Abstract
Until recently, influenza A viruses from wild waterfowl in South America were rarely isolated and/or characterized. To explore the ecology of influenza A viruses in this region, a long-term surveillance program was established in 2006 for resident and migratory water birds in Argentina. We report the characterization of 5 avian influenza viruses of the H6 hemagglutinin (HA) subtype isolated from rosy-billed pochards (Netta peposaca). Three of these viruses were paired to an N2 NA subtype, while the other two were of the N8 subtype. Genetic and phylogenetic analyses of the internal gene segments revealed a close relationship with influenza viruses from South America, forming a unique clade and supporting the notion of independent evolution from influenza A viruses in other latitudes. The presence of NS alleles A and B was also identified. The HA and NA genes formed unique clades separate from North American and Eurasian viruses, with the exception of the HA gene of one isolate, which was more closely related to the North American lineage, suggesting possible interactions between viruses of North American and South American lineages. Animal studies suggested that these Argentine H6 viruses could replicate and transmit inefficiently in chickens, indicating limited adaptation to poultry. Our results highlight the importance of continued influenza virus surveillance in wild birds of South America, especially considering the unique evolution of these viruses.
INTRODUCTION
Extensive epidemiological studies have revealed that influenza viruses can infect a wide range of hosts, including many types of birds, humans, pigs, horses, dogs, cats, and other mammals (16). Of these species, aquatic birds are considered the natural reservoir of avian influenza A viruses (AIVs), as all 16 known hemagglutinin (HA) subtypes (H1 to H16) and all 9 known neuraminidase (NA) subtypes (N1 to N9) have been recognized in these hosts (7). Influenza viruses have been isolated from different parts of the world. Phylogenetic analyses have revealed that influenza viruses in Eurasia and Australia are genetically distinct from those in North America, which is consistent with the notion of the confinement of birds to distinct flyways in each hemisphere (16).
In contrast to the extensive animal influenza virus surveillance activities carried out in Southern China, East Asia, Northern Europe, Australia, and North America (23), such efforts have been limited in South America. The first reported outbreak of avian influenza virus in South America occurred in commercial chickens and turkeys in Chile in 2002. This outbreak was caused by an H7N3 virus (21) that was later demonstrated to be closely related to an AIV from a wild duck from Bolivia [A/Cinnamon teal/Bolivia/4537/01 (H7N3)] (18, 19).
Since 2006, in Argentina, a partnership between the National Institute of Agricultural Technology (INTA), the Argentinean National Animal Health Service (SENASA), the Wildlife Conservation Society (WCS), and the University of Maryland, College Park, in the United States has implemented a long-term AIV surveillance program. AIV surveillance is carried out for resident and migratory water birds, seabirds, and shorebirds along the Argentine Atlantic coast and in freshwater wetlands formed by two of the major river systems in South America, the Paraná and Uruguay rivers. Surveillance efforts are aimed at exploring the ecology of AIVs in wild aquatic birds and to generate epidemiological data for an early warning of the introduction of AIV into commercial poultry. A/kelp gull/Argentina/LDC4/06 (H13N9) was isolated from these initial surveillance studies (17). As the surveillance efforts continued, additional AIVs were isolated from different species of wild aquatic birds (Table 1). In the present study, we focus on the characterization and phylogenetic analysis of H6 AIVs isolated from rosy-billed pochards (Netta peposaca) in Argentine wetlands because they were the subtype most consistently isolated. We investigated the replication and transmission of these H6 Argentine AIVs in chickens and compared these strains to prototypic H6 strains from North America and Eurasia. Our studies suggest the existence of an H6 South American influenza virus lineage in which viruses do not appear to be mixing with and are evolving independently from influenza viruses in other latitudes. Interestingly, one of the HA genes appears to have been derived from a virus of the North American lineage, indicating the presence of more than one H6 subtype cluster. Our studies also suggest a limited adaptation of these viruses to poultry and that additional molecular changes would be required for these viruses to replicate and transmit efficiently in chickens. Our study highlights the importance of the continuous surveillance of AIVs in South America as an integral part of worldwide efforts to better understand the ecology of these viruses and to prevent the emergence of novel, potentially pandemic strains.
Table 1.
Bird species, numbers of samples, and numbers of isolates obtained between 2007 and 2010
| Species | Common name | No. of samples | No. of RRT-PCR-positive samples | Isolate subtype(s) (no. of isolates)a |
|---|---|---|---|---|
| Amazonetta brasiliensis | Brazilian teal | 144 | 1 | |
| Anas cyanoptera | Cinnamon teal | 19 | 1 | H7N9 (1) |
| Anas platalea | Red shoveler | 279 | 2 | |
| Anas versicolor | Silver teal | 479 | 10 | H5N3 (1) |
| Callonetta leucophrys | Ringed teal | 157 | ||
| Dendrocygna bicolor | Fulvous whistling duck | 258 | ||
| Dendrocyna viduata | White-faced whistling duck | 218 | ||
| Larus dominicanus | Kelp gull | 104 | ||
| Netta peposaca | Rosy-billed pochard | 1,679 | 14 | H6N2 (3), H6N8 (2), H9N2 (1) |
| Phalacrocorax atriceps | Imperial shag | 207 | ||
| Spheniscus magellanicus | Magellanic penguin | 457 | ||
| Sterna hirundinacea | South American tern | 139 | ||
| Others | 364 | |||
| Total | 4,504 | 28 |
The total number of isolates was 8.
MATERIALS AND METHODS
Sample collection.
Sampling activities were performed by trained biologists and veterinarians. Cloacal swabs were collected from carcasses of hunter-killed ducks donated by licensed hunters during the hunting seasons of 2007 and 2010. Hunter-killed ducks were processed within 2 to 4 h postmortem. Cloacal swabs were collected by using single-use polyester sterile swabs and then stored separately in single plastic cryovials containing 2 ml of phosphate buffer solution (PBS) with 50% glycerol and 10,000 IU/ml penicillin, 5 mg/ml streptomycin, 1 mg/ml gentamicin sulfate, 700 μg/ml kanamycin sulfate, and 10 μg/ml amphotericin B (Sigma Chemical Co., St. Louis, MO). Samples were frozen in liquid nitrogen and transported on dry ice. Once in the laboratory, all samples were stored at −80°C until they were processed for molecular diagnosis and virus isolation.
Virus detection.
Viral RNA was extracted from 140 μl of a PBS suspension from cloacal swabs by use of a QIAamp viral RNA minikit (Qiagen Inc., Valencia, CA) in accordance with the manufacturer's instructions. RNA was eluted in a final volume of 60 μl and stored at −80°C. Viral cDNA was prepared by using 30 μl of viral RNA and random hexamers in a final volume of 60 μl by use of a High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). The cDNA was tested for avian influenza virus (AIV) by real-time reverse transcription-PCR (RRT-PCR) using TaqMan Universal PCR master mix (Applied Biosystems) directed to the matrix (M) gene. This system detects all type A influenza viruses (20). PCR was performed with an ABI Prism 7500 SDS system (Applied Biosystems).
Virus isolation.
All swab samples determined to be positive by RRT-PCR were inoculated into 9- to 11-day-old specific-pathogen-free (SPF) embryonated chicken eggs. Briefly, 200 μl of PBS suspension from the cloacal swab samples was injected into the allantoic cavity of the eggs, and the eggs were incubated for 72 h and harvested in accordance with standardized protocols described in the WHO Manual on Animal Influenza Diagnosis and Surveillance (25) and according to Argentine regulations.
Phylogenetic and molecular analysis.
Viral RNA was extracted from infected allantoic fluid by using an RNeasy minikit (Qiagen). Reverse transcription followed by PCR was performed by using specific primers for each gene segment as described previously (9). PCR products were purified with a QIAquick PCR purification kit (Qiagen). Sequencing was performed by using the BigDye Terminator v3.1 cycle sequencing kit on an ABI Prism 3700 DNA analyzer (Applied Biosystems) according to the manufacturer's instructions. The consensus amino acid and nucleotide sequences for all 8 gene segments of the viruses were generated by using Megalign (DNASTAR, Madison, WI). Phylogenetic analyses were performed by using additional influenza virus sequence data available in GenBank. Sequences were assembled and edited with Lasergene 8.1 (DNASTAR); BioEdit 7 was used for alignments and residue analyses. Neighbor-joining (NJ) trees were constructed by using PAUP* 4.0. Estimates of phylogenies were calculated by performing 1,000 NJ bootstrap replicates. The TREEVIEW program, version 1.6.6, was used for the visualization and printing of phylogenetic trees.
Animal studies.
Two-week-old SPF White Leghorn chickens (Charles River Laboratories, Wilmington, MA) were used throughout the studies. Groups of 3 chickens housed in HEPA-filtered isolator cages were inoculated intraocularly, intranasally, and intratracheally with 1 ml of virus containing 106 50% tissue culture infective doses (TCID50s). Virus transmission was monitored by introducing 3 direct-contact chickens into each group at 1 day postinoculation (dpi). Tracheal and cloacal swabs were collected at 1, 3, 5, and 7 dpi in 1 ml freezing medium (50% glycerol in PBS containing 1% antibiotics) and stored at −80°C until use for the titration of virus in MDCK cells. The inoculated chickens were boosted with the same amount of the corresponding viruses at 14 dpi, and serum samples were collected at 28 dpi. Chickens were observed daily for 28 days for signs of disease. Birds were monitored for appetite, activity, fecal output, and signs of distress, including cyanosis of the tongue or legs, ruffled feathers, and respiratory distress. Experiments were carried out under animal biosafety level 2+ (ABSL2+) conditions with investigators wearing appropriate protective equipment and compliant with all University of Maryland Institutional Animal Care and Use Committee (IACUC)-approved protocols and under Animal Welfare Act (AWA) regulations. Animal experiments with strains A/mallard/Alberta/206/1996 (H6N8) (206/H6N8), A/teal/Hong Kong/W312/97 (H6N1) (W312/H6N1), A/rosy-billed pochard/Argentina/CIP051-557/2007 (H6N2) (557/H6N2), A/rosy-billed pochard/Argentina/CIP051-575/2007 (H6N8) (575/H6N8), and A/rosy-billed pochard/Argentina/CIP051-925/2008 (H6N2) (925/H6N2) were performed twice by different laboratory personnel.
HI assay.
Serum samples were collected to determine the antigenic relatedness between these H6 viruses at the end of the experiment. Sera were treated with receptor-destroying enzyme (Accurate Chemical and Scientific Corp., Westbury, NY) to remove sialic acid receptors. The antiviral antibody titers were evaluated by using a hemagglutination inhibition (HI) assay system outlined in the WHO Manual on Animal Influenza Diagnosis and Surveillance (25). HI assays were performed by using homologous and heterologous viruses. The serum with highest HI titer was used to perform HI assays for antigenic analysis.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study are available in the GenBank database under accession numbers CY067691 to CY067714 and CY096050 to CY096065.
RESULTS
Geographic location of sampling site.
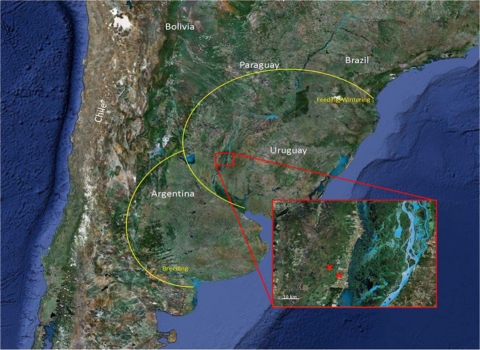
Sampling activities were performed in the Lower Paraná River Valley, along the Paraná River, one of the major river systems in South America. The valley is composed of a mosaic of rice fields, natural wetlands and marshes, native forests, and patches of land within the floodplain of the Parana River (30°41′S, 60°02′W). The Paraná River is also within an important migratory bird flyway and serves as a reservoir for multiple populations of resident waterfowl species (Fig. 1). In contrast to the Northern Hemisphere, South American duck populations have a more homogeneous distribution of the number of birds throughout the year, as winter migrations are not predominant. From May to August, the wetlands are used for game bird hunting focusing mainly on ducks, tinamous, and wild pigeons. Samples were collected between January 2007 and July 2010 at different hunting lodges in the Lower Paraná River Valley in Argentina. A total of 4,504 cloacal swabs representing 39 different bird species were collected and tested for the presence of AIV (Table 1). Twenty-eight of these samples from five different bird species were positive for AIV by RRT-PCR (0.62%). Fourteen positive samples were obtained from Netta peposaca (rosy-billed pochard), 10 were obtained from Anas versicolor (silver teal), two were obtained from Anas platalea (red shoveler), 1 was obtained from Amazoneta brasiliensis (Brazilian teal), and 1 was obtained from Anas cyanoptera (cinnamon teal). Viable AIVs were isolated from only eight RRT-PCR-positive samples, and six out from these eight viruses were from N. peposaca. In winter, N. peposaca birds migrate to the Rio Grande Do Sul state in Brazil and then return to Argentina in the summer during the breeding season. This pattern is also shared with a variety of other duck species, including Anas georgica (yellow-billed pintail) and Anas versicolor (silver teal).
Fig. 1.
Geographical location of the area under surveillance. Crosses indicate the exact sampling locations. The area surrounded in yellow indicates the breeding and the feeding/wintering areas for N. peposaca. (Reprinted from Google Images [copyright 2011 NASA, TerraMetrics].)
Since isolates belonging to the H6 subtype were those most frequently identified, they were subjected to a more detailed characterization. Of the 5 H6 isolates, 3 were of the H6N2 subtype, A/rosy-billed pochard/Argentina/CIP051-557/2007 (H6N2), A/rosy-billed pochard/Argentina/CIP051-925/2008 (H6N2), and A/rosy-billed pochard/Argentina/CIP051-1977/2010 (H6N2) (herein referred to as 557/ H6N2, 925/H6N2, and 1977/H6N2, respectively). The other 2 isolates were of the H6N8 subtype, A/rosy-billed pochard/Argentina/CIP051-575/2007 (H6N8) and A/rosy-billed pochard/Argentina/CIP051-269/2007 (H6N8) (herein referred to as 575/H6N8 and 269/H6N8, respectively) (Table 2).
Table 2.
Viruses in GenBank with the highest sequence similarity to the H6 Argentine AIVsa
| Virus | PB2 |
PB1 |
PA |
HA |
NP |
NA |
M |
NS |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain with highest nucleotide similarity | % similarity | Strain with highest nucleotide similarity | % similarity | Strain with highest nucleotide similarity | % similarity | Strain with highest nucleotide similarity | % similarity | Strain with highest nucleotide similarity | % similarity | Strain with highest nucleotide similarity | % similarity | Strain with highest nucleotide similarity | % similarity | Strain with highest nucleotide similarity | % similarity | |
| 557/H6N2 | rwt/Arg/08 H1N1 | 96 | rwt/Arg/08 H1N1 | 94 | rwt/Arg/08 H1N1 | 97 | mal/Alb/96 H6N8 | 91 | blwt/Alb/78 H6N2 | 94 | mtldk/LA/87 H6N2 | 88 | rwt/Arg/08 H1N1 | 98 | rwt/Arg/08 H1N1 | 99 |
| 575/H6N8 | rwt/Arg/08 H1N1 | 96 | rwt/Arg/08 H1N1 | 94 | rwt/Arg/08 H1N1 | 96 | mal/Alb/96 H6N8 | 91 | ct/Bolivia/01 H7N3 | 96 | blwt/Alb/91 H3N8 | 92 | rwt/Arg/08 H1N1 | 98 | rwt/Arg/08 H1N1 | 98 |
| 925/H6N2 | rwt/Arg/08 H1N1 | 96 | rwt/Arg/08 H1N1 | 94 | rwt/Arg/08 H1N1 | 97 | mal/Alb/96 H6N8 | 91 | ct/Bolivia/01 H7N3 | 96 | mtldk/LA/87 H6N2 | 88 | rwt/Arg/08 H1N1 | 98 | klpg/Arg/06 H13N9 | 96 |
| 269/H6N8 | rwt/Arg/08 H1N1 | 96 | rwt/Arg/08 H1N1 | 94 | rwt/Arg/08 H1N1 | 98 | mal/Alb/96 H6N8 | 91 | ct/Bolivia/01 H7N3 | 96 | blwt/Alb/91 H3N8 | 92 | rwt/Arg/08 H1N1 | 98 | ty/Chile/02(H7N3) | 97 |
| 1977/H6N2 | rwt/Arg/08 H1N1 | 95 | rwt/Arg/08 H1N1 | 94 | rwt/Arg/08 H1N1 | 98 | sb/DE/2004 H6N8 | 97 | ct/Bolivia/01 H7N3 | 96 | mtldk/LA/87 H6N2 | 88 | rwt/Arg/08 H1N1 | 98 | klpg/Arg/06 H13N9 | 96 |
rwt/Arg/08 H1N1, A/red-winged tinamou/Argentina/MP1/2008 (H1N1); mal/Alb/96 H6N8, A/Mallard/Alberta/206/96 (H6N8); sb/DE/2004 H6N8, A/shorebird/Delaware/12/2004 (H6N8); blwt/Alb/78 H6N2, A/blue-winged teal/Alberta/651/1978 (H6N2); ct/Bolivia/01 H7N3, A/cinnamon teal/Bolivia/4537/2001 (H7N3); mtldk/LA/87 H6N2, A/mottled duck/LA/32 M/1987 (H6N2); blwt/Alb/91 H3N8, A/blue-winged teal/Alberta/120/1991 (H3N8); klpg/Arg/06 H13N9, A/kelp gull/Argentina/LDC4/2006 (H13N9); ty/Chile/02(H7N3), A/turkey/Chile/4418/2002 (H7N3).
Genetic and phylogenetic analysis.
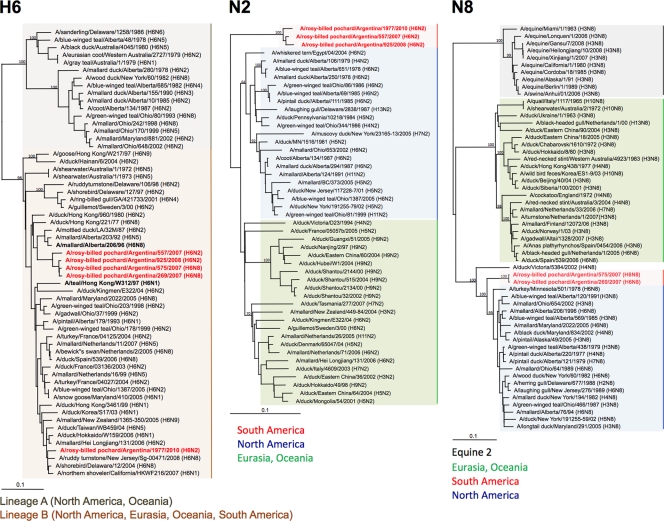
To better understand the evolutionary relationship of the Argentine H6 subtype influenza viruses with other avian influenza viruses, we performed full-length sequencing of the open reading frames followed by phylogenetic analysis. The internal genes of the Argentine H6 viruses were closely related to avian influenza viruses from South America, e.g., Argentina, Bolivia, and Chile (Table 2). Due to the lack of sequence information on H6 HA genes from South America, their closest relative is A/mallard/Alberta/206/1996 (H6N8) from the North American lineage, with only 91% nucleotide identity, except for the 1977/H6N2 isolate, which shares 97% homology with A/shorebird/DE/12/2004 (H6N8) (Table 2). Phylogenetic analyses of the HAs of H6 subtype viruses revealed two major distinct lineages (4a). Lineage A contains H6 viruses almost exclusively from North America and Oceania, whereas lineage B contains H6 viruses from Eurasia, North America, and Oceania and the 5 strains from Argentina (Fig. 2). Four of the H6 HA genes from the Argentine influenza viruses clustered together and formed a unique clade in lineage B, suggesting an independent evolution of the HA H6 genes of these viruses with respect to other influenza viruses. In contrast, 1977/H6N2 grouped with other viruses from North America, suggesting the presence of two independent H6 HA populations in South America and/or the potential for the exchange of gene segments from distinct lineages. Likewise, due to the lack of N2 and N8 NA genes from South America, their closest relatives shared only 88% and 92% sequence identities with viruses from Eurasia, A/whiskered tern/Egypt/04/2004 (H6N2), and North America, A/blue-winged teal/Alberta/120/1991 (H3N8), respectively. The N2 and N8 genes of the Argentine viruses also formed unique clades compared to the NA genes of other avian influenza viruses from other continents (Fig. 2).
Fig. 2.
Phylogenetic trees of H6 HA and N2 and N8 NA genes. Trees were generated by the neighbor-joining method with the PAUP* program. Numbers above branches indicate neighbor-joining bootstrap values. Not all supports are shown because of space constraints. Analysis was based on nucleotides 22 to 1036 (HA), 249 to 1303 (N2), and 41 to 1341 (N8). The H6 tree was rooted to A/mallard/Potsdam/178-4/83 (H2N2). The N2 tree was rooted to A/equine/Prague/1/1956 (H7N7). The N8 tree was rooted to A/ruddy turnstone/Delaware/97/2000 (H12N5). Viruses characterized in this study are highlighted in red. Scale bar, 0.1 substitutions per site.
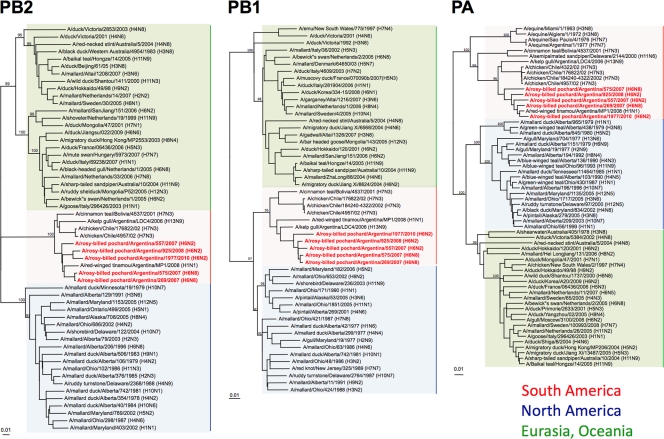
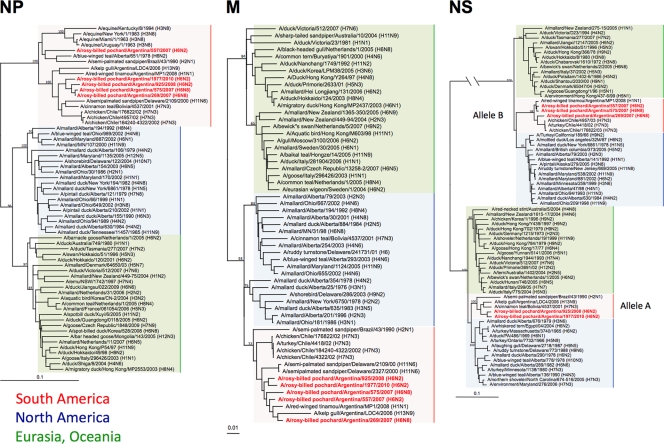
Consistent with previously reported observations, phylogenetic analyses of the internal genes (PB2, PB1, PA, NP, M, and NS) suggested that the H6 Argentine viruses followed an evolutionary pathway along with other influenza viruses in South America that was distinct from that of other avian influenza viruses (Fig. 3 and 4). In the phylogenetic trees of PB2, PB1, PA, and M, the 5 H6 viruses clustered together into the South American clade and were most closely related to A/red-winged tinamou/Argentina/MP1/2008 (H1N1) (Fig. 3 and 4). A phylogenetic analysis of the NP genes revealed that they are closely related to viruses from South America, although there was a more appreciable divergence than that for the other internal genes (Fig. 4). The NP gene of the 557/H6N2 virus was more closely related to A/semipalmated sandpiper/Brazil/43/1990 (H2N1) and, interestingly, belonged to a phylogenetic cluster that contains equine influenza viruses as well as an old (H6N2) isolate from a blue-winged teal from Alberta, Canada. These observations make it tempting to speculate on the potential for a South American origin of the so-called equine 2 influenza viruses, like A/equine/Uruguay/1/1963 (H3N8), for at least the NP gene. The NP genes of the other 4 viruses were grouped among typical avian influenza viruses from South America. Phylogenetic relationships among the PA genes revealed some similarities to the NP trees, in a clade more closely related to equine influenza viruses (Fig. 3 and 4). Phylogenetic analyses of the NS genes showed that these Argentine H6 viruses were more diversified than the other internal genes (Fig. 4). They belonged to alleles A (925/H6N2 and 1977/H6N2) and B (557/H6N2, 575/H6N8, and 269/H6N8). In each allele, the NS genes of the Argentine H6 viruses were most closely related to AIVs from South America, forming a unique clade.
Fig. 3.
Phylogenetic trees of PB2, PB1, and PA genes. Trees were generated as described in the legend of Fig. 2 and in Materials and Methods. Analysis was based on nucleotides 1079 to 2138 (PB2), 42 to 1217 (PB1), and 251 to 2049 (PA). The PB2 tree was rooted to A/equine/Prague/1/1956 (H7N7), and the PB1 and PA trees were rooted to A/Brevig Mission/1/1918 (H1N1). Viruses characterized in this study are highlighted in red. Scale bar, 0.1 substitutions per site (0.01 substitutions for PA).
Fig. 4.
Phylogenetic trees of NP, M, and NS genes. Trees were generated as described in the legend of Fig. 2 and in Materials and Methods. Analysis was based on nucleotides 31 to 917 (NP), 49 to 864 (M), and 88 to 815 (NS). The NP, M, and NS trees were rooted to A/equine/Prague/1/1956 (H7N7). Viruses characterized in this study are highlighted in red. Scale bar, 0.1 substitutions per site.
Compared with consensus sequences from North American and Eurasian AIVs from wild birds, the amino acid sequence homology between the Argentine H6 viruses and other AIVs was conserved across all gene segments. The amino acid identity of most of the gene segments was more than 98% compared to the North American or Eurasian consensus strains (Table 3). These results suggest that although there is an independent evolution of South American AIVs, they share significant amino acid identity, consistent with evolutionary stasis in the natural reservoir (25a).
Table 3.
Homology of Argentine H6 AIVs with consensus amino acid sequences of North American and Eurasian AIVs
| Strain | % sequence consensusa |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2 |
PB1 |
PA |
NP |
HA |
NA |
M1 |
NS1 |
|||||||||
| NA | EA | NA | EA | NA | EA | NA | EA | NA-A | EA | NA | EA | NA | EA | NA | EA | |
| 557/H6N2 | 98.3 | 98.2 | 99.2 | 99.2 | 97.9 | 97.9 | 98.4 | 98.0 | 97.5 | 97.3 | 94.0 | 93.6 | 100 | 100 | 98.7 | 99.6 |
| 575/H6N8 | 98.6 | 98.4 | 99.5 | 99.5 | 98.2 | 98.0 | 98.6 | 98.4 | 97.0 | 96.8 | 97.4 | 87.2 | 100 | 100 | 97.8 | 98.7 |
| 925/H6N2 | 98.4 | 98.3 | 99.3 | 99.3 | 98 | 98.2 | 98.6 | 98.4 | 97.0 | 96.8 | 93.8 | 93.4 | 100 | 100 | 97.4 | 97.4 |
| 269/H6N8 | 98.8 | 98.7 | 99.3 | 99.3 | 98.0 | 98.0 | 98.4 | 98.2 | 97.2 | 97.0 | 93 | 87.0 | 100 | 100 | 97.0 | 97.8 |
| 1977/H6N2 | 98.4 | 98.3 | 99.2 | 99.2 | 98.2 | 98.2 | 98.6 | 98.4 | 98.1 | 97.9 | 97 | 92.8 | 100 | 100 | 97.0 | 97.0 |
Consensus in amino acids. NA, North American lineage; EA, Eurasian lineage; NA-A, North American lineage, clade A.
An analysis of deduced amino acids revealed that these H6 viruses were typical low-pathogenic avian influenza (LPAI) viruses. The 5 Argentine H6 viruses have a PQIETRG motif at the HA cleavage site and retain avian-like amino acids at the receptor binding site (226Q and 228G). No mutations associated with mammalian adaptation were found, e.g., PB2-627K or PB2-701N. Also, no M2-31N mutations, responsible for amantadine resistance, were found (data not shown).
Virus replication and transmission in chickens.
Since H6 viruses have established stable lineages in terrestrial poultry in Eurasia (5, 6, 10–12, 14) and North America (24), we wanted to determine whether the Argentine H6 viruses represent a risk to local poultry. Thus, the replication and transmission of the H6 Argentine viruses were investigated in SPF White Leghorn chickens (n = 3/virus + n = 3 direct contacts) (Table 4). Two prototypic H6 viruses, one from Eurasia, A/teal/Hong Kong/W312/97 (H6N1), and one from North America A/mallard/Alberta/206/96 (H6N8), were included for comparison (herein referred to as W312/H6N1 and 206/H6N8), respectively. The 5 Argentine viruses showed very limited replication and transmission in chickens. Tracheal swabs showed the presence of virus above the limit of detection in one chicken inoculated with the 575/H6N8 strain (1.7 log10 50% egg infective doses [EID50]/ml at 7 dpi). In contrast, prototypic strain W312/H6N1, which is commonly found in live-bird poultry markets in Southeast Asia, showed more consistent respiratory virus replication in the inoculated group (5 out 6 chickens were positive). Tracheal swabs for the direct-contact groups were below the limit of detection irrespective of the strain used. Cloacal swabs of inoculated chickens showed discernible virus titers only for one chicken inoculated with W312/H6N1 (1.7 log10 EID50/ml at 7 dpi). The results with the W312/H6N1 strain are consistent with data from previous reports (11, 12). The prototypic North American strain, 206/H6N8, did not show signs of replication in inoculated chickens, although it showed the highest level of seroconversion, as measured by HI. In fact, directly inoculated chickens showed clear seroconversion regardless of the strain used, indicating that chickens had been infected. Transmission to direct-contact chickens resulted in no detectable virus in either tracheal or cloacal swabs, except for one direct-contact chicken in the 557/H6N2 group (3.2 log10 EID50/ml at 5 dpi). However, evidence of partial transmission was observed through seroconversion. Three out of six direct-contact chickens showed seroconversion in the W312/H6N1 group, whereas 2 out of 6 showed discernible HI titers in the 557/H6N2 and the 575/H6N8 groups.
Table 4.
Replication, transmission, and seroconversion of chickens infected with representative H6 AIVs
| Virus | No. of positive chickens/total no. of chickens (peak virus titer [log10 EID50/ml])a |
Mean HI titer (log2 serum dilution) (no. of positive chickens/total no. of chickens) |
||||
|---|---|---|---|---|---|---|
| Inoculated |
Direct contact |
|||||
| T | C | T | C | Inoculated | Contact | |
| 206/H6N8 | 0/6 | 0/6 | 0/6 | 0/6 | 475 (6/6) | 0/6 |
| W312/H6N1 | 5/6 (3.7)b | 1/6 (1.7)c | 0/6 | 0/6 | 217 (6/6) | 17 (3/6) |
| 557/H6N2 | 0/6 | 0/6 | 0/6 | 1/6 (3.2)d | 189 (6/6) | 40 (2/6) |
| 575/H6N8 | 1/6 (1.7)e | 0/6 | 0/6 | 0/6 | 133 (6/6) | 15 (2/6) |
| 925/H6N2 | 0/6 | 0/6 | 0/6 | 0/6 | 384 (6/6) | (0/6) |
| 269/H6N8 | 0/3 | 0/3 | 0/3 | 0/3 | 133 (3/3) | (0/3) |
| 1977/H6N2 | 0/3 | 0/3 | 0/3 | 0/3 | 73 (3/3) | (0/3) |
T, trachea; C, cloaca.
Virus shedding for 5 days and peak virus titer at 1 dpi.
Titer of 1.7 log10 EID50/ml at 7 dpi.
Titer of 3.2 log10 EID50/ml at 5 dpi.
Titer of 1.7 log10 EID50/ml at 7 dpi.
Antigenic profile of Argentine H6 AIVs.
The antigenic properties of the Argentine H6 viruses were investigated by using chicken antisera generated from the replication studies mentioned above. Antisera against the W312/H6N1 virus showed significant HI titers against the homologous viruses but significantly lower titers against the Argentine viruses (≥4-fold). Likewise, antisera against the other H6 strains showed low-level reactions against the W312/H6N1 virus (Table 5). The HI pattern followed the phylogenetic profile, with strains 557/H6N2, 575/H6N8, 925/H6N2, and 269/H6N8 being closer antigenically to each other than to the 1977/H6N2 strain or the 206/H6N8 strain.
Table 5.
Antigenic profiles of representative H6 AIVs
| Virus | HI titer (log2 serum dilution) for sera againsta: |
||||||
|---|---|---|---|---|---|---|---|
| 206/H6N8 | W312/H6N1 | 557/H6N2 | 575/H6N8 | 925/H6N2 | 269/H6N8 | 1977/H6N2 | |
| 206/H6N8 | 320 | 480 | 320 | 960 | 480 | 240 | 120 |
| W312/H6N1 | 160 | 3,840 | 160 | 320 | 160 | 40 | 40 |
| 557/H6N2 | 160 | 320 | 640 | 640 | 1,280 | 80 | 160 |
| 575/H6N8 | 240 | 960 | 480 | 1,280 | 1,280 | 160 | 160 |
| 925/H6N2 | 160 | 1,280 | 640 | 1,280 | 1,280 | 160 | 160 |
| 269/H6N8 | 160 | 640 | 320 | 640 | 960 | 320 | 120 |
| 1977/H6N2 | 120 | 640 | 640 | 320 | 640 | 240 | 160 |
HI titers for homologous viruses are in boldface type.
DISCUSSION
Improved AIV surveillance efforts in wild water birds in Argentina resulted in the isolation of strains with unique evolutionary pathways (4, 17). In this study, we report the first H6 subtype AIV isolated from 5 N. peposaca ducks from South America.
The present study further supports the notion of a unique South American lineage with probable independent evolution for all viral gene segments. The internal segments of the Argentine H6 viruses were closely related to those characterized in viruses previously isolated in this region (4, 17, 19, 21). Particularly, we observed the presence of the NS gene alleles A and B but forming distinct South American clades with a high level of bootstrap support. This observation emphasizes the possible independent evolution of South American AIVs. The relationship between the NP and PA genes from AIVs isolated in South America and equine influenza viruses is also remarkable, suggesting a probable common ancestor (18). Independent evolution is also obvious for the HA gene, although both major lineages have representative strains from North America and Oceania. More divergent evolution is evident for the N2 and N8 NA genes, which form clusters that lie outside other strains from either Eurasia or North America. This observation is particularly remarkable for the N2 NA subtype, which forms a unique branch completely detached from the Eurasian and the North American lineages. Given our findings, it appears that migration patterns are responsible for the independent evolution of these viruses. However, while it is relatively simple to understand the significant geographic barriers that limit the mixing of viruses between the Eurasian and North American lineages, those barriers are less obvious between viruses from North and South America. Further studies are needed to determine these potential boundaries and to fully comprehend the ecology of AIV on the American continent. Interestingly, the HA gene of one virus (1977/H6N2) clustered with viruses from North America, suggesting the presence of more than one AIV phylogenetic lineage in South America.
Importantly, AIVs of the H6 subtype are some of the most abundant AIVs detected in wild bird populations worldwide (13, 15, 22). H6 AIVs have crossed the species barrier and caused outbreaks in domestic poultry in Eurasia, North America, and Africa (1, 2, 11, 12, 24). In Southeast Asia, H6 AIVs have established stable lineages, which are hard to eradicate and likely contribute to the genetic diversity of influenza viruses in local domestic poultry (6, 10). In our animal studies with White Leghorn chickens, the Argentine H6 viruses showed evidence of limited replication and transmission reflected mainly by seroconversion, suggesting a potential risk to local poultry. Seroconversion may be the only sign of a previous AIV infection in poultry (3); therefore, the continuing evolution and movement of these unique Argentine H6 viruses should be carefully monitored. Our results are also consistent with a previous study that showed that the W312/H6N1 virus could replicate in chickens, although transmission was limited (8). Furthermore, long-term AIV surveillance in southern China has found that W312/H6N1-like viruses have established a stable lineage in live-poultry markets, especially in minor species such as quail, chukkar, and guinea fowl (6). In summary, we describe the presence of H6N2 and H6N8 AIVs isolated from wild ducks in Argentina, their evolutionary pattern, and their potential for infection of domestic poultry. Further studies are needed to better elucidate the ecology of AIVs in South America and to establish their host range and pandemic potential.
ACKNOWLEDGMENTS
We thank Theresa Wolter-Marth for her technical and administrative support.
This work was partially supported by the USDA grant no. 1865-05523 and by the NIAID Center for Research on Influenza Pathogenesis (CRIP) through University of Maryland, College Park, contract no. HHSN266200700010C. This work was also supported by P. E. INTA Exoticas y Emergentes and by the European Community (Proyecto Integrado Cadena Carne Aviar-BiotecSur) and was partially supported by the Global Avian Influenza Network for Surveillance (GAINS) program, funded in part by USAID grant no. LAG-A-00-99-00047-00.
The opinions expressed herein are those of the author(s) and do not necessarily reflect the views of the U.S. Agency for International Development.
Footnotes
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Abolnik C., Bisschop S., Gerdes T., Olivier A., Horner R. 2007. Outbreaks of avian influenza H6N2 viruses in chickens arose by a reassortment of H6N8 and H9N2 ostrich viruses. Virus Genes 34:37–45 [DOI] [PubMed] [Google Scholar]
- 2. Abolnik C., Bisschop S. P., Gerdes G. H., Olivier A. J., Horner R. F. 2007. Phylogenetic analysis of low-pathogenicity avian influenza H6N2 viruses from chicken outbreaks (2001–2005) suggest that they are reassortants of historic ostrich low-pathogenicity avian influenza H9N2 and H6N8 viruses. Avian Dis. 51:279–284 [DOI] [PubMed] [Google Scholar]
- 3. Alexander D. J. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74:3–13 [DOI] [PubMed] [Google Scholar]
- 4. Alvarez P., et al. 2010. First isolation of an H1N1 avian influenza virus from wild terrestrial non-migratory birds in Argentina. Virology 396:76–84 [DOI] [PubMed] [Google Scholar]
- 4a. Bahl J., Vijaykrishna D., Holmes E. C., Smith G. J., Guan Y. 2009. Gene flow and competitive exclusion of avian influenza A virus in natural reservoir hosts. Virology 390:289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown I. H. 2010. Summary of avian influenza activity in Europe, Asia, and Africa, 2006-2009. Avian Dis. 54:187–193 [DOI] [PubMed] [Google Scholar]
- 6. Chin P. S., et al. 2002. Molecular evolution of H6 influenza viruses from poultry in Southeastern China: prevalence of H6N1 influenza viruses possessing seven A/Hong Kong/156/97 (H5N1)-like genes in poultry. J. Virol. 76:507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fouchier R. A., Munster V. J. 2009. Epidemiology of low pathogenic avian influenza viruses in wild birds. Rev. Sci. Tech. 28:49–58 [DOI] [PubMed] [Google Scholar]
- 8. Gillim-Ross L., et al. 2008. Avian influenza H6 viruses productively infect and cause illness in mice and ferrets. J. Virol. 82:10854–10863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 10. Hoffmann E., et al. 2000. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J. Virol. 74:6309–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang K., et al. 2010. Establishment of an H6N2 influenza virus lineage in domestic ducks in southern China. J. Virol. 84:6978–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim H. R., et al. 2010. Genetic relatedness of H6 subtype avian influenza viruses isolated from wild birds and domestic ducks in Korea and their pathogenicity in animals. J. Gen. Virol. 91:208–219 [DOI] [PubMed] [Google Scholar]
- 13. Krauss S., et al. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 4:177–189 [DOI] [PubMed] [Google Scholar]
- 14. Lee H. J., et al. 2010. Continuing evolution and interspecies transmission of influenza viruses in live bird markets in Korea. Avian Dis. 54:738–748 [DOI] [PubMed] [Google Scholar]
- 15. Munster V. J., et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Munster V. J., Fouchier R. A. 2009. Avian influenza virus: of virus and bird ecology. Vaccine 27:6340–6344 [DOI] [PubMed] [Google Scholar]
- 17. Pereda A. J., et al. 2008. Avian influenza virus isolated in wild waterfowl in Argentina: evidence of a potentially unique phylogenetic lineage in South America. Virology 378:363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spackman E., McCracken K. G., Winker K., Swayne D. E. 2007. An avian influenza virus from waterfowl in South America contains genes from North American avian and equine lineages. Avian Dis. 51:273–274 [DOI] [PubMed] [Google Scholar]
- 19. Spackman E., McCracken K. G., Winker K., Swayne D. E. 2006. H7N3 avian influenza virus found in a South American wild duck is related to the Chilean 2002 poultry outbreak, contains genes from equine and North American wild bird lineages, and is adapted to domestic turkeys. J. Virol. 80:7760–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spackman E., et al. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suarez D. L., et al. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg. Infect. Dis. 10:693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wallensten A., et al. 2007. Surveillance of influenza A virus in migratory waterfowl in northern Europe. Emerg. Infect. Dis. 13:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Webby R. J., Webster R. G., Richt J. A. 2007. Influenza viruses in animal wildlife populations. Curr. Top. Microbiol. Immunol. 315:67–83 [DOI] [PubMed] [Google Scholar]
- 24. Webby R. J., Woolcock P. R., Krauss S. L., Webster R. G. 2002. Reassortment and interspecies transmission of North American H6N2 influenza viruses. Virology 295:44–53 [DOI] [PubMed] [Google Scholar]
- 25. Webster R. G., Cox N. J., Sthor K. 2002. WHO manual on animal influenza diagnosis and surveillance. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf [Google Scholar]
- 25a. Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]