Abstract
Innate immune deficiencies result in a spectrum of severe clinical outcomes following infection. In particular, there is a strong association between loss of the signal transducer and activator of transcription (Stat) pathway, breach of the blood-brain barrier (BBB), and virus-induced neuropathology. The gene signatures that characterize resistance, disease, and mortality in the virus-infected nervous system have not been defined. Herpes simplex virus type 1 (HSV-1) is commonly associated with encephalitis in humans, and humans and mice lacking Stat1 display increased susceptibility to HSV central nervous system (CNS) infections. In this study, two HSV-1 strains were used, KOS (wild type [WT]), and Δvhs, an avirulent recombinant lacking the virion host shutoff (vhs) function. In addition, two mouse strains were used: strain 129 (control) and a Stat1-deficient (Stat1−/−) strain. Using combinations of these virus and mouse strains, we established a model of infection resulting in three different outcomes: viral clearance without neurological disease (Δvhs infection of control mice), neurological disease followed by viral clearance (Δvhs infection of Stat1−/− mice and WT infection of control mice), or neurological disease followed by death (WT infection of Stat1−/− mice). Through the use of functional genomics on the infected brain stems, we determined gene signatures that were representative of the three infection outcomes. We demonstrated a pathological signature in the brain stem of Stat1-deficient mice characterized by upregulation of transcripts encoding chemokine receptors, inflammatory markers, neutrophil chemoattractants, leukocyte adhesion proteins, and matrix metalloproteases. Additionally, there was a greater than 100-fold increase in the inflammatory markers interleukin 1β (IL-1β) and IL-6. Consistent with this gene signature, we demonstrated profound CNS inflammation with a concomitant lethal breach of the BBB. Taken together, our results indicated an essential role for normal Stat1-dependent signaling in mediating a nonpathological immune response to viral CNS infection.
INTRODUCTION
Virus infection of the human central nervous system (CNS) is a significant source of morbidity and mortality worldwide. While both innate and adaptive immunity are essential for protection of the CNS, the immune response is also capable of causing significant collateral damage during and after clearance of infection (6, 27). As with other tissues, viral infection of the CNS is recognized by Toll-like receptors (TLRs) and other pattern recognition receptors (PRRs) that activate antiviral host defenses. Multiple signaling pathways are activated, including NF-κB, MAPK, and interferon (IFN), which initiate expression of inflammatory cytokines and chemokines (reviewed in references 1 and 62). In response, immune cells, including monocytes, neutrophils, natural killer cells, dendritic cells, and T cells, chemotactically migrate into the infected brain. Migration of cells into the CNS is controlled by the blood-brain barrier (BBB), a highly selective structure composed of endothelial cells, neurons, pericytes, and astrocytes that controls the migration and diffusion of molecules and cells from the vasculature into the parenchyma (reviewed in reference 17). CNS control and clearance of a viral infection can depend on the ability of immune cells and molecules to cross this barrier (47, 49). A pivotal aspect of a successful immune response is the balance between efficacy and pathology, and we hypothesize that the achievement of such a balance is critical in nonregenerative organs, such as the brain.
Herpes simplex virus type 1 (HSV-1) is a ubiquitous, neurotropic human pathogen that causes mostly self-limiting diseases in immunocompetent hosts but can cause encephalitis, meningitis, and hepatitis in immune-suppressed hosts (36, 42). Herpes simplex encephalitis (HSE) is the most common form of viral sporadic encephalitis (25) and, when left untreated, has a mortality rate of 70% (3, 22). HSE may result from primary infection or reactivation from latency, and it frequently involves frontal or temporal lobes, as well as the brain stem. HSE has a strong association with immune pathology; hence, steroids have been proposed as a treatment adjunct to antivirals (14). Nucleoside analogs are effective against HSE, but delayed diagnosis is frequent, and delayed treatment results in increased morbidity and mortality. Even with treatment, the 1-year mortality rate from HSE is 14%, with frequent neurological sequelae in survivors.
The IFN signaling pathway provides an effective and rapid antiviral host response to viral infection. Type I IFN (IFN-β and IFN-α) is produced by infected cells and establishes the antiviral state. Type II IFN (IFN-γ) also has multiple activities, and it serves to activate macrophages and NK cells and ultimately stimulate T-cell-mediated adaptive immunity. IFN receptor (IFNR) engagement leads to phosphorylation of associated Janus and tyrosine kinases, which in turn phosphorylate and activate Stat (signal transduction and transcription factor) proteins (45). The Stat proteins then translocate to the nucleus and initiate IFN-dependent transcription programs. While type I IFN stimulates the phosphorylation of Stat1, Stat2, and Stat3, type II IFN selectively induces phosphorylation of Stat1. Stat1 is therefore critical in both type I and type II IFN signaling, as it mediates expression from both IFN-stimulated response elements (ISREs) and gamma-activated sequence (GAS) motifs. ISRE and GAS-dependent genes are critical for establishing both antiviral and adaptive immune responses in response to infection, as shown by the increased susceptibility of Stat1-deficient mice to various pathogens (10, 11, 16, 21, 28, 30, 32, 35, 48). Previously, we demonstrated that Stat1−/− mice show significantly increased HSV-driven immune pathology in the cornea, independent of viral titer (32). Moreover, immune pathology was apparent even following infection with a highly attenuated HSV strain lacking the virion host shutoff (vhs) protein, a viral RNase that degrades both host and viral mRNA (32, 34, 41, 54). These findings, together with the immunopathological nature of HSE, led us to hypothesize that Stat1 may play a role in balancing the immune response to HSV infections in the brain.
To define gene signatures representative of a spectrum of HSV CNS infection, we utilized infection modalities to generate three infection outcomes. We used two viruses, wild-type HSV-1 KOS (WT) and an attenuated recombinant virus lacking the vhs function (Δvhs), and two mouse strains, control mice and Stat1−/− mice. The three outcomes were subclinical infection (by Δvhs infection of control mice), infection followed by neurological disease and recovery (by Δvhs infection of Stat1−/− mice and WT infection of control mice), and infection followed by neurological disease and death (by WT infection of Stat1−/− mice). We examined gene profiles in both diseased and protected CNS tissues and generated pathological and protective gene signatures that represent causation or prevention of neurological disease. We directly correlated neurological disease and gene signatures with in vivo data demonstrating increased immune cell infiltrates, BBB leakage, and changes in cytokine expression profiles. These data therefore revealed a critical dependence upon fully functional Stat1 to specifically protect the nervous system from neurotropic virus infection and associated immune pathology. In addition, this work provides a gene signature framework for diagnosis and prediction of clinical outcomes following virus infection of the brain.
MATERIALS AND METHODS
Ethics statement.
Animal treatment was consistent with the National Institutes of Health guidelines (30a). All experimental procedures were approved by the Washington University and Dartmouth Medical School Institutional Animal and Care Use Committees.
Cells, viruses, and mice.
Vero cells were used for amplification and titering of viral stocks as previously described (40). All viruses used in this study were of the KOS background (50). Construction of the vhs-null HSV-1 (Δvhs) virus has been previously described (54). Mock-treated animals were inoculated with uninfected Vero cell lysates prepared in a manner parallel to that used for infected Vero lysates. Mouse strains used were 129S6 (as control mice) (Taconic Farms, Germantown, NY) and 129S6 Stat1 knockout mice (Stat1−/−) (28). Mice were housed in the Washington University School of Medicine enhanced barrier facility and were infected in the Washington University School of Medicine biohazard facility. At both facilities, sentinel mice were screened every 3 months and determined to be negative for adventitious mouse pathogens, in particular mouse norovirus. Mice were infected between 6 and 8 weeks of age. Mice were housed, treated, and euthanized when necessary in accordance with all federal and university policies. All animal studies were reviewed and approved by the Washington University Animal Studies (protocol number 2005-0234).
Animal infection procedures.
For corneal infection, equivalent numbers of male and female mice were anesthetized intraperitoneally (i.p.) with ketamine (87 mg/kg body weight) and xylazine (13 mg/kg). Corneas were bilaterally scarified with a 25G syringe needle, and virus was inoculated by adding 2 × 106 PFU in a 5-μl volume per eye. Brain stem and liver were harvested from sacrificed mice on the indicated day postinfection, mechanically disrupted with 3-mm glass beads, and sonicated before tissue titers were determined on Vero cells (as PFU/tissue). For both viruses and each time point, at least eight animals were measured, with samples collected from at least 3 independent groups of the 2 different strains of mice. All titer values are expressed as mean ± standard error of the mean (SEM). Statistical significance was determined using the nonparametric Mann-Whitney U test, with values of P < 0.05 considered statistically significant.
Histological analysis.
At the specified day postinfection, brain stems were harvested from sacrificed mice and immediately fixed in 10% buffered neutral formalin. Paraffin-embedded brain stems were sagitally sectioned and counterstained with hematoxylin. HSV antigen was detected with an anti-HSV-1 rabbit polyclonal antibody (Dako, Denmark). Primary antibodies were detected with streptavidin-horseradish peroxidase (HRP) and diaminobenzidine (DAB). Sections were imaged at 20×.
Bead-based multiplex cytokine analysis.
Brain stems were harvested from mock-treated or infected mice at the indicated day postinfection and stored at −80°C in PBS until processing. Thawed samples were mechanically disrupted with 3-mm glass beads and sonicated. Large debris was removed by centrifugation for 5 min at 500 × g at 4°C. The resulting supernatant was clarified by centrifugation for 5 min at 14,000 × g at 4°C. An aliquot of the supernatant was mixed 1:1 with the Bio-Rad serum diluent solution (Bio-Rad, Hercules, CA). A Bradford assay was utilized to normalize protein levels. The bead-based cytokine BioPlex assay was performed according to the kit protocol (Bio-Rad). Cytokine concentrations were quantified by comparison to a standard curve and are reported as pg/ml. Results shown are the averages from at least two experiments, with each experiment containing 2 or more mice per data point (n ≥ 4).
EB dye uptake assay to measure blood-brain barrier integrity.
On the indicated days postinfection, 1 h prior to sacrifice, 300 μl of 2% Evans blue (EB) in saline (wt/vol) was administered i.p. Extremities were examined for coloration to confirm proper EB delivery. After 1 h, mice were euthanized with CO2 and immediately perfused with 50 ml phosphate-buffered saline (PBS). Mice not fully perfused were not included in the results. Brain and brain stem were removed and homogenized in 1 ml PBS. EB dye was precipitated with an equal volume of 100% trichloroacetic acid (TCA) on ice for 30 min. Samples were centrifuged at 2,800 × g for 30 min at 4°C, and the optical density at 620 nm (OD620) absorbance was measured on supernatants (64). Uptake of dye, as indicated by an increase in A620, correlates with increased blood-brain barrier permeability.
RNA preparation and oligonucleotide microarray processing.
At the indicated day postinfection, mice were sacrificed, brain stems harvested and homogenized, and RNA was extracted as previously described (32). RNA samples were spectroscopically verified for purity, and the quality of the intact RNA was assessed using an Agilent 2100 bioanalyzer. cRNA probes were generated from each sample by the use of an Agilent one-color LowInput quick amp labeling kit (Agilent Technologies, Santa Clara, CA). Individual cRNA samples were hybridized to Agilent mouse whole-genome oligonucleotide 4-by-44 microarrays (approximately 39,000 unique mouse genes) according to the manufacturer's instructions. Samples from individual animals were not pooled to enable examination of animal-to-animal variation as part of the data analysis. These included two animals per time point for each virus (73 animals total). Select samples were hybridized a second time (n = 2 technical replicates) to verify the quality of the process. Slides were scanned with an Agilent DNA microarray scanner, and the resulting images were analyzed using Agilent Feature Extractor (AFE), version 8.1.1.1. The AFE software was used to perform image analysis, including significance of signal and spatial detrending, and to apply a universal error model. For these hybridizations, the most conservative error model was applied. Raw data were then loaded into a custom-designed laboratory information management system (LIMS). Data were warehoused in a Labkey system (Labkey, Inc., Seattle, WA) and analyzed using Rosetta Resolver 7.2 (Rosetta Biosoftware, Seattle, WA) and Spotfire DecisionSite for Functional Genomics 9.1.1 software (Tibco Spotfire, Somerville, MA).
Microarray analysis and bioinformatics.
Global gene expression in infected brain stems was compared to mock control RNA prepared from a pool of equal masses of total RNA from brain stem tissue of two uninfected mice per time point. Probe labeling and microarray slide hybridization were performed as described elsewhere (5). The Resolver system creates ratio profiles by combining replicates while applying error weighting (termed a “squeeze operation”). The error weighting consists of adjusting for additive and multiplicative noise. A P value is generated that represents the probability that a gene is differentially expressed. In this study, a threshold P value of 0.01 was used to identify genes that were significantly differentially expressed. The Resolver system then combines ratio profiles to create ratio experiments using an error-weighted average as described elsewhere (52). Spotfire Decision Site 9.1.1 (Spotfire, Somerville, MA), and Ingenuity Pathways Analysis (IPA) 8.0 (Ingenuity Systems, Redwood City, CA) were also used for data analysis and mining. Primary gene expression data are available at http://viromics.washington.edu in accordance with proposed MIAME standards. Functional and network analysis of statistically significant gene expression changes was performed using IPA. Analysis considered all genes from the data set that met the 2-fold (P < 0.01) change cutoff and which were associated with biological functions in the Ingenuity Pathways knowledge base. For all gene set enrichment analyses, Fisher's exact test was used to determine the probability that each biological function assigned to the genes within each statistical analysis was due to chance alone.
qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed on samples from individual animals to validate specific cellular gene expression changes (CXCL10 and IFN-γ) detected by microarray. The QuantiTect reverse transcription kit (Qiagen Inc., Valencia, CA) was used to generate cDNA. qRT-PCR was run on an ABI 7500 PCR system, using TaqMan chemistry (Applied Biosystems, Foster City, CA). Gene expression assays specific to mouse cellular genes were purchased from Applied Biosystems. Differences in gene expression are represented as log10 of the fold change of the calibrator group relative to that of the target group (log10RQ) relative to a calibrator and normalized to a reference, using the 2−ΔΔCT method (24, 46).
RESULTS
Virological analysis of brain stems and livers.
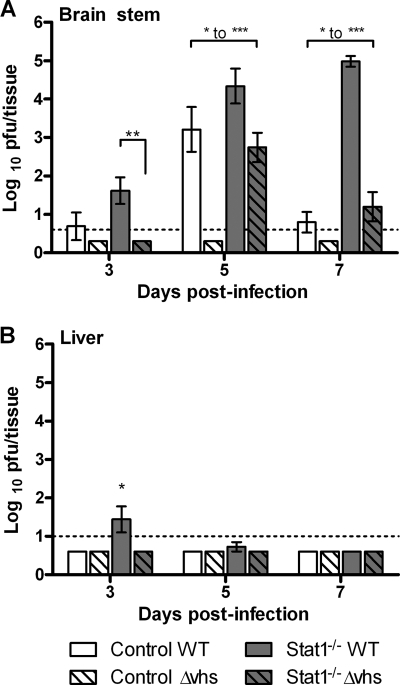
Previous studies have shown that while KOS (WT) grows robustly in corneas of control and Stat1−/− mice, viruses lacking vhs show profoundly attenuated replication relative to WT virus in both mouse strains (35, 53, 54). To better define infection kinetics and related gene expression changes in different tissues, we determined the titers of virus from the brain stems and livers of infected mice. Viral titers in the brain stems of WT-infected control mice peaked at 5 days postinfection (dpi) and decreased by 7 dpi. In contrast, Δvhs-infected control mice had undetectable levels of virus (Fig. 1A). Brain stems from WT-infected Stat1−/− mice showed steadily increasing viral titers out to 7 dpi, shortly after which these mice began to die. In contrast, virus titers in Δvhs-infected Stat1−/− mice peaked on day 5 and decreased thereafter, correlative with their survival. This is consistent with previous results showing that viral brain stem titers in Δvhs-infected Stat1−/− mice were significantly higher than those in control mice but significantly lower than those in WT-infected Stat1−/− mice (35). Notably, the WT virus-infected control and Stat1−/− mice and the Δvhs virus-infected Stat1−/− mice all had significant virus titers at 5 dpi, yet these mice presented with dramatic differences in disease.
Fig. 1.
Virological analysis of brain stems and livers. Control or Stat1−/− mice were inoculated on the cornea with 2 × 106 PFU/eye of HSV-1 WT or HSV-1 Δvhs. At the indicated time postinfection, brain stems (A) and livers (B) were harvested and mechanically disrupted for a plaque assay (n ≥ 8). Titers are presented as PFU/tissue and are the average of results for at least 8 animals. The dotted line indicates threshold of detection (LOD). Where average values fall below the LOD and error bars are shown, virus was detected in some samples and not in others. Where no error bars are shown, virus in all samples was below the LOD. Asterisks indicate ranges of statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001) for differences between bracketed bars. All bars on days 3 and 5 showed statistically significant differences from each other, with the exception of results for WT-infected control mice and Δvhs-infected Stat1−/− mice, which were not significantly different.
Like the brain stem, the liver is highly susceptible to HSV infection in mice and humans with compromised IFN signaling pathways (26, 31, 42) (Fig. 1B). We therefore examined the livers of Stat1−/− mice to test whether increased susceptibility extended beyond the brain stem. Virus was not detectable in the livers of control mice or following infection of either mouse strain by Δvhs. Livers of WT virus-infected Stat1−/− mice showed detectable virus at 3 dpi, but in contrast to infection in the brain, liver infection was undetectable by 5 dpi. Consistent with this, WT virus was similarly cleared from the lymph nodes and spleens of Stat1−/− mice (data not shown). These results demonstrate a difference in the ability of Stat1−/− mice to control HSV-1 infections in the liver and the brain stem.
Molecular signature associated with host protection.
The disease and virulence patterns of control and Stat1−/− mice infected with WT and Δvhs viruses have been previously described. Briefly, following corneal inoculation of WT virus, immunocompetent control mice show no neurological disease, while infected Stat1−/− mice exhibit neurological symptoms (hunched posture, lack of controlled movement, and weight loss) and die between 7 and 10 days postinfection (dpi) (32). In contrast, inoculation of Δvhs into control mice causes little to no disease, with significantly reduced virulence (53, 54). In Stat1−/− mice, however, Δvhs shows enhanced virulence (35). To address the pathways that protect the control mice, we performed functional genomics analysis on infected brain stems. The goal was to identify differences in gene expression between the nondiseased model (WT virus-infected control mice) and the diseased models (WT or Δvhs virus-infected Stat1−/− mice), with the aim of finding protective gene signatures. We performed a two-way analysis of variance (ANOVA) on the gene array data comparing nondiseased and diseased groups at 3 and 5 days postinfection. We used samples from 3 dpi because this represents the previously observed onset of robust viral infection in the brain and samples from 5 dpi because it corresponds to the previously observed peak viral titer in the WT virus-infected control mice and the onset of disease in the Stat1−/− mice (35).
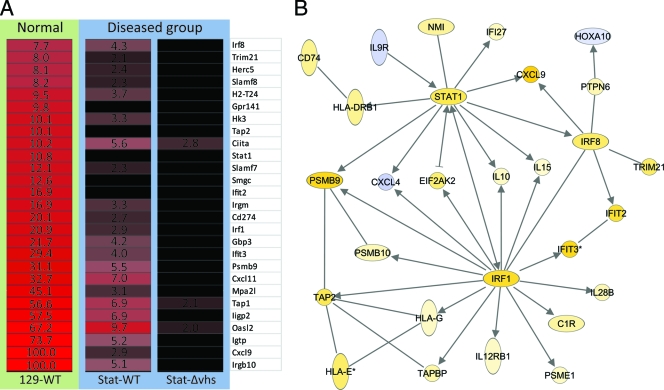
The majority of differentially expressed highly upregulated genes in the WT virus-infected control mice were IFN dependent (including Irgb10, Cxcl9, Igtp [Irgm3], Oasl2, Iigp2 [Irgm2 or Gtpi], Tap1, Cxcl11, Psmb9, Ifit3, Gbp3, Irf1, Irgm, Stat1, and Irf8) (Fig. 2A). While both type I and type II IFN can induce most of these genes, many have been shown to be preferentially or strongly induced by type II IFN. These include, but are not limited to, the GTPases Igtp, Iigp2, and Gbp3 (reviewed in reference 55), Cxcl9 and Cxcl11 (12, 13), interferon transcription factors Irf1 and Irf8 (7, 8, 19, 23), major histocompatibility complex (MHC) class II transactivator CIIta (57), Psmb9 (7), Tap 1 (7), and Stat1 (57). Cxcl10, encoding an IFN-γ inducible chemokine closely related to Cxcl9 and Cxcl11, is upregulated via IFN-independent mediators, which is likely why it is expressed in both control and Stat1−/− mice (reviewed in reference 29).
Fig. 2.
Protective signature genes. Control or Stat1−/− mice were inoculated on the cornea with 2 × 106 PFU/eye of HSV-1 WT or HSV-1 Δvhs or treated with a mock lysate. At the indicated time postinfection, RNA was harvested and processed for array analysis. (A) Heat map showing ANOVA results for differentially expressed genes within the cutoff values of ≥2-fold change and ANOVA P ≤ 0.01 in brain stems comparing nondiseased (WT virus-infected strain 129 control mice) and diseased (WT virus-infected Stat1−/− mice and Δvhs virus-infected Stat1−/− mice) models at 5 dpi. Red and black colors indicate upregulated and unchanged genes, respectively, compared to a mock-treated sample. The numbers indicate fold changes compared to results for a mock-treated sample. (B) Functional network analysis of 5 dpi ANOVA results determined by Ingenuity Pathways Analysis. Arrows indicate functional interactions between genes. Yellow and blue colors indicate up- and downregulated genes, respectively.
The genes upregulated in the control mice were further analyzed by Ingenuity Pathways Analysis (IPA) to identify functionally related gene networks (Fig. 2B). This led to identification of gene nodes for three IFN-γ-inducible transcription factors acting downstream of IFN receptors: Stat1, Irf8, and Irf1 (7, 8, 19, 23). These data indicate a distinct protective signature in the brain stems of the infected control mice that centers around IFN-stimulated genes. From these results, we conclude that the lack of Stat1 impacts not only the antiviral response but also the establishment of a nonpathological immune response in the brain stem.
Transcriptional profiling of liver tissues reveals differential regulation of IFN-responsive genes in brain stem and liver.
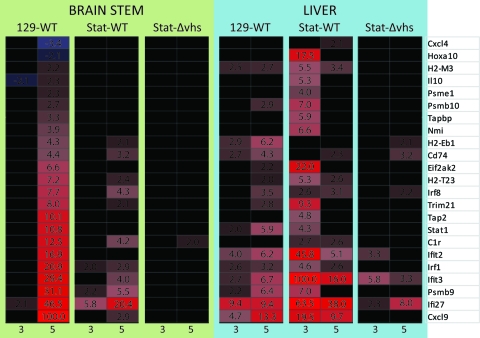
Compromised IFN responses have been previously shown to result in severe and lethal infection of the liver (26, 33). In this study, there was significant infection of the liver of WT-infected Stat1−/− mice on day 3 (Fig. 1B), yet these mice clear HSV from liver and subsequently succumb to CNS infection. These divergent outcomes suggested a differential role for Stat1 in protection of the CNS and the liver. We therefore did transcriptional profiling on livers from control and Stat1−/− mice (Fig. 3). Although WT virus titers were below the threshold of detection in the livers of control mice, our analysis showed upregulation at 5 dpi of many IFN-dependent genes, indicative of an effective immune response. More unexpectedly, there was upregulation of IFN-dependent genes in the livers of the Stat1−/− mice infected with WT virus. This was seen at 3 dpi, coincident with the peak of viral titer (Fig. 1A), and included Cxcl9, Ifi27, Ifit3, Psmb9, Ifit2, and Eif2ak2 (PKR). Many of these genes are distinct from those strongly upregulated in the brain stems of the same mice (Fig. 2A). Mice lacking Stat1 differed in their ability to express IFN-stimulated genes in the liver versus the brain stem, and this pattern of expression directly correlated with viral replication and clearance in each tissue (Fig. 1). These results suggest a specific requirement of Stat1 in the CNS for the generation of a proper response to IFN signaling. To explore this further, we examined pathological signatures of mice that exhibit severe disease and death from HSV infection.
Fig. 3.
Comparison of protective responses in brain stem and liver. Heat map comparing gene expression changes in the brain stems and livers of WT virus-infected control mice and WT or Δvhs virus-infected Stat1−/− mice at 3 and 5 dpi. Differentially expressed genes identified in the protective gene signature analysis are shown. Red and black colors indicate upregulated or unchanged genes, respectively, compared to a mock-treated sample. The numbers indicate fold changes compared to results for a mock-treated sample.
Molecular signature associated with pathogenicity.
Stat1−/− mice infected with the attenuated Δvhs virus exhibited disease similar to that found in WT virus-infected Stat1−/− mice, despite lower viral titers and eventual viral clearance (Fig. 1A). The similarity in disease course between the WT and Δvhs virus-infected Stat1−/− mice suggested a common disease process with divergent outcomes. We had thus far identified gene expression changes that correlated with protection, so we next examined the transcriptional profile with the goal of identifying broad pathogenic signatures that correlated with disease. Based on the known disease pathogenesis and the time course of viral replication (Fig. 1A) (35), we continued to examine infection models in which substantial viral replication and disease were observed. These infection models were control mice infected with the WT virus and Stat1−/− mice infected with either the WT or Δvhs virus.
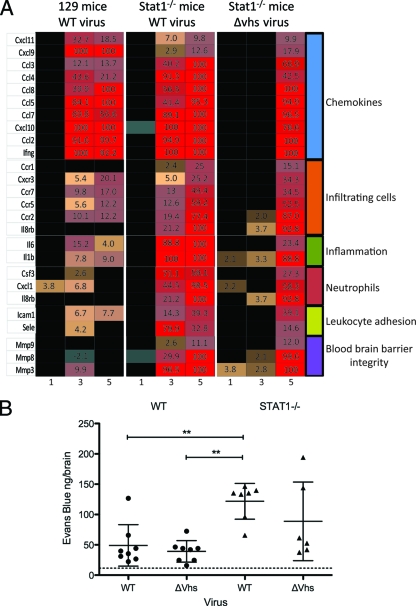
Although WT virus-infected control mice lacked behavioral changes indicative of CNS infection, there was substantial upregulation at 5 and 7 dpi of chemokines and immunostimulatory cytokines, including monocyte chemoattractant protein (MCP)-1 (CCL2), RANTES (CCL5), MCP-3 (CCL7), MCP-2 (CCL8), CXCL9, CXCL10, and IFN-γ (Fig. 4A). Of particular note was the strong upregulation of CXCL9 and CXCL11 in the control mice. Common to control and Stat1−/− mice was a remarkable upregulation of both CXCL10 and IFN-γ, which by qRT-PCR were induced by 1,000- to 10,000-fold (data not shown).
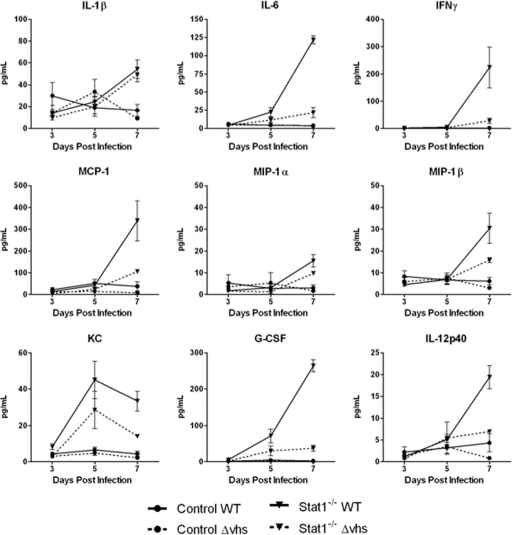
Fig. 4.
Pathogenicity genes and BBB permeability. (A) Heat map showing selected, differentially expressed genes within the cutoff values of ≥2-fold change and ANOVA P ≤ 0.01 in brain stems from WT virus-infected control mice, WT virus-infected Stat1−/− mice, or Δvhs virus-infected Stat1−/− mice at 5 dpi. The numbers indicate fold changes compared to results for mock-infected mice. (B) Quantification of Evans blue dye in brain tissue of infected mice. WT or Δvhs virus-infected control and Δvhs virus-infected Stat1−/− mice are shown at 7 dpi. WT virus-infected Stat1−/− mice were moribund at 7 dpi, and so data are shown for 6 dpi. The amount of dye is reported in ng/brain. The dotted line indicates the background level of Evans blue dye. *, P < 0.01.
In contrast to control mice, the brains of Stat1−/− mice showed significant upregulation of chemokine receptors, inflammatory markers, neutrophil chemoattractants, leukocyte adhesion proteins, and matrix metalloproteases. Upregulation of these factors in concert could attract leukocytes and control BBB permeability. The upregulation of chemokine receptors in the brain is especially indicative of increased immune infiltration through the BBB. Consistent with this, the observed increases included CCR1, CCR2, and CCR5 (chemokine receptors found on monocytes, macrophages, T cells, and dendritic cells), as well as CXCL1 and CXCR2 (neutrophil chemoattractants and receptors). Similar gene expression changes were found in the Δvhs virus-infected Stat1−/− mice, although with delayed or muted expression compared to that of the WT virus-infected Stat1−/− mice. Together, these gene products are strong chemotactic signals that mediate immune cell infiltration and are predictive of significant CNS inflammation. Several other upregulated genes in the Stat1−/− mice included the cytokines interleukin 1β (IL-1β) and IL-6, which influence the integrity of the BBB (2, 38), the chemokine MCP-1, which opens endothelial cell tight junctions (15, 20, 51), and the enzymes MMP-3, -8, and -9, which degrade extracellular matrix proteins at the BBB (43, 56). The altered expression of these genes suggested that the integrity of the BBB was being disrupted in the infected Stat1−/− mice. To assess this, we employed an Evans blue dye uptake assay in which dye uptake into the CNS correlates with increased BBB permeability (Fig. 4B). Coincident with the upregulation of cytokines known to affect BBB permeability, we found increased uptake of the dye in only the infected Stat1−/− mice, regardless of the viral strain and viral titer. Statistically significant changes were seen in the WT virus-infected Stat1−/− mice (P < 0.01). Increased uptake in the Δvhs-infected Stat1−/− mice occurred but was variable and not statistically significant. These more variable increases correlate with the variable viral titers seen at this time point (Fig. 1A) and the delayed/muted increases in expression of BBB breach-related genes (Fig. 4). Increased BBB permeability was not seen in infected mice lacking IFN receptors (P. C. Rosato and D. A. Leib, unpublished data), indicating that the mutation of Stat1, not the absence of IFN responses, was responsible for the BBB breach.
Multiplex cytokine and chemokine analysis.
A bead-based cytokine analysis was performed to confirm the inflammatory and chemoattractant array profile in the brain stems. Both WT and Δvhs virus-infected control mice showed little if any cytokine or chemokine expression following infection (Fig. 5), and results for them were similar to those for mock-treated tissues. In contrast, the infected Stat1−/− mice had sharp increases in the cytokines IL-6, granulocyte colony-stimulating factor (G-CSF), and IFN-γ, as well as the chemoattractants macrophage inflammatory protein (MIP)-1β and MCP-1 by 7 dpi. We also found modestly increased expression of IL-1β, MIP-1α, and IL-12p40 in the WT virus-infected Stat1−/− mice at 7 dpi. Expression of CXCL1 (also known as KC and IL-8, a neutrophil chemoattractant), peaked at 5 dpi in the Stat1−/− mice. The most significant changes in expression were found in the WT virus-infected Stat1−/− mice at 7 dpi, with elevated levels of all cytokines and chemokines shown. Cytokine expression was not as dramatic in the Δvhs virus-infected Stat1−/− mice at 5 and 7 dpi, with the exception of IL-1β expression. The chemokines MIP-1β, MCP-1, and KC are potent chemoattractants for natural killer cells, monocytes, memory T cells, dendritic cells, and neutrophils. The upregulation of these chemokines is consistent with the array data, with appearance of behavioral changes predictive of CNS disease and also predictive of immune cell migration and BBB breach in the Stat1−/− mice.
Fig. 5.
Bead-based cytokine analysis of brain stems. Control or Stat1−/− mice were inoculated with 2 × 106 PFU/eye of HSV-1 WT or HSV-1 Δvhs. At the indicated time postinfection, protein was extracted from brain stem tissue and analyzed by bead-based cytokine analysis (n ≥ 4). Cytokine concentrations are presented as pg/ml.
Histological analysis of infected brain stems.
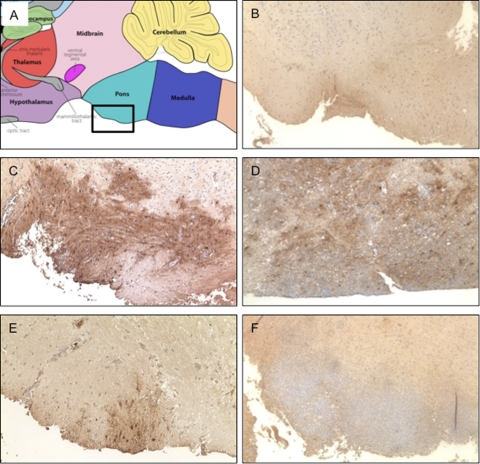
We evaluated histological sections of infected brain tissues to overview the location of viral antigen, immune cell infiltration, and the extent of tissue damage. We focused on the pons in the brain stem directly adjacent to the trigeminal ganglia (Fig. 6A). Tissue from the WT virus-infected control mice showed little if any viral antigen in the brain stem and was indistinguishable from mock-infected tissues (Fig. 6B and data not shown). In contrast, viral antigen staining in the WT virus-infected Stat1−/− brain stems showed viral invasion as early as 5 dpi (Fig. 6C). At 7 dpi, shortly before such mice succumb to infection, the virus had penetrated deeper into the brain stem and a significant immune cell infiltrate containing neutrophils was readily apparent (Fig. 6D). The Δvhs virus was not as invasive as the WT virus at 5 dpi (Fig. 6E), and by 7 dpi viral antigen was mostly cleared. Despite this clearance, we observed a persistent and massive immune cell infiltrate containing neutrophils (Fig. 6F). In the Δvhs virus-infected Stat1−/− mice, disease symptoms and the immune cell infiltrate persisted to at least 13 dpi (data not shown), long after Δvhs clearance from the CNS (Fig. 1A) (35). Importantly, however, neurological disease correlated with increased immune cell infiltrates and not with viral antigen or CNS viral titer, as both WT and Δvhs virus-infected Stat1−/− mice exhibited similarly severe and progressive patterns of neurological disease. This observation is consistent with the upregulation of BBB permeability-related genes in the Δvhs-infected brains (Fig. 4). Furthermore, Δvhs still induced severe inflammation beyond the clearance of infection, showing a pathological response to HSV-1 infection in the brains of Stat1−/− mice that was independent of viral virulence or titer.
Fig. 6.
Histological analysis of brain stem tissue. (A) Map taken from the Gene Expression Nervous System Atlas (GENSAT) Project (www.gensat.org) shows the region of the brain examined in the subsequent panels. (B) Control mice infected with WT virus, 5 dpi. (C) Stat1−/− brain infected with WT virus, 5 dpi. (D) Stat1−/− mice infected with WT virus, 7 dpi. (E) Stat1−/− mice infected with Δvhs virus, 5 dpi. (F) Stat1−/− mice infected with WT virus, 7 dpi. Mice were inoculated with 2 × 106 PFU/eye of virus per eye, and brain stems were harvested and fixed for histological analysis at times indicated. Tissue sections were stained for HSV-1 antigen (brown) and counterstained with hematoxylin. Representative images are shown of the pons area in the brain stem where the trigeminal ganglia connect to the brain stem.
DISCUSSION
The efficacy of innate immunity and the early phase response in the clearance of pathogens is dependent on several factors, including efficient detection of pathogen-associated molecular patterns (PAMPs), a measured IFN response, and appropriate expression of chemoattractant cytokines. In this study, we demonstrated the pivotal requirement for fully functional Stat1 protein in the protection of the central nervous system from an acute neurotropic virus infection. The increased susceptibility of Stat1-deficient mice to CNS pathology was mirrored in gene signatures that predicted significant permeability of the blood-brain barrier and dramatic increases in expression of proinflammatory cytokines. These signatures were subsequently authenticated through a series of biological assays. Through parallel infection of control mice or the use of an attenuated Δvhs virus in the Stat1−/− mice, we were also able to demonstrate protective signatures that resulted either from subclinical infection or from disease and recovery. While these infection scenarios resulted in a continuum of disease determined by viral virulence and host susceptibility, these gene signatures nevertheless demonstrate the delicate balance between recovery, disease, and death during viral infection in the CNS. Moreover, these signatures likely have strong correlates in human disease, especially since defects in Stat1 are correlated with susceptibility to HSV encephalitis (44).
It is notable in these data that while brain stems from both control and Stat1−/− mice upregulate chemokines in response to infection, it is only the Stat1−/− mice that show strong upregulation of inflammatory markers, leukocyte adhesion molecules, MMPs, and neutrophil chemotaxis genes. Notably these increases are independent of viral antigen and titer. Moreover, this profile is consistent with the previous finding that type I IFN inhibits dendritic cell migration into the CNS through Stat1-dependent suppression of MMP-9 and CCR7 expression (63). This previous finding provided mechanistic insight into the established use of IFN-β as a therapy for multiple sclerosis by reducing dendritic cell migration into the CNS, thereby helping to prevent demyelination. This previous study, along with the central findings of this study, points to the feasibility and potential efficacy of use of IFN-β as a therapeutic agent in combination with nucleoside analogs for treatment of HSE.
The two major organs affected following generalized HSV infection of immune-compromised mice and humans are the brain and the liver. This analysis revealed dramatic differences in IFN-dependent gene expression between the brain stems and livers of the Stat1−/− mice. Stat1−/− mice clear the liver infection, but not the CNS infection, with Stat1−/− livers showing a profile of IFN-dependent genes comparable to that found in the CNS and liver of control mice. A potential explanation for the expression of IFN-dependent genes in Stat1−/− livers is that Stat2-dependent signaling can be a source of interferon-stimulated gene (ISG) expression in Stat1−/− macrophages, although this observation cannot explain the lack of signaling in the CNS (37). Additionally, there could be a specific requirement for Stat1 in the CNS or in neuronal cells or perhaps differential effects of Stat1 and IFN in the CNS compared to that in non-CNS tissues (59–61). Another possibility is that there is heightened activation of Stat3 in Stat1−/− cells, and this may lead to pathological signaling in certain tissues (18, 32, 39, 58). Other research from our laboratory has revealed that in a distinct Stat1-deficient mouse line (10), HSV follows a more viscerotropic/hepatotropic pattern of spread. This implies that the different domains or levels of activity of Stat1 may be influencing tropism (33). It is possible that the N-terminal deleted Stat1−/− strain used in this study (28) has sufficient residual activity to protect the liver, but not the CNS, from lethal infection. Notably, Stat1-deficient humans are more susceptible to HSV encephalitis (9), implying that the mouse strain used in this study (lacking the N terminus of Stat1) is a better model for human disease than the mouse strain lacking the DNA-binding domain (10). A further comparison of these two mouse lines will likely reveal important details of Stat1 domains that determine host resistance to infection.
A comparison of data from the present study with those from a study of 129S6/SvEv mice infected with the highly pathogenic avian H5N1 virus revealed that some of the genes identified in the protective signature elicited in the HSV-infected mice were components of a lethality signature elicited during avian influenza virus infection (5). In contrast, some of the genes identified as part of the molecular pathogenicity signature associated with lethality in this study were also part of a functional network associated with Ebola virus lethality in other studies (4). Although these various studies differ in terms of mice, viruses, inocula, and time points, an overarching pattern is that interferon-dependent genes and inflammation-related genes play critical roles during lethal viral infection in determining the delicate homeostatic balance of the immune response and ultimately profoundly affect the fate of the host. These findings may have therapeutic implications, suggesting that compounds targeting these pathways may ameliorate severity and outcomes of infections but only in a virus-specific fashion. Study of virus- and tissue-specific protective and pathogenicity signatures will therefore have great value in fashioning predictions for therapeutic intervention in viral disease.
ACKNOWLEDGMENTS
The study was supported by National Institutes of Health grants to D.A.L. (EY 10707 and 09083) with a subaward to M.G.K. from EY 10707. The project was also supported by P20RR016437 from the National Center for Research Resources to Dartmouth.
We acknowledge Belinda McMahon and Bob Schmidt for assistance with the histology and Robyn Klein and the reviewers of manuscript version 1 for helpful and detailed comments on the manuscript.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Aravalli R. N., Hu S., Rowen T. N., Palmquist J. M., Lokensgard J. R. 2005. Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J. Immunol. 175:4189–4193 [DOI] [PubMed] [Google Scholar]
- 2. Argaw A. T., et al. 2006. IL-1beta regulates blood-brain barrier permeability via reactivation of the hypoxia-angiogenesis program. J. Immunol. 177:5574–5584 [DOI] [PubMed] [Google Scholar]
- 3. Baringer J. R. 2008. Herpes simplex infections of the nervous system. Neurol. Clin. 26:657–674, viii [DOI] [PubMed] [Google Scholar]
- 4. Cilloniz C., et al. 2011. Functional genomics reveals the induction of inflammatory response and metalloproteinase gene expression during lethal Ebola virus infection. J. Virol. 85:9060–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cilloniz C., et al. 2010. Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J. Virol. 84:7613–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conrady C. D., Drevets D. A., Carr D. J. 2010. Herpes simplex type I (HSV-1) infection of the nervous system: is an immune response a good thing? J. Neuroimmunol. 220:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Der S. D., Zhou A., Williams B. R., Silverman R. H. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Driggers P. H., et al. 1990. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. U. S. A. 87:3743–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dupuis S., et al. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391 [DOI] [PubMed] [Google Scholar]
- 10. Durbin J. E., Hackenmiller R., Simon M. C., Levy D. E. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450 [DOI] [PubMed] [Google Scholar]
- 11. Durbin J. E., et al. 2002. The role of IFN in respiratory syncytial virus pathogenesis. J. Immunol. 168:2944–2952 [DOI] [PubMed] [Google Scholar]
- 12. Ellis S. L., et al. 2010. The cell-specific induction of CXC chemokine ligand 9 mediated by IFN-gamma in microglia of the central nervous system is determined by the myeloid transcription factor PU.1. J. Immunol. 185:1864–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farber J. M. 1990. A macrophage mRNA selectively induced by gamma-interferon encodes a member of the platelet factor 4 family of cytokines. Proc. Natl. Acad. Sci. U. S. A. 87:5238–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fitch M. T., van de Beek D. 2008. Drug insight: steroids in CNS infectious diseases—new indications for an old therapy. Nat. Clin. Pract. Neurol. 4:97–104 [DOI] [PubMed] [Google Scholar]
- 15. Gourmala N. G., Buttini M., Limonta S., Sauter A., Boddeke H. W. 1997. Differential and time-dependent expression of monocyte chemoattractant protein-1 mRNA by astrocytes and macrophages in rat brain: effects of ischemia and peripheral lipopolysaccharide administration. J. Neuroimmunol. 74:35–44 [DOI] [PubMed] [Google Scholar]
- 16. Halford W. P., et al. 2006. ICP0 antagonizes Stat 1-dependent repression of herpes simplex virus: implications for the regulation of viral latency. Virol. J. 3:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hawkins B. T., Davis T. P. 2005. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol. Rev. 57:173–185 [DOI] [PubMed] [Google Scholar]
- 18. Hong F., et al. 2002. Opposing roles of STAT1 and STAT3 in T cell-mediated hepatitis: regulation by SOCS. J. Clin. Invest. 110:1503–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horiuchi M., Itoh A., Pleasure D., Ozato K., Itoh T. 2011. Cooperative contributions of interferon regulatory factor 1 (IRF1) and IRF8 to interferon-γ-mediated cytotoxic effects on oligodendroglial progenitor cells. J. Neuroinflammation 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang D. R., Wang J., Kivisakk P., Rollins B. J., Ransohoff R. M. 2001. Absence of monocyte chemoattractant protein 1 in mice leads to decreased local macrophage recruitment and antigen-specific T helper cell type 1 immune response in experimental autoimmune encephalomyelitis. J. Exp. Med. 193:713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karst S. M., Wobus C. E., Lay M., Davidson J., Virgin IV H. W. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578 [DOI] [PubMed] [Google Scholar]
- 22. Kennedy P. G., Chaudhuri A. 2002. Herpes simplex encephalitis. J. Neurol. Neurosurg. Psychiatry 73:237–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ko J., Gendron-Fitzpatrick A., Splitter G. A. 2002. Susceptibility of IFN regulatory factor-1 and IFN consensus sequence binding protein-deficient mice to brucellosis. J. Immunol. 168:2433–2440 [DOI] [PubMed] [Google Scholar]
- 24. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Livorsi D., et al. 2010. Brainstem encephalitis: an unusual presentation of herpes simplex virus infection. J. Neurol. 257:1432–1437 [DOI] [PubMed] [Google Scholar]
- 26. Luker G. D., Prior J. L., Song J., Pica C. M., Leib D. A. 2003. Bioluminescence imaging reveals systemic dissemination of herpes simplex virus type 1 in the absence of interferon receptors. J. Virol. 77:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lundberg P., et al. 2008. The immune response to herpes simplex virus type 1 infection in susceptible mice is a major cause of central nervous system pathology resulting in fatal encephalitis. J. Virol. 82:7078–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meraz M. A., et al. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431–442 [DOI] [PubMed] [Google Scholar]
- 29. Muller M., Carter S., Hofer M. J., Campbell I. L. 2010. Review: the chemokine receptor CXCR3 and its ligands CXCL9, CXCL10 and CXCL11 in neuroimmunity—a tale of conflict and conundrum. Neuropathol. Appl. Neurobiol. 36:368–387 [DOI] [PubMed] [Google Scholar]
- 30. Mumphrey S. M., et al. 2007. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J. Virol. 81:3251–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a. National Institutes of Health 2000. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD [Google Scholar]
- 31. Norvell J. P., Blei A. T., Jovanovic B. D., Levitsky J. 2007. Herpes simplex virus hepatitis: an analysis of the published literature and institutional cases. Liver Transpl. 13:1428–1434 [DOI] [PubMed] [Google Scholar]
- 32. Pasieka T. J., et al. 2009. Host responses to wild-type and attenuated herpes simplex virus infection in the absence of Stat1. J. Virol. 83:2075–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pasieka T. J., et al. 2011. Bioluminescent imaging reveals divergent viral pathogenesis in two strains of Stat1-deficient mice, and in αβγ interferon receptor-deficient mice. PLoS One 6(9):e24018 doi:10.1371/journal.pone.0024018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pasieka T. J., et al. 2008. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J. Virol. 82:5527–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pasieka T. J., Lu B., Leib D. A. 2008. Enhanced pathogenesis of an attenuated herpes simplex virus for mice lacking Stat1. J. Virol. 82:6052–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perez de Diego R., et al. 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity 33:400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perry S. T., Buck M. D., Lada S. M., Schindler C., Shresta S. 2011. STAT2 mediates innate immunity to dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 7:e1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poller B., Drewe J., Krahenbuhl S., Huwyler J., Gutmann H. 2010. Regulation of BCRP (ABCG2) and P-glycoprotein (ABCB1) by cytokines in a model of the human blood-brain barrier. Cell. Mol. Neurobiol. 30:63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qing Y., Stark G. R. 2004. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J. Biol. Chem. 279:41679–41685 [DOI] [PubMed] [Google Scholar]
- 40. Rader K. A., Ackland-Berglund C. E., Miller J. K., Pepose J. S., Leib D. A. 1993. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. J. Gen. Virol. 74(Pt 9):1859–1869 [DOI] [PubMed] [Google Scholar]
- 41. Read G. S., Karr B. M., Knight K. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Riediger C., et al. 2009. Herpes simplex virus sepsis and acute liver failure. Clin. Transplant. 23(Suppl. 21):37–41 [DOI] [PubMed] [Google Scholar]
- 43. Rosenberg G. A. 2002. Matrix metalloproteinases in neuroinflammation. Glia 39:279–291 [DOI] [PubMed] [Google Scholar]
- 44. Sancho-Shimizu V., et al. 2007. Genetic susceptibility to herpes simplex virus 1 encephalitis in mice and humans. Curr. Opin. Allergy Clin. Immunol. 7:495–505 [DOI] [PubMed] [Google Scholar]
- 45. Schindler C., Levy D. E., Decker T. 2007. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 282:20059–20063 [DOI] [PubMed] [Google Scholar]
- 46. Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 47. Shresta S., et al. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 78:2701–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shresta S., et al. 2005. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 175:3946–3954 [DOI] [PubMed] [Google Scholar]
- 49. Shrestha B., Diamond M. S. 2004. Role of CD8+ T cells in control of West Nile virus infection. J. Virol. 78:8312–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith K. O. 1964. Relationship between the envelope and the infectivity of herpes simplex virus. Proc. Soc. Exp. Biol. Med. 115:814–816 [DOI] [PubMed] [Google Scholar]
- 51. Song L., Pachter J. S. 2004. Monocyte chemoattractant protein-1 alters expression of tight junction-associated proteins in brain microvascular endothelial cells. Microvasc. Res. 67:78–89 [DOI] [PubMed] [Google Scholar]
- 52. Stoughton R., Dai H. February 2002. Statistical combining of cell expression profiles. U.S. patent 6,351,712
- 53. Strelow L. I., Leib D. A. 1996. Analysis of conserved domains of UL41 of herpes simplex virus type 1 in virion host shutoff and pathogenesis. J. Virol. 70:5665–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strelow L. I., Leib D. A. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taylor G. A., Feng C. G., Sher A. 2004. p47 GTPases: regulators of immunity to intracellular pathogens. Nat. Rev. Immunol. 4:100–109 [DOI] [PubMed] [Google Scholar]
- 56. Toft-Hansen H., Nuttall R. K., Edwards D. R., Owens T. 2004. Key metalloproteinases are expressed by specific cell types in experimental autoimmune encephalomyelitis. J. Immunol. 173:5209–5218 [DOI] [PubMed] [Google Scholar]
- 57. Waddell S. J., et al. 2010. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS One 5:e9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Walters D. M., et al. 2005. Susceptibility of signal transducer and activator of transcription-1-deficient mice to pulmonary fibrogenesis. Am. J. Pathol. 167:1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang J., Campbell I. L. 2005. Innate STAT1-dependent genomic response of neurons to the antiviral cytokine alpha interferon. J. Virol. 79:8295–8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang J., Campbell I. L., Zhang H. 2008. Systemic interferon-alpha regulates interferon-stimulated genes in the central nervous system. Mol. Psychiatry 13:293–301 [DOI] [PubMed] [Google Scholar]
- 61. Wang J., Schreiber R. D., Campbell I. L. 2002. STAT1 deficiency unexpectedly and markedly exacerbates the pathophysiological actions of IFN-alpha in the central nervous system. Proc. Natl. Acad. Sci. U. S. A. 99:16209–16214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wuest T. R., Carr D. J. 2008. The role of chemokines during herpes simplex virus-1 infection. Front. Biosci. 13:4862–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yen J. H., Kong W., Ganea D. 2010. IFN-beta inhibits dendritic cell migration through STAT-1-mediated transcriptional suppression of CCR7 and matrix metalloproteinase 9. J. Immunol. 184:3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou J., Stohlman S. A., Hinton D. R., Marten N. W. 2003. Neutrophils promote mononuclear cell infiltration during viral-induced encephalitis. J. Immunol. 170:3331–3336 [DOI] [PubMed] [Google Scholar]