Abstract
The highly pathogenic severe acute respiratory syndrome coronavirus (SARS-CoV) poses a constant threat to human health. The viral spike protein (SARS-S) mediates host cell entry and is a potential target for antiviral intervention. Activation of SARS-S by host cell proteases is essential for SARS-CoV infectivity but remains incompletely understood. Here, we analyzed the role of the type II transmembrane serine proteases (TTSPs) human airway trypsin-like protease (HAT) and transmembrane protease, serine 2 (TMPRSS2), in SARS-S activation. We found that HAT activates SARS-S in the context of surrogate systems and authentic SARS-CoV infection and is coexpressed with the viral receptor angiotensin-converting enzyme 2 (ACE2) in bronchial epithelial cells and pneumocytes. HAT cleaved SARS-S at R667, as determined by mutagenesis and mass spectrometry, and activated SARS-S for cell-cell fusion in cis and trans, while the related pulmonary protease TMPRSS2 cleaved SARS-S at multiple sites and activated SARS-S only in trans. However, TMPRSS2 but not HAT expression rendered SARS-S-driven virus-cell fusion independent of cathepsin activity, indicating that HAT and TMPRSS2 activate SARS-S differentially. Collectively, our results show that HAT cleaves and activates SARS-S and might support viral spread in patients.
INTRODUCTION
The severe acute respiratory syndrome coronavirus (SARS-CoV) is the causative agent of a novel lung disease, SARS, which was first observed in Guangdong Province, southern China, in 2002 (32, 33). Subsequently, SARS spread to more than 30 countries with 8,096 cases. About 10% of the afflicted individuals died from the disease, with elderly patients being disproportionally affected (32, 33). The emergence of the SARS-CoV was traced back to palm civets and other animals sold in Chinese wet markets (15). It is believed that these animals serve as intermediate hosts, while Chinese horseshoe bats, which harbor SARS-CoV-related viruses, constitute a natural reservoir (27, 29). The circulation of SARS-CoV in an animal reservoir poses the continuous threat of reintroduction of the virus into the human population. Therefore, the development of preventive and therapeutic measures is required, and host cell factors essential for spread of SARS-CoV and potentially other respiratory viruses are attractive targets.
The SARS-CoV spike protein (SARS-S), jointly with the viral M and E proteins, is incorporated into the viral membrane, and SARS-S mediates infectious viral entry into target cells (20). For this, SARS-S needs to bind to a receptor on the host cell surface and to fuse the viral membrane with a host cell membrane, thereby allowing delivery of SARS-CoV proteins and genomic information into the host cell cytoplasm, the location of SARS-CoV replication (20, 43). Angiotensin-converting enzyme 2 (ACE2) has been identified to be the SARS-CoV receptor (28) and was found to be expressed on type II pneumocytes and enterocytes, major viral target cells (16, 31, 46, 47). ACE2 expression protects against experimentally induced lung disease, and viral interference with receptor expression might contribute to SARS pathogenesis (22, 25).
Receptor engagement and membrane fusion are accomplished by two separate subunits in SARS-S, the N-terminal surface unit S1 (receptor binding) and the C-terminal transmembrane unit S2 (membrane fusion). In order to transit into an active state, viral glycoproteins with an architecture similar to that of SARS-S, termed class I fusion proteins, frequently depend on cleavage by host cell proteases (20). An initial study indeed suggested that efficient SARS-CoV spread might depend on SARS-S activation by furin (3), but these findings were not substantiated by subsequent work. A seminal study by Simmons and colleagues revealed that cathepsins, pH-dependent, endo-/lysosomal cysteine proteases, activate SARS-S (40). Thus, binding of SARS-CoV to ACE2 is thought to trigger receptor-mediated endocytosis and transport of virions into host cell endosomes (48), where SARS-S is activated for membrane fusion upon cleavage by cathepsin L (40). Consequently, it was proposed that cathepsin inhibitors could be developed for therapy of SARS-CoV infection (40), and such efforts are under way (36).
Type II transmembrane serine proteases (TTSPs) play an important role in development and homeostasis, and dysregulated TTSP expression is a hallmark of different cancers (10). Human influenza viruses parasitize transmembrane protease, serine 2 (TMPRSS2), and human airway trypsin-like protease (HAT), members of the TTSP family, to facilitate their activation (5, 7, 9). TMPSS2 has also recently been shown to cleave and activate SARS-S for cell-cell and virus-cell fusion, thereby allowing cathepsin-independent host cell entry (14, 30, 38). Whether HAT plays a role in SARS-S activation is at present unknown.
Here, we report that HAT cleaves SARS-S at position R667 and activates SARS-S for cell-cell fusion, while TMPRSS2 has multiple cleavage sites in SARS-S and activates SARS-S for cell-cell and virus-cell fusion. HAT was found to be coexpressed with ACE2 in human lung epithelium, indicating that this protease could promote viral spread in patients.
MATERIALS AND METHODS
Plasmids.
Expression plasmids pCAGGS-SARS-S, encoding the spike protein of the Frankfurt strain of SARS-CoV, and pcDNA3-hACE2, encoding human ACE2 (hACE2), have been described previously (18, 19). The plasmids encoding human TMPRSS2, TMPRSS3, TMPRSS4, TMPRSS6, and hepsin have also been described previously (9, 24, 35, 37). HAT was PCR amplified from cDNA from human bronchus, employing primers p5_EcoRI_HAT (GCGAATTCACCATGTATAGGCCAGCACGTGTAACTTCG) and p3_NheI_HAT (GCGCTAGCGCCTAGATCCCAGTTTGTTGCCTAATCC). The PCR product was inserted into pCAGGS via EcoRI and NheI and controlled by sequencing.
Cell culture.
293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin and were grown in a humidified atmosphere containing 5% CO2. 293T cells stably expressing ACE2, 293T-hACE2, were generated by transfection of plasmid pcDNA3.1zeo-hACE2 (18) into 293T cells, followed by selection of resistant cells with zeocin (Invitrogen) at 50 μg/ml, as previously reported (13). Homogeneous surface expression of ACE2 on stably transfected cells was confirmed by flow cytometry.
Cleavage of cellular SARS-S by HAT.
For detection of cleavage of SARS-S by HAT and other TTSPs, 293T cells were cotransfected with SARS-S expression plasmid jointly with a TTSP expression plasmid or empty plasmid. At 6 to 8 h posttransfection, the medium was changed, and at 48 h posttransfection, the cells were washed once with 1× phosphate-buffered saline (PBS) and lysed in 2× sodium dodecyl sulfate (SDS) loading buffer. For immunoblotting, the lysates were separated by SDS-gel electrophoresis and transferred onto nitrocellulose membranes. SARS-S protein was detected by staining with a rabbit serum specific for the S1 subunit, generated by immunization with a peptide comprising SARS-S amino acids 19 to 48 (17). For loading control, the stripped membranes were incubated with an anti-β-actin antibody (Sigma).
VLPs.
For production of virus-like particles (VLPs), 293T cells were cotransfected with the HIV-1 Gag (p55)-encoding plasmid p96ZM651gag-opt (12), SARS-S expression plasmid, TTSP expression plasmid, or empty vector. The cellular supernatants were harvested at 48 h posttransfection and concentrated by ultrafiltration employing VivaSpin centrifugal concentrators (Sartorius). Subsequently, VLPs were purified from concentrated supernatants by ultracentrifugation through a 20% sucrose cushion for 2 h at 25,000 rpm and 4°C. Finally, supernatants and pellets of the ultracentrifuge reactions were treated with PBS or trypsin, followed by addition of soybean trypsin inhibitor (Sigma) and analysis by Western blotting, employing a commercially available antibody directed against the S2 subunit of SARS-S (Imgenex). In addition, the presence of HIV Gag in pellets and supernatants was detected using anti-p24 hybridoma supernatant 183-H12-5C.
HAT-dependent activation of SARS-S for cell-cell fusion.
The cell-cell fusion assay was carried out essentially as described previously (14, 21, 39). For analysis of SARS-S-driven cell-cell fusion in the absence of TTSPs, 293T effector cells seeded in 6-well plates at 1.2 × 105/well were CaPO4 transfected with either SARS-S expression plasmid or empty plasmid (negative control) in combination with plasmid pGAL4-VP16, which encodes the herpes simplex virus VP16 trans activator fused to the DNA binding domain of the Saccharomyces cerevisiae transcription factor GAL4 (21). In parallel, 293T target cells were seeded in 48-well plates at 0.8 × 105/well and transfected with either hACE2-encoding plasmid or empty vector (negative control) together with plasmid pGal5-luc, which encodes the luciferase reporter gene under the control of a promoter containing five GAL4 binding sites. For analysis of SARS-S activation by HAT and other TTSPs in cis, the fusion assay was carried out as described above, but effector cells were additionally transfected with a TTSP expression plasmid. In order to determine SARS-S activation by HAT and other TTSPs in trans, the fusion assay was carried out as described above, but target cells were additionally transfected with a TTSP expression plasmid. At 24 h posttransfection, the effector cells were detached by pipetting, resuspended in fresh medium, and added to the target cells. At 6 h postcocultivation, medium supplemented with trypsin (final concentration, 100 ng/ml; Sigma) or PBS was added to the samples. At 16 h after trypsin treatment, medium was completely removed and fresh culture medium without trypsin was added. Finally, at 24 h after medium change, the cell-cell fusion was quantified by determination of luciferase activities in cell lysates using a commercially available kit (Promega).
Activation of SARS-S-bearing lentiviral pseudotypes by HAT.
Lentiviral pseudotypes bearing SARS-S were essentially generated as described previously (14, 21, 39, 41). In brief, 293T cells were transiently cotransfected with pNL4-3 E-R-Luc (11), SARS-S expression plasmid, and either TTSP expression plasmid or empty plasmid. At 16 h posttransfection, the culture medium was replaced by fresh medium, and at 48 h posttransfection, culture supernatants were harvested. The supernatants were passed through 0.45-μm-pore-size filters, aliquoted, and stored at −80°C. In order to analyze cis activation of virus-cell fusion, pseudotypes generated in TTSP-expressing cells were used for infection experiments. Specifically, equal volumes of pseudotypes normalized for comparable infectivity for 293T-hACE2 cells were employed for infection of 293T-hACE2 cells pretreated for 30 min at 37°C with the indicated concentrations of ammonium chloride or the cathepsin inhibitor MDL28170 (Calbiochem). At 16 h postinfection, the infection medium was replaced by fresh culture medium without inhibitor, and at 72 h postinfection, luciferase activities in cell lysates were determined employing a commercially available kit (Promega). For the assessment of trans activation of virus-cell fusion, the assay was carried out as described for the cis setting, except that pseudotypes produced in the absence of TTSP expression were used for infection of 293T-hACE2 target cells transfected to express TTSPs.
HAT-dependent syncytium formation.
293T-hACE2 cells (2 × 105 cells per ml) were seeded on cover slides in 24 wells and were transfected with 1 μg HAT, TMPRSS2, or TMPRSS4 expression plasmids or transfected with empty control plasmid (pCAGGS) using X-tremeGENE reagent (Roche), according to the manual instructions. After 24 h, cells were infected with SARS-CoV (Frankfurt-1; multiplicity of infection, 0.1) for 30 min at 4°C, and slides were fixed with paraformaldehyde (8%) after another 24 h. Cells were permeabilized with 0.1% Triton X-100, and SARS-CoV antigen was detected by immunostaining by applying a SARS patient antiserum diluted 1:100. In parallel, proteases were detected with mouse anti-HAT (R&D Systems), mouse anti-TMPRSS2 (Santa Cruz), or mouse anti-Myc (for Myc-tagged TMPRSS4; Life Technologies), all diluted 1:100, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Secondary detection was performed with Cy3-conjugated goat anti-human (1:200) and Cy2-labeled goat anti-mouse (1:200) antibodies (Dianova). All pictures were taken with the help of an Axio Imager.M1 fluorescence microscope (Zeiss).
Digest of recombinant SARS-S by recombinant HAT.
Eight micrograms of recombinant SARS-S (rSARS-S [Beijing 02]), produced in 293T cells and obtained from a commercial source (Geneimmune), was incubated with 200 ng HAT (R&D Systems) in assay buffer, 50 mM Tris, 0.05% (wt/vol) Brij 35, pH 9.5, for 2 h at 37°C in a total volume of 25 μl. Subsequently, the reactions were stopped by addition of SDS-loading buffer and the reaction products were analyzed by 12.5% SDS-PAGE and Western blotting. Alternatively, the reaction products were separated on NuPAGE gradient gels and analyzed by mass spectrometry (MS), as described below.
Mass spectrometric analysis of SARS-S cleavage products.
SARS-S cleavage products were separated on precast NuPAGE bis-Tris 4 to 12% gradient gels using a morpholinepropanesulfonic acid buffer system according to the manufacturer (Invitrogen). After colloidal Coomassie staining, gel plugs were excised and subjected to tryptic in-gel digest using our automated platform for the identification of gel-separated proteins by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS and Mascot database search (23). The remainders of the bands were subjected to manual in-gel digest with sequencing-grade endoproteinase Asp-N (Roche) under standard conditions (including prior reduction and carboxamidomethylation of Cys residues). Extracted peptides were dried, redissolved in 0.5% trifluoroacetic acid–0.1% octyl-glucopyranoside, and prepared for MALDI MS by thin-layer affinity preparation on an alpha-cyano-4-hydroxycinnamic acid matrix (23). Peptide survey and fragment ion mass spectra were acquired on an Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics).
Immunostaining of tissue sections.
Formalin-fixed paraffin-embedded lung tissue was obtained with full ethical approval from the National Research and Ethics Service (Oxfordshire Research and Ethics Committee A, reference 04/Q1604/21) and immunostained for HAT and ACE2. Antigen retrieval was performed by pressure cooking in different antigen retrieval solutions. Slides were mounted in Aquatex mounting medium (Merck, United Kingdom). ACE2 immunostaining (affinity-purified goat polyclonal serum; R&D Systems, Abingdon, United Kingdom) was performed, and ACE2 was detected using a mouse anti-goat Ig (GTI-75) (16) and a Novolink max polymer detection system (Leica Microsystems, Newcastle, United Kingdom), according to the manufacturer's instructions, after antigen retrieval in citrate, pH 6.0. HAT immunostaining (mouse monoclonal antibody 337029; R&D Systems, Abingdon, United Kingdom) was performed using the Novolink max polymer detection system, after antigen retrieval in Dako Target retrieval solution, pH 6.0 (Dako, Cambridge, United Kingdom). Alveolar macrophages in lung tissue were used as an internal positive control for ACE2 immunostaining (45), while bronchus tissue was used as a positive control for HAT immunostaining (44). As negative controls for immunostaining, normal goat polyclonal serum was substituted for the anti-ACE2 primary antibody and an irrelevant mouse monoclonal (anti-ALK1 antibody, clone ALK1 [34]) was substituted for the anti-HAT primary antibody. Stained sections were photographed with a Nikon DS-FI1 camera with a Nikon DS-L2 control unit (Nikon United Kingdom Limited, Kingston-upon-Thames, United Kingdom) and an Olympus BX40 microscope (Olympus United Kingdom Limited, Watford, United Kingdom).
RESULTS
HAT cleaves SARS-S.
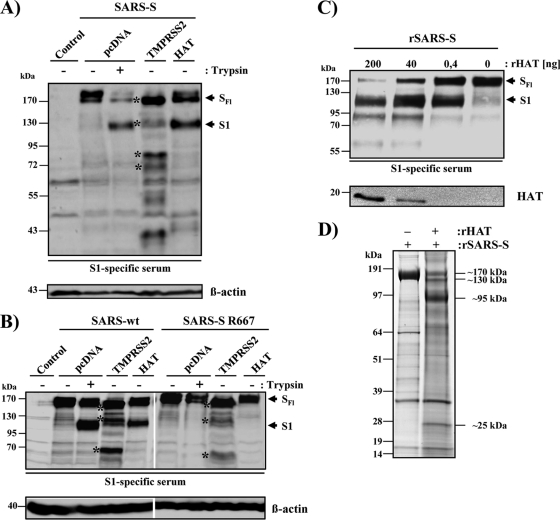
We first investigated whether HAT cleaves SARS-S. For this, the protease and the S protein were coexpressed in 293T cells and the cell lysates were analyzed by Western blotting for cleavage products. As a positive control for SARS-S cleavage, lysates from cells transfected with SARS-S alone were treated with trypsin, which cleaves SARS-S at the boundary between the S1 and S2 subunits (2, 39). In the absence of TTSP coexpression and trypsin treatment of cell lysates, only full-length SARS-S (180 kDa) was detected (Fig. 1 A). Trypsin digestion generated the S1 subunit (120 kDa). Coexpression of TMPRSS2 resulted in SARS-S cleavage at multiple sites, as expected (14), while coexpression of HAT produced a cleavage pattern indistinguishable from the one observed upon trypsin treatment (Fig. 1A). Thus, SARS-S is a substrate for HAT and trypsin and HAT cleave SARS-S at similar or identical sites.
Fig. 1.
HAT cleaves SARS-S. (A) Cleavage of SARS-S by HAT in cotransfected 293T cells. Expression plasmids coding for SARS-S and the proteases indicated or empty vector (pcDNA) were transiently cotransfected into 293T cells, which were then treated with trypsin or PBS. Subsequently, S-protein cleavage was detected by Western blot analysis of cell lysates using a serum specific for the S1 subunit of SARS-S. Detection of β-actin served as a loading control. SFl, full-length SARS-S; S1, S1 subunit of SARS-S; asterisks, SARS-S cleavage fragments generated by TMPRSS2. (B) HAT cleaves SARS-S at arginine 667. Plasmids encoding wild-type SARS-S or the SARS-S R667A mutation were transfected into 293T cells jointly with TMPRSS2, HAT expression plasmids, or empty vector (pcDNA). Subsequently, the cells were treated with PBS or trypsin, and SARS-S cleavage was analyzed by Western blot analysis of cell lysates, using an S1-specific antiserum. Expression of β-actin in cell lysates was assessed as a loading control. (C) Cleavage of recombinant SARS-S (rSARS-S) by recombinant HAT. Recombinant SARS-S was incubated with the indicated concentrations of recombinant HAT, and cleavage products were analyzed by Western blot analysis employing a SARS-S1-specific serum. Recombinant HAT was also detected. (D) Separation of SARS-S cleavage products for mass spectrometric analysis. SARS-S cleavage products were separated by gel electrophoresis and visualized by colloidal Coomassie staining. Major differential bands appearing upon HAT treatment (see apparent molecular mass annotations on the right) were subjected to in-gel digest with trypsin or Asp-N, followed by mass spectrometric analysis. Thereby, we identified the 130-kDa band to be S2 with its N terminus starting at S668 and the 95-kDa band to be S1 with its C terminus ending at R667. Untreated SARS-S (170-kDa band in the left lane) was processed in parallel as a control.
HAT cleaves SARS-S at arginine 667.
Trypsin and HAT generated indistinguishable SARS-S cleavage fragments, and the cleavage site of trypsin has previously been mapped to arginine 667 (2, 39). Therefore, we investigated whether this amino acid is also a target for SARS-S proteolysis by HAT. While coexpression of wild-type (wt) SARS-S and HAT in 293T cells or trypsin treatment of wt SARS-S-expressing cells resulted in comparable SARS-S cleavage, no cleavage of the SARS-S mutant with the R667A mutation by trypsin or HAT was observed (Fig. 1B). In contrast, mutation R667A had little impact on SARS-S processing by TMPRSS2 (Fig. 1B). These results indicate that R667 is essential for SARS-S proteolysis by trypsin and HAT but not TMPRSS2.
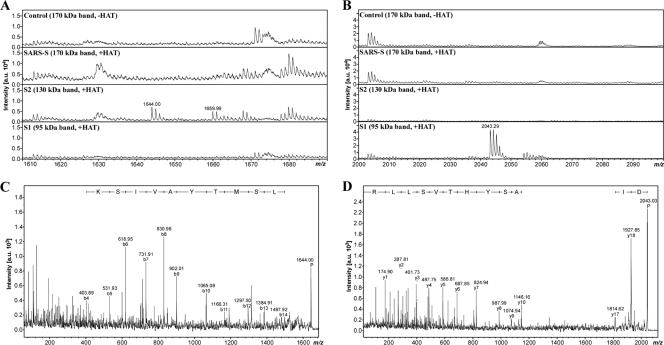
We next sought to provide direct proof that HAT indeed cleaves SARS-S at R667. To this end, we digested soluble SARS-S with recombinant HAT and analyzed the cleavage products by gel electrophoresis and mass spectrometry. Digest of SARS-S with HAT generated the S1 subunit in a concentration-dependent manner (Fig. 1C), indicating that the recombinant proteins used adequately model SARS-S processing by HAT in cells. Next, we repeated the digest and visualized the cleavage fragments by gel electrophoresis, followed by colloidal Coomassie staining. As expected, two prominent bands (∼130 kDa, ∼95 kDa), potentially corresponding to the S1 and S2 subunits, were detected upon SARS-S incubation with HAT and were absent in reactions carried out in the absence of HAT (Fig. 1D). These bands, together with the bands most likely representing intact SARS-S (∼170 kDa) and recombinant HAT (∼25 kDa), were subjected to tryptic in-gel digest and mass spectrometric protein identification. By database search against the Swiss-Prot database, all four bands analyzed were identified to be spike glycoprotein (Swiss-Prot database accession number P59594). However, closer inspection of the sequence coverage (i.e., the distribution of the detected tryptic peptides) revealed that the 170-kDa band represents full-length SARS-S, while the 130-kDa and 95-kDa bands correspond to S2 and S1, respectively. The slower migration of the S2 fragment relative to the S1 fragment, which was confirmed by Western blot analysis (data not shown), might be due to its more extensive glycosylation and/or the particular gel electrophoresis conditions used. In the 25-kDa band, a C-terminal fragment of SARS-S (matching amino acids 800 to 1000) and HAT (identified to be transmembrane protease serine 11D, Swiss-Prot database accession number O60235) were detected. If HAT indeed cleaves SARS-S C terminally of R667, a digest of the S2 subunit with endoproteinase Asp-N (which cleaves N terminally of Asp) should yield the peptide 668-STSQKSIVAYTMSLGA-683 (M + H+ calculated = 1,642.8 Da) as a cleavage product from the N terminus of S2. By in-gel digest with Asp-N and mass spectrometry, the respective mass signal was exclusively detected from the 130-kDa S2 band but not from the bands corresponding to S1 and intact SARS-S, and its identity was further confirmed by mass spectrometric sequencing (Fig. 2 A and C). Consistently, we detected and sequenced the C-terminal Asp-N fragment of S1 (649-DIPIGAGICASYHTVSLLR-667, M + H+ calculated = 2,042.1 Da with carboxamidomethylated Cys) only from the 95-kDa S1 band (Fig. 2B and D). Thus, our mass spectrometric data unambiguously demonstrated that HAT generates a SARS-S cleavage fragment compatible only with cleavage at R667, confirming the results of our mutagenic analysis.
Fig. 2.
Mass spectrometric analysis of Asp-N-digested SARS-S cleavage products. (A and B) Zoomed-in peptide survey spectra of the Asp-N digests. The mass signals at m/z 1,644.00 (A) and m/z 2,043.29 (B) represent the N-terminal Asp-N fragment of S2 (668-STSQKSIVAYTMSLGA-683, M + H+ calculated = 1,643.83 Da) and the C-terminal Asp-N fragment of S1 (649-DIPIGAGICASYHTVSLLR-667, M + H+ calculated = 2,043.069 Da with carboxamidomethylated Cys), respectively, as they were exclusively detected in the corresponding fractions. Note that the mass signal at m/z 1,659.99 in panel A represents the Met-oxidized variant of 668-STSQKSIVAYTMSLGA-683 (+16 mass units). (C and D) Mass spectrometric sequencing of the terminal Asp-N fragments. (C) The fragment ion mass spectrum of m/z 1,644 clearly confirmed the identity of the peptide to be 668-STSQKSIVAYTMSLGA-683 through a conclusive N-terminal b-ion series. The predominant occurrence of b-type ions is in agreement with the charge localization at Lys in position 5, and only this ion series is annotated for the sake of clarity (P, precursor). (D) The fragment ion mass spectrum of m/z 2,043 clearly confirmed the identity of the peptide to be 649-DIPIGAGICASYHTVSLLR-667 through a conclusive C-terminal y-ion series. The predominant occurrence of y-type ions is in agreement with the charge localization at the C-terminal Arg, and only this ion series is annotated for the sake of clarity. Mascot database searches of both fragment ion mass spectra against the Swiss-Prot database (with enzyme set to Arg-C plus Asp-N to mimic the cleavage scenario) identified spike glycoprotein (Swiss-Prot database accession number P59594) with significant MS/MS ion scores. a.u., arbitrary units.
SARS-S cleaved by HAT is incorporated into virus-like particles.
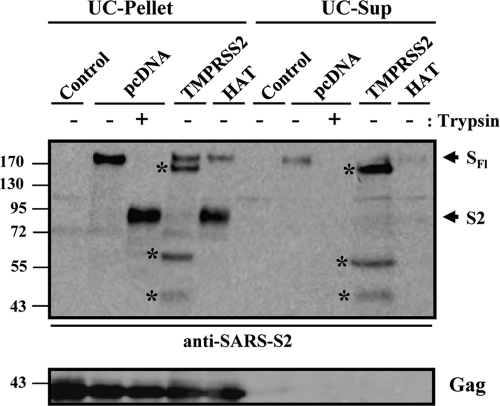
Coexpression of SARS-S and TMPRSS2 produces several S-protein fragments, the largest of which is shed into culture supernatants (14). Therefore, we next examined whether the SARS-S cleavage products generated by HAT remained particle associated or were shed into culture supernatants. For this, HIV Gag protein-derived VLPs carrying the SARS-S protein were produced in the absence or presence of HAT and TMPRSS2. Subsequently, particles were pelleted through a 20% sucrose cushion, and pellets as well as supernatants were treated with trypsin or PBS, followed by Western blot analysis for the presence of S protein (Fig. 3). Particles produced in the presence of TMPRSS2 were found to harbor SARS-S fragments of different sizes (pellet), including the full-length spike protein of 180 kDa. Upon coexpression of HAT, full-length SARS-S and its S2 subunit (90 kDa) were detected in the pellet, similarly to the cleavage products resulting from trypsin treatment of particles produced in the absence of TMPRSS2 or HAT (Fig. 3). The supernatants of particle preparations produced in TMPRSS2-expressing cells contained the 150-kDa subunit of SARS-S as well as smaller cleavage products (Fig. 3), indicating that these fragments were released into culture supernatants. In contrast, SARS-S was largely absent from supernatants produced in control or HAT-expressing cells. Thus, SARS-S cleaved by HAT retains its particle association, while a substantial proportion of the SARS-S fragments generated by TMPRSS2 is shed into culture supernatants.
Fig. 3.
Expression of HAT does not induce SARS-S shedding. VLPs were produced in 293T cells by coexpression of HIV p55-Gag and SARS-S in the absence and presence of coexpressed TMPRSS2, HAT, or cotransfected empty vector. Supernatants were collected and subjected to ultrafiltration followed by ultracentrifugation through a 20% sucrose cushion. Afterwards, pellets and supernatants of the ultracentrifuge reactions were treated with trypsin or PBS and analyzed for the presence of S protein, using a serum specific for the S2 subunit of SARS-S. In parallel, the presence of HIV p55-Gag was determined. UC pellet, VLP preparation subjected to ultrafiltration followed by ultracentrifugation and analysis of the pellets; UC-Sup, VLP preparation subjected to ultrafiltration followed by ultracentrifugation and analysis of the supernatants of ultracentrifuge reactions; SFl, full-length SARS-S; S2, S2 subunit of SARS-S; asterisks, SARS-S cleavage fragments generated by TMPRSS2.
HAT activates SARS-S in cis and trans for cell-cell fusion.
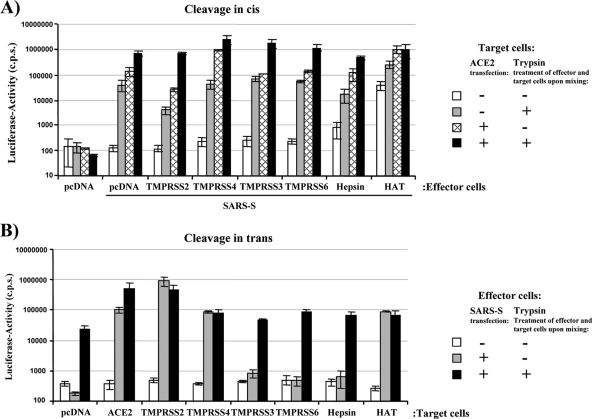
We next investigated if cleavage of SARS-S by HAT activates the S protein for cell-cell fusion. Our previous studies demonstrated that in the 293T cell system, both receptor expression and proteolytic activation limit the rate of SARS-S-driven cell-cell fusion (14, 39). Thus, S-protein-mediated fusion with 293T cells, which express small amounts of endogenous ACE2 (28), is detectable but inefficient and can be rescued by ACE2 overexpression on target cells (14, 39). Under these conditions, fusion is driven by an as yet unidentified leupeptin-sensitive protease endogenously expressed by 293T cells (39). Alternatively, fusion can be rescued by expression of TMPRSS2 on target cells or by trypsin treatment (14, 39), which both ensure efficient SARS-S activation.
Employing this cell-cell fusion assay, we first tested if coexpression of HAT in S-protein-expressing effector cells enhances cell-cell fusion activity (cis activation). For this, effector cells were transfected with SARS-S expression plasmid and a TTSP expression plasmid or empty plasmid. Effector cells transfected with empty plasmid alone served as negative controls. Target cells were transfected with an ACE2 expression plasmid or empty plasmid. Subsequently, effector and target cells were mixed and treated with trypsin or PBS, and the rate of efficiency of cell-cell fusion was determined at 48 h after cell mixing. The presence of SARS-S in effector cells was critical for cell-cell fusion, as expected (Fig. 4 A, pcDNA). In the absence of exogenous ACE2 expression on target cells and trypsin treatment or expression of TTSPs, no notable S-protein-driven cell-cell fusion was observed (pcDNA/SARS-S, white bar). Trypsin treatment overcame low-level receptor expression on target cells and led to a substantial increase in the fusogenic activity of SARS-S (pcDNA/SARS-S, gray bar), as expected (39) (Fig. 4A). Similarly, ACE2 overexpression augmented cell-cell fusion, which was further enhanced by trypsin treatment (pcDNA/SARS-S, lined bar and black bar, respectively), in agreement with our recent findings (14, 39). Expression of TMPRSS2, TMPRSS3, TMPRSS4, TMPRSS6, and hepsin, TTSPs previously examined for their ability to activate SARS-S (14) and influenza virus hemagglutinin (4), in SARS-S-transfected effector cells had no marked influence on the fusogenic activity of SARS-S (Fig. 4A). In contrast, HAT efficiently activated SARS-S for fusion with target cells expressing endogenous ACE2, indicating that SARS-S cleavage in cis by HAT but not TMPRSS2 activates SARS-S for cell-cell fusion.
Fig. 4.
HAT activates SARS-S for cell-cell fusion in cis and in trans. (A) cis activation of SARS-S by HAT. 293T effector cells were cotransfected with pGAL4-VP16 expression plasmid, SARS-S plasmid, and plasmids encoding the indicated proteases or empty plasmid (pcDNA) and mixed with target cells cotransfected with ACE2 expression plasmid or empty plasmid and a plasmid encoding luciferase under the control of a promoter with five GAL4 binding sites. The cell mixtures were then treated with PBS or trypsin, and the luciferase activities in cell lysates were quantified at 48 h after cell mixing. The results of a representative experiment performed in triplicate are shown; error bars indicate standard deviations (SDs). Similar results were observed in an independent experiment. (B) trans activation of SARS-S by HAT. The cell-cell fusion assay was performed as described in the legend for panel A, but proteases were expressed in target cells. The results of a representative experiment performed in triplicate are shown and were confirmed in two separate experiments. Error bars indicate SDs.
To address whether HAT can also activate SARS-S in trans, the cell-cell fusion assay was carried out as described above, but TTSPs were expressed in target instead of effector cells. Expression of TMPRSS3, TMPRSS6, and hepsin in target cells had no effect on SARS-S-driven cell-cell fusion, while expression of TMPRSS2 or TMPRSS4 markedly increased the fusogenic activity of SARS-S (Fig. 4B), in agreement with our published results (14). The same observation was made for expression of HAT, which induced efficient SARS-S-driven cell-cell fusion with target cells expressing endogenous, low levels of ACE2 (Fig. 4B). Thus, HAT activates the SARS spike protein both in cis and in trans, while activation of SARS-S by TMPRSS2 occurs only in trans.
Cleavage activation of SARS-S by HAT does not bypass the need for cathepsin activity.
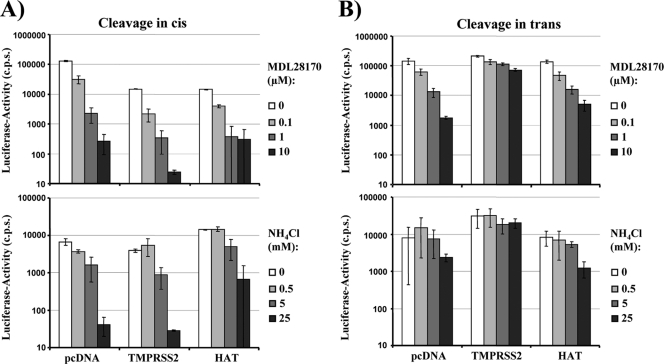
The pH-dependent protease cathepsin L can activate SARS-S for virus-cell fusion (40). Accordingly, SARS-S-driven virus-cell fusion can be inhibited by the lysosomotropic agent ammonium chloride (NH4Cl) and the cathepsin inhibitor MDL28170 (40, 42). Since HAT activated SARS-S for cell-cell fusion in cis, we next investigated whether viruses produced in HAT-expressing cells and thus bearing activated SARS-S were no longer dependent on cathepsin activity for infection of target cells. Infectious entry of pseudotypes bearing SARS-S was efficiently inhibited by both NH4Cl and MDL28170 in a concentration-dependent manner independent of the expression of HAT or TMPRSS2 in virus-producing cells (Fig. 5 A). Thus, cis cleavage of SARS-S by TMPRSS2 or HAT does not render virions independent of cathepsin activity for infectious host cell entry. In contrast, expression of TMPRSS2 on target cells (trans cleavage) allowed efficient infection of NH4Cl- or MDL28170-treated cells by SARS-S-bearing pseudotypes (Fig. 5B), in accordance with our previous results (14). Notably, this observation did not hold true for infection of HAT-expressing target cells, which was efficiently blocked by both agents (Fig. 5B). These results suggest that HAT activates cell- but not virus-associated SARS-S in trans and emphasize that TMPRSS2 and HAT activate SARS-S differentially.
Fig. 5.
Activation of SARS-S by HAT does not bypass the requirement for cathepsin activity for virus-cell fusion. (A) cis cleavage of SARS-S by HAT does not rescue SARS-S-driven infectious entry from blockade by cathepsin inhibitors. Lentiviral pseudotypes bearing SARS-S were generated in 293T cells coexpressing the proteases indicated or cotransfected with empty plasmid (pcDNA). The pseudotypes were used to infect 293T cells engineered to express high levels of ACE2 and preincubated with the indicated concentrations of the cathepsin B/L inhibitor MDL28170 or the lysosomotropic agent NH4Cl. Luciferase activities in cell lysates were determined at 72 h postinfection. The results of a representative experiment performed in triplicate are shown; error bars indicate SDs. Comparable results were obtained in two independent experiments. (B) trans cleavage of SARS-S by HAT does not rescue SARS-S-driven infectious entry from blockade by cathepsin inhibitors. The experiment was carried out as described in the legend for panel A, but proteases were expressed in viral target cells. The results of a representative experiment performed in triplicate are shown and were confirmed in two separate experiments. Error bars indicate SDs.
ACE2 is coexpressed with HAT in human lung tissue and activates the SARS coronavirus for cell-cell fusion.
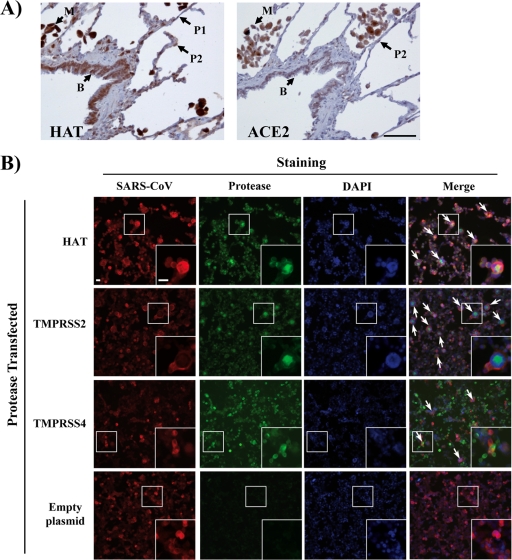
Finally, we explored if activation of SARS-S by HAT could modulate viral spread in infected patients. Immunostaining for HAT demonstrated strong expression by extra- and intrapulmonary bronchial epithelial cells and alveolar macrophages (Fig. 6 A and data not shown). Weaker positive immunostaining suggested expression at a lower level by type I and type II pneumocytes (Fig. 6A). Analysis of a serial section of lung demonstrated coexpression of ACE2 with HAT by intrapulmonary bronchial epithelial cells, alveolar macrophages, and type II pneumocytes, although no expression of ACE2 by type I pneumocytes was detected (Fig. 6A). Thus, HAT is expressed in or near potential SARS-CoV target cells and might modulate viral spread in infected humans. Indeed, expression of HAT and TMPRSS2 promoted formation of syncytia in ACE2-transfected and SARS-CoV-infected 293T cells; syncytia were absent in uninfected cells and much less frequent in infected cells transfected with empty plasmid (Fig. 6B). Similarly, syncytium formation was inefficient in TMPRSS4-transfected cells (Fig. 6B), in agreement with our previous finding that TMPRSS4 does not activate SARS-CoV (14). In summary, HAT is expressed in viral target cells in human lung and can promote fusion of SARS-CoV-infected cells with neighboring permissive cells.
Fig. 6.
HAT and ACE2 are coexpressed in human lung, and HAT activates the SARS coronavirus for cell-cell fusion. (A, left) Section of lung immunostained for HAT, using the peroxidase technique (brown), demonstrating strong expression by intrapulmonary bronchial epithelial cells (B) and alveolar macrophages (M). Weaker positive immunostaining suggested expression at a lower level by type 1 pneumocytes (P1; thin flat cells) and type 2 pneumocytes (P2; plumper cells). (A, right) Serial section of lung immunostained for ACE2, using the peroxidase technique (brown), demonstrating expression by intrapulmonary bronchial epithelial cells (B), alveolar macrophages (M), and type 2 pneumocytes (P2), although no expression of ACE2 by type 1 pneumocytes is seen. Bar, 50 μm (the bar pertains to both panels). (B) ACE2-expressing 293T cells were transfected with HAT, TMPRSS2, or TMPRSS4 expression plasmids or transfected with empty plasmid (control) and infected with SARS-CoV (Frankfurt-1; multiplicity of infection, 0.1). At 24 h postinfection, the cells were fixed with paraformaldehyde (8%) and permeabilized, and SARS-CoV antigen was detected by immunostaining using a human antiserum and a goat anti-human Cy3-labeled secondary antibody (red). In parallel, proteases were detected with mouse anti-HAT, mouse anti-TMPRSS2, or mouse anti-Myc (for Myc-tagged TMPRSS4) and a Cy2-conjugated goat anti-mouse antibody (green). Nuclei were stained with DAPI (blue). Arrows, examples of syncytium formation that were partially magnified (white squares). Bars, 25 μm. Similar results were obtained in an independent experiment.
DISCUSSION
TMPRSS2 activates influenza virus (4, 7, 8), human metapneumovirus (37), and SARS-CoV (14, 30, 38) in cell culture and is coexpressed with ACE2 in type II pneumocytes, major SARS-CoV target cells (14, 30). It is thus likely that this protease modulates SARS-CoV spread in infected humans. The related protease HAT, which can regulate the structure and function of the urokinase receptor and potentially other receptors (1), also activates influenza viruses (7), but its potential role in other viral infections has not been analyzed. Here, we show that HAT, like TMPRSS2, cleaves and activates SARS-S, but the mode and outcome of SARS-S activation by these proteases are notably different.
First, HAT and TMPRSS2 cleave different motifs in SARS-S. Mutagenic analysis and mass spectrometry revealed that HAT, like trypsin, cleaves SARS-S at arginine 667. In contrast, TMPRSS2 cleaves SARS-S at multiple, at present unidentified sites, and it is unknown which of the cleavage events is important for SARS-S activation. A recent study suggests that activation of SARS-S involves two cleavage events, one at arginine 667 and one at arginine 797 (2), and one can speculate that the latter amino acid residue defines the cleavage site recognized by TMPRSS2. Alternatively, TMPRSS2 might recognize the same motif as cathepsin L, which has been shown to cleave at R678 (6). Differential cleavage of SARS-S by HAT and TMPRSS2 most likely reflects differential substrate specificity, and only the substrate specificity of HAT has been determined (49). Alternatively, SARS-S sequences distant from the cleavage sites might modulate recognition by TTSPs, a possibility that remains to be investigated.
Second, HAT activates cellular SARS-S in cis and in trans, while TMPRSS2 is active only in trans. The inability of TMPRSS2 to activate SARS-S in cis could be explained by aberrant S-protein cleavage or lack of cleavage under these conditions. Our previous analysis of cleavage fragments argues against but does not exclude these possibilities (14). A more likely explanation could be that activation of SARS-S by TMPRSS2 is a spatially and temporally restricted process which can proceed only once SARS-S makes contact with ACE2 and TMPRSS2 on target cells. The recent finding that ACE2 binds to and is cleaved by TMPRSS2 lends support to such a scenario (38).
Third, TMPRSS2 but not HAT expression allows SARS-S-driven virus fusion with target cells treated with a cathepsin inhibitor. This observation is particularly striking, since trypsin treatment of cell-bound, SARS-S-bearing virions bypasses the need for cathepsin activity for infectious entry (2, 39, 40). A possible explanation for this apparent discrepancy is that trypsin sequentially cleaves SARS-S at arginines 667 and 797, while cleavage by HAT might be limited to arginine 667 and might be sufficient to activate SARS-S for cell-cell but not virus-cell fusion. Alternatively, we have previously provided evidence for a requirement for conformational rearrangements (induced upon ACE2 binding) within SARS-S for proteolysis and infectious cell entry (40). These conformational changes may not proceed to completion in the relatively short time that the virus has available to make contact with HAT during virus-cell fusion, precluding proteolytic activation of SARS-S. In contrast, substantial amounts of S protein, receptor, and protease are stably available at the cell surface for prolonged time periods during cell-cell fusion, potentially allowing conformational changes and subsequent proteolysis to occur.
Regardless of the differences in mode and outcome of SARS-S activation by HAT and TMPRSS2, both proteases were able to activate SARS-S for cell-cell fusion in the context of authentic SARS-CoV. The resulting syncytia were readily detectable in the infected cultures and, as determined by immunostaining, were positive for both SARS-CoV and protease, as expected. Numeric analysis revealed that syncytium formation was about 15-fold more frequent in TMPRSS2- and HAT-transfected cells than control cells. Given that HAT was detected in ACE2-positive bronchial epithelial cells, in agreement with a report describing HAT expression in the bronchus (44), it is conceivable that HAT contributes to the previously documented syncytium formation in SARS-CoV-infected patients (26). Similarly, the documented findings that TMPRSS2 is expressed on SARS-CoV target cells and activates the virus in cell culture suggest that this protease might promote viral spread in humans (14, 30). In sum, HAT and TMPRSS2 activate SARS-S by different mechanisms and might support viral spread in infected humans.
ACKNOWLEDGMENTS
We thank T. F. Schulz, S. Treue, and T. Liepold for support.
Hayley Lavender was funded by the NIHR Biomedical Research Centre Programme. S.P. and C.D. were funded by BMBF (SARS-Verbund, 01KI1005C). Y.H. was supported by China 973 program (2010CB530100). G.S. was supported by grant R01AI074986 from the NIAID. C.D. was funded by DFG Africa (DR 772/3-1) and EMPERIE (project code 223498).
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Beaufort N., et al. 2007. The human airway trypsin-like protease modulates the urokinase receptor (uPAR, CD87) structure and functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L1263–L1272 [DOI] [PubMed] [Google Scholar]
- 2. Belouzard S., Chu V. C., Whittaker G. R. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 106:5871–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bergeron E., et al. 2005. Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem. Biophys. Res. Commun. 326:554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertram S., et al. 2010. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 84:10016–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bertram S., Glowacka I., Steffen I., Kuhl A., Pöhlmann S. 2010. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 20:298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosch B. J., Bartelink W., Rottier P. J. 2008. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82:8887–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Böttcher E., et al. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Böttcher-Friebertshauser E., Stein D. A., Klenk H. D., Garten W. 2011. Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J. Virol. 85:1554–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaipan C., et al. 2009. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 83:3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi S. Y., Bertram S., Glowacka I., Park Y. W., Pöhlmann S. 2009. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol. Med. 15:303–312 [DOI] [PubMed] [Google Scholar]
- 11. Connor R. I., Chen B. K., Choe S., Landau N. R. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944 [DOI] [PubMed] [Google Scholar]
- 12. Gao F., et al. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retroviruses 19:817–823 [DOI] [PubMed] [Google Scholar]
- 13. Glowacka I., et al. 2010. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 84:1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glowacka I., et al. 2011. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85:4122–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guan Y., et al. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278 [DOI] [PubMed] [Google Scholar]
- 16. Hamming I., et al. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He Y., et al. 2004. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 173:4050–4057 [DOI] [PubMed] [Google Scholar]
- 18. Hofmann H., et al. 2004. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 319:1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hofmann H., et al. 2004. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 78:6134–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann H., Pöhlmann S. 2004. Cellular entry of the SARS coronavirus. Trends Microbiol. 12:466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hofmann H., et al. 2006. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80:8639–8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imai Y., et al. 2005. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436:112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jahn O., Hesse D., Reinelt M., Kratzin H. D. 2006. Technical innovations for the automated identification of gel-separated proteins by MALDI-TOF mass spectrometry. Anal. Bioanal. Chem. 386:92–103 [DOI] [PubMed] [Google Scholar]
- 24. Jung H., et al. 2008. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene 27:2635–2647 [DOI] [PubMed] [Google Scholar]
- 25. Kuba K., et al. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11:875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuiken T., et al. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lau S. K., et al. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li W., et al. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li W., et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 30. Matsuyama S., et al. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84:12658–12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mossel E. C., et al. 2008. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology 372:127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peiris J. S., Guan Y., Yuen K. Y. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peiris J. S., Yuen K. Y., Osterhaus A. D., Stohr K. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431–2441 [DOI] [PubMed] [Google Scholar]
- 34. Pulford K., et al. 1997. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood 89:1394–1404 [PubMed] [Google Scholar]
- 35. Scott H. S., et al. 2001. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 27:59–63 [DOI] [PubMed] [Google Scholar]
- 36. Shah P. P., et al. 2010. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and Ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol. Pharmacol. 78:319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shirogane Y., et al. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 82:8942–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shulla A., et al. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simmons G., et al. 2011. Different host cell proteases activate the SARS-coronavirus spike-protein for cell-cell and virus-cell fusion. Virology 413:265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simmons G., et al. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 102:11876–11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simmons G., et al. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115–123 [DOI] [PubMed] [Google Scholar]
- 42. Simmons G., et al. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U. S. A. 101:4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stadler K., et al. 2003. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 1:209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takahashi M., et al. 2001. Localization of human airway trypsin-like protease in the airway: an immunohistochemical study. Histochem. Cell Biol. 115:181–187 [DOI] [PubMed] [Google Scholar]
- 45. Tedoldi S., et al. 2006. Jaw1/LRMP, a germinal centre-associated marker for the immunohistological study of B-cell lymphomas. J. Pathol. 209:454–463 [DOI] [PubMed] [Google Scholar]
- 46. To K. F., Lo A. W. 2004. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J. Pathol. 203:740–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. To K. F., et al. 2004. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 202:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang H., et al. 2008. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 18:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wysocka M., et al. 2010. Substrate specificity and inhibitory study of human airway trypsin-like protease. Bioorg. Med. Chem. 18:5504–5509 [DOI] [PubMed] [Google Scholar]