Abstract
RNA silencing provides protection against RNA viruses by targeting both the helper virus and its satellite RNA (satRNA). Virus-derived small interfering RNAs (vsiRNAs) bound with Argonaute (AGO) proteins are presumed participants in the silencing process. Here, we show that a vsiRNA targeted to virus RNAs triggers the host RNA-dependent RNA polymerase 6 (RDR6)-mediated degradation of viral RNAs. We confirmed that satRNA-derived small interfering RNAs (satsiRNAs) could be associated with different AGO proteins in planta. The most frequently cloned satsiRNA, satsiR-12, was predicted to imperfectly match to Cucumber mosaic virus (CMV) RNAs in the upstream area of the 3′ untranslated region (3′ UTR). Moreover, an artificial satsiR-12 (asatsiR-12) mediated cleavage of a green fluorescent protein (GFP) sensor construct harboring the satsiR-12 target site. asatsiR-12 also mediated reduction of viral RNAs in 2b-deficient CMV (CMVΔ2b)-infected Nicotiana benthamiana. The reduction was not observed in CMVΔ2b-infected RDR6i plants, in which RDR6 was silenced. Following infection with 2b-containing CMV, the reduction in viral RNAs was not observed in plants of either genotype, indicating that the asatsiR-12-mediated reduction of viral RNAs in the presence of RDR6 was inhibited by the 2b protein. Our results suggest that satsiR-12 targeting the 3′ UTR of CMV RNAs triggered RDR6-dependent antiviral silencing. Competition experiments with wild-type CMV RNAs and anti-satsiR-12 mutant RNA1 in the presence of 2b and satRNA demonstrate the inhibitory effect of the 2b protein on the satsiR-12-related degradation of CMV RNAs, revealing a substantial suppressor function of the 2b protein in native CMV infection. Our data provide evidence for the important biological functions of satsiRNAs in homeostatic interactions among the host, virus, and satRNA in the final outcome of viral infection.
INTRODUCTION
Diverse RNA silencing processes exist in plants, including silencing by microRNAs (miRNAs) and several classes of endogenous small interfering RNAs (siRNAs) involved in pathways with multiple Dicer-like proteins (DCLs) and Argonaute proteins (AGOs) (3, 4). There are four specialized DCLs (DCL1 to DCL4) that produce different kinds of small RNAs (sRNAs; 21 to 25 nucleotides [nt] in length) from double-stranded RNAs or stem-loop precursors in Arabidopsis thaliana. The resulting sRNAs bind to an AGO protein to form effector complexes, termed RNA-induced silencing complexes (RISCs), to target the cleavage or translational suppression of cRNA based on sequence complementarity (1). There are three clades of AGOs, including 10 subfamily members in Arabidopsis (50). Recent studies reveal distinct binding affinities of sRNAs with different 5′-terminal nucleotides targeted to AGO proteins (31). AGO1 displays a strong bias for sRNAs with a 5′-terminal uridine, while AGO2 and AGO4 are associated mainly with sRNA sequences that are initiated with a 5′ adenosine, and AGO5 is highly enriched for sRNA sequences that are initiated with a 5′ cytosine (31).
RNA silencing also provides protection against diverse RNA viruses in many eukaryotic organisms (28, 38, 52). Two classes of viral siRNAs are generated in virus infections: the primary siRNAs, derived from the DCL-mediated cleavage of a viral RNA precursor, and the secondary siRNAs, whose biogenesis requires host RNA-dependent RNA polymerases (RDRs) (12, 16, 20, 52) in a process likely resembling the production of secondary siRNAs in plants, which is triggered by an AGO1-containing 22-nt miRNA or siRNA (7, 9, 42). To counter or escape from host antiviral silencing, many viruses have evolved viral suppressors of RNA silencing (VSR) to interfere with host RNA silencing at different stages (11, 14, 27). For example, the 2b protein, a VSR encoded by a Cucumber mosaic virus (CMV) (5, 21, 29), perturbs RNA silencing by directly interacting with AGO1, which results in a block of the loading of sRNAs into the AGO1 RISC and, thereby, interferes with the Slicer activity of AGO1 (55).
CMV is one of the best-characterized tripartite positive-sense single-stranded RNA viruses, with genes encoding five proteins distributed on three genomic and two subgenomic RNAs. The 1a and 2a proteins encoded by genomic RNA1 and RNA2, respectively, are RDRs involved in viral replication (24, 35, 36). Genomic RNA3 encodes the 3a protein, or movement protein (MP) (13). The 2b and cost protein (CP) are expressed from subgenomic RNA4A and RNA4, which are transcribed from genomic RNA2 and RNA3, respectively (15, 44). High nucleotide sequence similarity is present within the 3′ untranslated regions (3′ UTRs) of the three genomes and the two subgenomes (18, 41). A tRNA-like structure (TLS) located at the 3′ terminus of the 3′ UTR is required for viral RNA replication, interacting with the replicases (1a and 2a) and other host factors (19). 3′ UTRs are effectively targeted by host antiviral silencing (18).
Plants infected with some isolates of CMV also contain high levels of satellite RNA (satRNA), which requires the helper virus CMV to supply proteins for replication in the cytoplasm (17, 46). These approximately 340- to 400-nt-long linear RNAs with highly conserved secondary structures can also be targeted by the host silencing machinery and processed into siRNAs, yielding satRNA-derived siRNAs (satsiRNAs) (17, 51). We have previously characterized satsiRNAs in Arabidopsis infected with a severe field Shan-Dong strain of CMV (SD-CMV) (17). satsiRNAs with lengths of both 21 and 22 nt were identified. It has been shown previously that specific nucleotide residues or satRNA structures determine the capability for modulating symptoms caused by helper viruses (8, 30, 37, 47). It was also suggested that the effect of satRNA on helper viruses could be related to the competition of the replication between the helper virus and satRNA (40, 46). We have recently found that SD-CMV satRNA reduces the accumulation of the 2b protein and attenuates the yellowing caused by SD-CMV infection (23). Recently, it has been reported that satsiRNA derived from CMV Y-satRNA silences the endogenous chlorophyll biosynthetic gene CHL1 and results in yellowing symptoms in infected plants (45, 48). However, it is not known whether satsiRNAs play a role(s) in pathogenicity in the trilateral interactions among host plants, helper viruses, and satRNAs relevant to RNA silencing. Functional analyses of a particular pathogen-derived siRNA targeting viral genomes in plants have not been reported so far. In this study, we confirmed that satsiRNAs could be associated with AGO proteins in planta. We found that satsiR-12 imperfectly matched with CMV RNAs at the 3′ UTR near the 5′-terminal upstream region and that expression of artificial satsiR-12 (asatsiR-12) downregulated the expression of a sensor construct, green fluorescent protein (GFP) fused to an upstream sequence (180 bp) of an SD-CMV RNA 3′ UTR (3′UTR1). Transgenic expression of asatsiR-12 triggered RDR6-dependent degradation of CMV RNAs following 2b-deficient CMV (CMVΔ2b) infection. The contribution of satsiR-12 to the satRNA-related reduction of CMV RNAs in natural infection was further supported by a competition experiment with wild-type CMV RNA and anti-satsiR-12 mutant RNA1 (RNA1m) in the presence of satsiR-12. Together, our results reveal that satsiR-12 targets the 3′ UTRs of CMV RNAs and triggers the RDR6-dependent amplification of antiviral silencing.
MATERIALS AND METHODS
Plasmid construction.
The asatsiR-12-containing artificial miRNA precursor was amplified with the Arabidopsis pre-miR159a sequence as a backbone as described previously (34). The asatsiR-12 primers were as follows: 5′-ACTAGTGGCCGGGTTCTGCGTGCAAACTGATGAGTTGAGCAGGGTAAAG-3′ (forward) and 5′-GGCCGGGTTCTGCTAGCAAACTGGAAGAGTAAAAGCCATTAAAG-3′ (reverse). The forward and reverse primers contain the asatsiR-12* sequence (underlined) and the mature asatsiR-12 reverse complementary sequence (underlined), respectively. The resulting pre-miR159a/asatsiR-12 fragment was cloned into pCAMBIA-1300221 to generate 35S–asatsiR-12.
For 35S-GFP sensor constructs, upstream sequences (180 bp) of SD-CMV RNA 3′ UTRs (3′UTR1, 3′UTR2, and 3′UTR3) were amplified and cloned into the pGEM-T vector (Tiangen). SpeI-3′UTR1/2/3-SacI fragments were each inserted into SpeI/SacI-digested pCAMBIA-1300221–GFP to make 35S–GFP-3′UTR1/2/3. For mutant constructs, oligonucleotide-directed mutagenesis was introduced into p35S-GFP-3′UTR2 and p35S-GFP-3′UTR3 to form p35S-GFP-3′UTR2m and p35S-GFP-3′UTR3m, respectively. The TaKaRa MutanBest kit was used according to the manufacturer's instructions. The primers for p35S-GFP-3′UTR2m and p35S-GFP-3′UTR3m were as follows: for p35S-GFP-3′UTR2m, primer 1, 5′-AACTGCCAACTCAGCTCCCGCCTC-3′, and primer 2, 5′-CTGCTACAAACTGTCTGAAGTCAC-3′, and for p35S-GFP-3′UTR3m, primer 1, 5′-GAACTGCCAACTCAGCTCCCGCCT-3′, and primer 2, 5′-TGCTACAAACTGTCTGAAGTCACT-3′.
For 35S-β-glucuronidase (GUS) sensor construction, the asatsiR-12 target sequence of 3′UTR1 was fused to the GUS sequence in plasmid pCAMBIA-1300221 (18). A 426-bp fragment of the 3′ terminus of the GUS gene was created by PCR amplification using a pair of primers as follows: forward, 5′-TGTACAGCGAAGAGGCAGTC-3′, and reverse, 5′-TGTACACAGTTTGTAGCAGAACTGCCAACTTGAGTGCAGCCCGGCTAAC-3′. The reverse primer contains a 23-nt asatsiR-12 target sequence, corresponding to the 50 to 72 nt of the SD-CMV RNA1 3′ UTR (underlined). The generated GUS sensor fragment was digested with BsrGI and replaced the corresponding GUS 3′-terminal sequence in pCAMBIA-1300221, resulting in the 35S-GUS sensor.
For the 35S-satRNA construct, a 334-nt fragment was amplified by reverse transcription-PCR (RT-PCR) from sap prepared from tobacco leaves with native SD-CMV infection, and the amplified fragment was finally cloned downstream of the 35S promoter in the binary vector pCAMBIA-1300221. The primers were as follows: forward, 5′-TCTAGAGTTTTGTTTGATGGAGAA-3′, and reverse, 5′-GAGCTCGGGTCCTGCAGAGGAATGA-3′; italics indicate the introduced restriction cleavage sites of XbaI and SacI. For 35S-mutant satRNA (satRm) constructs, oligonucleotide-directed mutagenesis was introduced into p35S-satR to form p35S-satRm by using a site-directed mutagenesis kit (NEB) according to the manual. The primers for p35S-satRm were as follows: primer A, 5′-aAGAACCCGGCACATGGTTCGC-3′, and primer B, 5′-gatGgAAACTGAGCGGGGGCTCAAATG-3′. Lowercase letters indicate the mutated nucleotides. For 35S-RNA1m constructs, oligonucleotide-directed mutagenesis was introduced into 35S-RNA1-pBI121 to form p35S-RNA1m using the same method. The primers for p35S-RNA1m were as follows: primer A, 5′-GTTGGCAGTTCTcacctAAACTGTCTGAAG-3′, and primer B, 5′-TCAGCTCCCGCCTCAGAAAACTGG-3′. Lowercase letters indicate the mutated nucleotides.
Plant transformation, virus inoculation, and Agrobacterium infiltration.
Nicotiana benthamiana plants (wild-type plants or plants in which RDR6 was silenced [RDR6i plants]) were transformed with 35S–asatsiR-12 or the pCAMBIA-1300221 control vector by Agrobacterium-mediated leaf disc transformation (22). The positive transformants (T0) were verified with RNA blots for the presence of the transgene. The T1 transgenic plants were selected on Murashige and Skoog (MS) medium with hygromycin (20 mg/liter). Rooted plants were transferred into soil. Leaves of 4-week-old Arabidopsis and transgenic tobacco plants were inoculated with fresh sap prepared from SD-CMV-infected tobacco leaves and SD-CMV infectious cDNA clones (23).
The EHA105 strain of Agrobacterium tumefaciens was transformed with the 35S-GFP sensor, 35S–asatsiR-12, and pCAMBIA-1300221 (vector control) constructs by electroporation, and transformants were selected on Luria-Bertani medium containing rifampin at 10 mg/liter and kanamycin at 50 mg/liter. For Agrobacterium infiltration experiments, equal volumes of Agrobacterium cultures containing 35S–asatsiR-12 or pCAMBIA-1300221 (optical density at 600 nm [OD600], 1.5) and the 35S-GFP sensor were mixed (yielding an OD600 of 0.5) before infiltration, and wild-type or RDR6i N. benthamiana leaves were infiltrated with the mixture. For SD-CMV infectious clone inoculation, equal volumes of Agrobacterium cultures containing RNA1, RNA2 (or RNA2Δ2b, which does not express 2b), and RNA3 cDNAs were mixed together at an OD600 of 1.0 and wild-type or RDR6i plants were infiltrated with the mixture.
AsatsiR-12 cloning and sequencing.
Small RNAs were extracted from the injected N. benthamiana plants transiently expressing 35S–asatsiR-12 and separated by a 15% denaturing polyacrylamide gel. The small RNAs, with sizes ranging from 18 to 24 nt, were gel extracted as reported previously (17). Cloning and expression of mature asatsiR-12 by miRNA rapid amplification of cDNA ends (miR-RACE) was performed as described previously (49). 5′ miR-RACE reactions were performed with the miRRacer 5′ primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) and a satsiR-12-specific forward primer (GSP1; 5′-TTTTTTTTTTGCCGGGTTCTGCTAGCA-3′), and the 3′ miR-RACE reactions were carried out with the miRRacer 3′ primer (5′-ATTCTAGAGGCCGAGGCGGCCGACATG-3′) and a satsiR-12-specific reverse primer (GSP2; 5′-GGAGTAGAAACAGTTTGCTAGCAGAAC-3′). Both GSP1 and GSP2 contained 17-nucleotide sequences (underlined) corresponding to satsiR-12. The 5′ RACE and 3′ RACE PCR products were separated on a 2.5% agarose gel and visualized by ethidium bromide (EtBr) staining. The expected DNA fragments were purified and directly cloned into the pGEM-T vector (Tiangen), and positive clones were subjected to DNA sequencing.
RNA ligase-mediated (RLM)-5′ RACE assay.
Mapping of the cleavage sites for the GUS sensor of the 3′UTR1 was carried out by 5′ RACE. The 5′ RACE assay was performed using the FirstChoice RLM-RACE kit (Ambion). About 2 μg of total RNA was used for direct ligation to the 5′ RACE RNA adapter, and subsequent steps were done according to the manufacturer's instructions. The PCR products obtained were cloned into the pGEM-T vector (Tiangen), and individual clones were selected for DNA sequencing.
RNA blot and immunoblot analyses.
Total RNA was extracted from plant issue using the TRIzol reagent (Invitrogen). For high-molecular-weight RNA blots, 5 μg of total RNA was separated on 1.2% agarose gels containing 6% formaldehyde and transferred onto nylon N+ membrane. DNA probes were labeled with [α-32P]dCTP using the Rediprime II system (Amersham). For low-molecular-weight RNA blots, 20 μg of total RNA was separated by electrophoresis on a 17% PAGE gel and electrically transferred onto nylon N+ membrane. [α-32P]UTP-labeled transcript or [γ-32P]ATP-labeled specific oligonucleotide probe sequences were used.
Leaf tissues from Agrobacterium-infiltrated N. benthamiana were ground with liquid nitrogen, homogenized in 2 ml/g lysis buffer (50 mM Tris-HCl at pH 7.4, 100 mM KCl, 2.5 mM MgCl2, 0.1% NP-40, and 2× complete protease inhibitor cocktail; Roche), and then centrifuged at 12,000 × g for 20 min at 4°C. Proteins were separated on an SDS–10% polyacrylamide gel and transferred onto polyvinylidene fluoride membranes with a semidry transfer cell (Bio-Rad). Bovine serum albumin (BSA; 5%) in phosphate-buffered saline–Tween (PBS-T) was used to block the membrane at 4°C overnight. A commercial mouse monoclonal GFP antibody (Abmart) was used for immunoblot analyses.
Co-IP.
The coimmunoprecipitation (co-IP) assay was performed as reported previously (6) with minor modifications. In brief, aerial or inflorescence tissues from SD-CMV-infected A. thaliana at 10 days postinoculation were ground under liquid nitrogen and homogenized in 3 ml/g lysis buffer. After centrifugation for 20 min at 9,500 × g, the supernatant was transferred into a new tube and centrifuged again. The lysates were precleared for 30 min at 4°C with a 10-μl bed volume of protein A-agarose (Santa Cruz). Precleared lysates were treated with 4 μg of anti-Myc (Santa Cruz) or anti-AGO2/anti-AGO5 (anti-AGO2/5; Abmart) antibodies per ml for 1 h at 4°C and then with a 50-μl bed volume of protein A-agarose per ml for 3 h at 4°C. Precipitates were washed three times in lysis buffer and divided for protein and nucleic acid analyses. Nucleic acids were extracted using the TRIzol reagent (Invitrogen), separated by 17% PAGE, and blotted. Membranes were hybridized with [γ-32P]ATP-labeled DNA oligonucleotide probes complementary to the satsiRNA, miR159, or miR390a. Part of the coimmunoprecipitated protein complex was subjected to immunoblotting with anti-Myc or anti-AGO2/5 antibodies as described above.
RESULTS
SatsiRNAs could be loaded onto the AGO RISC.
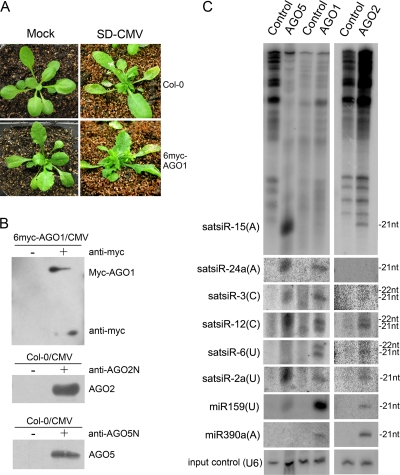
Our previous study reported that the cloned satsiRNAs are primarily 21 nt (64.7%) or 22 nt (22%) in length (17). To ascertain that these satsiRNAs possess biological functions, we first examined whether satsiRNAs are associated with the AGO proteins that mediate silencing. Wild-type Arabidopsis (Col-0) and 6-Myc–AGO1 transgenic Arabidopsis, constructed by transforming the ago1-27 mutant (33) with 35S–6-Myc–AGO1 plasmids, were infected with SD-CMV (Fig. 1 A). Tissues collected from infected plants were subjected to immunoprecipitation to pull down protein complexes containing AGO1 (with anti-Myc antibody [α-Myc]), AGO2 (with α-AGO2), or AGO5 (with α-AGO5). The copurified small RNAs were then isolated from the immunoprecipitates and evaluated by RNA blot hybridization. miR159 and miR390a, whose 5′-terminal nucleotides are U and A, respectively, were used as controls. Consistent with the data in a previous report (31), miR159 was detected predominantly in the AGO1 co-IP complex whereas miR390a was detected mostly in the AGO2 and weakly in the AGO1 but not in the AGO5 co-IP complex (Fig. 1C). Several satsiRNAs identified in our previous study with different 5′-terminal nucleotides (17) were examined. satsiR-15, having a 5′-terminal A, was detected in the AGO2 and strongly in the AGO5 but not in the AGO1 co-IP complex (Fig. 1C). satsiR-24a, also beginning with a 5′-terminal A, was detected with AGO5 and AGO1 but not AGO2 (Fig. 1C). satsiR-3, starting with a 5′-terminal C, was detected mainly with AGO1 and AGO5 (Fig. 1C). satsiR-12 also contains a 5′-terminal C and could be associated with the three AGOs (Fig. 1C). Consistent with the data in a previous report (17), two bands of signals were detected, suggesting that both the 21- and 22-nt versions of satsiR-12 were loaded onto AGO proteins. satsiR-6 has a 5′-terminal U and was associated with AGO1 and weakly associated with AGO2 but not with AGO5 (Fig. 1C). However, satsiR-2a, which also has a 5′-terminal U, was detected in the three AGO co-IP complexes (Fig. 1C). Nevertheless, these data indicated that satellite RNA-derived siRNAs could be sorted into AGO-containing effector components. In addition to the 5′-terminal nucleotide bias (31), the relative strand stability of satsiRNA duplexes or other factors may also determine the fate of satsiRNAs for binding certain AGO proteins (25).
Fig. 1.
Analysis of satsiRNA association with AGO-containing complexes. (A) SD-CMV infection symptoms in wild-type Arabidopsis (Col-0) and 6-Myc-AGO1 ago1-27 (6myc-AGO1) transgenic Arabidopsis plants. Photographs were taken at 10 dpi. (B) AGO-containing complexes were immunopurified from total protein extracted from SD-CMV-infected Col-0 and 6myc-AGO1 plants by using specific antibodies as indicated. Total protein extracts incubated with protein A were used as negative controls (−). (C) Small RNA detection of copurified satsiRNAs and miRNAs. Small RNAs were extracted from immunoprecipitates of AGO-containing complexes and protein A controls as indicated at the top. Membranes were hybridized with 32P-labeled specific oligodeoxynucleotide probes for each satsiRNA and miR-159 and miR-390a. U6 RNA hybridization is shown as an input sample control.
Bioinformatics analysis of potential targets of satsiRNAs.
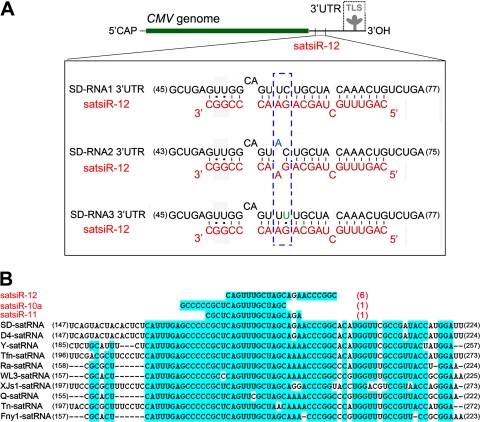
The ability of satsiRNAs to be loaded onto the AGO RISC prompted us to seek potential targets in both viral and host plant genomic sequences. Via bioinformatic predictions, we found that several satsiRNAs could potentially target A. thaliana genome sequences, including some genes encoding transcription factors. However, a 5′ RACE analysis showed that none of the cleavage sites were located in or near the satsiRNA/target complementary regions. By searching for complementation between satsiRNAs and CMV genomic sequences, we found that satsiR-12 imperfectly matched to a region upstream of the TLS in the SD-CMV RNA 3′ UTR (Fig. 2 A). There are three mismatches between satsiR-12 and SD-CMV RNA1 and RNA3 and four mismatches between satsiR-12 and RNA2 (Fig. 2A). Sequence alignments with different satRNA strains and variants indicated that satsiR-12 originated from a highly conserved region of CMV satRNA (Fig. 2B). Together with the fact that satsiR-12 is the most frequently cloned satsiRNA (17), these data suggest that satsiR-12 may not be an occasional by-product and that it probably plays a role in interacting with the helper virus CMV by targeting viral RNAs.
Fig. 2.
Imperfect complementarity between satsiR-12 and the 3′ UTR of SD-CMV RNA and alignment of representative CMV satRNA strains with satsiRNAs. (A) Sequence alignment among satsiR-12 and the SD-CMV RNA1, RNA2, and RNA3 3′ UTRs upstream of the TLS region. 5′CAP represents the 5′-terminal cap structure. The three mismatches between satsiR-12 and SD-CMV RNA1 and RNA3 and four mismatches between satsiR-12 and RNA2 are shown. The differences among the three target regions are indicated in the dashed box. (B) Alignment of representative CMV satRNA isolates/strains with satsiRNAs. Three cloned satsiRNAs (17) mapped to the conserved region (highlighted with a blue background) are shown. Red numbers in parentheses indicate the clone frequencies for each satsiRNA in this conserved region (17). Black numbers at the ends of the sequences indicate nucleotide positions.
Artificial satsiR-12 downregulated the expression of the GFP sensor construct.
To examine the potential function of satsiR-12, we expressed artificial satsiR-12 (asatsiR-12) using an A. tumefaciens MIR159a precursor backbone as described previously (18). N. benthamiana plants were infiltrated with Agrobacterium containing 35S–asatsiR-12, and the expression of asatsiR-12 was confirmed (Fig. 3 B). Sequencing analysis using miR-RACE (49) PCR products confirmed that asatsiR-12 molecules were produced with lengths of both 21 and 22 nt. The 21-nt asatsiR-12 was identical to the first 21 nt of the natural satsiR-12.
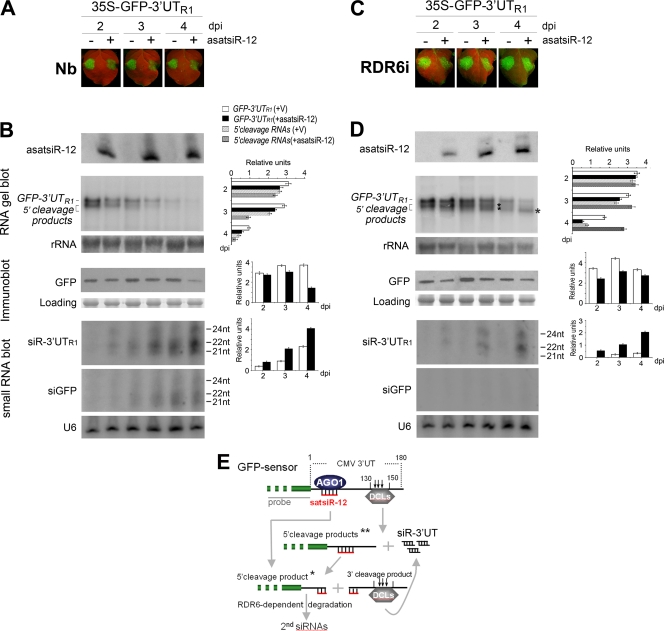
Fig. 3.
Analysis of the effect of asatsiR-12 on regulating expression of GFP-3′UTR1. (A and C) Observation of GFP fluorescence in samples from wild-type N. benthamiana plants (Nb) (A) and RDR6i plants (C) coexpressing GFP-3′UTR1 with asatsiR-12 or a vector control (−). Photographs were taken under UV light at 2, 3, and 4 dpi. (B and D) Detection of the expression of asatsiR-12 and the accumulation of GFP-3′UTR1 RNA, 5′ cleavage products, 3′UTR1-derived siRNAs (siR-3′UTR1), and GFP-derived siRNAs (siGFP), as well as the accumulation of GFP, in wild-type N. benthamiana plants (B) and RDR6i plants (D). 32P-labeled GFP DNA probes and GFP-specific antibodies were used. For small RNA blots, 32P-labeled in vitro transcripts from RNA1 construct 3′UTR1 and from GFP were used as probes. Methylene blue-stained rRNA, U6 RNA hybridization, and Coomassie blue-stained total proteins are shown as RNA and protein loading controls. Pools of samples collected from four plants were analyzed for each time point. Quantification of GFP-3′UTR1, 5′ cleavage products, GFP, and siGFP relative to the loading control are shown to the right of each panel using ImageQuant TL (GE Healthcare Life Sciences). +V, in the presence of the control vector. (E) A sketch map for the possible cleavage of GFP sensor RNA mediated by asatsiR-12 and DCLs (18). * indicates an asatsiR-12-mediated 5′ cleavage product, and ** indicates DCL-mediated cleavage products (18).
To investigate the effects of satsiR-12 on CMV RNAs, we linked the CMV RNA1 3′ UTR (3′ UTR1) to the 3′ terminus of the GFP coding sequence. However, this transiently expressed GFP-CMV 3′ UTR1 sequence was rapidly degraded. This was consistent with our previous report that the highly structured 3′ UTRs of CMV RNAs are effectively targeted by the plant RNA silencing system (18). We therefore constructed a series of GFP sensor plasmids with only the 5′-terminal sequences (∼180 bp each) of three CMV genomic RNA 3′ UTRs without the downstream TLS sequence (referred to herein as 3′UTs); these constructs contained satsiR-12 target sites (Fig. 2A) and were named GFP-3′UTR1, GFP-3′UTR2, and GFP-3′UTR3. Transiently expressed GFP-3′UTR1 sequences accumulated to levels comparable to that of the control 35S-GFP transcript. 5′ terminus-related cleavage products were seen in samples from plants infiltrated with GFP-3′UTR1, due presumably to DCL-mediated cleavage of hot spot regions located in the 130 to 150 bp of 3′UTR1 downstream of the satsiR-12 target site (18) (Fig. 3E). However, the fluorescence of GFP-3UTR1 was similar to that of the control 35S-GFP construct, suggesting that the GFP sensor plasmids were suitable for further analysis. Therefore, we coinfiltrated N. benthamiana plants with GFP sensors and 35S–asatsiR-12 or vector controls, and GFP fluorescence was monitored at 2, 3, and 4 days postinfiltration (dpi). The fluorescence of GFP-3′UTR1, but not that of GFP-3′UTR2 or GFP-3′UTR3, was reduced in leaves coexpressing asatsiR-12 compared to that in leaves coexpressing vector controls at each time point (Fig. 3A and 4A and B). The effects of asatsiR-12 on the expression of GFP-3′UTR1 were also observed by using GFP mRNA blot and protein immunoblot analyses with pools of samples collected from four plants for each time point (Fig. 3B). Consistent with the GFP fluorescence intensity, GFP and GFP mRNA accumulation was generally reduced in samples with GFP-3′UTR1/asatsiR-12 coexpression, especially at 4 dpi, compared to samples with GFP-3′UTR1/control vector coexpression.
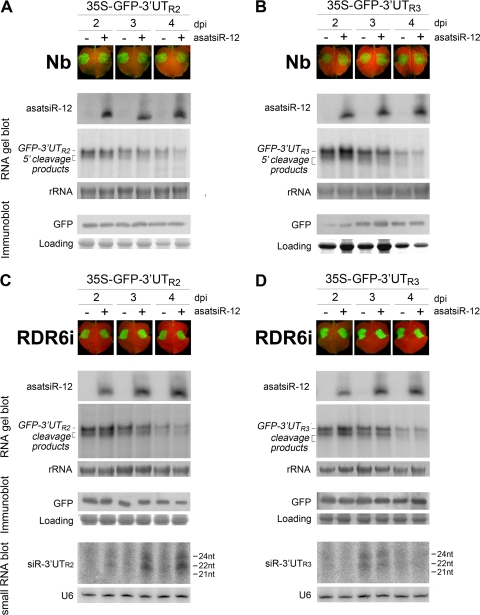
Fig. 4.
Analysis of the effect of asatsiR-12 on regulating expression of GFP-3′UTR2. (A and C) and GFP-3′UTR3 (B and D) in wild-type N. benthamiana plants (A and B) and RDR6i plants (C and D). Photographs were taken under UV light at 2, 3, and 4 dpi. The expression of asatsiR-12 and the accumulation of GFP-3′UTR2 and GFP-3′UT3 RNAs, 5′ cleavage products, and 3′UTR2- and 3′UTR3-derived siRNAs (siR-3′UTR2 and siR-3′UTR3), as well as the expression of GFP, were detected as described in the legend to Fig. 3.
RNA gel blot results also showed that 5′ terminus-related cleavage products of GFP sensors were generated in all coinfiltration samples despite the presence or absence of asatsiR-12 (Fig. 3B and 4A and B). These data further suggested that the GFP sensor RNA, containing the 3′UT at the 3′ terminus, was targeted for cleavage independently of asatsiR-12. A small RNA blot was used to detect the accumulation of CMV 3′UTR1-derived siRNAs (siR-3′UTR1). Hybridization results indicated that siR-3′UTR1 indeed accumulated in all GFP-3′UTR1 infiltration samples, with higher levels when plants were coinfiltrated with asatsiR-12 and lower levels when were plants were coinfiltrated with the vector control (Fig. 3B). These results supported the idea that the upstream 180-bp sequences of CMV RNA 3′ UTRs were indeed targeted by plant RNA silencing to be processed into siRNAs. The higher level of siR-3′UTR1 detected with the GFP-3′UTR1/asatsiR-12 coexpression indicated the contribution of asatsiR-12 in increasing GFP-3′UTR1 degradation.
GFP-specific siRNA (siGFP) was also examined and clearly detected in GFP-3′UTR1 coinfiltration samples at 3 and 4 dpi (Fig. 3B), suggesting the involvement of transitive siRNAs synthesized from the 3′UTR1 terminus to the GFP coding sequence, a process dependent on RDR6 (32, 54). This indicated that blocking the RDR6-dependent degradation might provide us with the possibility of distinguishing different 5′ cleavage products that were derived from asatsiR-12-mediated or asatsiR-12-independent cleavage. We therefore coexpressed GFP-3′UTR1 with asatsiR-12 or a vector control in N. benthamiana plants in which RDR6 had been silenced (RDR6i plants) (43, 54). Accumulation of different 5′ cleavage products was indeed observed in GFP-3′UTR1/asatsi-R-12 and GFP-3′UTR1/vector samples (Fig. 3D). Steady accumulation of a shorter asatsiR-12-mediated 5′ cleavage product was obviously detected at 3 and especially at 4 dpi (Fig. 3D and E). There was no RDR6-dependent transitive siGFP detected (Fig. 3D), whereas siR-3′UTR1 was clearly detected in GFP-3′UTR1/asatsiR-12 samples but levels were very low in GFP-3′UTR1/vector samples (Fig. 3D). These data clearly showed the effect of asatsiR-12 in the mediation of GFP-3′UTR1 cleavage. The resulting short 5′ cleavage product (Fig. 3E) steadily accumulated without subsequent RDR6-dependent degradation, whereas the 3′ cleavage products were further targeted probably for dicing in production of siR-3′UTR1 (Fig. 3D and E). Similarly, smear hybridization signals of asatsiR-12-independent 5′ cleavage products (Fig. 3E) were also detected at 4 dpi in GFP-3′UTR1/vector samples from RDR6i plants (Fig. 3D). However, the accumulation of the asatsiR-12-mediated cleavage product of GFP-3′UTR1 RNA without subsequent RDR6-dependent degradation was remarkable. Although the accumulation of the full-length GFP-3′UTR1 RNA was reduced, steady-state levels of 5′ cleavage products in RDR6i plants may maintain the ability of the GFP coding sequence to be translated for the production and maintenance of the protein (Fig. 3C). Together with the accumulation of similar levels of GFP in samples coexpressing asatsiR-12 and those coexpressing the vector control (Fig. 3D), our results suggest that the asatsiR-12-mediated downregulation of GFP-3′UTR1 likely does not act at the translational level.
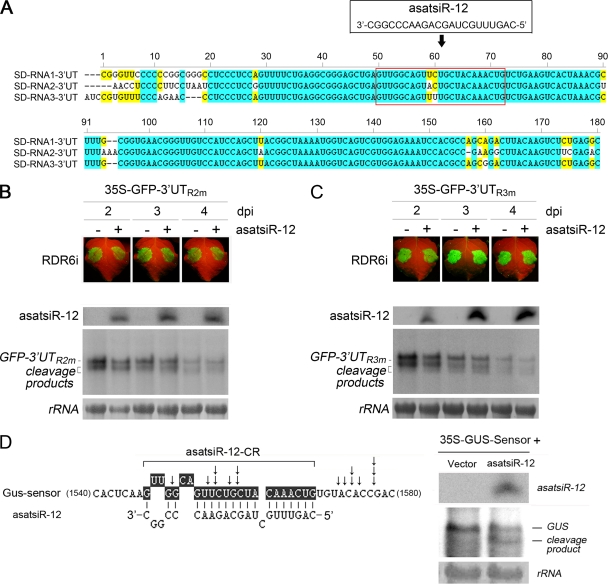
As in wild-type N. benthamiana plants, there were no obvious differences between the expression of GFP-3′UTR2 or GFP-3′UTR3 in the presence of asatsiR-12 and that in the presence of the vector control in RDR6i plants (Fig. 4 C and D). Small RNA detection showed slight increases in siR-3′UTR2 but not siR-3′UTR3 in the asatsiR-12 coexpression sample (Fig. 4C and D). These results further implied that asatsiR-12 might exhibit low or no activity for targeting GFP-3′UTR2 or GFP-3′UTR3. The similar degrees of complementary mismatch between asatsiR-12 and 3′UTR1, asatsiR-12 and 3′UTR2, and asatsiR-12 and 3′UTR3 (Fig. 2A and 5 A) suggested that the spatial structure of the flanking sequence around the satsiR-12 target site of 3′UTR1 may be different from that of 3′UTR2 and 3′UTR3, resulting in different accessibilities for the asatsiR-12–AGO complex. Results obtained from nucleotide substitution in GFP sensor constructs and coinfiltration experiments supported this idea (Fig. 5B and C).
Fig. 5.
Detection of asatsiR-12-mediated cleavage of nucleotide substitution mutant GFP sensors (GFP-3′UTR2m and GFP-3′UTR3m) and GUS sensor cleavage. (A) Alignment of 5′-terminal sequences (∼180 bp) of three SD-CMV genomic RNA 3′ UTRs (3′UT) containing the satsiR-12 target site. satsiR-12 and its target region are marked in the red box. Identical and conserved nucleotides are highlighted in blue and yellow, respectively. (B and C) GFP-3′UTR2m and GFP-3′UTR3m, in which the satsiR-12 target site in 3′UTR2 or 3′UTR3 was replaced by that of 3′UTR1, were constructed by oligonucleotide-directed mutagenesis. The bases A57 in 3′UTR2 and U60 in 3′UTR3 are changed to U and C. Coexpression of GFP-3′UTR2m (B) or GFP-3′UTR3m (C) with asatsiR-12 or a vector control (−) in RDR6i plants was examined. GFP fluorescence was photographed under UV light at 2, 3, and 4 dpi. There were no obvious differences in accumulation of GFP-3′UTR2m, GFP-3′UTR3m, and their 5′ cleavage products between samples in which the construct was coexpressed with asatsiR-12 and those in which it was coexpressed with the vector control at each time point. (D) Analysis of the asatsiR-12-mediated cleavage of the GUS sensor. The GUS sensor construct contains the exact satsiR-12 target site sequence of 3′UTR1 (highlighted in black). The arrows indicate the cleavage sites of the GUS sensor RNA detected by 5′ RACE. RDR6i plants were coinfiltrated with the GUS sensor and a vector or the GUS sensor and asatsiR-12, and samples were subjected to RNA blot analysis of GUS RNA at 4 dpi. A 32P-labeled DNA probe specific for GUS mRNA was used. The 5′ cleavage product is indicated.
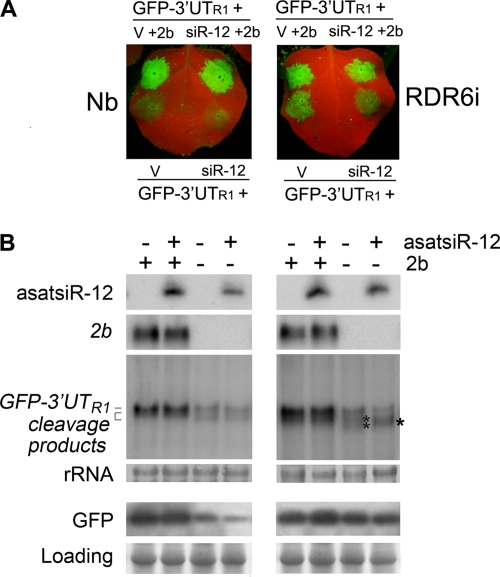
Artificial satsiR-12-mediated cleavage of the GFP sensor was inhibited by the CMV 2b suppressor.
The location of the satsiR-12 target site in the 3′ UTR made it less likely that satsiR-12 guided the cleavage of mRNA targets as most plant miRNAs do (39). Despite this, the RNA gel blot analysis of the coinfiltrations (Fig. 3) suggested that asatsiR-12 guided the target for cleavage. To examine the asatsiR-12-mediated cleavage of GFP-3′UTR1, 5′ RACE was performed with samples collected at 4 dpi from RDR6i plants coinfiltrated with GFP-3′UTR1 and asatsiR-12. However, the silencing of cleavage hot spots in the downstream region of the satsiR-12 target site (18) (Fig. 3E) obstructed the detection of asatsiR-12 cleavage sites via the 5′ RACE approach. Therefore, we constructed a GUS sensor that contained the exact satsiR-12 target site sequence of 3′UTR1 for the coinfiltration assay with RDR6i plants. Cleavage sites mapped by 5′ RACE confirmed that asatsiR-12 could mediate the cleavage of the GUS sensor in the transcribed region, and the cleavage sites at or near the complementary region of the siRNA/target pairing area were mapped (Fig. 5D). RNA gel blot analysis showed that a specific 5′ cleavage fragment was produced in coinfiltration with the GUS sensor and asatsiR-12 (Fig. 5D).
To further verify that satsiR-12 also mediated cleavage at the 3′ terminus region, we introduced the CMV 2b-silencing suppressor protein into the coinfiltration assay. The Fny 2b protein, encoded by a CMV subgroup IA Fny strain, has been reported previously to interfere with the Slicer activity of AGO1 (55) but not affect plant miRNA-mediated translational repression (26). Coexpression of GFP-3′UTR1 with asatsiR-12 or the vector control in the presence or absence of the SD-CMV 2b protein in both wild-type N. benthamiana and RDR6i plants was carried out. Consistent with the results of the above-described coinfiltration assay, in the absence of the SD-CMV 2b protein, the differences in the levels of accumulation of GFP-3′UTR1 and the cleavage products when the construct was coexpressed with asatsiR-12 versus with the vector control were more evident in RDR6i plants than in wild-type N. benthamiana plants (Fig. 6). Steady accumulation of asatsiR-12-mediated 5′ cleavage products in RDR6i plants was observed when the construct was coexpressed with asatsiR-12 (Fig. 6B). However, in the presence of the SD-CMV 2b protein, the steady accumulation of asatsiR-12-mediated 5′ cleavage products in RDR6i plants was not detected (Fig. 6B), indicating that the SD-CMV 2b protein interfered with the asatsiR-12-mediated cleavage of GFP-3′UTR1 RNA and supporting the idea that asatsiR-12 mediated cleavage at the 3′ terminus. Consistent with SD-CMV 2b suppression of silencing activity, intensive GFP fluorescence was observed in the presence of SD-CMV 2b in plants of either genotype (Fig. 6A). In wild-type N. benthamiana plants, the accumulation of GFP-3′UTR1 RNA was higher in the presence of SD-CMV 2b but lower in the absence of SD-CMV 2b with or without asatsiR-12 (Fig. 6B).
Fig. 6.
Analysis of the inhibitory effect of SD-CMV 2b on asatsiR-12-mediated cleavage of GFP-3′UTR1. (A) Coexpression of GFP-3′UTR1 with asatsiR-12 (siR-12) or a vector control (V) in the absence or presence of SD-CMV 2b protein in wild-type N. benthamiana plants (left panel) and RDR6i plants (right panel). GFP fluorescence was photographed under UV light at 4 dpi. (B) Expression of asatsiR-12 and accumulation of SD-CMV 2b and GFP-3′UTR1 RNA and 5′ cleavage products, as well as expression of GFP, were detected as described in the legend to Fig. 3B.
Effect of artificial satsiR-12 on viral RNA accumulation in SD-CMV infection.
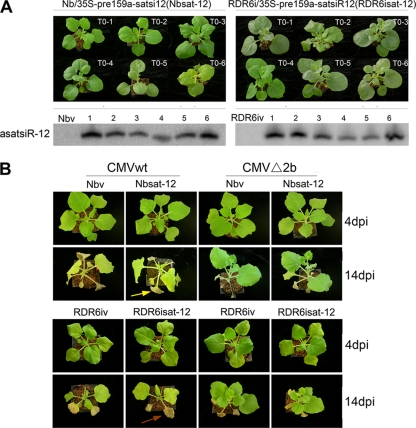
To examine whether asatsiR-12 would downregulate the accumulation levels of SD-CMV RNAs in native CMV infection, we generated asatsiR-12 transgenic N. benthamiana (Nbsat-12) and RDR6i (RDR6isat-12) plants. The expression of asatsiR-12 was confirmed by a small RNA gel blot analysis (Fig. 7 A). To avoid disturbances by satsiRNAs during native infection with SD-CMV containing SD-CMV satRNA (17), two infectious clones, CMVwt, containing SD-CMV RNA1, RNA2, and RNA3, and CMVΔ2b, containing CMV RNA1, RNA2Δ2b, and RNA3, in which 2b protein expression was abolished (23), were inoculated into Nbsat-12, RDR6isat-12, and vector transgenic control plants.
Fig. 7.
Morphology and infection symptoms of transgenic Nbsat-12 and RDR6isat-12 plants carrying 35S–asatsiR-12. (A) Morphology of transgenic Nbsat-12 and RDR6isat-12 T0 plants and detection of asatsiR-12 expression levels in these transgenic plants. The corresponding vector transgenic plants (Nbv and RDR6iv plants) were used as controls. A 32P-labeled oligodeoxynucleotide probe specific for asatsiR-12 was used. (B) Infection symptoms of Nbsat-12 and RDR6isat-12 plants and corresponding control (Nbv and RDR6iv) plants inoculated with an SD-CMV infectious clone: CMVwt, containing SD-CMV RNA1, RNA2, and RNA3, or CMVΔ2b, containing CMV RNA1, RNA2Δ2b, and RNA3, in which 2b protein expression is abolished (23). Photographs were taken at 4 and 14 dpi, respectively. Leaves of RDR6isat-12 plants with wild-type CMV-related yellowing turned brown (brown arrow) and dried rapidly compared to leaves of the wild-type CMV-infected Nbsat-12 plants (yellow arrow) at 14 dpi.
Typical leaf mosaic and curling symptoms appeared in the newly growing leaves at 4 dpi, and no significant difference was observed between CMVwt-inoculated Nbsat-12 plants and the corresponding control transgenic plants (Fig. 7B). At 14 dpi, noninoculated leaves of systemically CMVwt-infected Nbsat-12 and control plants exhibited severe developmental defects, and CMVwt-related yellowing symptomatic phenotypes were also observed (Fig. 7B) (23). Symptoms were greatly attenuated in CMVΔ2b-infected Nbsat-12 and control plants, consistent with our previous report (23). These results revealed that the expression of asatsiR-12 in N. benthamiana did not cause visible alterations of the symptomatic phenotype following CMVwt and CMVΔ2b infection.
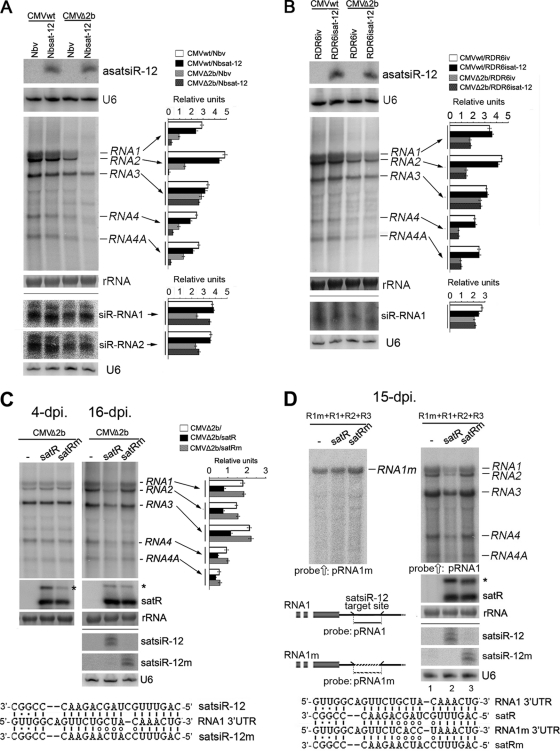
We then examined the accumulation of viral RNAs in locally inoculated leaves at 4 dpi. An RNA gel blot analysis showed similar accumulation levels of CMV RNAs in CMV wild type-infected Nbsat-12 and vector control plants (Fig. 8A). However, accumulation of CMV genomic RNA1 and RNA2 in CMVΔ2b-infected Nbsat-12 plants was greatly reduced compared to that in infected control plants (Fig. 8A), indicating that asatsiR-12 may mediate CMV RNA1 and RNA2 degradation in the absence of the SD-CMV 2b protein. Accumulation of subgenomic RNA4A, which is transcribed from genomic RNA2 and has the same 3′ UTR as RNA2, was also reduced (Fig. 8A), suggesting that asatsiR-12 also targeted CMV RNA4A for degradation. However, we could not rule out that asatsiR-12-mediated CMV RNA1 degradation might also indirectly cause the reduced accumulation of CMV RNA2 and RNA4A. Viral siRNA detection supported this supposition. There was an increased level of viral siR-RNA1 in CMVΔ2b-infected Nbsat-12 plants compared to that in CMVΔ2b-infected vector control plants (Fig. 8A); however, there was no obvious increase in viral siR-RNA2 in CMVΔ2b-infected Nbsat-12 plants compared to that in CMVΔ2b-infected control plants (Fig. 8A). This finding supports the asatsiR-12-mediated degradation of CMV RNA1 but shows less of an effect on RNA2/RNA4A. The similar accumulation levels of RNA3 in CMVΔ2b-infected Nbsat-12 plants and infected control plants (Fig. 8A) suggested that asatsiR-12 might not target CMV RNA3 and its subgenomic RNA4 region in native CMV infection. The reduced accumulation of subgenomic RNA4 might also result from the reduction in RNA1 and RNA2, which encode the 1a and 2a RDRs, respectively. A reduction in viral RNAs was not observed following wild-type CMV infection (Fig. 8A), indicating that the asatsiR-12-mediated downregulation of CMV RNAs was inhibited in the presence of the 2b protein.
Fig. 8.
Analysis of the effect of asatsiR-12 and satsiR-12 mutant satRNAm on accumulation of wild-type viral RNAs and anti-satsiR-12 mutant RNA1 (RNA1m) in viral infection. (A and B) Detection of expression of asatsiR-12 and accumulation of CMV RNAs, siR-RNA1, and siR-RNA2 in leaves of Nbsat-12, Nbv, RDR6isat-12, and RDR6iv plants with local CMVwt or CMVΔ2b infection at 4 dpi. siR-RNA1 and siR-RNA2 represent CMV RNA1- and RNA2-derived vsiRNAs respectively. (C) Detection of accumulation of CMV RNAs in wild-type N. benthamiana leaves in systemic CMVΔ2b coinfection with wild-type satRNA and satsiR-12 mutant satRNAm at 4 and 16 dpi. An asterisk denotes a replication form of satRNA. Alignment of satsiR-12 and mutant satsiR-12m with CMV RNA1 3′-UTR RNA is shown under the blot. The satsiR-12/RNA1 3′-UTR pair contains three mismatches in base pairing, whereas there are eight mismatches in the satsiR-12m/RNA1 3′-UTR pair. (D) Detection of accumulation of CMV RNAs and RNA1m at 15 dpi in plants inoculated with a mixture of RNA1m, RNA1, RNA2, and RNA3 with or without satRNA or satRNAm. Membrane for detection of viral RNAs was first hybridized with an RNA1m-specific oligodeoxynucleotide probe (pRNA1m) (left panel) and then stripped and rehybridized with an RNA1-specific oligodeoxynucleotide probe (pRNA1), which can also detect other CMV RNAs (right panel), but not RNA1m. Alignment of satRNA (satR) and mutant satRNA (satRm) with CMV RNA1 and RNA1m 3′-UTR RNA at the satsiR-12 complementation region is shown under the blot. The satR/RNA1m and satRm/RNA1m pairs each contain eight mismatches in base pairing in the 3′ UTRs. Pools of samples collected from four plants were used. Hybridizations were performed with 32P-labeled oligodeoxynucleotide probes specific for asatsiR-12, satsiR-12m, RNA1, or RNA1m (D) or a mix of probes for 32P-labeled SD-CMV genomic RNA 3′ UTRs (A to C) or satRNA or 32P-labeled in vitro transcripts from RNA1 and RNA2. Methylene blue-stained rRNA and the U6 RNA hybridization are shown as loading controls. Quantification of each CMV RNA and siRNA relative to the loading control is shown to the right of each panel.
Similar leaf mosaic and curling symptoms were also observed in the newly growing leaves in CMVwt-infected RDR6isat-12 plants and corresponding vector control transgenic plants (RDR6iv plants) (Fig. 7B). However, the leaves with CMVwt-related yellowing turned brown and dried rapidly compared to those of CMVwt-infected Nbsat-12 and control plants (Fig. 7B), consistent with the lower resistance capacity of RDR6i plants (43, 54). CMVΔ2b infection caused a less severe curling phenotype than that observed following CMVwt infection, and similar symptomatic phenotypes in RDR6isat-12 and RDR6iv plants were observed (Fig. 7B). These results revealed that the expression of asatsiR-12 in RDR6i plants did not cause visible alterations in symptomatic phenotypes following infection with either CMVwt or CMVΔ2b. However, in contrast to the reduction of CMV RNAs in CMVΔ2b-infected Nbsat-12 plants (Fig. 8A), an RNA gel blot analysis showed that the accumulation levels of CMV RNAs did not change in CMVwt- and CMVΔ2b-infected RDR6isat-12 plants compared to those in infected RDR6iv plants at 4 dpi (Fig. 8B). Together with the above-mentioned observation for the CMVΔ2b-infected Nbsat-12 plants (Fig. 8A), our results indicate that asatsiR-12-mediated degradation of viral RNAs requires the RDR6-dependent silencing pathway. In other words, asatsiR-12 targeting the 3′ UTR of CMV RNAs triggered the host RDR6-dependent degradation of CMV RNAs. The fact that the asatsiR-12-mediated GFP-3′UTR1 cleavage product steadily accumulated in the absence of the 2b protein (Fig. 3 and 6) suggested that in CMVΔ2b-infected RDR6isat-12 plants, asatsiR-12-mediated cleavage alone without RDR6-dependent degradation had little effect on the accumulation of CMV RNAs.
We previously detected a reduction in CMV RNAs following CMVΔ2b infection in the presence of replicating satRNA at 20 dpi compared to those in CMVΔ2b-infected wild-type plants (23). To examine whether satsiR-12 derived from satRNA replication contributed to the reduction in CMV RNAs, we constructed a satsiR-12 substitution mutant satRNA (satRNAm), the replication of which produced satsiR-12m but not satsiR-12 (Fig. 8C), for coinfection with CMVΔ2b. Reduced accumulation of the replication form of satRNAm was detected at 4 dpi, but not at 16 dpi (Fig. 5C), compared to wild-type satRNA. This finding suggests that the substitution mutation of satRNAm may affect its replication capacity at an early stage. Production of satsiR-12 and satsiR-12m was also confirmed at 16 dpi (Fig. 8C). Similar accumulation levels of CMV RNAs were observed with coexpression of satRNA and satRNAm at 4 dpi (Fig. 8C), while accumulation of the CMV RNAs was reduced in coinfection with wild-type satRNA but not obviously changed in coinfection with satRNAm at 16 dpi (Fig. 8C). This result clearly indicated that degradation of CMV RNAs resulted, at least in part, from the action of satsiR-12.
To further examine the effect of satsiR-12-mediated degradation of CMV RNAs in the presence of the 2b protein, mutated CMV RNA1 (RNA1m), in which the 3′ UTR was unable to be targeted by either satsiR-12 or satsiR-12m (Fig. 8D), was created and a competition experiment was performed. Wild-type plants were coinoculated with a mixture of RNA1m, RNA1, RNA2, and RNA3 with or without satRNA or satRNAm. Figure 8D shows that there was no significant difference observed in accumulation levels of RNA1m in coinfection with or without satRNA or satRNAm (Fig. 8D, left panel). Similar accumulation levels of wild-type CMV RNAs were also observed in coinfection with or without satRNAm (Fig. 8D, right panel, lanes 3 and 1). However, accumulation levels of wild-type CMV RNAs were clearly lower in coinfection with wild-type satRNA (Fig. 8D, right panel, lane 2). These results clearly indicate that the satRNA did affect RNA1, but not RNA1m, through satsiR-12 during wild-type CMV infection.
Taken together, our data for wild-type N. benthamiana plants indicate that asatsiR-12 has a role in interacting with helper SD-CMV by targeting viral RNAs at the 3′ UTR and triggering the host RDR6-dependent degradation of viral RNAs. Reduction in the CMV RNA1, but not the mutant RNA1m, in the competition infection with wild-type satRNA suggests that satsiR-12-related degradation of viral RNAs was partially inhibited by the 2b protein following wild-type CMV infection. In other words, the inhibitory effect of the 2b protein could not completely compensate for the satsiR-12-mediated RDR6-dependent degradation, resulting in a reduction in CMV RNAs in coinfection with satRNA but not with or without satRNAm. Nevertheless, our results also demonstrate the suppressor activity of SD-CMV 2b in the context of native CMV infection. In the absence of the 2b protein, the asatsiR-12-mediated downregulation of viral RNAs was not observed in RDR6i plants, suggesting that the inhibitory effect of 2b on asatsiR-12 function counteracted the RDR6-dependent antiviral silencing.
DISCUSSION
In this study, we first confirmed that natural satRNA-derived siRNAs could be associated with AGO proteins in planta. The AGO-binding abilities further strengthened the idea that satsiRNAs play an important role in pathogenicity by interfering with either endogenous or helper virus gene expression. The study of asatsiR-12 in the downregulation of the expression of the sensor construct GFP-3′UTR1 (Fig. 3) and CMV RNA(s) in native infections (Fig. 8) revealed that satsiR-12 interacted with helper virus genomic sequences via targeting of the 3′-UTR sequence.
Infection of wild-type (Nbsat-12) and RDR6i (RDR6isat-12) plants (Fig. 8) revealed that asatsiR-12 targeting the 3′ UTR of CMV RNAs triggers RDR6-dependent degradation of viral RNAs. It has been found previously that RDR6i plants infected with CMV in combination with the Y satellite display more severe symptoms than infected control plants but that the symptoms of RDR6i and control plants infected with CMV without the Y satellite are similar (43). One of the possible explanations for the weak symptoms in CMV/Y satellite-infected wild-type plants could be the involvement of the RDR6-dependent degradation of viral RNAs in the presence of Y satellite-derived siRNAs. Together with the asatsiR-12-mediated reduction of CMV RNAs (Fig. 8A) and the detection of AGO-binding activity of the natural satsiR-12 (Fig. 1), our results support the contribution of satsiR-12 to the silencing of CMV RNAs. The constitutive expression of asatsiR-12, without being affected by other satRNA-derived siRNAs, may explain the effect of asatsiR-12 on reducing CMV RNAs detected at earlier time points, whereas the reduction of CMV RNAs by satRNA was detected at later time points (23) (Fig. 8).
Consistent with a recent report that shows an overall reduction in the density of CMV siRNAs targeting both RNA1 and RNA2 in rdr6 plants compared to wild-type plants infected with CMVΔ2b (52), extensive degradation of RNA1 and RNA2, but not RNA3, in Nbsat-12 plants infected with CMVΔ2b (Fig. 8A) further supports the role of satsiR-12 in triggering the production of RDR6-dependent secondary CMV siRNAs. Furthermore, our competition experiment shows that in wild-type plants, the wild-type CMV RNA1 and the anti-satsiR-12 mutant RNA1m accumulated to similar amounts in competition with each other in the absence or presence of the mutant satRNAm. In contrast, when in competition with wild-type satRNA, CMV RNA1, but not RNA1m, was reduced. Our results clearly indicated that the satRNA was affecting the wild-type virus through satsiR-12. Our data also reveal that the effect of the SD-CMV 2b protein in interfering with the asatsiR-12-mediated cleavage of wild-type CMV RNAs protected CMV RNAs from subsequent RDR6-mediated degradation. However, during native viral infection, the inhibitory effect of the SD-CMV 2b protein may not completely overcome RDR6-dependent degradation. More severe symptoms were observed in CMV/Y satellite-infected RDR6i plants than in infected wild-type plants (43). Similarly, the CMV-related yellowing of leaves was less severe in Nbsat-12 plants than in CMV-infected RDR6isat-12 plants, where leaves turned brown and dried rapidly in comparison with the control (Fig. 7B). Collectively, these results reveal that RDR6-dependent anti-CMV activities in the presence of the Y satellite or asatsiR-12 were not completely overcome by the inhibitory effect of 2b proteins. In this scenario, the satsiR-12 targeting of the CMV 3′ UTR strengthened the host antiviral silencing during the defense and counterdefense arms race between the host and virus in native CMV infection.
The diverse RNA silencing pathways and involvement of large numbers of silencing proteins and their multiple functions, in addition to the various evolving functions of viral silencing suppressors, complicate host-pathogen interactions. Some effects of antiviral or silencing suppression may not be readily shown during normal infections. For example, Turnip crinkle virus (TCV)-derived DCL4-dependent 21-nt siRNAs were detected only following infection with TCVΔP38, which lacks the viral suppressor P38. In normal TCV infection, DCL2-dependent 22-nt viral siRNAs accumulate, suggesting that P38 interferes with DCL4 function during normal infections and that DCL2 takes over the antiviral defense (10). By analysis of infection with the CMV 2b deletion mutant CMVΔ2b in the Arabidopsis rdr mutant, Diaz-Pendon and colleagues showed that the production of CMV-derived siRNAs is largely RDR1 dependent (12). However, in the presence of the 2b protein, the RDR1-dependent production of CMV siRNAs was undetectable. Thus, their study shows the inhibitory effects of 2b in the RDR1-dependent production of viral siRNAs (12). The antiviral functions of RDR1 and RDR6 are further indicated by the finding that each RDR exhibits specificity in targeting the tripartite CMV RNA genome in the production of secondary viral siRNAs (52). The increased production of viral siRNAs targeting the 3′ half of RNA3 detected in rdr1 mutant plants was suggested to be RDR6 dependent (52). We have reported previously that CMV RNA3-derived siRNAs accumulate at very low levels during wild-type CMV infection but at high levels during CMVΔ2b infection (23), suggesting that the 2b protein suppresses the silencing processes that specifically target some region of CMV RNA3, probably for RDR6-dependent secondary siRNA production, which may be triggered following viral RNA replication-triggered biogenesis of primary siRNAs (52). Interestingly, satellite RNA itself is not targeted by either RDR for amplification of secondary siRNAs (52). This highlights the role that satsiR-12 plays in triggering the RDR-dependent silencing for the amplification of anti-CMV effectors specifically against helper virus RNA1 and RNA2, but not satellite RNAs.
We have previously described the trilateral interactions among host plant, helper virus, and satRNA relevant to host RNA silencing (23). SD-CMV satRNA reduces the accumulation of SD-CMV subgenomic RNA4A, which encodes the 2b protein (23), presumably as a result of the natural satRNA-derived satsiR-12-mediated degradation, as found for asatsiR-12 (Fig. 8A). A reduction in the silencing suppressor 2b protein by SD-CMV satRNA relieves normal host regulatory silencing processes influenced by the 2b protein. The high level of satRNA-derived siRNA accumulation (17, 23) also reveals that the recruitment of silencing components to the abundant satRNA itself partially compensates for the increased silencing due to the reduced accumulation of the 2b suppressor protein caused by satRNA/satsiRNAs.
By using bioinformatic prediction, several satsiRNAs were found that potentially target sequences in the Arabidopsis genome. Although a 5′ RACE analysis of samples from SD-CMV-infected Arabidopsis plants showed that none of the cleavage sites were located in or near the satsiRNA/target complementary region, we could not rule out the possibility that satsiRNAs, like siRNAs from CMV Y-satRNA (48), may have a role in interacting with host genes. This may be because the satsiRNA-mediated cleavage of host target genes is inhibited during normal infections with 2b-containing wild-type CMV. Further investigation with an infectious clone of CMVΔ2b will be able to address these issues.
It was suggested previously that the effect of satRNA on helper viruses could be related to the competitiveness of the replication between the helper virus and satRNA (40, 46). Our data show a new mechanism of satRNA in reducing helper virus RNAs via targeting of viral RNA by satRNA-derived siRNA, which triggers subsequent host RDR6-dependent antiviral silencing. satsiR-12 is the most frequently cloned satsiRNA (17) and originates from a region of CMV satRNA highly conserved in different satRNA strains and variants (Fig. 2B and data not shown). Together with the high nucleotide sequence similarity present within the 3′ UTRs among different CMV subgroups (18, 41), our results suggest that the function of satsiR-12 in reducing helper virus RNAs may evolve de novo to strengthen the host RDR6-dependent amplification of anti-CMV effectors. Our findings provide evidence that satsiRNAs play an important biological function in homeostatic interactions among the host, virus, and satellite RNA to determine the final outcome of viral infection. Suppression of satsiR-12-mediated cleavage also demonstrates a concrete biological function for the 2b protein as a silencing suppressor in native CMV infections.
ACKNOWLEDGMENTS
We thank David Baulcombe for RDR6i seeds, Steve E. Jacobsen for ago1-27 seeds, and Yijun Qi for antibodies to AGO2 and AGO5.
This research was supported by grants from the Ministry of Science and Technology (grant 2011CB100700), the National Science Foundation of China (grants 31030009, 30900057, and 30940003), and the Chinese Academy of Sciences (Knowledge Innovation Program [KSCX2-EW-N-06]).
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 2. Reference deleted.
- 3. Baulcombe D. 2004. RNA silencing in plants. Nature 431:356–363 [DOI] [PubMed] [Google Scholar]
- 4. Baumberger N. 2005. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 102:11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brigneti G., et al. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Chapman E. J. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H. M., et al. 2010. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U. S. A. 107:15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collmer C. W., Howell S. H. 1992. Role of satellite RNA in the expression of symptoms caused by plant viruses. Annu. Rev. Phytopathol. 30:419–442 [DOI] [PubMed] [Google Scholar]
- 9. Cuperus J. T., et al. 2010. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17:997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deleris A., et al. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68–71 [DOI] [PubMed] [Google Scholar]
- 11. Díaz-Pendón J. A., Ding S.-W. 2008. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46:303–326 [DOI] [PubMed] [Google Scholar]
- 12. Diaz-Pendon J. A., Li F., Li W.-X., Ding S.-W. 2007. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19:2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding B., Li Q., Nguyen L., Palukaitis P., Lucas W. J. 1995. Cucumber mosaic virus 3a protein potentiates cell-to-cell trafficking of CMV RNA in tobacco plants. Virology 207:345–353 [DOI] [PubMed] [Google Scholar]
- 14. Ding S.-W., Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding S. W., Anderson B. J., Haase H. R., Symons R. H. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198:593–601 [DOI] [PubMed] [Google Scholar]
- 16. Donaire L., et al. 2008. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 82:5167–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Du Q. S., et al. 2007. DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J. Virol. 81:9142–9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duan C. G., Wang C. H., Fang R. X., Guo H. S. 2008. Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 82:11084–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fernandez-Cuartero B., et al. 1994. Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology 203:373–377 [DOI] [PubMed] [Google Scholar]
- 20. Garcia-Ruiz H., et al. 2010. Arabidopsis RNA-dependent RNA polymerases and Dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 22:481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo H. S., Ding S. W. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horsch R., et al. 1985. A simple and general method for transferring genes into plants. Science 227:1229–1231 [DOI] [PubMed] [Google Scholar]
- 23. Hou W.-N., Duan C.-G., Fang R.-X., Zhou X.-Y., Guo H.-S. 2011. Satellite RNA reduces expression of the 2b suppressor protein resulting in the attenuation of symptoms caused by Cucumber mosaic virus infection. Mol. Plant Pathol. 12:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishihama A., Barbier P. 1994. Molecular anatomy of viral RNA-directed RNA polymerases. Arch. Virol. 134:235–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khvorova A., Reynolds A., Jayasena S. D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216 [DOI] [PubMed] [Google Scholar]
- 26. Lanet E., et al. 2009. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21:1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li F., Ding S.-W. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Llave C. 2010. Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 15:701–707 [DOI] [PubMed] [Google Scholar]
- 29. Lucy A. P., Guo H. S., Li W. X., Ding S. W. 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19:1672–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Masuta C., Takanami Y. 1989. Determination of sequence and structural requirements for pathogenicity of a cucumber mosaic virus satellite RNA (Y-satRNA). Plant Cell 1:1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mi S., et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moissiard G., Parizotto E. A., Himber C., Voinnet O. 2007. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 13:1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morel J. B., et al. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14:629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niu Q. W., et al. 2006. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24:1420–1428 [DOI] [PubMed] [Google Scholar]
- 35. O'Reilly E. K., Kao C. C. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287–303 [DOI] [PubMed] [Google Scholar]
- 36. Palukaitis P., Garcia-Arenal F. 2003. Cucumoviruses. Adv. Virus Res. 62:241–323 [DOI] [PubMed] [Google Scholar]
- 37. Palukaitis P., Roossinck M. J. 1996. Spontaneous change of a benign satellite RNA of cucumber mosaic virus to a pathogenic variant. Nat. Biotechnol. 14:1264–1268 [DOI] [PubMed] [Google Scholar]
- 38. Qu F. 2010. Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. Mol. Plant Microbe Interact. 23:1248–1252 [DOI] [PubMed] [Google Scholar]
- 39. Rhoades M. W., et al. 2002. Prediction of plant microRNA targets. Cell 110:513–520 [DOI] [PubMed] [Google Scholar]
- 40. Rizzo T. M., Palukaitis P. 1990. Construction of full-length cDNA clones of cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol. Gen. Genet. 222:249–256 [DOI] [PubMed] [Google Scholar]
- 41. Roossinck M. J., Zhang L., Hellwald K.-H. 1999. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol. 73:6752–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwab R., Voinnet O. 2010. RNA silencing amplification in plants: size matters. Proc. Natl. Acad. Sci. U. S. A. 107:14945–14946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwach F., Vaistij F. E., Jones L., Baulcombe D. C. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138:1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwinghamer M. W., Symons R. H. 1975. Fractionation of cucumber mosaic virus RNA and its translation in a wheat embryo cell-free system. Virology 63:252–262 [DOI] [PubMed] [Google Scholar]
- 45. Shimura H., et al. 2011. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simon A. E., Roossinck M. J., Havelda Z. 2004. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu. Rev. Phytopathol. 42:415–437 [DOI] [PubMed] [Google Scholar]
- 47. Sleat D. E., Palukaitis P. 1992. A single nucleotide change within a plant virus satellite RNA alters the host specificity of disease induction. Plant J. 2:43–49 [PubMed] [Google Scholar]
- 48. Smith N. A., Eamens A. L., Wang M. B. 2011. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 7:e1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Song C., et al. 2010. MiR-RACE, a new efficient approach to determine the precise sequences of computationally identified trifoliate orange (Poncirus trifoliata) microRNAs. PLoS One 5:e10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vaucheret H. 2008. Plant ARGONAUTES. Trends Plant Sci. 13:350–358 [DOI] [PubMed] [Google Scholar]
- 51. Wang M. B., Wesley S. V., Finnegan E. J., Smith N. A., Waterhouse P. M. 2001. Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA 7:16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X. B., et al. 2010. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 107:484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reference deleted.
- 54. Ying X. B., et al. 2010. RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell 22:1358–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang X., et al. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20:3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]