Abstract
Cytomegalovirus (CMV) infection is the most common opportunistic infection in immunosuppressed individuals, such as transplant recipients or people living with HIV/AIDS, and congenital CMV is the leading viral cause of developmental disabilities in infants. Due to the highly species-specific nature of CMV, animal models that closely recapitulate human CMV (HCMV) are of growing importance for vaccine development. Here we present the genomic sequence of a novel nonhuman primate CMV from cynomolgus macaques (Macaca fascicularis; CyCMV). CyCMV (Ottawa strain) was isolated from the urine of a healthy, captive-bred, 4-year-old cynomolgus macaque of Philippine origin, and the viral genome was sequenced using next-generation Illumina sequencing to an average of 516-fold coverage. The CyCMV genome is 218,041 bp in length, with 49.5% G+C content and 84% protein-coding density. We have identified 262 putative open reading frames (ORFs) with an average coding length of 789 bp. The genomic organization of CyCMV is largely colinear with that of rhesus macaque CMV (RhCMV). Of the 262 CyCMV ORFs, 137 are homologous to HCMV genes, 243 are homologous to RhCMV 68.1, and 200 are homologous to RhCMV 180.92. CyCMV encodes four ORFs that are not present in RhCMV strain 68.1 or 180.92 but have homologies with HCMV (UL30, UL74A, UL126, and UL146). Similar to HCMV, CyCMV does not produce the RhCMV-specific viral homologue of cyclooxygenase-2. This newly characterized CMV may provide a novel model in which to study CMV biology and HCMV vaccine development.
INTRODUCTION
Human cytomegalovirus (HCMV), also known as human herpesvirus 5 (HHV-5), is a member of the Betaherpesvirus family (including HHV-6 and HHV-7). Cytomegalovirus (CMV) is a double-stranded DNA virus with the largest genome of any herpesvirus. The virus is transmitted horizontally through bodily secretions and can cross the placental barrier to facilitate vertical transmission (reviewed in reference 44). CMV results in a lifelong infection characterized by the establishment of latency in myeloid progenitor cells, followed by periodic reactivation. CMV elicits a strong cellular immune response, and the CMV-specific T cells of some individuals can account for greater than 10% of the total T-cell population (16, 22, 64). In immunocompetent individuals, CMV infection is generally asymptomatic and controlled by the cell-mediated immune response; however, in immunocompromised individuals (i.e., neonates, transplant patients, and AIDS patients), it can cause severe diseases, such as congenital disorders, CMV retinitis, and a variety of opportunistic infections.
Various lab-adapted and clinical strains of HCMV have been isolated and sequenced; most notable are AD169 (13), Toledo (46), Towne (17), and Merlin (15). Furthermore, there are a number of clinical strains that have been cloned as bacterial artificial chromosomes, such as TB40/E (62), TR, PH, and FIX (VR1814) (46). The full-length genomes of CMVs from a number of different animal species, including mice (54), rats (68), guinea pigs (33, 59), and tree shrews (6), have been isolated and sequenced. Given their high degree of genetic relatedness to humans, nonhuman primates (NHPs) likely represent the best animal model to study HCMV biology. A variety of CMVs from Old and New World primates have also been described (37), including chimpanzee CMV (14, 63), rhesus CMV strains 68.1 and 180.92 (28, 57), cercopithecine herpesvirus 5 (CeHV-5) strains GR2715 and Colburn (accession no. FJ483968 and FJ483969, respectively), squirrel monkey CMV (SsciCMV-1; accession no. FJ483967), and owl monkey CMV (AtriCMV-1; accession no. FJ483970). CMVs are highly species-specific viruses (32, 44) and are consequently incapable of infecting even closely related species (A. P. N. Ambagala et al., unpublished data). This specificity restricts the study of CMV to its target species and reiterates the importance of developing animal models that are closely related to humans in an effort to study HCMV pathogenesis.
Animal models to study CMV biology have been largely limited to mice, guinea pigs, and rhesus macaques. As an alternative, cynomolgus macaques (Macaca fascicularis) are a species of Old World monkeys that have the potential to serve as a novel NHP model to study CMV pathogenesis. Cynomolgus macaques are extensively used as an animal model for infectious disease research (12, 23, 36, 67) and transplant research (21, 34, 60) and are becoming an increasingly popular NHP model for human immunodeficiency virus (HIV) vaccine development (7, 47, 66). Within the field of HIV vaccine design, there is a real need to diversify the pool of vectors undergoing testing. Recent studies using rhesus macaque CMV (RhCMV) as a simian immunodeficiency virus (SIV) vaccine vector have shown much promise in the ability of the vaccine to mount a robust effector memory response, thus providing vaccinated macaques with long-term protection from SIV disease progression (26, 29). CMV strains are not conserved even between closely related NHPs, and our recent experience suggests that cynomolgus macaques are not readily infected with RhCMV (Ambagala et al., unpublished). In order to overcome this strong host restriction and evaluate CMV as an HIV vector in a cynomolgus macaque-SIV model, we must use a cynomolgus macaque CMV (CyCMV). We have recently isolated and characterized a novel CyCMV (Ottawa strain) (3). Here we describe the complete genomic sequence and organization of the CyCMV genome for its use as an alternative NHP model to evaluate CMV pathogenesis and vaccine strategies. We compare and contrast the structural and functional genes of CyCMV with those of HCMV and RhCMV with respect to pathogenesis, immune evasion, and species specificity.
MATERIALS AND METHODS
CyCMV viral DNA isolation.
Ottawa strain CyCMV was isolated from catheter-derived urine samples from a healthy, captive-bred, 4-year-old cynomolgus macaque of Philippine origin as described previously (3). A cynomolgus macaque fibroblast cell line was not available at the time of isolation. Initial attempts were made to grow CyCMV in telomerase-immortalized rhesus macaque fibroblast cells (Telo-RF) (35); however, given the rapid growth properties of Telo-RF cells, this cell line could not support the slow growth kinetics of the CyCMV clinical isolate (3). To circumvent this, we propagated the virus in human fetal lung fibroblast cells (MRC-5) (30), which have slower growth properties and have been used extensively to propagate CMVs (9, 18, 63). CyCMV was passaged 16 times in MRC-5 cells to obtain high-titer virus stocks. The virus was not plaque purified, and thus, the sequence likely represents a consensus of one or more strain variants. In order to isolate viral DNA, CyCMV-infected cells were lysed with Hirt extraction buffer (2× Tris-EDTA, 1.2% SDS) for 20 min at room temperature, treated with 1 M sodium chloride overnight at 4°C, and subsequently centrifuged at 27,000 × g for 35 min at 4°C to precipitate the cellular DNA and proteins. The supernatant containing viral DNA was treated with an RNase cocktail (60 μg/ml RNase A and 160 U/ml RNase T1; Fermentas) for 2 h at 37°C and with pronase (1 mg/ml; Roche) for 2 h at 37°C. The supernatant was deproteinized with three phenol-chloroform extractions, and the viral DNA was precipitated with 0.3 M sodium acetate and 2 volumes of absolute ethanol overnight at −20°C. The sample was centrifuged at 17,000 × g for 30 min at 4°C, and the viral DNA was overlaid on a discontinuous 5 to 20% sucrose gradient containing ethidium bromide (2 μg/ml). Following centrifugation at 200,000 × g for 2.5 h at 4°C, the viral DNA was visualized by UV illumination, collected and diluted in 1.5 volumes of water, and precipitated with 0.3 M sodium acetate and 2.5 volumes of absolute ethanol overnight at −20°C. The sample was centrifuged at 15,000 × g for 30 min at 4°C and washed once with 70% ethanol, and the viral DNA was resuspended in water. CyCMV viral DNA (1 μg) was digested with 20 U of HindIII or BamHI at 37°C overnight and fractionated by gel electrophoresis on a 0.8% agarose gel.
Next-generation DNA sequencing.
Using 9.4 μg of CyCMV DNA, a paired-end library with a 500-bp insert size was prepared to generate read lengths of 72 bp. To sequence the complete CyCMV genome, high-throughput Illumina Genome Analyzer II paired-end sequencing was performed at The Centre of Applied Genomics, Toronto, Ontario, Canada.
Bioinformatic assembly.
The CyCMV genome was assembled de novo from 18,205,114 paired 72-bp reads (∼6,000-fold coverage) derived from a run of the Illumina Genome Analyzer II platform. Isolated paired ends were filtered to match the barcode (3,391,350 paired reads, ∼1,120-fold coverage) and were assembled using Velvet (version 0.7.55) (73). The best results were obtained using a kmer length of 39, the shortPaired mode, an insert length of 500 bp, and an expected coverage of 242 to yield a single large contig of 220 kbp.
Gap closing.
The resulting Velvet assembly had 11 gaps (runs of Ns), with lengths of 9 to 124. We implemented a simple greedy assembly program that started from a seed sequence, identifying all possible overlapping reads and extended the seed until no further extension was possible. This process is analogous to those described previously (40, 72). By providing the Velvet program with the areas close to gaps as seeds, we were able to generate sequence and close 10 of the 11 gaps in the initial Velvet assembly.
PCR sequencing.
To confirm the integrity of the sequence, areas of low coverage from the next-generation sequencing data were verified by Sanger sequencing. PCR amplicons were gel purified with GeneClean II (MP Biomedicals) if necessary, cloned into the pCR-Blunt II-TOPO vector (Invitrogen), and transformed with chemically competent cells (Invitrogen). The full-length gene inserts were confirmed by Sanger sequencing using standard sequencing primers [M13 For (UP), 5′-CACGACGTTGTAAAACGAC-3′; M13 Rev (−27), 5′-CAGGAAACAGCTATGAC-3′ (Invitrogen)]. Any remaining sequence was determined by primer walking. Sequences were aligned by ClustalW using Geneious Pro 5.1.7 (Biomatters Ltd., Auckland, New Zealand).
Error correction.
In order to identify additional assembly errors, we aligned all of the Illumina reads with the finished assembly and corrected 64 positions (out of a total assembly size of 220 kbp) where the base pair present in the reference genome occurred 7 times less frequently than an alternative base and replaced each such base with the alternative. Furthermore, for regions of the assembly with low coverage or with conflicting base calls (including all 11 of the gaps initially identified as described above), we generated 71 Sanger sequences. By analyzing the Sanger sequences together with the base qualities in the aligned Illumina data, we were able to correct 24 additional assembly errors (14 single base pair modifications and 10 insertions/deletions of 1 to 23 bp). Finally, the CyCMV sequence was aligned with that of RhCMV 68.1 (accession no. AY186194) to confirm the orientation of the sequence.
ORF assignment.
Open reading frames (ORFs) were identified on both strands with Geneious Pro 5.1.7 (Biomatters Ltd., Auckland, New Zealand) by using the following criteria: (i) the ORF began with a start codon (ATG) and ended with a stop codon, (ii) the ORF was a minimum of 100 amino acids (aa) long (including the start and stop codons), and (iii) the ORF did not overlap another ORF on the same strand within the same reading frame (13). The majority (83%) of the genes were annotated by using these criteria, and the remaining genes were identified using a smaller size criterion of ≥30 aa in an effort to identify all possible ORFs. Homology for each ORF was determined using the BLASTP program (NCBI) with >20% identity. BLAST scores (2) and homology details were obtained from NCBI.
The genome homology of CyCMV with other CMVs was determined by Geneious alignment using a global alignment with free and end gaps and a cost matrix of 65% similarity (5.0/−4.0), a gap open penalty of 12, and a gap extension penalty of 3 (Geneious Pro 5.1.7; Biomatters Ltd., Auckland, New Zealand).
Nucleotide sequence accession number.
The fully annotated and complete nucleotide sequence of the Ottawa strain of CyCMV has been submitted to GenBank and assigned accession number JN227533.
RESULTS
Genomic analysis of CyCMV.
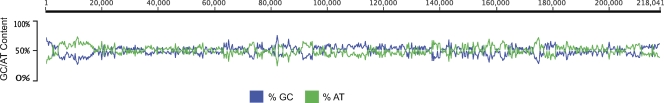
CyCMV was isolated from a cynomolgus macaque of Philippine origin and sequenced using paired-end sequencing (Illumina) to an average of 516-fold coverage/nucleotide. The CyCMV genome is 218,041 bp in length with a 49.5% G+C content and 84% protein-coding density. CyCMV is shorter and less GC rich than HCMV AD169 (229,354 bp, 57.2%) (13) or chimpanzee CMV (CCMV) (241,087 bp, 61.7%) (14) and more comparable to both strains of RhCMV (68.1, 221,459 bp; 180.92, 215,678 bp, 49% G+C content) (28, 57) (Fig. 1). Similar to other CMV genomes, CyCMV has a low G+C content at the beginning of the genome (bp 4559 to 17305), in various regions across the genome (bp 72074 to 74174, 91077 to 94827, 137006 to 140096, 161230 to 163989, and 172919 to 175284), and at the end of the genome (bp 205859 to 211243). Consistent with other CMV genomes, CyCMV is organized with a unique long region followed by a unique short region. When the genome sequence and gene products of CyCMV are compared to those of other CMVs, CyCMV most closely resembles RhCMV. The CyCMV genome is 54.8% identical to that of HCMV AD169 (13), 53.6% identical to that of CCMV (14), 54.2% identical to that of AtriCMV-1 (accession no. FJ483970), 57.5% identical to that of SsciCMV-1 (accession no. FJ483967), 67.5% identical to that of CeHV-5 strain GR2715 (accession no. FJ483968), 67.8% identical to that of CeHV-5 strain Colburn (accession no. FJ483969), 89.8% identical to that of RhCMV 68.1 (28), and 88.2% identical to that of RhCMV 180.92 (57) at the nucleotide level. The first base call of CyCMV corresponds to bp 3996 of HCMV (AD169), is −490 bp from the first nucleotide of the CCMV genome, and is −50 bp from beginning of the RhCMV genome.
Fig. 1.
G+C content of the CyCMV genome. The base composition across the CyCMV genome is represented by percent GC (blue) and AT (green) contents. Window size, 50 bp.
Restriction digestion.
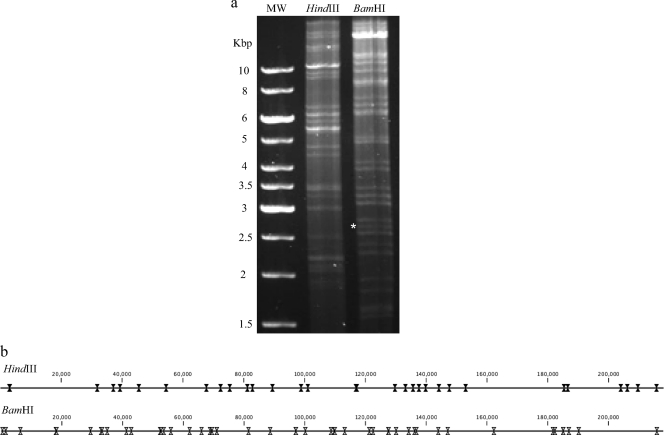
To assess gross viral genome structure, a restriction digest analysis was performed and digested bands were confirmed based on predicted fragment sizes (Geneious Pro 5.1.7; Biomatters Ltd., Auckland, New Zealand). The CyCMV genome was digested with restriction enzymes HindIII and BamHI and fractionated on an agarose gel (Fig. 2a). The digested CyCMV fragments were compared with the predicted fragments generated from the sequence data using bioinformatic software (Fig. 2b). All fragments were present at the expected size, with the exception of an additional band running at approximately 2.7 kbp upon digestion with BamHI (Fig. 2a). It is possible that there is an extra BamHI site located between cy92 and cyUL69 that is not accurately represented in the final sequence data. This region encompasses the putative origin of lytic replication, a region known for structural complexity (10). This region is inherently challenging to sequence due the presence of inverted and repeated sequence motifs (10) and proved difficult to sequence in our study by both next-generation and Sanger sequencing. The RhCMV genome does indeed contain a BamHI restriction site in the origin of lytic replication. We predict that the additional CyCMV band is the result of a missing restriction site in the sequence, although the restriction site may be present in the viral DNA.
Fig. 2.
Restriction enzyme digestion of CyCMV genome. To assess gross viral genome structure, a restriction digest analysis was performed. CyCMV viral DNA was digested with the HindIII and BamHI restriction enzymes. DNA fragments (900 ng) were separated by electrophoresis on a 0.8% agarose gel (a). The digested CyCMV DNA has an additional BamHI band (*) at approximately 2.7 kbp. A map of the CyCMV genome digested with HindIII (31 sites) and BamHI (49 sites) was generated using CLC Main Workbench (v 6.1) (b). Lane MW, 10-kbp ladder.
Gene assignment.
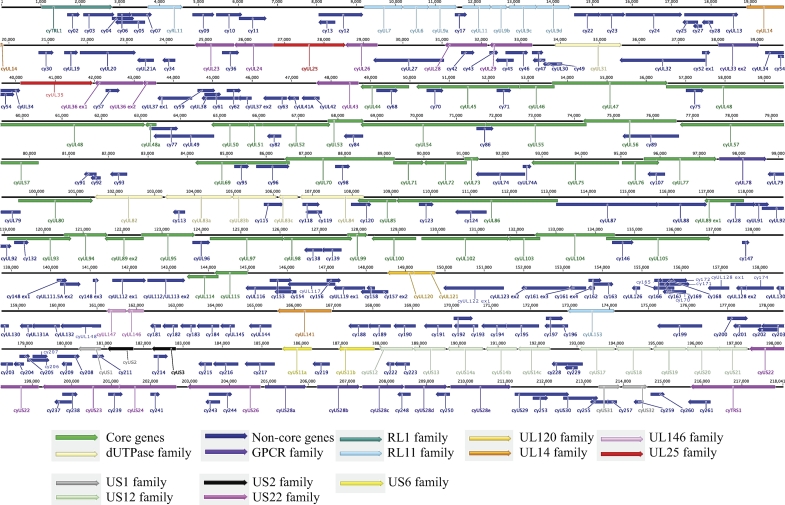
We have identified 262 putative ORFs (Table 1) with a mean coding length of 789 bp. The genes were numbered starting at the left of the genome and continuing to the right with nomenclature similar to that used in annotating the ORFs in other NHP CMVs. The genomic organization of CyCMV is largely colinear with that of RhCMV. The CyCMV gene arrangement with color-coded herpesvirus core genes and gene families is shown in Fig. 3. All alpha-, beta-, and gammaherpesviruses have 40 conserved genes known as core genes (44). CyCMV contains 39 of the 40 core genes with no homologue to HCMV UL108. The function of UL108 is not known, and its deletion from the HCMV genome results in only moderate growth defects (17).
Table 1.
CyCMV gene products
| ORFa | Translation |
Strand | Size (aa) | Mol mass (kDa) | Function/gene family(ies)b | HCMVc homologue (%)d | RhCMV homologue (%)d |
||
|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | 68.1 | 180.92 | ||||||
| cyTRL1 | 1040 | 2776 | + | 579 | 63.6 | RL1 | TRL1 (37) | Rh01 (84.1) | rhRL1 (84.9) |
| cy02 | 1693 | 1953 | + | 87 | 9.9 | rh02 (76.8) | rh2 (78) | ||
| cy03 | 2097 | 2831 | + | 245 | 26.2 | rh2.1 (62) | rh2.1 (65.1) | ||
| cy04 | 2828 | 3325 | + | 166 | 18.2 | rh03 (92.7) | rh3 (92) | ||
| cy05 | 2913 | 3611 | − | 233 | 30.0 | rh04 (88.4) | rh4 (87.9) | ||
| cy06 | 2961 | 3263 | + | 101 | 11.5 | rh3.1 (97) | rh3.1 (96) | ||
| cy07 | 3298 | 3783 | + | 162 | 18.2 | rh3.2 (89) | rh3.2 (89.6) | ||
| cyRL11 | 3738 | 4559 | + | 274 | 30.1 | IgG Fc-binding membrane glycoprotein/RL11 | RL11 (31.3) | Rh05 (94.9) | rh5 (95.2) |
| cy09 | 4873 | 5358 | + | 162 | 18.6 | rh06 (47.6) | rh6 (46.8) | ||
| cy10 | 5475 | 6074 | + | 200 | 22.7 | rh07 (91.5) | rh7 (91.5) | ||
| cy11 | 6049 | 6690 | + | 214 | 24.3 | rh08 (88.4) | rh8 (82.4) | ||
| cy12 | 8067 | 9140 | + | 358 | 39.9 | Rh17 (85.2) | rh17 (86.3) | ||
| cy13 | 8075 | 8437 | − | 121 | 13.1 | rh18 (71.9) | rh18 (72.5) | ||
| cyUL7 | 9222 | 10166 | + | 315 | 35.6 | Putative membrane glycoprotein/RL11 | UL7 (34.7) | Rh19 (87.6) | rhUL7 (87.6) |
| cyUL6 | 10224 | 10817 | + | 198 | 22.1 | Putative membrane glycoprotein/RL11 | UL6 (27.9) | Rh20 (84.8) | rhUL6 (84.1) |
| cyUL9a | 10844 | 11527 | + | 228 | 26.4 | Putative membrane glycoprotein/RL11 | UL9 (28.5) | Rh21 (62.2) | rh21 (66) |
| cy17 | 11527 | 11772 | + | 82 | 10.0 | Rh22 (42.9) | rh22 (56.8) | ||
| cyUL11 | 11724 | 12404 | + | 227 | 25.4 | Membrane glycoprotein/RL11 | UL11 (33.3) | Rh23 (80.6) | rhUL11 (81) |
| cyUL9b | 12455 | 12817 | + | 121 | 13.7 | Putative membrane glycoprotein/RL11 | UL9 (25.3) | rh24 (87.5) | rh24 (87.5) |
| cyUL9c | 12896 | 13570 | + | 225 | 24.7 | Putative membrane glycoprotein/RL11 | UL9 (28.5) | Rh25 (92.9) | rh26 (29.5) |
| cyUL9d | 13590 | 14408 | + | 273 | 31.0 | Putative membrane glycoprotein/RL11 | UL9e (31.9) | Rh26 (77.7) | rh26 (78.1) |
| cy22 | 14556 | 15164 | + | 203 | 23.0 | rh27 (94.1) | rh27 (90.1) | ||
| cy23 | 15166 | 15789 | + | 208 | 23.8 | rh28 (73.4) | rh28 (77.3) | ||
| cy24 | 15865 | 17235 | + | 457 | 49.6 | Rh29 (85.5) | rh29 (91) | ||
| cy25 | 17311 | 17634 | − | 108 | 11.5 | rh30 (94.4) | rh30 (93.2) | ||
| cyUL13 | 17351 | 18658 | + | 436 | 50.3 | Putative secreted protein | UL13 (34.2) | Rh31 (92.9) | rh31 (93.6) |
| cy27 | 17573 | 17743 | − | 57 | 6.3 | rh32 (89.3) | rh32 (87.5) | ||
| cy28 | 17798 | 18025 | − | 76 | 8.7 | rh32 (85.3) | rh32 (86.7) | ||
| cyUL14 | 18934 | 19845 | + | 304 | 35.3 | Putative membrane glycoprotein/UL14 | UL14 (32.8) | Rh33 (97.7) | rhUL14 (97.4) |
| cy30 | 20786 | 21109 | + | 108 | 12.7 | Rh35 (95.4) | |||
| cyUL19 | 21438 | 21725 | + | 96 | 10.9 | UL19 (46.2) | rhUL19 (97) | rhUL19 (95.8) | |
| cyUL20 | 21834 | 23189 | + | 452 | 51.0 | T-cell receptor γ chain homologue | UL20 (36.7) | Rh36 (94.5) | rhUL20 (94.7) |
| cyUL21A | 23294 | 23656 | − | 121 | 13.9 | CC chemokine-binding protein | UL21A (41.6) | rh37 (97.5) | rhUL21a (98.3) |
| cy34 | 23932 | 24195 | − | 88 | 9.2 | rh39 (56) | |||
| cyUL23 | 24752 | 25690 | − | 313 | 35.9 | TP/US22 | UL23 (44.4) | Rh40 (95.2) | |
| cy36 | 25437 | 25817 | + | 127 | 14.7 | rh41 (95.2) | rh41 (94.4) | ||
| cyUL24 | 25747 | 26673 | − | 309 | 35.1 | TP/US22 | UL24 (53.3) | Rh42 (98.1) | rhUS22 (28) |
| cyUL25 | 26740 | 28509 | + | 590 | 67.4 | Tegument phosphoprotein/UL25 | UL25 (40.4) | Rh43 (95.1) | rhUL25 (95.2) |
| cyUL26 | 28573 | 29328 | − | 252 | 28.2 | TP; transcriptional activator of major immediate-early promoter/US22 | UL26 (46.7) | Rh44 (97.1) | rhUL26 (97.1) |
| cyUL27 | 29282 | 31015 | − | 578 | 65.7 | UL27 (55.8) | Rh46 (97.8) | rhUL27 (97.8) | |
| cyUL28 | 31100 | 32113 | − | 338 | 38.7 | US22 | UL28 (67.1) | Rh47 (97.6) | rhUL28 (97.9) |
| cy42 | 31514 | 31786 | + | 91 | 10.2 | rh48 (94.4) | |||
| cy43 | 32224 | 32388 | + | 55 | 6.3 | Rh49 (88.7) | rh49 (90.6) | ||
| cyUL29 | 32242 | 33252 | − | 337 | 38.9 | US22 | UL29 (62.1) | Rh50 (99.1) | rhUL29 (98.8) |
| cy45 | 32385 | 32789 | − | 135 | 15.2 | rh51 (93.3) | rh51 (94) | ||
| cy46 | 32638 | 33150 | + | 171 | 19.3 | rh52 (93.7) | |||
| cy47 | 33304 | 33537 | + | 78 | 9.0 | rh52 (100) | |||
| cyUL30 | 33326 | 33592 | − | 89 | 10.3 | UL30 (37.7) | |||
| cy49 | 33582 | 34016 | − | 145 | 17.4 | rh53 (95.5) | |||
| cyUL31 | 33899 | 35524 | + | 542 | 61.0 | dUTPase | UL31 (58.6) | Rh54 (98.2) | |
| cyUL32 | 35535 | 37667 | − | 711 | 79.5 | Major TP (pp150) | UL32 (54.1) | Rh55 (93) | rhUL32 (93.1) |
| cy52 ex1 | 37621 | 37773 | + | 51 | 6.0 | UL33 (94) | |||
| cyUL33 ex2 | 38035 | 39024 | + | 330 | 37.2 | Virion envelope protein/GPCR | UL33 (56.2) | Rh56 (92.4) | rhUL33 ex2 (95.7) |
| cyUL34 | 39229 | 40086 | + | 286 | 32.9 | Repression of US3 transcription | UL34 (64.4) | Rh57 (96.6) | rhUL34 (98.9) |
| cy54 | 39543 | 39929 | + | 129 | 13.9 | rh58 (92.2) | rh58 (93) | ||
| cyUL35 | 40149 | 41921 | + | 591 | 66.9 | Tegument phosphoprotein; interaction with UL82 protein/UL25 | UL35 (43.9) | Rh59 (98.5) | rhUL25 (22.3) |
| cyUL36 ex1 | 42038 | 43201 | − | 388 | 44.7 | Immediate-early TP; inhibitor of caspase-8-induced apoptosis (vICA)/US22 | UL36 (44) | rhUL36 (96.9) | rhUL36 (97.2) |
| cy57 | 42308 | 42607 | + | 100 | 11.5 | rh59.1 (92) | rh59.1 (91.9) | ||
| cyUL36 ex2 | 43248 | 43529 | − | 94 | 10.5 | Immediate-early TP; inhibitor of caspase-8-induced apoptosis (vICA)/US22 | UL36 (58.4) | rhUL36 (96.6) | rhUL36 (97.4) |
| cyUL37 ex1 | 43627 | 44445 | − | 273 | 31.2 | Immediate-early glycoprotein; mitochondrial inhibitor of apoptosis (vMIA) | UL37 (30.5) | rhUL37 (96) | rhUL37 (96) |
| cy59 | 44646 | 45029 | + | 128 | 13.5 | rh63 (96.9) | |||
| cyUL38 | 44750 | 45631 | − | 294 | 33.2 | Virion envelope glycoprotein | UL38 (54.7) | Rh64 (96.6) | rhUL38 (96.9) |
| cy61 | 44816 | 45181 | + | 122 | 13.8 | rh65 (93.4) | rh65 (92.6) | ||
| cy62 | 45377 | 45676 | + | 100 | 11.3 | rh65.1 (89) | rh65.1 (87.9) | ||
| cyUL37 ex2 | 45673 | 45963 | − | 97 | 11.0 | Immediate-early glycoprotein | UL37 (36.6) | rhUL37 (90.6) | rhUL37 (90.3) |
| cy63 | 46313 | 46843 | − | 177 | 19.0 | rh67 (93.2) | |||
| cyUL41A | 46921 | 47160 | − | 80 | 9.4 | Virion envelope protein | UL41Af (36.7) | rhUL41a (99) | rhUL41a (98.7) |
| cyUL42 | 47291 | 47680 | − | 130 | 14.3 | Putative MP | UL42 (45.9) | Rh68 (96.8) | rhUL42 (96.1) |
| cyUL43 | 47664 | 48665 | − | 334 | 38.5 | TP/US22 | UL43 (46.3) | Rh69 (97.9) | rhUS22 (26.1) |
| cyUL44 | 48784 | 49956 | − | 391 | 44.0 | DNA polymerase processivity factor | UL44 (69.2) | Rh70 (99) | |
| cy68 | 49186 | 49662 | + | 159 | 18.0 | rh71 (92.4) | |||
| cyUL45 | 50197 | 52749 | − | 851 | 97.0 | TP; ribonucleotide reductase subunit 1 | UL45 (61.2) | Rh72 (98.5) | |
| cy70 | 50458 | 50817 | + | 120 | 13.4 | rh73 (92.4) | |||
| cy71 | 52235 | 52537 | + | 101 | 11.3 | rh74 (100) | |||
| cyUL46 | 52768 | 53640 | − | 291 | 33.1 | Capsid triplex subunit 1 | UL46f (72.1) | Rh75 (98.6) | |
| cyUL47 | 53639 | 56515 | + | 959 | 110.6 | TP | UL47 (41.8) | Rh76 (97.3) | rhUL47 (97.2) |
| cyUL48 | 56536 | 63069 | + | 2,178 | 246.9 | Large TP | UL48 (41.3) | Rh78 (98.3) | rhUL48 (98.4) |
| cy75 | 57035 | 57427 | + | 131 | 15.1 | rh78.1 (89) | rh78.1 (89.2) | ||
| cyUL48a | 63141 | 63362 | − | 74 | 8.4 | Small capsid protein | UL48a (65.3) | rhUL48a (100) | rhUL48a (100) |
| cy77 | 63241 | 63906 | + | 222 | 23.8 | rh79 (98.6) | |||
| cyUL49 | 63355 | 64824 | − | 490 | 56.2 | UL49 (71.5) | Rh80 (99.2) | rhUL49 (99.4) | |
| cyUL50 | 64814 | 65695 | − | 294 | 32.4 | Inner nuclear MP; nuclear egress MP | UL50 (80.6) | rhUL50 (99.3) | |
| cyUL51 | 65721 | 66056 | − | 112 | 12.4 | DNA-packaging protein; terminase component | UL51 (83.1) | Rh82 (99.1) | rhUL51 (97.3) |
| cyUL52 | 66116 | 67774 | + | 553 | 62.5 | UL52 (55.9) | Rh83 (98.2) | rhUL52 (98.4) | |
| cy82 | 66226 | 66534 | − | 103 | 10.8 | rh84 (97.6) | rh84 (97.6) | ||
| cyUL53 | 67767 | 68630 | + | 288 | 32.9 | TP; nuclear matrix protein; nuclear egress lamina protein | UL53 (75.4) | Rh85 (98.3) | |
| cy84 | 68175 | 68606 | − | 144 | 16.0 | rh86 (100) | |||
| cyUL54 | 68608 | 71715 | − | 1,036 | 116.6 | DNA polymerase | UL54 (62.2) | Rh87 (99) | rhUL54 (98.9) |
| cy86 | 71536 | 71913 | + | 126 | 14.1 | Rh88 (97.6) | rh88 (98.4) | ||
| cyUL55 | 71734 | 74283 | − | 850 | 97.1 | Glycoprotein B | UL55 (58.4) | Rh89 (79.5) | rhUL55 (76.8) |
| cyUL56 | 74249 | 76558 | − | 770 | 88.3 | DNA-packaging terminase component | UL56 (73.2) | Rh91 (98.6) | rhUL56 (98.7) |
| cy89 | 75252 | 76622 | − | 457 | 48.4 | rh91.1 (97) | rh91.1 (97.6) | ||
| cyUL57 | 76705 | 80196 | − | 1,164 | 129.3 | Single-stranded DNA-binding protein | UL57 (74.8) | Rh92 (99.7) | rhUL57 (99.6) |
| cy91 | 81413 | 81661 | − | 83 | 9.3 | rh93 (86.6) | rh93 (87.8) | ||
| cy92 | 81579 | 81770 | − | 64 | 6.7 | rh93 (50) | |||
| cy93 | 82075 | 82446 | − | 124 | 12.6 | rh95 (97.4) | rh95 (98.3) | ||
| cyUL69 | 84231 | 86564 | − | 778 | 87.4 | TP; multiple regulatory protein | UL69f (49.5) | Rh97 (96) | rhUL69 (96.1) |
| cy95 | 85222 | 85530 | + | 103 | 11.8 | rh98 (99) | |||
| cy96 | 85773 | 86561 | + | 263 | 28.4 | rh99 (94.7) | rh99 (95) | ||
| cyUL70 | 86498 | 89236 | − | 913 | 105.5 | DNA helicase primase subunit | UL70 (65.3) | Rh100 (98.5) | rhUL70 (98.5) |
| cy98 | 87773 | 88096 | + | 108 | 11.5 | rh99.1 (94) | rh99.1 (94.4) | ||
| cyUL71 | 89249 | 89968 | + | 240 | 26.4 | TP | UL71 (59.5) | rhUL71 (95) | rhUL71 (95.4) |
| cyUL72 | 90036 | 91067 | − | 344 | 39.2 | dUTPase/dUTPase | UL72 (58.7) | Rh101 (98) | rhUL72 (97.7) |
| cyUL73 | 91062 | 91373 | + | 104 | 11.8 | Glycoprotein N | UL73 (60.8) | Rh102 (97.1) | |
| cyUL74 | 91354 | 92535 | − | 394 | 45.9 | Glycoprotein O | UL74 (43.7) | Rh103 (94.7) | rhUL74 (94.4) |
| cyUL74A | 92534 | 92704 | + | 57 | 6.4 | Envelope glycoprotein 24 | UL74A (55.3) | ||
| cyUL75 | 92761 | 94923 | − | 721 | 81.6 | Glycoprotein H | UL75 (49.1) | Rh104 (98.3) | rhUL75 (98.2) |
| cyUL76 | 95056 | 95940 | + | 295 | 32.8 | Virion-associated regulatory protein | UL76 (55.4) | Rh105 (98.6) | rhUL76 (98.3) |
| cyUL77 | 95609 | 97396 | + | 596 | 67.4 | Portal-capping protein; DNA packaging | UL77 (64.5) | Rh106 (99.5) | rhUL77 (99.3) |
| cy107 | 95705 | 96088 | − | 128 | 14.4 | rh106.1 (96.1) | |||
| cyUL78 | 97523 | 98662 | + | 380 | 41.9 | Putative chemokine receptor/GPCR | UL78 (30.4) | Rh107 (91.6) | rhUL78 (91.6) |
| cyUL79 | 98758 | 99558 | − | 267 | 30.5 | UL79 (71) | Rh108 (98.9) | rhUL79 (99.2) | |
| cyUL80 | 99557 | 101392 | + | 612 | 66.4 | Capsid maturation protease | UL80 (44.1) | Rh109 (97.9) | rhUL80 (97.9) |
| cyUL82 | 101507 | 103156 | − | 550 | 61.6 | Tegument phosphoprotein pp71 (upper matrix protein)/UL82, dUTPase | UL82 (42.3) | Rh110 (95.3) | rhUL83b (26) |
| cyUL83a | 103286 | 104911 | − | 542 | 62.2 | Major tegument phosphoprotein pp65 (lower matrix protein)/UL82, dUTPase | UL83 (35.3) | Rh111 (95.6) | rhUL83a (95.4) |
| cy113 | 103486 | 103737 | − | 84 | 9.2 | R83a (89.5) | |||
| cyUL83b | 104980 | 106050 | − | 357 | 40.4 | Major tegument phosphoprotein pp65 (lower matrix protein)/UL82, dUTPase | UL83 (38.9) | Rh112 (94.7) | rhUL83b (94.4) |
| cy115 | 105787 | 106218 | + | 144 | 15.8 | rh113 (87.6) | rh113 (86.9) | ||
| cyUL83c | 106169 | 106594 | − | 142 | 15.9 | Major tegument phosphoprotein pp65 (lower matrix protein)/UL82, dUTPase | UL83 (45.2) | Rh112 (97.1) | rhUL83b (97.1) |
| cyUL84 | 106714 | 108258 | − | 515 | 57.6 | Role in organizing DNA replication/UL82, dUTPase | UL84 (50) | Rh114 (98.8) | rhUL84 (98.6) |
| cy118 | 106775 | 107149 | + | 125 | 13.5 | rh115 (96) | rh115 (95.2) | ||
| cy119 | 106850 | 107209 | − | 120 | 13.3 | rh115.1 (90.8) | |||
| cy120 | 108000 | 108458 | + | 153 | 16.1 | rh116 (97.4) | |||
| cyUL85 | 108173 | 109099 | − | 309 | 34.7 | Capsid triplex subunit 2 | UL85f (74) | Rh117 (99.4) | |
| cyUL86 | 109160 | 113191 | − | 1,344 | 151.3 | Major capsid protein | UL86 (76) | Rh118 (98.9) | rhUL86 (98.8) |
| cy123 | 109725 | 110039 | + | 105 | 11.8 | rh119 (98.1) | |||
| cy124 | 110652 | 111386 | − | 245 | 28.7 | rh120 (97.1) | |||
| cyUL87 | 113206 | 115758 | + | 851 | 96.5 | UL87 (67.7) | Rh122 (99.4) | ||
| cyUL88 | 115771 | 116973 | + | 401 | 45.5 | TP | UL88g (53.7) | Rh123 (98) | |
| cyUL89 ex1 | 116970 | 117917 | − | 316 | 35.8 | DNA-packaging terminase component | UL89 (84.8) | UL89 (100) | rhUL89 (100) |
| cy128 | 117633 | 118202 | + | 190 | 20.8 | rh125 (94.7) | rh125 (94.2) | ||
| cyUL91 | 118234 | 118545 | + | 104 | 10.9 | UL91 (56.3) | Rh126 (92.3) | rhUL91 (94.2) | |
| cyUL92 | 118430 | 119143 | + | 238 | 26.5 | UL92h (90.5) | UL92 (100) | rhUL92 (98.7) | |
| cyUL93 | 119109 | 120671 | + | 521 | 59.7 | DNA-packaging; TP | UL93 (48.2) | Rh128 (96.5) | rhUL93 (96.5) |
| cy132 | 119270 | 119590 | + | 107 | 11.1 | rh128.1 (94) | rh128.1 (93.4) | ||
| cyUL94 | 120547 | 121587 | + | 347 | 37.8 | TP; binds single-stranded DNA | UL94 (58.8) | Rh129 (98.5) | rhUL94 (96.2) |
| cyUL89 ex2 | 121576 | 122508 | − | 311 | 36.2 | DNA-packaging terminase component | UL89 (86.5) | UL89 (99.7) | rhUL89 (100) |
| cyUL95 | 122507 | 123802 | + | 432 | 47.1 | UL95 (66.3) | Rh130 (99.5) | rhUL95 (99.3) | |
| cyUL96 | 123799 | 124188 | + | 130 | 14.8 | TP | UL96 (59.8) | Rh131 (97.7) | rhUL96 (96.9) |
| cyUL97 | 124245 | 126071 | + | 609 | 67.9 | TP; viral serine-threonine protein kinase | UL97 (66.5) | Rh132 (96.4) | UL97 (96.9) |
| cyUL98 | 126122 | 127792 | + | 557 | 63.4 | DNase | UL98 (66) | Rh134 (99.5) | |
| cy138 | 126634 | 127035 | − | 134 | 14.6 | rh135 (98.5) | |||
| cy139 | 127073 | 127522 | − | 150 | 16.9 | Rh136 (99.3) | rh136 (98.7) | ||
| cyUL99 | 127729 | 128181 | + | 151 | 16.5 | Myristylated tegument phosphoprotein (pp28) | UL99 (57.1) | rh137 (94) | |
| cyUL100 | 128354 | 129424 | − | 357 | 41.0 | Glycoprotein M | UL100 (58.1) | Rh138 (98.3) | |
| cyUL102 | 129613 | 131790 | + | 726 | 80.5 | DNA helicase primase subunit | UL102 (67) | Rh139 (97.8) | rhUL102 (97.7) |
| cyUL103 | 131812 | 132567 | − | 252 | 28.9 | TP | UL103f (55.1) | Rh140 (97.2) | |
| cyUL104 | 132494 | 134464 | − | 657 | 75.5 | Capsid portal protein | UL104 (68.3) | Rh141 (99.4) | rhUL104 (99.2) |
| cyUL105 | 134301 | 136880 | + | 860 | 97.6 | DNA helicase primase subunit | UL105 (71.9) | Rh142 (99.2) | |
| cy146 | 134450 | 134929 | + | 160 | 18.0 | rh142.1 (92) | rh142.1 (93) | ||
| cy147 | 137750 | 137881 | + | 44 | 5.2 | rh142.3 (97.6) | |||
| cy148 ex1 | 140183 | 140380 | + | 66 | 7.1 | Interleukin-10-like protein precursor | Ul11A (90.6) | rhUL111a (93.5) | |
| cyUL111.5A ex2 | 140268 | 140729 | + | 154 | 17.7 | Latency-associated viral interleukin-10 | UL111.5A (37) | Rh143 (92.8) | rhUL111a (91) |
| cy148 ex3 | 141097 | 141186 | + | 30 | 3.3 | Interleukin-10-like protein precursor | Ul11A (91.3) | rhUL111a (91.3) | |
| cyUL112 ex1 | 141591 | 142385 | + | 265 | 28.3 | Early phosphoprotein (p50) | UL112 (54.3) | rhUL112 (91.4) | |
| cyUL112/UL113 ex2 | 142484 | 143374 | + | 297 | 30.7 | Early phosphoprotein (p84) | UL112/UL113 (34.8) | rhUL112 (94.6) | rhUL112 (94.6) |
| cyUL114 | 143485 | 144228 | − | 248 | 28.3 | Uracil-DNA glycosylase | UL114 (69) | Rh146 (99.6) | |
| cyUL115 | 144191 | 144967 | − | 259 | 29.2 | Glycoprotein L | UL115 (50.6) | Rh147 (98.8) | rhUL115 (98.4) |
| cyUL116 | 144978 | 146060 | − | 361 | 38.2 | Putative membrane glycoprotein | UL116 (26.7) | Rh148 (92.5) | rhUL116 (82.5) |
| cy153 | 145585 | 146082 | + | 166 | 16.8 | rh147.1 (97) | rh147.1 (86.7) | ||
| cy154 | 145714 | 146199 | − | 162 | 20.1 | rh149 (92.9) | rh149 (90.7) | ||
| cyUL117 | 146042 | 147190 | − | 383 | 42.6 | UL117 (47.8) | Rh150 (99.7) | rhUL117 (99.5) | |
| cy156 | 147026 | 147328 | + | 101 | 11.1 | rh149.1 (97) | rh149.1 (98) | ||
| cyUL119 ex1 | 147215 | 147946 | − | 244 | 28.4 | Virion envelope glycoprotein; IgG Fc-binding glycoprotein | UL119 (35.1) | rhUL119 (96.5) | rhUL119 (96.1) |
| cy157 ex2 | 147894 | 148532 | − | 213 | 21.3 | rhUL119 (79.8) | rhUL119 (65.8) | ||
| cy158 | 148043 | 148195 | − | 51 | 5.4 | rh153 (76.9) | rh153 (87.5) | ||
| cyUL120 | 148581 | 149177 | − | 199 | 22.6 | Putative membrane glycoprotein/UL120 | UL120 (44.1) | Rh154 (92.4) | |
| cyUL121 | 149179 | 149727 | − | 183 | 21.0 | Putative membrane glycoprotein/UL120 | UL121 (26.9) | Rh155 (96.7) | rh155 (96.2) |
| cyUL122 ex1 | 149990 | 151474 | − | 495 | 53.6 | Immediate-early 2 transactivator | UL122 (58.7) | IE (89.9) | rhUL122 (88.7) |
| cyUL123 ex2 | 151977 | 153122 | − | 382 | 43.1 | Major immediate-early 1 cotransactivator | UL123 (23.7) | IE 1 (59.4) | rhUL123 (60.7) |
| cy161 ex3 | 153182 | 153559 | − | 126 | 14.4 | Immediate-early protein | IE (63.3) | rhUL122 (63.3) | |
| cy161 ex4 | 153665 | 153784 | − | 40 | 4.1 | Immediate-early protein | IE 1 (87.2) | rhUL123 (84.6) | |
| cy162 | 153674 | 153937 | + | 88 | 10.4 | rh156.1 (92) | rh156.1 (90.8) | ||
| cy163 | 153783 | 154238 | + | 152 | 15.8 | rh156.2 (93) | rh156.2 (94) | ||
| cyUL126 | 154779 | 154925 | − | 49 | 5.7 | UL126 (55.8) | |||
| cy165 | 155301 | 155795 | − | 165 | 18.6 | rh157.1 (87) | rh157.1 (87.8) | ||
| cy166 | 155342 | 155491 | + | 50 | 5.8 | rh157.1 (61) | rh157.1 (61.4) | ||
| cy167 | 155476 | 155679 | + | 68 | 7.1 | rh157.3 (55.1) | |||
| cy168 | 155547 | 155780 | + | 78 | 8.4 | rh157.1 (47) | rh157.1 (53.6) | ||
| cy169 | 155555 | 155704 | − | 50 | 5.2 | rh157.3 (91.8) | |||
| cy170 | 155717 | 156094 | − | 126 | 14.8 | rh157 (70.2) | rh157.3 (79.6) | ||
| cy171 | 155719 | 156183 | + | 155 | 18.3 | rh157 (75.8) | rh157 (80.9) | ||
| cy172 | 155823 | 156128 | + | 102 | 11.6 | rh157.2 (82) | rh157.2 (82) | ||
| cyUL128 ex1 | 156700 | 157182 | − | 161 | 18.3 | Putative secreted protein; putative CC chemokine | UL128 (37.1) | rhUL128 (96.7) | |
| cyUL128 ex2 | 157420 | 157608 | − | 63 | 7.1 | Putative secreted protein; putative CC chemokine | UL128 (55.2) | rhUL128 (93.6) | |
| cy174 | 157610 | 158317 | − | 236 | 25.5 | rh157.4 (76.2) | |||
| cyUL130 | 158438 | 158731 | − | 98 | 11.5 | Putative secreted protein | UL130 (40.7) | rhUL130 (85.6) | |
| cyUL131A | 159065 | 159322 | − | 86 | 9.7 | Putative secreted protein | UL131A (35.3) | rh131a (96.5) | |
| cyUL132 | 159366 | 160025 | − | 220 | 24.1 | Envelope glycoprotein | UL132 (32.1) | Rh160 (94.9) | rhUL132 (94.4) |
| cyUL148 | 160091 | 161071 | − | 327 | 37.0 | Putative membrane glycoprotein | UL148 (30.5) | Rh159 (89) | rhUL148 (89.1) |
| cyUL147 | 161273 | 161707 | − | 145 | 16.5 | Chemokine vCXCL2/UL146 | UL147 (43.3) | Rh158 (88.2) | |
| cyUL146 | 161786 | 162148 | − | 121 | 13.5 | Chemokine vCXCL1/UL146 | UL146i (28.1) | ||
| cy181 | 162372 | 162590 | − | 73 | 8.2 | Alpha-chemokine-like protein | |||
| cy182 | 162689 | 163015 | − | 109 | 12.1 | Alpha-chemokine-like protein | |||
| cy183 | 163138 | 163470 | − | 111 | 12.5 | Alpha-chemokine-like protein | rh161 (35.2) | ||
| cy184 | 163554 | 164072 | − | 173 | 19.3 | Alpha-chemokine-like protein | rh161 (97.3) | ||
| cyUL145 | 164157 | 164462 | − | 102 | 11.3 | UL145 (65.6) | Rh162 (99) | ||
| cyUL144 | 164880 | 165395 | − | 172 | 18.7 | Membrane glycoprotein; tumor necrosis factor receptor homologue | UL144f (29.6) | Rh163 (98.8) | |
| cyUL141 | 165611 | 166909 | − | 433 | 49.1 | Membrane glycoprotein/UL14 | UL141 (39.4) | Rh164 (97) | |
| cy188 | 167408 | 167854 | − | 149 | 17.1 | rh165 (94.6) | |||
| cy189 | 167899 | 168426 | − | 176 | 19.4 | Rh166 (96) | |||
| cy190 | 168557 | 169060 | − | 168 | 18.2 | rh167 (95.2) | rh167 (95.9) | ||
| cy191 | 169318 | 169977 | − | 220 | 24.7 | rh168 (94.1) | rh168 (93.6) | ||
| cy192 | 170074 | 170505 | + | 144 | 16.3 | rh168.1 (79) | rh168.1 (79.7) | ||
| cy193 | 170080 | 170643 | − | 188 | 20.8 | rh169 (90.4) | rh169 (88.2) | ||
| cy194 | 170777 | 171343 | − | 189 | 21.2 | rh170 (96.8) | rh170 (97.3) | ||
| cy195 | 171346 | 172185 | − | 280 | 30.7 | Rh171 (91.4) | rh171 (90.7) | ||
| cy196 | 172385 | 172915 | − | 177 | 19.9 | rh172 (96) | rh172 (97.2) | ||
| cy197 | 172418 | 172792 | + | 125 | 13.9 | rh171.1 (90) | rh171.1 (91.9) | ||
| cyUL153 | 172967 | 174088 | − | 374 | 40.6 | MP RL13/RL11 | UL153h (32.8) | rh173 (57.3) | rh173 (93) |
| cy199 | 175240 | 176325 | − | 362 | 39.9 | rh174 (92.2) | rh174 (92.2) | ||
| cy200 | 177022 | 177480 | + | 153 | 16.5 | rh175 (92.8) | rh175 (93.4) | ||
| cy201 | 177171 | 177824 | − | 218 | 23.7 | rh176 (92.2) | rh176 (93.1) | ||
| cy202 | 177790 | 178263 | − | 158 | 17.8 | rh177 (78.8) | rh177 (78.2) | ||
| cy203 | 177884 | 178657 | − | 258 | 28.2 | rh178 (86.1) | rh178 (84.9) | ||
| cy204 | 178734 | 178931 | − | 66 | 7.0 | rh178.2 (82.8) | |||
| cy205 | 178882 | 179079 | + | 66 | 6.9 | rh178.1 (51) | rh178.1 (47.8) | ||
| cy206 | 179072 | 179575 | + | 168 | 18.2 | rh178.3 (64.1) | rh178.3 (71.7) | ||
| cy207 | 179212 | 179535 | + | 108 | 11.8 | rh178.3 (78) | rh178.3 (80.8) | ||
| cy208 | 179835 | 180350 | + | 172 | 18.6 | rh179 (92.4) | rh179 (95.3) | ||
| cy209 | 179951 | 180157 | − | 69 | 6.8 | rh180 (91.2) | rh180 (95.6) | ||
| cyUS1 | 180377 | 180883 | − | 169 | 19.3 | US1 | US1 (49.4) | Rh181 (97.6) | rhUS1 (98.8) |
| cy211 | 180732 | 180965 | + | 78 | 8.3 | rh180.1 (91) | rh180.1 (96.1) | ||
| cyUS2 | 181118 | 181708 | − | 197 | 23.3 | Membrane glycoprotein/US2 | US2 (21) | Rh182 (76.9) | rh182 (78.5) |
| cyUS3 | 182241 | 182786 | − | 182 | 20.7 | Immediate-early glycoprotein/US2 | US3 (26.4) | Rh184 (92.7) | rh184 (92.7) |
| cy214 | 182286 | 182579 | + | 98 | 10.8 | Rh183 (90.7) | rh183 (90.7) | ||
| cy215 | 183413 | 183712 | − | 100 | 11.1 | rh184.1 (96) | rh184.1 (96) | ||
| cy216 | 183794 | 184366 | − | 191 | 21.4 | Rh185 (95.3) | rh185 (94.7) | ||
| cy217 | 184596 | 185300 | − | 235 | 27.8 | rh186 (82.9) | rh186 (82.5) | ||
| cyUS11a | 185545 | 186228 | − | 228 | 25.7 | Membrane glycoprotein/US6 | US11 (24) | Rh187 (92.1) | rh187 (91.6) |
| cy219 | 186324 | 186698 | − | 125 | 14.6 | rh188 (94.4) | |||
| cyUS11b | 186981 | 187823 | − | 281 | 32.8 | Membrane glycoprotein/US6 | US11 (29.1) | Rh189 (87.6) | rhUS11 (87.6) |
| cyUS12 | 188025 | 188807 | − | 261 | 30.0 | Putative multiple-transmembrane protein/US12 | US12 (37.2) | Rh190 (98.5) | |
| cy222 | 188182 | 188334 | − | 51 | 5.4 | rh191 (84) | |||
| cy223 | 188395 | 188724 | − | 110 | 12.0 | rh191 (87.2) | |||
| cyUS13 | 188865 | 189629 | − | 255 | 29.7 | Putative multiple-transmembrane protein/US12 | US13f (25.9) | Rh192 (99.6) | rhUS13 (99.2) |
| cyUS14a | 189742 | 190575 | − | 278 | 31.4 | Putative multiple-transmembrane protein/US12 | US14 (25.3) | Rh194 (97.8) | rh194 (98.2) |
| cyUS14b | 190706 | 191434 | − | 243 | 27.4 | Putative multiple-transmembrane protein/US12 | US14 (25.3) | Rh195 (98.8) | rhUS14 (26.4) |
| cyUS14c | 191527 | 192285 | − | 253 | 29.5 | Putative multiple-transmembrane protein/US12 | US14 (29.7) | Rh196 (98.4) | rhUS14 (98.8) |
| cy228 | 192391 | 193116 | − | 242 | 28.0 | Rh197 (96.3) | rh197 (96.3) | ||
| cy229 | 192716 | 192985 | − | 90 | 10.8 | rh196.1 (92.1) | |||
| cyUS17 | 193094 | 193918 | − | 275 | 30.4 | Putative multiple-transmembrane protein/US12 | US17 (42.9) | Rh198 (98.2) | rhUS17 (97.4) |
| cyUS18 | 194024 | 194824 | − | 267 | 30.1 | Putative multiple-transmembrane protein/US12 | US18f (28.6) | rhUS18 (98.1) | |
| cyUS19 | 194944 | 195729 | − | 262 | 30.0 | Putative multiple-transmembrane protein/US12 | US19 (27.8) | Rh200 (95) | rh200 (95) |
| cyUS20 | 195790 | 196551 | − | 254 | 28.6 | Putative multiple-transmembrane protein/US12 | US20 (43.9) | Rh201 (99.6) | rhUS13 (27) |
| cyUS21 | 196599 | 197285 | − | 229 | 26.1 | Putative multiple-transmembrane protein/US12 | US21 (61.1) | Rh202 (98.2) | |
| cyUS22 | 197407 | 199131 | − | 575 | 65.8 | TP/US22 | US22 (46.3) | Rh203 (96.9) | rhUS22 (96.7) |
| cyUS23 | 199290 | 201161 | − | 624 | 72.7 | TP/US22 | US23 (51.8) | Rh204 (98.2) | rhUS23 (97.9) |
| cy237 | 199589 | 200002 | − | 138 | 16.5 | rh206 (90.5) | rh206 (89.7) | ||
| cy238 | 199788 | 200117 | + | 110 | 12.9 | rh207 (92.5) | rh205 (92.5) | ||
| cy239 | 200946 | 201263 | + | 106 | 11.5 | rh208 (89.5) | rh208 (90.5) | ||
| cyUS24 | 201185 | 202615 | − | 477 | 56.6 | TP/US22 | US24 (66.2) | rh209 (98.7) | |
| cy241 | 202022 | 202207 | + | 62 | 7.0 | rh210 (90) | |||
| cyUS26 | 202978 | 204759 | − | 594 | 67.3 | US22 | US26 (46.8) | rh211 (96.8) | rhUS26 (96.6) |
| cy243 | 203421 | 203735 | − | 105 | 11.6 | rh212 (98.1) | |||
| cy244 | 203512 | 204027 | + | 172 | 19.0 | rh213 (94.7) | |||
| cyUS28a | 204931 | 205917 | + | 329 | 37.5 | MP; CC and CX3C chemokine receptor; mediates cellular activation and migration; virion envelope glycoprotein/GPCR | US28 (29.1) | Rh214 (97.9) | rh214 (98.2) |
| cyUS28b | 206261 | 207274 | + | 338 | 38.7 | MP; CC and CX3C chemokine receptor; mediates cellular activation and migration; virion envelope glycoprotein/GPCR | US28 (25.4) | Rh215 (93.5) | rh218 (38.1) |
| cyUS28c | 207400 | 208401 | + | 334 | 38.1 | MP; CC and CX3C chemokine receptor; mediates cellular activation and migration; virion envelope glycoprotein/GPCR | US28 (24.5) | rhUS28.2 (97.3) | rh218 (37.8) |
| cy248 | 208296 | 208568 | − | 91 | 10.2 | Rh217 (88.9) | rh217 (91.1) | ||
| cyUS28d | 208474 | 209496 | + | 341 | 39.1 | MP; CC and CX3C chemokine receptor; mediates cellular activation and migration; virion envelope glycoprotein/GPCR | US28 (26.2) | Rh218 (96.2) | rh218 (97.1) |
| cy250 | 209266 | 209571 | − | 102 | 11.4 | rh219 (94.1) | rh219 (93.1) | ||
| cyUS28e | 209641 | 211104 | + | 488 | 54.0 | MP; CC and CX3C chemokine receptor; mediates cellular activation and migration; virion envelope glycoprotein/GPCR | US28 (40) | Rh220 (85.3) | rhUS28 (89.2) |
| cyUS29 | 211265 | 212581 | + | 439 | 49.3 | Putative membrane glycoprotein | US29 (38.1) | Rh221 (95) | rhUS29 (94.8) |
| cy253 | 211728 | 212051 | + | 108 | 12.7 | rh222 (97.2) | |||
| cyUS30 | 212499 | 213320 | + | 274 | 30.8 | Putative membrane glycoprotein | US30 (22.2) | Rh223 (96) | rh223 (96) |
| cy255 | 213182 | 213544 | − | 121 | 12.6 | rh224 (95.8) | rh224 (95.8) | ||
| cyUS31 | 213396 | 213881 | + | 162 | 18.5 | US1 | US31 (42.7) | Rh225 (94.4) | rhUS31 (93.2) |
| cy257 | 213572 | 213799 | − | 76 | 8.5 | rh224 (95.2) | rh224 (93.7) | ||
| cyUS32 | 214008 | 214568 | + | 187 | 22.2 | US1 | US32 (44.7) | Rh226 (96.8) | rhUS32 (96.2) |
| cy259 | 214288 | 214482 | − | 65 | 7.2 | rh227 (93.7) | |||
| cy260 | 214712 | 215017 | + | 102 | 10.9 | rh228 (93.1) | rh228 (88.3) | ||
| cy261 | 215664 | 216176 | − | 171 | 18.6 | rh229 (86.2) | rh229 (86.8) | ||
| cyTRS1 | 215743 | 217818 | − | 692 | 77.3 | TP; immediate-early protein/US22 | TRS1 (37.4) | Rh230 (92.4) | rhTRS1 (92.2) |
The CyCMV genes with an HCMV homologue are annotated as “cy” followed by the HCMV name.
Functions and gene families were assigned based on studies of HCMV (44).
HCMV homologues are from strains AD169 and Toledo unless otherwise specified.
Percent identity based on a BLASTP search conducted in June 2011.
HCMV strain 3301.
HCMV strain Merlin.
HCMV strains CINCY and Towne.
HCMV strain Towne.
HCMV strain NT.
Fig. 3.
Map of ORFs in the CyCMV genome. CyCMV encodes 262 putative ORFs that are annotated by gene name and color coded based on gene families. The genomic organization of CyCMV is largely colinear with that of RhCMV. The CyCMV genes with an HCMV homologue are annotated by “cy” followed by the HCMV name, and the arrowheads indicate the directions of the ORFs. Core genes are herpesvirus core genes.
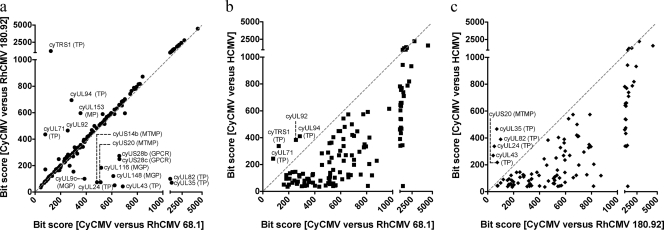
Of the 262 CyCMV genes, 137 are homologous to HCMV genes, 243 are homologous to RhCMV 68.1 genes, and 200 are homologous to RhCMV 180.92 genes. With respect to the RhCMV genomes, CyCMV encodes homologues for 230 (89%) of the 260 RhCMV 68.1 genes and 180 (70%) of the 258 RhCMV 180.92 genes. CyCMV gene homologues were compared between HCMV and both strains of RhCMV based on their alignment bit scores to determine if a particular CyCMV gene is more closely related to a particular strain of CMV (Fig. 4). The majority of the CyCMV genes are biased toward the RhCMV genomes; however, there are some exceptions in which the HCMV gene homologues have the higher bit scores. The outliers are mainly membrane proteins (MP) and tegument proteins (TP).
Fig. 4.
CyCMV gene similarities between RhCMV strains and HCMV. A bit score was calculated for each CyCMV gene versus its putative homologue in RhCMV or HCMV. The data points would be expected to be distributed along the line x = y (gray dashed line), where CyCMV is no more closely related to HCMV than to RhCMV 68.1 or 180.92, respectively. The graphs represent comparisons of the CyCMV gene homologue bit scores versus the two RhCMV strains (a), RhCMV 68.1 versus HCMV (b), and RhCMV 180.92 versus HCMV (c). Outlier genes are annotated according to their CyCMV names and putative functions. MTMP, multiple-transmembrane protein; MGP, membrane glycoprotein.
Genes missing from other macaque CMVs.
In the 262 CyCMV ORFs, four genes show homology to HCMV genes that are not present in either strain of the RhCMV sequenced genomes (68.1 or 180.92). These genes include UL30, UL74A, UL126, and UL146. It should be noted that a wild-type isolate of RhCMV (RhCMVCNPRC) does contain the HCMV homologue of UL146 (48) and cyUL146 has 76.1% identity with its RhCMVCNPRC counterpart. The functions of UL30 (cyUL30) and UL126 (cyUL126) have yet to be elucidated; however, it is known that UL74A (cyUL74A) encodes an envelope glycoprotein and UL146 (cyUL146) contains an alpha-chemokine homologue, vCXCL1, belonging to the UL146 gene family (50). UL146 exhibits a high degree of sequence variability between HCMV strains and among species (5). Notably, cyUL146 has retained the chemokine motif, ELRCXC (not shown), that is required for alpha-chemokines to recruit neutrophils (5). During CMV infection, vCXCL1 plays a role in neutrophil attraction and degranulation, resulting in increased viral dissemination both within and between hosts (50).
Tropism genes.
CyCMV encodes a number of HCMV homologues for tropism genes that have been shown to be essential for HCMV propagation in various cell types. The HCMV homologues (UL128, UL130, and UL131A) that have been shown to be associated with endothelial cell, macrophage, and dendritic cell tropism (25, 61) have been retained in CyCMV. The CyCMV genes that show homology with the HCMV UL128-131 region include cyUL128 ex1 (37.1%) and ex2 (55.2%), cyUL130 (40.7%), and cyUL131A (35.3%). An additional HCMV gene known to be required for viral replication in human microvascular endothelial cells (HMVEC) is UL24 (17), for which CyCMV encodes cyUL24 as a homologue with 53.3% identity. Similarly, HCMV UL64 and US29 were shown to be required for growth in human retinal pigment epithelial (RPE) cells (17). CyCMV encodes a US29 homologue (cyUS29) with 38.1% identity but does not encode a UL64 homologue. Functional studies are required to determine if CyCMV can replicate in epithelial cells in the absence of a UL64 homologue. Furthermore, in the functional profiling of HCMV, it was determined that the deletion of UL10 and UL16 increases the viral titer in RPE cells and the deletion of US16 and US19 also results in a higher viral titers in HMVEC (17). CyCMV does not encode the above-listed HCMV homologues, with the exception of cyUS19, which is an HCMV homologue of US19. With respect to RhCMV-specific tropism genes, four genes (Rh01, Rh159, Rh160, and Rh203) have been shown to be tropism determinants for RhCMV (strain 68.1) replication in rhesus RPE cells (38). The CyCMV homologues of these RhCMV tropism genes include cyTRL1 (84.1%), cyUL148 (89%), cyUL132 (94.9%), and cyUS22 (96.9%). CyCMV deletion studies are required to determine if CyCMV exhibits the same impaired viral replication in epithelial cells. The CyCMV tropism genes (cyUL24, cyUL131A, cyUL148, and cyUS22) encode full-length homologues of their respective HCMV and/or RhCMV counterparts. However, the homologues for cyTRL1, cyUL128 ex1/ex2, cyUL130, cyUL132, cyUS19, and cyUS29 represent only partial alignments with the intact CyCMV ORF due to N- and/or C-terminal truncations.
Functional genes.
Although CyCMV has homology with a number of HCMV MP and TP, the genes that have a functional role in DNA replication, packaging, and egress are the most conserved. These proteins include DNA-packaging terminase components (cyUL89 ex1, 84.8%; cyUL89 ex2, 86.5%; cyUL56, 73.2%), a DNA-packaging protein (cyUL51, 83.1%), nuclear egress membrane and lamina proteins (cyUL50, 80.6%; cyUL53, 75.4%), a major capsid protein (cyUL86, 76%), a single-stranded DNA-binding protein (cyUL57, 74.8%), capsid triplex subunits 1 and 2 (cyUL46, 72.1%; cyUL85, 74%), a DNA polymerase processivity factor (cyUL44, 69.2%), a uracil-DNA glycosylase (cyUL114, 69%), DNA helicase primase subunits (cyUL70, 65.3%; cyUL102, 67%; cyUL105, 71.9%), a capsid portal protein (cyUL104, 68.3%), a viral serine-threonine protein kinase (cyUL97, 66.5%), a DNase (cyUL98, 66%), a DNA polymerase (cyUL54, 62.2%), a small capsid protein (cyUL48a, 65.3%), a portal-capping/DNA-packaging protein (cyUL77, 64.5%), and ribonucleotide reductase subunit 1 (cyUL45, 61.2%). The percent identities to HCMV genes are described, and it should be noted that the RhCMV complements for these same genes are even more conserved, with an average of 99% identity to their CyCMV counterparts.
Surface glycoproteins.
CMV surface glycoproteins are commonly used for identification and classification purposes and to assess phylogenetic relationships between CMVs (3). CyCMV encodes HCMV homologues for glycoproteins B (cyUL55), N (cyUL73), O (cyUL74), H (cyUL75), M (cyUL100), and L (cyUL115). Glycoprotein N (UL73) is a highly variable HCMV glycoprotein (51); however, its CyCMV homologue (cyUL73) exhibits the highest degree of homology (60.8%) compared to the remaining HCMV glycoprotein homologues. Another highly polymorphic HCMV glycoprotein is glycoprotein O (UL74) (51), which is the least conserved of the glycoprotein homologues in CyCMV, with 43.7% identity.
Viral homologues of chemokine receptor and GPCR proteins.
Chemokine receptor (CXCL) and G protein-coupled receptor (GPCR) gene homologues are carried by CMVs from various species. These receptor homologues are organized in gene clusters, and the number of repeated genes in a cluster differs between species and between isolates, given that these genes are dispensable for growth in fibroblast cells (1). CyCMV has retained six alpha-chemokine receptor homologues that are clustered together in a 3.98-kbp coding region encompassing cyUL147 to cy184. Likewise, CyCMV contains a cluster of five GPCR homologues (cyUS28a, cyUS28b, cyUS28c, cyUS28d, and cyUS28e) that encode the HCMV homologue of US28, a GPCR known to bind chemokines (69). CyCMV encodes seven genes (cyUL33 ex2, cyUL78, and cyUS28a to cyUS28e) that are homologous to three of the four GPCR family genes (UL33, UL78, and US28). The only GPCR homologue absent from the CyCMV genome is US27, a virion envelope glycoprotein (44).
Immunomodulatory genes.
CMVs contain a number of genes that function to evade the immune response of the infected host. CyCMV encodes HCMV homologues for major histocompatibility complex class I (MHC-I) downregulation genes (US2, US3, and US11), viral interleukin-10 (UL111.5A), a tumor necrosis factor receptor homologue (UL144), and antiapoptotic genes (UL36, UL37 ex1, and UL38). In addition to the HCMV genes, cy203 carries a homologue of the RhCMV-specific gene (rh178) involved in MHC-I downregulation by interference with the translation of the heavy chain portion of the MHC-I molecule (53, 56). Although it was originally thought to be unique to RhCMV (53), it appears that this immunomodulatory gene may in fact be an NHP-specific immunevasin.
The HCMV immunomodulatory genes β2.7, UL16, UL18, UL142, US6, US8, and US10 are not present in CyCMV. Although these genes are important for evading the immune system, their deletion does not have any effect on viral growth in vitro (17, 42). According to the criterion (>20% identity) used to assign CyCMV homologues, CyCMV does not contain a homologue of the HCMV US6 gene. The RhCMV gene Rh185 has a low degree of sequence homology with US6; however, it has been shown to be functionally similar to US6 and therefore has been assigned as a putative homologue (49). Given that cy216 shows significant homology with Rh185 (95.3%) from the RhCMV 68.1 genome, we propose that this CyCMV gene may also function to downregulate MHC-I and may be considered a US6 homologue. We have previously shown that CyCMV downregulates MHC-I expression on the surface of CyCMV-infected cells (3). Further functional studies are needed to determine if CyCMV has equivalent or uncharacterized homologues of the missing immunomodulatory genes to evaluate the effects of these deletions on immunomodulation.
Antiapoptotic genes.
The antiapoptotic genes carried by HCMV include a 2.7-kbp viral RNA (β2.7) and the UL36 to UL38 genes (41). It is likely that CyCMV does not transcribe a β2.7 gene equivalent, given than CyCMV does not encode an HCMV homologue of the predicted ORF (RL4) from which the β2.7 transcript is derived (42). The absence of the β2.7 gene does not affect HCMV growth kinetics in vitro (42). CyCMV contains homologues for the UL36 to UL38 genes (cyUL36 ex1 and ex2, cyUL37 ex1, and cyUL38, respectively). The function of UL36 is to inhibit caspase-8-induced apoptosis (vICA), and cyUL36 ex1 and ex2 have 44% and 58.4% identity with UL36. Similarly, UL37 is a mitochondrial inhibitor of apoptosis (vMIA) and cyUL37 ex1 has 30.5% identity with this HCMV gene. Human CMV UL38 (54.7% identity to cyUL38) is an antiapoptotic gene that blocks the cellular response pathway induced by stress, thus preventing cellular apoptosis; alternatively, when it is deleted from the genome, the target cells undergo apoptosis and HCMV exhibits viral replication defects (45, 65). It remains to be determined if these CyCMV genes have the same antiapoptotic roles as their HCMV homologues.
Latency genes.
Of the known CMV latency transcripts, CyCMV encodes only an HCMV homologue of UL111.5A (cyUL111.5A). The second exon of cy148 (cyUL111.5A ex2) has 37% identity with the latency-associated UL111.5A HCMV gene that encodes the viral interleukin-10 protein. Like HCMV, cy148 carries a spliced transcript with 3 exons, which is analogous to UL111.5A during productive infection (31) and differs from the RhCMV 180.92 homologue (RhUL111a), which contains 4 exons (57). Comparable to RhCMV, CyCMV does not contain an HCMV homologue of UL81 and thus does not contain the UL81-82 antisense transcript (LUNA) that is involved in latency (8, 55). Although CyCMV has retained a number of gene homologues from the HCMV ULb′ region, it is missing the HCMV homologue of UL138, a known latency gene that is also absent from the RhCMV genomes. Substitution studies have shown that removing UL138 from HCMV does not affect the in vitro growth kinetics of the virus when propagated in fibroblast cells, although they suggest that it could be cell type specific (24).
Spliced transcripts.
CyCMV encodes at least eight genes that are the products of spliced mRNA transcripts, and these include the commonly spliced CMV genes. The spliced transcripts that have two exons include a virion envelope protein/GPCR family protein (cy52 ex1 and cyUL33 ex2), tegument protein vICA (cyUL36), immediate-early glycoprotein vMIA (cyUL37), a DNA-packaging terminase component (cyUL89), an early phosphoprotein (cyUL112), IgG Fc-binding glycoprotein (cyUL119 ex1 and cy57 ex2), and a putative CC chemokine (cyUL128). Furthermore, cy148 contains 3 exons (cy148 ex1, cyUL111.5A ex2, and cy148 ex3) to produce the viral interleukin-10 protein. The cy161 ORF is spliced into 4 exons, where cyUL122 ex1 and cyUL123 ex2 produce immediate-early proteins 2 and 1, respectively, and cy161 ex3 and cy161 ex4 encode immediate-early proteins.
Missing genes.
Similar to HCMV, CyCMV does not contain the RhCMV-specific viral homologue of cyclooxygenase-2 (COX-2) (28). CyCMV lacks an approximately 6.7-kbp coding region equivalent to that of RhCMV rh9 to rh16, which encompasses the COX-2 gene homologue of rh10. In comparison to HCMV, CyCMV is lacking full complements for 84 HCMV genes, although the vast majority of these genes are uncharacterized and their deletion from the HCMV genome does not affect viral growth kinetics (Table 2). At the left terminus of the genome, CyCMV is lacking all of the RL genes except TRL1 and RL11 (cyTRL1 and cyRL11, respectively). These genes are generally present only in clinical isolates of CMV, as they are dispensable for growth in vitro (17). The RL11 family of genes is not present in mouse or rat CMV (54, 68). Of the absent HCMV genes, the only ones that have been reported to be required for viral replication are UL60 and UL90, both of which encode functionally uncharacterized proteins. It appears that CyCMV is lacking the HCMV homologues (UL58 to UL68) spanning the origin of lytic replication (oriLyt) that resides between cy92 and cyUL69. These HCMV genes (UL58 to UL68) are present in the AD169 strain; however, they are not present in the Toledo strain of HCMV or in either of the RhCMV strains. The only RhCMV homologues missing from the oriLyt area are rh94 and rh96, suggesting that CyCMV is not lacking any crucial genes in this region and has a gene allocation similar to that of HCMV (Toledo) and RhCMV.
Table 2.
HCMV genes not present in CyCMV
| Missing HCMV genea | Function/gene familyb | Effect of deletion on viral growth kineticsc |
|---|---|---|
| RL2 | − | |
| RL3 | ND | |
| RL4 | − | |
| RL5 | ND | |
| RL6 | RL11 family | − |
| RL7 | ND | |
| RL8 | ND | |
| RL9 | − | |
| RL10 | Envelope glycoprotein | − |
| RL12 | Putative membrane glycoprotein/RL11 family | − |
| RL13 | Putative membrane glycoprotein/RL11 family | − |
| RL14 | RL11 family | ND |
| UL1 | RL11 family | ND |
| UL2 | Putative MP | + |
| UL3 | − | |
| UL4 | Transcriptionally regulated envelope | − |
| glycoprotein/RL11 family | ||
| UL5 | RL11 family | − |
| UL8 | RL11 family | − |
| UL10 | Putative membrane glycoprotein/RL11 family | − |
| UL12 | + | |
| UL15 | Putative MP | − |
| UL16 | Membrane glycoprotein | − |
| UL17 | Seven-TM membrane glycoprotein | − |
| UL18 | Putative membrane glycoprotein; MHC-I | − |
| homologue/UL18 family | ||
| UL22 | Envelope glycoprotein; secreted glycoprotein | ND |
| UL39 | − | |
| UL40 | Membrane glycoprotein | ND |
| UL58 | ND | |
| UL59 | − | |
| UL60 | +++ | |
| UL61 | ND | |
| UL62 | − | |
| UL63 | ND | |
| UL64 | − | |
| UL65 | + | |
| UL66 | ND | |
| UL67 | − | |
| UL68 | ND | |
| UL80.5 | Capsid scaffold protein | ND |
| UL81 | ND | |
| UL90 | +++ | |
| UL101 | ND | |
| UL106 | ND | |
| UL107 | ND | |
| UL108 | + | |
| UL109 | − | |
| UL110 | − | |
| UL118 | ND | |
| UL124 | Putative membrane glycoprotein | ++d |
| UL125 | ND | |
| UL127 | − | |
| UL129 | + | |
| UL147A | Putative MP | ND |
| UL143 | ND | |
| UL142 | Putative membrane glycoprotein; MHC-I | ND |
| homologue/UL18 family | ||
| UL140 | Putative MP | ND |
| UL139 | Putative membrane glycoprotein | ND |
| UL138 | Putative MP | ND |
| UL137 | ND | |
| UL136 | Putative MP | ND |
| UL135 | Putative secreted protein | ND |
| UL134 | ND | |
| UL133 | Putative MP | ND |
| UL148A | Putative MP | ND |
| UL148B | Putative MP | ND |
| UL148C | Putative MP | ND |
| UL148D | Putative MP | ND |
| UL149 | ND | |
| UL150 | Putative secreted protein | ND |
| IRS1 | Immediate-early protein; TP/US22 family | − |
| US4 | ND | |
| US5 | ND | |
| US6 | Putative membrane glycoprotein/US6 family | − |
| US7 | Membrane glycoprotein/US6 family | − |
| US8 | Membrane glycoprotein/US6 family | − |
| US9 | Membrane glycoprotein/US6 family | − |
| US10 | Membrane glycoprotein/US6 family | − |
| US15 | Putative multiple-transmembrane | − |
| protein/US12 family | ||
| US16 | Putative multiple-transmembrane | − |
| protein/US12 family | ||
| US25 | − | |
| US27 | Virion envelope glycoprotein/GPCR family | − |
| US33 | − | |
| US34 | Putative secreted protein | − |
| US34A | Putative MP | ND |
Furthermore, CyCMV is also lacking complements of the UL2, UL12, UL65, UL108, and UL129 genes, all of which do not have a known function, with the exception of UL2 (putative MP). Only a modest effect on viral replication has been observed when these genes are deleted from the HCMV genome (17). There is ambiguity in the literature regarding the effect of knocking out the MP UL124 (17). The remaining HCMV gene deletions have yet to be examined for their effects on viral growth kinetics.
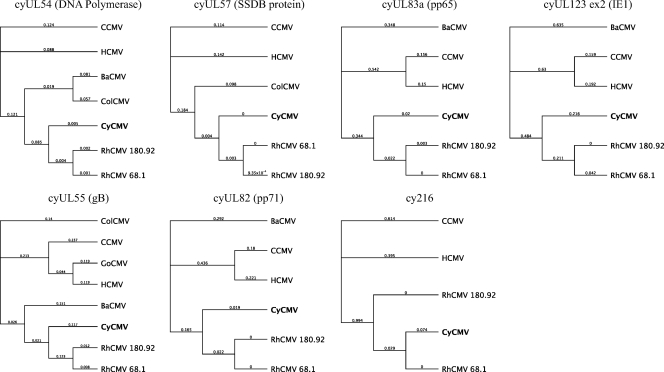
Phylogenetic analysis.
Phylogenetic trees have been generated to depict the evolutionary relatedness of genes common to different CMVs (Fig. 5). Genes that had the greatest number of sequenced strains were included in this analysis. As expected, these genes uniformly group more closely with RhCMV than with the other CMV strains. The cy216 gene was included in this analysis to clarify the discrepancy regarding the HCMV US6 homologue. For this gene, the number of substitutions per site for the branches separating CyCMV and RhCMV from HCMV is relatively high (0.994) in comparison to those of other genes represented by the phylogenetic trees. Furthermore of interest is the cy216 gene, in which the CyCMV complement is more closely related to RhCMV 68.1 than is RhCMV 180.92.
Fig. 5.
Phylogenetic analysis of CyCMV genes. Unrooted phylogenetic trees were created using Geneious Tree Builder with the protein sequences of various CMV strains. The relationships between strains are shown as cladograms, and the number of substitutions per site is listed on each branch. The CMV strains include CyCMV (bold), HCMV AD169, CCMV, BaCMV (baboon CMV), GoCMV (gorilla CMV), ColCMV (colobus guereza CMV), and RhCMV 68.1 and 180.92. SSDB, single-stranded DNA-binding protein.
DISCUSSION
The newly characterized CyCMV (Ottawa strain) is 218,041 bp in length, encodes 262 ORFs, and is most closely related to the two published genomes of RhCMV (strains 68.1 and 180.92). Although we have predicted 262 ORFs, we acknowledge that this may not be a complete representation of the CyCMV genome and that there may be additional ORFs carried in the genome that have yet to be elucidated. The virus was not plaque purified, and thus, the sequence likely represents a consensus of one or more strain variants. Our particular Illumina sequencing run had a calculated error frequency of 1% and acted as a baseline for substitution frequencies. In this manner, we determined that sequence calls other than the consensus represent, on average, only 1.9%. This determination does not allow us to calculate how many deviances from the consensus are contained simultaneously in a single genome sequence but does suggest that our current sequence likely represents a mixed population with only a diminutive portion of variants. With respect to interhost variability, we have previously examined the amino acid sequence of glycoprotein B (cyUL55) and have observed 99% identity between animals from the same geographic origin (3).
As the Ottawa strain of CyCMV is a multiply tissue culture-passaged virus, in vitro passage may have resulted in deletions impairing its coding capacity. However, in comparison to the multiple passages required to generate HCMV strains AD169 (54 passages) (19) and Towne (125 passages) (52), CyCMV (16 passages) would be considered only a moderately passaged isolate. Potential gene deletions could be further investigated by sequencing and characterizing a different isolate of CyCMV, specifically, a low-passage strain. The CyCMV genome is unique in that it contains four HCMV homologues (UL30, UL74A, UL126, and UL146) that are not present in either of the published RhCMV genomes (68.1 or 180.92), although an HCMV homologue of UL146 is present in a wild-type strain of RhCMV (RhCMVCNPRC) (48). There is no putative function for UL30 and UL126; however, it is known that UL74A is an envelope glycoprotein and UL146 is an alpha-chemokine homologue (vCXCL1). It has been suggested that vCXCL1 may act as a virulence determinant of CMV disease in individuals with a compromised adaptive immune system (43). Although CyCMV is a multipassaged derivative, with respect to the chemokine and GPCR gene clusters, CyCMV resembles a minimally passaged virus in that it has retained the clusters of six alpha-chemokine receptor homologues and five GPCR homologues. The wild-type isolates of RhCMV contain six CXCL and five GPCR gene clusters; however, in annotated RhCMV strains 68.1 and 180.92, half or all of the CXCL genes are deleted while all of the GPCR genes in the clusters are retained (1). CyCMV does not appear to have lost these genes in the same way that the RhCMV genomes have.
CyCMV does not contain a viral COX-2 gene that appears to be unique to RhCMV (28). Cellular COX-2 expression is induced upon HCMV infection and has been shown to play an important role in HCMV replication (74). Unlike HCMV infection, RhCMV infection does not induce cellular COX-2 expression in the presence of the viral COX-2 isoform in the RhCMV genome (rh10) (58). Further studies are required to determine if CyCMV infection induces cellular COX-2 expression in the same way as HCMV infection.
Given the importance of cynomolgus macaques as a widely utilized animal model for infectious disease and transplant research, the isolation and characterization of this highly prevalent endogenous virus may have a variety of applications. The seroprevalence of CyCMV in the cynomolgus macaque colony at the Public Health Agency of Canada in Ottawa, Ontario, is estimated to be 100% as measured by a CyCMV-specific enzyme-linked immunosorbent assay (3). In other studies, it has been observed that greater than 95% of NHPs bred in captivity are CMV seropositive (4). Fortunately, it has been shown that CMV-seropositive rhesus macaques can be superinfected with RhCMV (27). This has yet to be examined in cynomolgus macaques, although we believe that, as with RhCMV and HCMV, it would be possible achieve superinfection with CyCMV.
CMVs have evolved with their hosts over millions of years and have contained CMV-specific genes that are related to each host species. We have preliminary data suggesting that it may be inherently difficult to cross the species-specific barrier and infect cynomolgus macaques with RhCMV (Ambagala et al., unpublished). Although the CyCMV genome is nearly 90% identical to that of RhCMV (at the nucleotide level) and the genes are largely colinear with those of RhCMV, clearly there are factors influencing the host range specificity of the virus. The mechanism by which a host cell restricts viral replication from a different species has not been well elucidated. However, it is known that this restriction does not occur exclusively during the entry phase of CMV infection, as it has been shown that CMV has the capacity to infect a host cell from a distant species (20). One possible mechanism by which the host cell inhibits CMV replication from other species may be mediated by apoptosis, suggesting that the foreign virus cannot overcome the cellular innate immune defense of the host (32). CMVs contain antiapoptotic genes (β2.7, UL36, UL37 ex1, and UL38) that function to overcome the apoptosis response induced by the host innate immune response following CMV infection (44). The HCMV homologues of the UL36 to UL38 antiapoptotic genes are encoded by the CyCMV genome (cyUL36 ex1 and ex2, cyUL37 ex1, and cyUL38), and these CyCMV genes show high homology (∼96 to 97% identity) with their RhCMV counterparts (Table 1). Although it has been suggested that these antiapoptotic genes are important for host restriction in vivo, this does not appear to be the situation in vitro, where the species specificity is less restricted. CyCMV productively infects and replicates in human (MRC-5), rhesus macaque (Telo-RF), and cynomolgus macaque (MSFT) fibroblast cell lines (unpublished data). It is known that when CMV strains are grown in fibroblast cell lines, they classically eliminate the tropism genes required for replication in different cell types, most notably, endothelial cells (71). Although CyCMV has been propagated in fibroblast cells prior to sequencing, the genes that are required for endothelial cell tropism (UL128, UL130, and UL131A) have been retained in CyCMV (cyUL128 ex1/ex2, cyUL130, and cyUL131A, respectively). Furthermore, we have preliminary evidence demonstrating that CyCMV infects and efficiently replicates in human umbilical vein endothelial cells (data not shown). Endothelial cell tropism plays an important role in natural infection and viral transmission (70); thus, CyCMV may have utility for examining viral dissemination and pathogenesis in endothelial cells.
Congenital CMV remains the most common viral cause of birth defects in newborns, and yet there is still no vaccine (11). The burden of CMV disease is apparent not only in children but also in adults, specifically, those receiving solid organ or bone marrow transplants and those suffering from an immunocompromising disease such as HIV/AIDS. We have reason to be hopeful regarding the ability to make a CMV vaccine, given the success attained with another herpesvirus, varicella-zoster virus, in which the licensed vaccine has been highly effective in reducing the mortality associated with varicella infections in the United States (39). We hope this newly sequenced and characterized CyCMV genome will provide the necessary groundwork for further studies evaluating the utility of cynomolgus macaques as an alternative NHP model in which to study CMV biology, pathogenesis, and vaccine design.
ACKNOWLEDGMENTS
A.K.M. was supported by the Queen Elizabeth II/Community Health Graduate Scholarships in Science and Technology and the Bernhard Cinader Graduate Scholarship in Immunology. D.O.W. was supported by a Junior Investigator Development Award from the Ontario HIV Treatment Network (OHTN). A.P.N.A. and K.S.M. also received funding from the OHTN as a postdoctoral fellowship and a career scientist award, respectively. This research was funded in part by the Canadian Institutes of Health Research.
We sincerely thank the veterinary and technical staff at the NHP colony of Health Canada.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Alcendor D. J., et al. 2009. Patterns of divergence in the vCXCL and vGPCR gene clusters in primate cytomegalovirus genomes. Virology 395: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ambagala A. P., et al. 2011. Isolation and characterization of cynomolgus macaque (Macaca fascicularis) cytomegalovirus (CyCMV). Virology 412: 125–135 [DOI] [PubMed] [Google Scholar]
- 4. Andrade M. R., et al. 2003. Prevalence of antibodies to selected viruses in a long-term closed breeding colony of rhesus macaques (Macaca mulatta) in Brazil. Am. J. Primatol. 59: 123–128 [DOI] [PubMed] [Google Scholar]
- 5. Arav-Boger R., Foster C. B., Zong J. C., Pass R. F. 2006. Human cytomegalovirus-encoded alpha-chemokines exhibit high sequence variability in congenitally infected newborns. J. Infect. Dis. 193: 788–791 [DOI] [PubMed] [Google Scholar]
- 6. Bahr U., Darai G. 2001. Analysis and characterization of the complete genome of tupaia (tree shrew) herpesvirus. J. Virol. 75: 4854–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baroncelli S., Negri D. R., Michelini Z., Cara A. 2008. Macaca mulatta, fascicularis and nemestrina in AIDS vaccine development. Expert Rev. Vaccines 7: 1419–1434 [DOI] [PubMed] [Google Scholar]
- 8. Bego M., Maciejewski J., Khaiboullina S., Pari G., St. Jeor S. 2005. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J. Virol. 79: 11022–11034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blewett E. L., Lewis J., Gadsby E. L., Neubauer S. R., Eberle R. 2003. Isolation of cytomegalovirus and foamy virus from the drill monkey (Mandrillus leucophaeus) and prevalence of antibodies to these viruses amongst wild-born and captive-bred individuals. Arch. Virol. 148: 423–433 [DOI] [PubMed] [Google Scholar]
- 10. Borst E. M., Messerle M. 2005. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J. Virol. 79: 3615–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cannon M. J. 2009. Congenital cytomegalovirus (CMV) epidemiology and awareness. J. Clin. Virol. 46 (Suppl. 4): S6–S10 [DOI] [PubMed] [Google Scholar]
- 12. Capuano S. V., III, et al. 2003. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect. Immun. 71: 5831–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chee M. S., et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154: 125–169 [DOI] [PubMed] [Google Scholar]
- 14. Davison A. J., et al. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84: 17–28 [DOI] [PubMed] [Google Scholar]
- 15. Dolan A., et al. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85: 1301–1312 [DOI] [PubMed] [Google Scholar]
- 16. Dunn H. S., et al. 2002. Dynamics of CD4 and CD8 T cell responses to cytomegalovirus in healthy human donors. J. Infect. Dis. 186: 15–22 [DOI] [PubMed] [Google Scholar]
- 17. Dunn W., et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100: 14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eizuru Y., Tsuchiya K., Mori R., Minamishima Y. 1989. Immunological and molecular comparison of simian cytomegaloviruses isolated from African green monkey (Cercopithecus aethiops) and Japanese macaque (Macaca fuscata). Arch. Virol. 107: 65–75 [DOI] [PubMed] [Google Scholar]
- 19. Elek S. D., Stern H. 1974. Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet i: 1–5 [DOI] [PubMed] [Google Scholar]
- 20. Fioretti A., Furukawa T., Santoli D., Plotkin S. A. 1973. Nonproductive infection of guinea pig cells with human cytomegalovirus. J. Virol. 11: 998–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flores M. G., et al. 2005. A technique of bone marrow collection from vertebral bodies of cynomolgus macaques for transplant studies. J. Surg. Res. 124: 280–288 [DOI] [PubMed] [Google Scholar]
- 22. Gillespie G. M., et al. 2000. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8(+) T lymphocytes in healthy seropositive donors. J. Virol. 74: 8140–8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goff A. J., et al. 2011. A novel respiratory model of infection with monkeypox virus in cynomolgus macaques. J. Virol. 85: 4898–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goodrum F., Reeves M., Sinclair J., High K., Shenk T. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110: 937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn G., et al. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78: 10023–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen S. G., et al. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hansen S. G., et al. 2010. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328: 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen S. G., Strelow L. I., Franchi D. C., Anders D. G., Wong S. W. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J. Virol. 77: 6620–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hansen S. G., et al. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jacobs J. P., Jones C. M., Baille J. P. 1970. Characteristics of a human diploid cell designated MRC-5. Nature 227: 168–170 [DOI] [PubMed] [Google Scholar]
- 31. Jenkins C., Abendroth A., Slobedman B. 2004. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 78: 1440–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jurak I., Brune W. 2006. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 25: 2634–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanai K., et al. 2011. Re-evaluation of the genome sequence of guinea pig cytomegalovirus. J. Gen. Virol. 92: 1005–1020 [DOI] [PubMed] [Google Scholar]
- 34. Kawai T., et al. 2002. Effect of mixed hematopoietic chimerism on cardiac allograft survival in cynomolgus monkeys. Transplantation 73: 1757–1764 [DOI] [PubMed] [Google Scholar]
- 35. Kirchoff V., Wong S., St J. S., Pari G. S. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147: 321–333 [DOI] [PubMed] [Google Scholar]
- 36. Lawler J. V., et al. 2006. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. 3: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leendertz F. H., et al. 2009. Novel cytomegaloviruses in free-ranging and captive great apes: phylogenetic evidence for bidirectional horizontal transmission. J. Gen. Virol. 90: 2386–2394 [DOI] [PubMed] [Google Scholar]
- 38. Lilja A. E., Chang W. L., Barry P. A., Becerra S. P., Shenk T. E. 2008. Functional genetic analysis of rhesus cytomegalovirus: Rh01 is an epithelial cell tropism factor. J. Virol. 82: 2170–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marin M., Zhang J. X., Seward J. F. 2011. Near elimination of varicella deaths in the US after implementation of the vaccination program. Pediatrics 128: 214–220 [DOI] [PubMed] [Google Scholar]
- 40. Massouras A., et al. 2010. Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods 7: 485–486 [DOI] [PubMed] [Google Scholar]
- 41. McCormick A. L. 2008. Control of apoptosis by human cytomegalovirus. Curr. Top. Microbiol. Immunol. 325: 281–295 [DOI] [PubMed] [Google Scholar]
- 42. McSharry B. P., Tomasec P., Neale M. L., Wilkinson G. W. 2003. The most abundantly transcribed human cytomegalovirus gene (beta 2.7) is non-essential for growth in vitro. J. Gen. Virol. 84: 2511–2516 [DOI] [PubMed] [Google Scholar]
- 43. Mocarski E. S., Jr 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 10: 332–339 [DOI] [PubMed] [Google Scholar]
- 44. Mocarski E. S., Shenk T., Pass R. F. 2007. Cytomegaloviruses, p. 2702–2757 In Knipe D. M., et al. (ed.), Fields virology, 5th ed., vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 45. Moorman N. J., et al. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murphy E., et al. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100: 14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O'Connor D. H. 2006. Chinese rhesus and cynomolgus macaques in HIV vaccine and pathogenesis research. Future Virol. 1: 165–172 [Google Scholar]
- 48. Oxford K. L., et al. 2008. Protein coding content of the UL)b′ region of wild-type rhesus cytomegalovirus. Virology 373: 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pande N. T., Powers C., Ahn K., Fruh K. 2005. Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J. Virol. 79: 5786–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Penfold M. E., et al. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. U. S. A. 96: 9839–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pignatelli S., Dal Monte P., Rossini G., Landini M. P. 2004. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev. Med. Virol. 14: 383–410 [DOI] [PubMed] [Google Scholar]
- 52. Plotkin S. A., Furukawa T., Zygraich N., Huygelen C. 1975. Candidate cytomegalovirus strain for human vaccination. Infect. Immun. 12: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Powers C. J., Fruh K. 2008. Signal peptide-dependent inhibition of MHC class I heavy chain translation by rhesus cytomegalovirus. PLoS Pathog. 4: e1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rawlinson W. D., Farrell H. E., Barrell B. G. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70: 8833–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reeves M. B., Sinclair J. H. 2010. Analysis of latent viral gene expression in natural and experimental latency models of human cytomegalovirus and its correlation with histone modifications at a latent promoter. J. Gen. Virol. 91: 599–604 [DOI] [PubMed] [Google Scholar]
- 56. Richards R., Scholz I., Powers C., Skach W. R., Fruh K. 2011. The cytoplasmic domain of rhesus cytomegalovirus Rh178 interrupts translation of major histocompatibility complex class I leader peptide-containing proteins prior to translocation. J. Virol. 85: 8766–8776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rivailler P., Kaur A., Johnson R. P., Wang F. 2006. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J. Virol. 80: 4179–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rue C. A., et al. 2004. A cyclooxygenase-2 homologue encoded by rhesus cytomegalovirus is a determinant for endothelial cell tropism. J. Virol. 78: 12529–12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schleiss M. R., et al. 2008. Analysis of the nucleotide sequence of the guinea pig cytomegalovirus (GPCMV) genome. Virol. J. 5: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schnorr J., et al. 2002. Functional studies of subcutaneous ovarian transplants in non-human primates: steroidogenesis, endometrial development, ovulation, menstrual patterns and gamete morphology. Hum. Reprod. 17: 612–619 [DOI] [PubMed] [Google Scholar]
- 61. Sinzger C., Digel M., Jahn G. 2008. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 325: 63–83 [DOI] [PubMed] [Google Scholar]
- 62. Sinzger C., et al. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89: 359–368 [DOI] [PubMed] [Google Scholar]
- 63. Swinkels B. W., Geelen J. L., Wertheim-van Dillen P., van Es A. A., van der Noordaa J. 1984. Initial characterization of four cytomegalovirus strains isolated from chimpanzees. Brief report. Arch. Virol. 82: 125–128 [DOI] [PubMed] [Google Scholar]
- 64. Sylwester A. W., et al. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202: 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Terhune S., et al. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81: 3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Turbant S., Martinon F., Moine G., Le Grand R., Leonetti M. 2009. Cynomolgus macaques immunized with two HIV-1 Tat stabilized proteins raise strong and long-lasting immune responses with a pattern of Th1/Th2 response differing from that in mice. Vaccine 27: 5349–5356 [DOI] [PubMed] [Google Scholar]
- 67. Van Andel R., et al. 2008. Clinical and pathologic features of cynomolgus macaques (Macaca fascicularis) infected with aerosolized Yersinia pestis. Comp. Med. 58: 68–75 [PMC free article] [PubMed] [Google Scholar]
- 68. Vink C., Beuken E., Bruggeman C. A. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74: 7656–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vomaske J., Nelson J. A., Streblow D. N. 2009. Human cytomegalovirus US28: a functionally selective chemokine binding receptor. Infect. Disord. Drug Targets 9: 548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Waldman W. J., Knight D. A., Huang E. H., Sedmak D. D. 1995. Bidirectional transmission of infectious cytomegalovirus between monocytes and vascular endothelial cells: an in vitro model. J. Infect. Dis. 171: 263–272 [DOI] [PubMed] [Google Scholar]
- 71. Waldman W. J., et al. 1991. Preservation of natural endothelial cytopathogenicity of cytomegalovirus by propagation in endothelial cells. Arch. Virol. 117: 143–164 [DOI] [PubMed] [Google Scholar]
- 72. Warren R. L., Sutton G. G., Jones S. J., Holt R. A. 2007. Assembling millions of short DNA sequences using SSAKE. Bioinformatics 23: 500–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zerbino D. R., Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu H., Cong J. P., Yu D., Bresnahan W. A., Shenk T. E. 2002. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc. Natl. Acad. Sci. U. S. A. 99: 3932–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]