI] [PubMed] [Google Scholar]
- 25. Kiyosawa K., et al. 1990. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology 12:671–675 [DOI] [PubMed] [Google Scholar]
- 26. Koblinski J. E., et al. 2002. Interaction of human breast fibroblasts with collagen I increases secretion of procathepsin B. J. Biol. Chem. 277:32220–32227 [DOI] [PubMed] [Google Scholar]
- 27. Koike K., et al. 1997. Sialadenitis histologically resembling Sjogren syndrome in mice transgenic for hepatitis C virus envelope genes. Proc. Natl. Acad. Sci. U. S. A. 94:233–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koike K., Tsutsumi T., Yotsuyanagi H., Moriya K. 2010. Lipid metabolism and liver disease in hepatitis C viral infection. Oncology 78(Suppl. 1):24–30 [DOI] [PubMed] [Google Scholar]
- 29. Komatsu M., et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884 [DOI] [PubMed] [Google Scholar]
- 30. Komatsu M., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163 [DOI] [PubMed] [Google Scholar]
- 31. Lee J. H., et al. 2010. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141:1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 34. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 35. Manns M. P., et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965 [DOI] [PubMed] [Google Scholar]
- 36. McHutchison J. G., et al. 2009. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 360:1827–1838 [DOI] [PubMed] [Google Scholar]
- 37. Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873 [DOI] [PubMed] [Google Scholar]
- 38. Moradpour D., Penin F., Rice C. M. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 39. Moriishi K., Matsuura Y. 2007. Host factors involved in the replication of hepatitis C virus. Rev. Med. Virol. 17:343–354 [DOI] [PubMed] [Google Scholar]
- 40. Moriishi K., Matsuura Y. 2003. Mechanisms of hepatitis C virus infection. Antivir. Chem. Chemother. 14:285–297 [DOI] [PubMed] [Google Scholar]
- 41. Oda K., Nishimura Y., Ikehara Y., Kato K. 1991. Bafilomycin A1 inhibits the targeting of lysosomal acid hydrolases in cultured hepatocytes. Biochem. Biophys. Res. Commun. 178:369–377 [DOI] [PubMed] [Google Scholar]
- 42. Orvedahl A., et al. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–35 [DOI] [PubMed] [Google Scholar]
- 43. Poordad F., et al. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pua H. H., Dzhagalov I., Chuck M., Mizushima N., He Y. W. 2007. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204:25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramachandran N., et al. 2009. VMA21 deficiency causes an autophagic myopathy by compromising V-ATPase activity and lysosomal acidification. Cell 137:235–246 [DOI] [PubMed] [Google Scholar]
- 46. Saito K., et al. 2008. Hepatitis C virus inhibits cell surface expression of HLA-DR, prevents dendritic cell maturation, and induces interleukin-10 production. J. Virol. 82:3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmid D., Munz C. 2007. Innate and adaptive immunity through autophagy. Immunity 27:11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schutte K., Bornschein J., Malfertheiner P. 2009. Hepatocellular carcinoma—epidemiological trends and risk factors. Dig. Dis. 27:80–92 [DOI] [PubMed] [Google Scholar]
- 49. Sir D., et al. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sloane B. F., et al. 2005. Cathepsin B and tumor proteolysis: contribution of the tumor microenvironment. Semin. Cancer Biol. 15:149–157 [DOI] [PubMed] [Google Scholar]
- 51. Stankovic-Djordjevic D., et al. 2007. Hepatitis C virus genotypes and the development of hepatocellular carcinoma. J. Dig. Dis. 8:42–47 [DOI] [PubMed] [Google Scholar]
- 52. Strader D. B., Wright T., Thomas D. L., Seeff L. B. 2004. Diagnosis, management, and treatment of hepatitis C. Hepatology 39:1147–1171 [DOI] [PubMed] [Google Scholar]
- 53. Taguwa S., et al. 2009. Cochaperone activity of human butyrate-induced transcript 1 facilitates hepatitis C virus replication through an Hsp90-dependent pathway. J. Virol. 83:10427–10436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tal M. C., et al. 2009. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U. S. A. 106:2770–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tardif K. D., Siddiqui A. 2003. Cell surface expression of major histocompatibility complex class I molecules is reduced in hepatitis C virus subgenomic replicon-expressing cells. J. Virol. 77:11644–11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Walker C. M. 1997. Comparative features of hepatitis C virus infection in humans and chimpanzees. Springer Semin. Immunopathol. 19:85–98 [DOI] [PubMed] [Google Scholar]
- 58. Wasley A., Alter M. J. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1–16 [DOI] [PubMed] [Google Scholar]
- 59. Wong J., et al. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yoshimori T., Noda T. 2008. Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 20:401–407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.