Abstract
Previously, we have shown that horses could be divided into susceptible and resistant groups based on an in vitro assay using dual-color flow cytometric analysis of CD3+ T cells infected with equine arteritis virus (EAV). Here, we demonstrate that the differences in in vitro susceptibility of equine CD3+ T lymphocytes to EAV infection have a genetic basis. To investigate the possible hereditary basis for this trait, we conducted a genome-wide association study (GWAS) to compare susceptible and resistant phenotypes. Testing of 267 DNA samples from four horse breeds that had a susceptible or a resistant CD3+ T lymphocyte phenotype using both Illumina Equine SNP50 BeadChip and Sequenom's MassARRAY system identified a common, genetically dominant haplotype associated with the susceptible phenotype in a region of equine chromosome 11 (ECA11), positions 49572804 to 49643932. The presence of a common haplotype indicates that the trait occurred in a common ancestor of all four breeds, suggesting that it may be segregated among other modern horse breeds. Biological pathway analysis revealed several cellular genes within this region of ECA11 encoding proteins associated with virus attachment and entry, cytoskeletal organization, and NF-κB pathways that may be associated with the trait responsible for the in vitro susceptibility/resistance of CD3+ T lymphocytes to EAV infection. The data presented in this study demonstrated a strong association of genetic markers with the trait, representing de facto proof that the trait is under genetic control. To our knowledge, this is the first GWAS of an equine infectious disease and the first GWAS of equine viral arteritis.
INTRODUCTION
Equine arteritis virus (EAV) is a small enveloped virus with a positive-sense, single-stranded RNA genome of 12.7 kb and belongs to the family Arteriviridae (genus Arterivirus, order Nidovirales) (16, 64). EAV is the causal agent of equine viral arteritis (EVA), a disease of equids (12, 13). The vast majority of EAV infections are inapparent or subclinical (5, 6, 68). However, some acutely infected horses may develop any combination of the following clinical signs: pyrexia, depression, anorexia, dependent edema (scrotum, ventral trunk, and limbs), conjunctivitis, lacrimation and swelling around the eyes (periorbital or supraorbital edema), respiratory distress, urticaria, and leukopenia (5, 6). During natural outbreaks of the disease, the virus can cause abortion in pregnant mares, with abortion rates varying from 10 to 71%, depending on the virus strain (5, 6). Following EAV infection, a variable proportion of stallions (30 to 70%) can become persistently infected and continuously shed the virus in their semen (55, 68). The mechanism of persistence of EAV in the male reproductive tract is not clear. However, studies have established that persistence of EAV in stallions is testosterone dependent (42, 52). Moreover, the prevalences of EAV infection differ markedly among different breeds of horses (68), strengthening the assumption of genetic influence on susceptibility to infection. The seroprevalences of EAV infection of horses vary between countries and among horses of different breeds and ages, with marked disparity between the prevalence of infection among Standardbred (STB) horses and that among Thoroughbred (TB) horses (49). EAV infection is considered endemic in STB horses but not in TB horses in the United States, with 77.5% to 84.3% of all STB horses but only up to 5.4% of TB horses being seropositive for the virus (5, 32, 49). Furthermore, the seroprevalence of EAV infection among Warmblood stallions is also very high in a number of European countries, as about 55 to 93% of Austrian Warmblood stallions are positive for antibodies to EAV (56). Although many of these differences are likely attributable to circulating virus strains and environmental or management conditions, inherent genetic differences may also play a significant role in determining clinical outcome and influence severity of disease following exposure to EAV.
In a recent study using dual-color flow cytometry, we demonstrated that the virulent Bucyrus strain of EAV (EAV VBS) can infect a small population of CD3+ T lymphocytes in vitro from some but not all horses (28). The data suggested that the CD3+ T lymphocyte subpopulation of individual horses varied in their susceptibilities to in vitro EAV VBS infection and they could be divided into susceptible and resistant groups (28). The susceptible/resistant CD3+ T lymphocyte phenotypes were not associated with age, prior exposure to EAV, or presence of antibodies to EAV but appeared to show an association with breed in preliminary studies. Therefore, we hypothesized that susceptibility and resistance of equine CD3+ T lymphocytes to EAV reflect genetic differences between horses in their response to infection with the virus. The primary objective of this study was to identify chromosomal regions and candidate genes associated with susceptibility/resistance of CD3+ T lymphocytes to EAV infection in horses by using a genome-wide association study (GWAS). Genetic factors contribute to host susceptibility and progression of disease, but the genes responsible for disease development are largely unknown. Until now, there was no evidence to indicate what role genetic factors play in determining the susceptibility to and outcome of EAV infection in horses. Availability of the equine genome sequence and development of genome-wide screening technologies offer unparalleled opportunities to identify variants associated with increased susceptibility/resistance to infectious disease agents such as EAV (20, 24, 31, 67, 70). In this study, we describe the identification of a distinct phenotypic trait that can be used as a marker to divide equine populations, regardless of breed, into susceptible and resistant groups based on an in vitro assay system. Using GWAS in combination with biological pathway analysis, we have identified for the first time several cellular genes that may be associated with the trait responsible for in vitro CD3+ T lymphocyte susceptibility/resistance to EAV infection.
MATERIALS AND METHODS
Horses.
Horses from four different breeds, Thoroughbred (TB), American Saddlebred (ASB), Standardbred (STB), and Quarter Horse (QH), a total of 310 horses, were randomly selected from farms in central Kentucky for this study. Blood samples were collected by jugular venipuncture into 10-ml Vacutainer tubes containing 15% EDTA (Monoject; Tyco Healthcare Group LP, Mansfield, MA) with short-term (<5 min) physical restraint of horses.
Virus and antibodies.
The virulent Bucyrus strain of EAV (EAV VBS; ATCC VR-796, American Type Culture Collection, Manassas, VA) was used for in vitro infection of equine peripheral blood mononuclear cells (PBMCs) as previously described (28). The monoclonal antibody (MAb) to the equine CD3 surface molecule, UC F6G, was kindly provided by Jeff Stott, University of California, Davis. The R-phycoerythrin (R-PE)-conjugated F(ab′)2 fragment of goat anti-mouse IgG1 (Southern Biotech, Birmingham, AL) was used as the secondary antibody. To detect EAV antigen in infected cells, Alexa Fluor 488-labeled MAb against nonstructural protein 1 (NSP1; MAb 12A4) was used (28, 71).
Phenotypic trait.
The susceptible or resistant phenotype of each animal was defined by dual-color flow cytometric analysis of in vitro EAV-infected CD3+ T lymphocytes as described previously (28). The horses were classified as susceptible or resistant to in vitro EAV infection based on their susceptible/resistant CD3+ T lymphocyte phenotype.
DNA extraction.
Genomic DNA (gDNA) was obtained from PBMCs of each animal by using the Puregene whole-blood extraction kit (Qiagen, Valencia, CA) by following the manufacturer's instructions. DNA quality and concentration were assessed using Nanodrop (Thermo Scientific, Wilmington, DE) at an absorbance ratio of optical density at 260 nm/280 nm (OD260/280).
Genotyping and quality control.
Samples were genotyped using Equine SNP50 BeadChip (Illumina, San Diego, CA) at the core facility for the Mayo Clinic in Rochester, MN. This array contains 59,355 single nucleotide polymorphisms (SNPs) derived from the EquCab2.0 SNP database of the horse genome (http://www.broadinstitute.org/mammals/horse), with an average probe spacing of 43.2 kb between adjacent variants. For the initial GWAS analysis (n = 37 horses), DNA samples from 16 TB horses known to be susceptible and 21 TB horses known to be resistant for the in vitro EAV infectivity trait were used for the Equine SNP50 BeadChip genotyping analysis (Illumina, San Diego, CA) (Fig. 1). Subsequently, additional SNPs from the region (Table 1) were selected using the Broad Institute Equine SNP database (http://www.broadinstitute.org/ftp/distribution/horse_snp_release/v2/) for further testing along with some of the original SNPs. The second SNP assay was conducted at Geneseek, Inc. (Lincoln, NE) using the Sequenom MassARRAY system (Sequenom Inc., San Diego, CA). Horses tested in this second assay included a second population of TB horses (n = 57) as well as horses of several different breeds (60 ASB, 60 STB, and 53 QH horses) plus the 37 TB horses tested in the original Illumina assay.
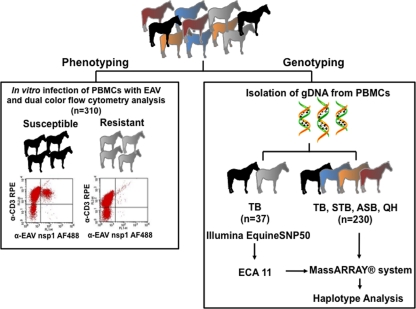
Fig. 1.
Schematic representation of the study design. A total of 310 horses of different breeds, American Saddlebred (ASB; n = 60), Standardbred (STB; n = 60), Thoroughbred (TB; n = 137), and Quarter Horse (QH n = 53), were used in the study. Horses were phenotyped first and grouped into the susceptible or resistant phenotype group based on their CD3+ T cell infectivity to EAV. To identify the region associated with EAV-susceptible/resistant phenotype, gDNA from 37 TB horses was isolated and analyzed using Illumina Equine SNP50. Results from the initial study were confirmed by genotyping 267 additional horses, including TB horses tested with Illumina Equine SNP50, with MassARRAY system technologies.
Table 1.
ECA11 SNPs selected for the MassARRAY system
| SNPc | Position on ECA11 (bp) |
|---|---|
| BIEC2-157847 | 49389493 |
| BIEC2-157867a | 49424839 |
| BIEC2-157892 | 49446386 |
| BIEC2-157899b | 49459247 |
| BIEC2-165951b | 49473877 |
| BIEC2-165967b | 49475165 |
| BIEC2-157944 | 49509504 |
| BIEC2-157945 | 49514091 |
| BIEC2-165988 | 49529433 |
| BIEC2-157947 | 49529680 |
| BIEC2-165991 | 49533085 |
| BIEC2-165992 | 49537432 |
| BIEC2-157953b | 49541824 |
| BIEC2-165997 | 49545584 |
| BIEC2-165998 | 49549767 |
| BIEC2-165999 | 49558533 |
| BIEC2-166001 | 49564266 |
| BIEC2-166003 | 49569444 |
| BIEC2-166005 | 49572804 |
| BIEC2-166006 | 49580531 |
| BIEC2-166009 | 49584513 |
| BIEC2-166011 | 49589952 |
| BIEC2-166013b | 49594275 |
| BIEC2-166014b | 49604594 |
| BIEC2-166015 | 49613954 |
| BIEC2-166016 | 49621050 |
| BIEC2-166018 | 49627471 |
| BIEC2-166020 | 49634340 |
| BIEC2-166021 | 49637772 |
| BIEC2-166027 | 49643932 |
| BIEC2-166028b | 49649380 |
| BIEC2-166029 | 49660962 |
| BIEC2-157989 | 49663560 |
| BIEC2-166033 | 49665617 |
| BIEC2-166034 | 49676359 |
| BIEC2-166036 | 49682023 |
| BIEC2-166040 | 49684089 |
| BIEC2-157998 | 49684299 |
| BIEC2-166042b | 49688617 |
| BIEC2-158000 | 49691181 |
| BIEC2-166044 | 49693957 |
| BIEC2-166047 | 49696679 |
| BIEC2-166049 | 49700501 |
| BIEC2-166050 | 49707607 |
| BIEC2-166052b | 49711761 |
| BIEC2-166056 | 49716340 |
| BIEC2-166060 | 49724419 |
| BIEC2-166065 | 49729958 |
| BIEC2-166066 | 49747467 |
| BIEC2-158024b | 49757978 |
| BIEC2-166449 | 50863263 |
| BIEC2-158587 | 51188148 |
| BIEC2-159231 | 52379156 |
Most highly associated SNP in the initial assay.
SNPs included on the assay but excluded due to failure to type or to low minor allele frequency.
The SNPs included on the Illumina Equine SNP50 chip are shown in bold font.
Data analyses.
Data from both assays were analyzed using the Golden Helix SNP & Variation Suite 7 (Bozeman, MT). Of the 59,355 SNPs, 53,747 had call rates above 95%. The rest were excluded from the analyses. Filtering for minor allele frequencies (removing SNPs with minor allele frequencies below 55%) and genotyping (excluding SNPs with genotypes involving less than 90% of the horses), 42,506 SNPs were evaluated. Data from both studies were evaluated using the association (chi-square) in the additive model and linkage disequilibrium (LD).
Gene ontology clustering and pathway analysis.
Unsupervised analysis was performed using the Ingenuity Pathways Analysis (IPA) tool (Ingenuity Systems Inc., Redwood City, CA) to investigate biological and molecular networks associated with genes identified within the selected region (3, 14). The IPA builds networks relevant to queried genes based on their functional annotation and their known molecular interactions supported by previously published peer-reviewed scientific articles which are then stored in Ingenuity Pathways Knowledge Base (IPKB). A list of interactions between genes identified in the GWAS (focus genes) and all other genes stored in the IPKB was obtained, with a maximum network size of 35 genes. Networks of direct or indirect interactions among genes are displayed graphically. In addition, IPA computes a score for each network, the −log (P value), which indicates the likelihood of the focus genes in a network from IPKB being found together due to random chance. To investigate the molecular and biological functions, genes identified within the region were categorized using the PANTHER classification system (www.pantherdb.org). In addition to the above-mentioned database analyses, functional information was also obtained from a literature search.
RESULTS
Phenotype analysis based on in vitro susceptibility of CD3+ T lymphocytes to EAV infection.
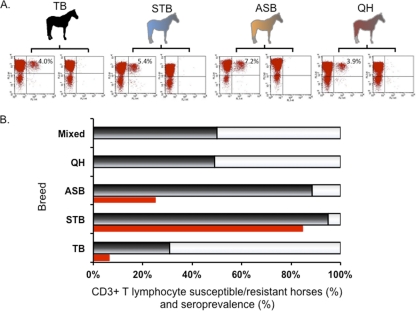
In an attempt to define the susceptible or resistant CD3+ T lymphocyte phenotype, blood was collected from each horse (n = 310) and PBMCs were isolated. PBMCs were infected with EAV VBS and subsequently subjected to dual-color flow cytometric analysis to identify horses with the susceptible or resistant CD3+ T lymphocyte phenotype to EAV infection (Fig. 1, left). Cells from horses with a susceptible CD3+ T lymphocyte phenotype were stained with antibodies to both the CD3+ T lymphocyte surface molecule and intracellular EAV antigen (NSP1). In contrast, cells from horses with a resistant CD3+ T lymphocyte phenotype were stained with antibody to the CD3+ T lymphocyte surface molecule but not to intracellular EAV antigen. Consistent with previously reported results, horses were divided into two distinct groups, those with CD3+ T cells susceptible and resistant to EAV infection. Of the 310 horses, 167 horses had the susceptible CD3+ T lymphocyte phenotype, and 143 horses had the resistant CD3+ T lymphocyte phenotype. Subsequently, data were analyzed by individual horse breed. In the susceptible phenotype group, there was no significant difference in the percentage of the EAV-infected CD3+ T lymphocyte subpopulation, regardless of breed. The percentages of the CD3+ T cell subpopulation susceptible to EAV were 4.0% ± 0.95% in TB horses, 5.4% ± 0.4% in STB horses, 7.2% ± 0.6% in ASB horses, and 3.9% ± 0.4% in QH horses (Fig. 2A). The results confirm that determination of the EAV-susceptible/resistant CD3+ T cell phenotype is consistent in horses, regardless of breed. Interestingly, there was a clear difference in prevalence of the susceptibility/resistance phenotype among breeds (Fig. 2B). Ninety-five percent of STB horses tested in this study were of the susceptible phenotype (57 out of 60). Similarly, 53 out of 60 ASB horses had the CD3+ T lymphocyte susceptible phenotype (∼90%). The lowest prevalence of the susceptible phenotype was in TB horses, 32 out of 137 (23%), and QH horses had both phenotypes evenly distributed, with approximately 50% prevalence of each phenotype (Fig. 2B).
Fig. 2.
Effect of breeds in prevalence of T cell susceptible/resistant phenotypes. (A) Representative dot plots from flow cytometry analysis for each breed are shown. The mean percentage of CD3+ T cells with intracellular EAV NSP1 antigen is indicated in the right upper quadrant for the susceptible phenotype. (B) The percentages of susceptible (black) and resistant (white) phenotypes for each breed are indicated in a bar graph. Seroprevalences of each breed (red) are indicated below the bar, representing phenotypic prevalence where available. TB, Thoroughbred (n = 137); STB, Standardbred (n = 60); ASB, American Saddlebred (n = 60); QH, Quarter Horse (n = 53).
GWAS.
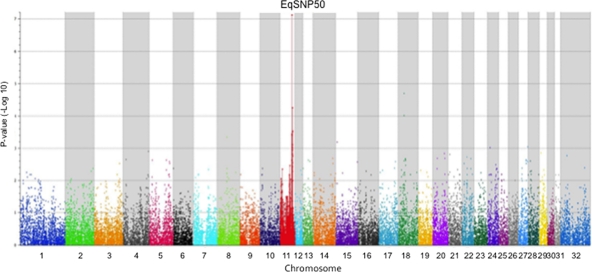
To investigate the possibility of genetic influence on susceptibility/resistance of CD3+ T lymphocyte to in vitro EAV infection, we applied the GWAS to identify the genetic element(s) associated with this phenotypic trait. In order to simplify the process of defining any genetic differences between positive and negative groups, we initially focused on TB horses since they have less diversity for genetic markers than most other breeds of horses. A group of 80 TB yearlings, serologically negative for EAV, were tested for the phenotype described above. Only 16 out of 80 TB yearlings had the susceptible CD3+ T lymphocyte phenotype. Thus, 37 horses were selected for the GWAS, including 16 with the susceptible phenotype and 21 with the resistant phenotype. DNA from these horses (Illumina TB horses) was genotyped for SNPs using the Equine SNP50 BeadChip (Illumina Inc., San Diego, CA) (Fig. 1, right). Using the option for association studies, the distributions of SNPs were compared between the susceptible horses and the resistant horses. The results from this study are represented graphically in a Manhattan plot (Fig. 3) showing the P values when comparing the distributions of markers for susceptible and resistant horses for each SNP. The values from a region of equine chromosome 11 (ECA11) showed a distinctive peak. The 10 highest associations that were found included 7 SNPs in one region on ECA11 (Table 2). The single highest association was found for SNP marker BIEC2-157867 from ECA11, which had statistical significances (P values) of 7.42 × 10−8 (uncorrected) and 0.003 (using the Bonferroni correction for the number of assays). None of the other markers achieved statistical significance when the correction to the probability value was factored in. These data clearly suggested that susceptibility/resistance of CD3+ T lymphocytes to in vitro EAV infection is highly associated with ECA11.
Fig. 3.
Manhattan plot showing the distribution of probability values (−log10 transformed) for the 42,506 SNPs investigated using the 37 Thoroughbred horses (16 were susceptible and 21 were resistant). Genomic positions are indicated by chromosomes with different colors.
Table 2.
Results from the GWAS study identifying the SNPs, chromosome location, and P values for the 10 highest associations found using the Illumina Equine SNP50 BeadChip based on 37 TB horses exhibiting the in vitro infection phenotype (n = 16 susceptible) and the in vitro resistance phenotype (n = 21 resistant)
| SNP no. | Chromosome no. | Position | Uncorrected P value |
|---|---|---|---|
| BIEC2_157867 | 11 | 49424839 | 7.42 × 10−8 |
| BIEC2_409206 | 18 | 24674961 | 2.00 × 10−5 |
| BIEC2_158932 | 11 | 51677777 | 5.42 × 10−5 |
| BIEC2_409219 | 18 | 24736336 | 9.69 × 10−5 |
| BIEC2_159231 | 11 | 52379156 | 2.90 × 10−4 |
| BIEC2_158587 | 11 | 51188148 | 3.06 × 10−4 |
| BIEC2_158588 | 11 | 51188190 | 3.06 × 10−4 |
| BIEC2_157160 | 11 | 48595739 | 3.63 × 10−4 |
| BIEC2_157720 | 11 | 49039853 | 3.98 × 10−4 |
| BIEC2_1046042 | 8 | 40043641 | 4.47 × 10−4 |
Study validation and haplotype analysis.
To confirm and extend the results from this initial study, additional SNPs were selected from the EquCab2.0 SNP database (Table 1) along with some of the original SNPs for the region on ECA11 and tested using the Sequenom MassARRAY system (Sequenom Inc., San Diego, CA). The Illumina-tested TB horses served as controls, being tested in both SNP assays, and had identical results for the 12 SNPs in common in the two assays. Table 3 provides the results from comparing the distributions of SNPs among susceptible and resistant horses for all horses (combined), TB horses alone, QH horses alone, ASB horses alone, and STB horses alone. The SNP showing the strongest association in the Illumina study, BIEC2-157867, had a significant and slightly higher P value of 5.84 × 10−5 for this new set of TB horses. The most significant value was found for BIEC2-166050 with the combined set of horses (P = 1.73 × 10−22). For the group of TB horses, the most significant P value was for BIEC2-166027 (P = 4.96 × 10−7). All breeds except STB horses showed statistically significant associations for the trait with SNPs in this region.
Table 3.
Probability values comparing cases to controls for the combined set of horses (n = 267) and for the individual groups of TB (n = 57), QH (n = 53), ASB (n = 60), and STB (n = 60) horses, showing the SNPs in ECA11 with the lowest 17 P values
| BIEC2 SNP no. | Position |
P value |
||||
|---|---|---|---|---|---|---|
| Combined | TB | QH | ASB | STB | ||
| 166050 | 49707607 | 1.73 × 10−22 | 6.11 × 10−4 | 3.00 × 10−3 | 6.18 × 10−11 | N/Ab |
| 166027 | 49643932 | 1.49 × 10−21 | 4.96 × 10−7 | 1.62 × 10−3 | 1.24 × 10−5 | 0.61 |
| 166011 | 49589952 | 1.17 × 10−19 | 1.46 × 10−4 | 1.07 × 10−3 | 1.74 × 10−6 | N/A |
| 165991 | 49533085 | 1.70 × 10−19 | 7.64 × 10−5 | 6.26 × 10−4 | 2.00 × 10−4 | 0.72 |
| 166065 | 49729958 | 3.44 × 10−19 | 7.02 × 10−5 | 9.40 × 10−4 | 9.48 × 10−5 | N/A |
| 166015 | 49613954 | 4.99 × 10−19 | 2.74 × 10−4 | 9.40 × 10−4 | 4.11 × 10−5 | N/A |
| 166021 | 49637772 | 5.97 × 10−19 | 1.87 × 10−4 | 9.40 × 10−4 | 4.11 × 10−5 | N/A |
| 166009 | 49584513 | 6.30 × 10−19 | 1.87 × 10−4 | 9.40 × 10−4 | 3.24 × 10−5 | N/A |
| 166047 | 49696679 | 3.23 × 10−18 | 7.02 × 10−5 | 9.40 × 10−4 | 9.48 × 10−5 | 0.61 |
| 166018 | 49627471 | 6.03 × 10−18 | 3.72 × 10−4 | 1.64 × 10−3 | 4.11 × 10−5 | N/A |
| 157944 | 49509504 | 5.75 × 10−17 | 5.78 × 10−4 | 3.02 × 10−3 | 2.00 × 10−4 | 0.56 |
| 166001 | 49564266 | 3.10 × 10−13 | 2.69 × 10−3 | 0.01 | 3.89 × 10−4 | 0.44 |
| 157867a | 49424839 | 1.12 × 10−9 | 5.84 × 10−5 | 0.50 | 1.66 × 10−5 | 0.48 |
| 157847 | 49389493 | 3.60 × 10−9 | 0.01 | 0.17 | 2.97 × 10−6 | 0.93 |
| 165999 | 49558533 | 5.45 × 10−9 | 0.10 | 0.18 | 0.10 | 0.34 |
| 165997 | 49545584 | 2.28 × 10−5 | 0.26 | 0.23 | 0.69 | 0.80 |
| 166056 | 49716340 | 6.52 × 10−5 | 3.93 × 10−3 | 0.07 | 0.02 | 0.46 |
The most highly associated SNP in the initial assay.
N/A represents no variation observed between the resistant and susceptible groups.
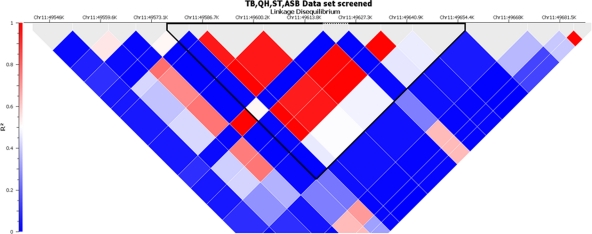
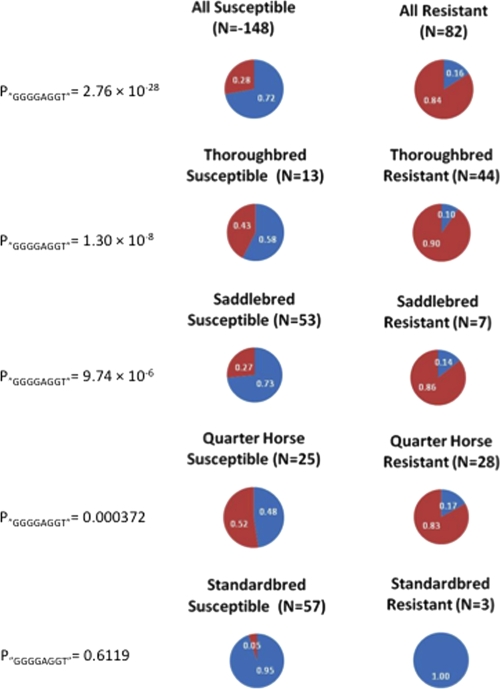
Next, linkage disequilibrium (LD) among the SNPs in this region was investigated and compared to the distribution of susceptible and resistant phenotypes in the different populations. A haplotype defined by 8 SNPs, BIEC2-166005, -166006, -166009, -166011, -166015, -166018, -166021, and -166027, spanning 71 kb from ECA11 positions 49572804 to 49643932, had the highest association across all breeds. The SNP BIEC2-166016 did not contribute to the definition of the haplotype and was therefore not included in the haplotype definition. The haplotype associated with the susceptibility phenotype was defined by the presence of nucleotides GGGGAGGT at those 8 SNP sites. Figure 4 shows a schematic representation of LD for the haplotype among all groups of horses tested. Figure 5 provides the frequencies for this haplotype among susceptible and resistant horses as well as the statistical significance for the differences in each group. In each breed, including STB horses, this haplotype was associated with susceptible horses. The haplotype was also present among the resistant horses but at a much lower frequency. Among the 13 TB horses with susceptible phenotype, all but one horse were heterozygous for the haplotype, possessing one copy of the haplotype; the remaining susceptible TB horses did not possess the haplotype (data not shown). Among the 44 resistant TB horses, 8 horses possessed one copy of the haplotype, while two horses were homozygotes (data not shown). Similar results were found for the distribution of the haplotype in other breeds.
Fig. 4.
Linkage disequilibrium (LD) plots for all breeds, showing the region used for defining the EAV susceptibility haplotype (ECA11 positions 49572804 to 49643932). The haplotype block is highlighted with a black line.
Fig. 5.
Frequency of the GGGGAGGT haplotype found for selected SNPs between ECA11 positions 49572804 to 49643932 among horses susceptible and resistant for the EAV in vitro infection phenotype. The blue area in the pie chart represents the proportion of the GGGGAGGT haplotype, and the red area represents the allelic haplotypes. The frequency represented by each section of the pie chart is shown on the pie chart in white. Data are represented for all horses together (All) and for the individual breeds (Thoroughbred, American Saddlebred, Quarter Horse, and Standardbred). The statistical significance for the frequency differences of the GGGGAGGT haplotype between susceptible and resistant horses is shown to the left of each set of pie charts (P“GGGGAGGT”).
Gene network analysis.
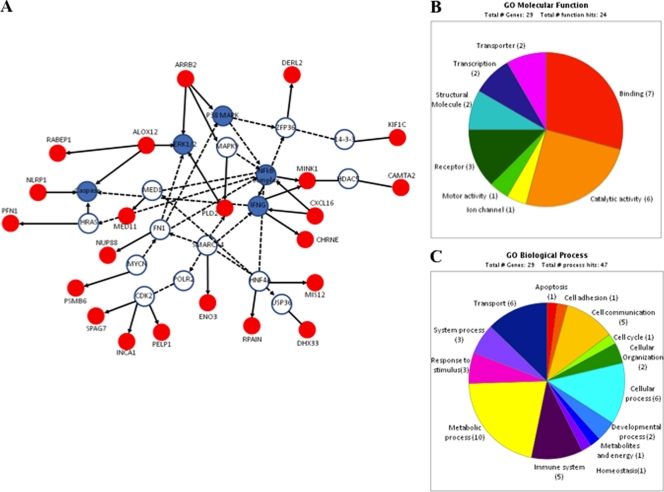
The EAV-associated haplotype is located in a block with strong LD, expanding ∼71 kb. This LD block includes five genes: the SPAG7, ENO3, PFN1, CHRNE, and MINK1 genes. For the purpose of analyzing possible functional relationship among genes using the IPA (Ingenuity Systems) and the PANTHER classification system, we have included those genes in high LD and genes located outside this particular LD block (500 kb upstream and downstream of the most significantly associated gene, the KIF1C gene). Unsupervised IPA analysis retrieved three top networks that were merged into a single interaction network (Fig. 6A). The network contained 22 genes showing significant correlation with networks implicated in cell morphology and cellular movement. Similarly, genes categorized by biological functions were mostly involved in cellular movement (P = 1.2 × 10−5), cell death (P = 1.45 × 10−3), cell morphology (P = 1.45 × 10−3), cellular assembly and organization (P = 1.45 × 10−3), and cellular function and maintenance (P = 1.45 × 10−3). We also classified the genes from the region by their molecular function and biological process categories using the PANTHER classification database. The enriched molecular function categories included binding, catalytic activities, and receptors, suggesting that these genes might mediate virus-cell interaction and facilitate viral entry (Fig. 6B). Furthermore, there was a predominance of genes regulating metabolic and cellular processes as well as cell communication and immune responses (Fig. 6C; Table 4).
Fig. 6.
(A) Three top networks retrieved using unsupervised IPA were merged into a single interaction network. Red nodes represent molecules encoded by genes found in the region of ECA11; white nodes are genes identified by IPA with direct or indirect interactions with genes of ECA11. The blue nodes represent molecules/complexes identified by IPA involved in canonical pathways. Classification of candidate molecules based on their molecular functions (B) and biological processes (C).
Table 4.
List of genes located in close vicinity with the highest associated haplotype block to the EAV-susceptible/resistant phenotypes
| Type | Gene symbola | NCBI gene ID | Description | Known molecular function | Virus (reference) |
|---|---|---|---|---|---|
| Receptor activity | CHRNE | 100146223 | Similar to nicotinic acetylcholine receptor epsilon | Acetylcholine receptor activity, ligand-gated ion channel activity | HIV-1 (8) |
| ARRB2 | 100072892 | Similar to beta-arrestin 2 | Receptor internalization, endocytosis | HIV-1 (4, 54) | |
| PELP1 | 100072897 | Similar to proline, glutamic acid, and leucine-rich protein 1 | Estrogen receptor coactivator, receptor activity | ||
| RABEP1 | 100072805 | RAB GTPase binding effector protein 1 | Endocytic pathway, fusion of early endosome, and regulation of recycling vesicle formation | ||
| CXCL16b | 100061442 | Similar to SR-PSOX | Scavenger receptor family, NKT cell, and macrophage (MØ) activation | HIV, severe acute respiratory syndrome coronavirus (SARS-CoV), Kaposi's sarcoma-associated herpesvirus (KSHV) (38, 76, 77) | |
| Intracellular vesicle trafficking | DERL2 | 100061027 | Derlin-2 (Der1-like protein 2) | Removal of misfolded protein from endoplasmic reticulum | Murine polyomavirus (41) |
| KIF1C | 100072836 | Kinesin family member 1C | Vesicle transport, redistribution of Golgi membrane, microtubule motor activity | ||
| Actin and cytoskeleton | SPAG7 | 100061215 | Similar to sperm-associated antigen 7 | Nucleic acid binding, cytoskeleton | Human parvovirus B19 (34, 35) |
| PFN1 | 100061295 | Similar to profilin 1 | Actin polymerization, cytoskeletal signaling pathway | Respiratory syncytial virus (RSV) (7) | |
| Innate and adaptive immunity | NLRP1 | NLR family, pyrin-containing domain 1 | Regulation of inflammatory response, induction of apoptosis | KSHV/human herpesvirus 8 (HHV-8), Myxoma virus, HIV-1 (30, 61) | |
| DHX33 | 100072797 | DEAH (Asp-Glu-Ala-His) box polypeptide 33 | Putative RNA helicase | HIV-1 (9) | |
| NUP88 | 100061117 | Nucleoporin, 88 kDa | Innate immunity, nuclear export | Hepatitis C and B viruses (36, 74) | |
| PSMB6 | 100072870 | Proteasome subunit beta type 6 precursor | Peptidase activity, involved in antigen presentation by major histocompatibility complex I (MHCI) | ||
| ALOX12 | 100072916 | Similar to 12-lipoxygenase | Oxidoreductase activity, MØ activation, B cell-mediated immunity | ||
| CXCL16c | 100061442 | Similar to SR-PSOX | Scavenger receptor family, NKT cell, and MØ activation | HIV, SARS-CoV, KSHV (38, 76, 77) | |
| Transcription factors | RPAIN | 100061089 | RNase protection assay (RPA)-interacting protein | Nuclear transporter of RPA | |
| ZFP3 | 100061153 | Zinc finger protein 3 homolog | DNA binding, transcription factor | ||
| CAMTA2 | 100072844 | Similar to calmodulin-binding transcription activator 2 | Transcriptional coactivator, DNA binding | ||
| MED11 | 100072890 | Mediator complex subunit 11; similar to HSPC296 | Component of pol II transcription machinery, transcription initiation complex stabilization | Measles virus (63) | |
| Enzyme activity | ENO3 | 100061187 | Hypothetical protein LOC100061187 | Lyase activity | |
| MINK1 | 100061370 | Misshapen-like kinase | Stress-activated protein kinase (SAPK)/Jun N-terminal protein kinase (JNK) signal pathway, kinase activity | ||
| PLD2 | 100072863 | Similar to phospholipase D2 | ERK1/2 signaling pathway, hydrolase activity | HIV-1 (59) | |
| Others | MIS12 | 100060987 | MIND kinetochore complex component | Chromosome segregation | |
| C17orf87 | Transmembrane protein C17orf87 | Transmembrane protein | |||
| L11_049426896 | 100072818 | Hypothetical protein | |||
| L11_049471914 | Hypothetical protein | ||||
| INCA1 | 100072840 | Similar to HSD45 | Regulation of cell proliferation, spermatogenesis | ||
| GLTPD2 | 100072874 | Glycolipid transfer protein domain-containing protein 2 | Lipid transport | ||
| VMO1 | 100072878 | Vitelline membrane outer layer protein 1 homolog precursor | |||
| TM4SF5 | 100072881 | Transmembrane 4 L6 family member 5 | |||
| ZMYND15 | 100061402 | Zinc finger MYND domain-containing protein 15 | |||
| L11_049822588 | Hypothetical protein |
Based on consensus equine protein-coding gene annotation (19).
Transmembrane form.
Secretory form.
DISCUSSION
The data presented in this study demonstrate a very strong association of genetic markers with the susceptible phenotype described for in vitro infection of CD3+ T lymphocytes with EAV VBS. Association of genetic markers with a trait is de facto proof that the trait is under genetic control. To our knowledge, this is the first GWAS of an equine infectious disease and the first GWAS of EVA. We have developed an easy and reliable in vitro assay based on dual-color flow cytometry to phenotype an equine population. The data presented in this study clearly demonstrated that there was a genetic difference within and among the horse populations based on the susceptibility of their CD3+ T lymphocytes to in vitro infection with EAV VBS. The in vitro assay described is currently the most precise method for determining the phenotype. However, it is expensive, time-consuming, and cumbersome to run on a routine or large-scale basis. The association of the ECA11 haplotype with the phenotype will allow us to develop a molecular test in large-scale studies to determine the impact of this phenotype on the consequences of infection.
The association of the haplotype with CD3+ T lymphocyte susceptibility in all four breeds of horses (TB, ASB, STB, and QH) is remarkable. This implies that the trait first appeared in a common ancestor of these breeds. Although these horses are products of four different breed registries, they are known to have common ancestors within the last 100 years (11). The use of the four breeds together also produced the strongest associations for the trait using both the haplotype and individual SNPs (Table 3). This was a product of the wide differences among the selected horse breeds for the in vitro susceptibility/resistance trait. The choice of TB horses for the initial stage of the study was fortuitous since the susceptibility/resistance trait is highly polymorphic among TB horses. Had the initial study been conducted among STB horses, the association would not have been uncovered, because almost all STB horses have the susceptibility phenotype. Indeed, STB horses appear almost fixed for the genes and the trait. Therefore, the results of this study suggest that preliminary studies of such traits might best be conducted using pools of different horse breeds when that trait is manifested across breed lines.
Interestingly, distinct phenotypic prevalence of each breed found in this study was similar to EAV seroprevalence that has been reported previously (Fig. 2B) (32, 49). Similar distributions of both seroprevalence and the susceptible CD3+ T lymphocyte trait strengthened our hypothesis that genetic factors might play a role in determining the CD3+ T lymphocyte susceptibility/resistance of horses to EAV infection. The haplotype associated with the trait was distributed among horses in a manner characteristic for that harboring a gene with a dominant mode of action. However, the association of the trait with the genetic markers was imperfect, since few susceptible horses were found without the implicated genetic markers implicated for susceptibility, and conversely, some resistant horses were found with susceptibility markers. However, this is not surprising since the 53 SNPs under study represented a very small proportion from the 3 million-bp-long region, and the chances of having selected the SNP defining the actual mutation defy expectations. Although additional work is needed to investigate candidate genes and resequencing of candidate regions, it is clear that genes within the region of ECA11 positions 49572804 to 49643932 play a major role in determining the CD3+ T lymphocyte susceptibility phenotype. In a parallel study, we have experimentally inoculated both groups of horses and assessed the severity of clinical disease, virus shedding, and production of proinflammatory/immunomodulatory cytokines in PBMCs using real-time quantitative reverse transcription (RT)-PCR (Y. Y. Go, R. F. Cook, J. Q. Fulgêncio, J. R. Campos, P. Henney, P. J. Timoney, and U. B. Balasuriya, submitted for publication). The data showed that there was a significant difference between the two groups of horses in terms of cytokine mRNA expression and increased clinical signs in horses possessing the in vitro resistant CD3+ T cell phenotype. Thus, there was a clear correlation between the in vitro susceptibility or resistance phenotype of CD3+ T lymphocytes and the different clinical/cytokine expression responses observed in horses following infection with EAV. Therefore, further studies on genes located in ECA11 will enhance our understanding of the pathogenesis of EAV infection and variation in susceptibility or resistance to establishment of the carrier state in stallions and will hopefully lead to the identification of a putative EAV cell receptor(s).
In this study, we have identified for the first time a significant number of candidate genes that might be associated with CD3+ T lymphocyte susceptibility to EAV infection, and these genes may also play a major role in pathogenesis and persistent infection in the stallion. We used IPA in an unsupervised manner, allowing identification of gene-gene relationships without a priori assumptions. This analysis linked 22 of 28 genes to other nodes in a highly significant network (Fig. 6A). These genes might be functionally interrelated and may play a direct or indirect role in susceptibility to EAV infection and pathogenesis of EVA. The proteins encoded by these genes are involved in virus attachment and entry pathway (putative cellular receptors for viral entry, molecules associated with cytoskeleton and endocytosis), various transcription factors, cell-signaling molecules, and molecules associated with inflammatory response and innate and adaptive immunity (Fig. 6A and B). Interestingly, some of these proteins are well-known cellular factors involved in other virus infections (Table 4) (4, 7–9, 30, 34–36, 38, 41, 54, 59, 61, 63, 74, 76, 77).
We found several proteins, such as CHRNE, CXCL16, RABEP1, ARRB2, and PELP1, that may be involved in EAV attachment and entry into the cell. Although the cellular receptor for EAV is not yet known, it is plausible that some of these molecules (e.g., CHRNE, CXCL16, and PELP1) may act as putative EAV receptors or serve as essential components in viral attachment and entry processes. In addition, proteins involved in the endocytic pathway, such as multifunctional proteins ARRB2 (17, 18, 60) and RABEP1 (69), may also play a critical role in the resistance or susceptibility of cells to EAV since it has been demonstrated that EAV is taken up via clathrin-dependent endocytosis and is delivered to acidic endosomal compartments during entry into the cell (57). Interestingly, the endocytic recycling receptor molecules, ASGPR1 and ASGPR2 (65), were located in the same chromosome approximately 800 kb downstream of RABEP1, making them attractive receptor candidates in line with clathrin-dependent endocytosis.
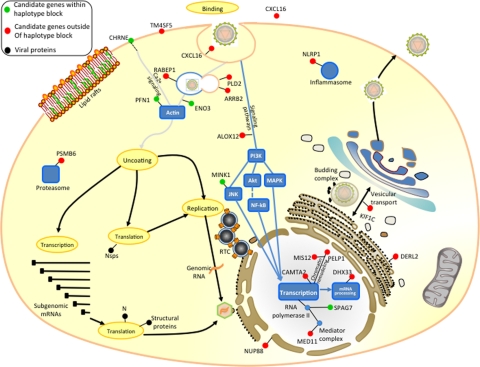
The molecules involved in intracellular trafficking, cell movements, cell-to-cell communication, as well as cell-substrate adhesion pattern and cell polarization are critical in relation to the viral life cycle (2, 10, 27, 33, 40, 47, 58). It has become apparent that numerous viruses interact with cytoskeletal components at various stages of their life cycles, disrupting or remodeling cytoskeletal networks to their own advantage (73). Some viruses can directly or indirectly hijack cytoskeletal cell component organization to their own advantage, often altering cell behavior and cell fate. In EAV-infected cells, cytopathic effect (CPE) is characterized by rounding, vacuolation, increased optical density, refraction, and detachment from the supporting surface, indicating viral modulations of the actin cytoskeleton (15, 37, 50, 51). In the region of ECA11 identified in this study, there are several genes that encode proteins determined to be involved in cellular movement function (e.g., ARRB2, PLD2, and PFN1 genes) as well as in actin cytoskeleton reorganization (Fig. 7) (7, 29).
Fig. 7.
Subcellular location of candidate genes and EAV life cycle. The function and subcellular location of candidate molecules were manually curated based on PANTHER, IPA, and published literature search. Subsequently, molecules were mapped along with stages of the EAV life cycle at the position most likely to interact with the virus life cycle. Green circles, molecules encoded by genes located in the haplotype block; red circles, putative molecules encoded by genes found within 500 kb upstream and downstream of the haplotype block; blue circles, molecules in the vicinity interacting with genes located in the region retrieved in the network analysis; black circles, viral proteins.
Finally, our network analysis also included some genes encoding proteins associated with apoptosis and modulating innate and adaptive immune responses to virus infections (Fig. 7) (25, 39, 62, 66). Proteins such as ARRB2 have been increasingly implicated in the modulation of nuclear factor kappa B (NF-κB) signaling and interacting with I kappa B alpha (IκBα) and extracellularly regulated kinases 1 and 2 (ERK1/2) (21–23, 26, 44–46, 53, 66, 72, 75). In addition, some genes in the region were associated with the host inflammatory response and innate immunity. For example, CXCL16, a member of the CXC chemokine family, is expressed as a soluble form and a transmembrane protein (38, 43). While the transmembrane form may have receptor activity, the soluble CXCL16 induces inflammatory responses (1, 38, 48). EAV may interact with some of these cellular proteins to evade the equine immune response and establish persistent infection in the cells of the reproductive tract of the stallion. However, further studies are needed to investigate potential mechanisms that involve the roles of proteins encoded by genes located in this region of ECA11 in the virus life cycle and host response to EAV infection, as well as to identify the mechanism of establishment of persistent infection in some but not all stallions infected with the virus. The findings from this study can help to develop working hypotheses to decipher novel mechanisms of viral immune evasion, establishment of persistent infection, and viral pathogenesis.
ACKNOWLEDGMENTS
We thank horse owners of various breeds who kindly provided samples for the study. We also thank Edward Squires at the Maxwell H. Gluck Equine Research Center for coordinating the collection of blood samples from various farms with TB, STB, ASB, and QH breeds.
This work was supported by the Morris Animal Foundation, Denver, CO, Maxwell H. Gluck Equine Research Center intramural research grant program, and the Frederick Van Lennep Chair endowment fund at the Maxwell H. Gluck Equine Research Center, University of Kentucky. Yun Young Go is supported by a Geoffrey C. Hughes Foundation graduate fellowship.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Abel S., et al. 2004. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J. Immunol. 172:6362–6372 [DOI] [PubMed] [Google Scholar]
- 2. Ait-Ali T., et al. 2009. Functional analysis of the porcine USP18 and its role during porcine arterivirus replication. Gene 439:35–42 [DOI] [PubMed] [Google Scholar]
- 3. Anselmo A., et al. 2011. Co-expression of host and viral microRNAs in porcine dendritic cells infected by the pseudorabies virus. PLoS One 6:e17374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aramori I., et al. 1997. Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signaling and internalization are dissociable from its role as an HIV-1 co-receptor. EMBO J. 16:4606–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balasuriya U. B., MacLachlan N. J. 2007. Equine viral arteritis, p. 153–164 In Sellon D. C., Long M. T.(ed.), Equine infectious diseases. Elsevier, St. Louis, MO [Google Scholar]
- 6. Balasuriya U. B., MacLachlan N. J. 2004. The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 102:107–129 [DOI] [PubMed] [Google Scholar]
- 7. Bitko V., Oldenburg A., Garmon N. E., Barik S. 2003. Profilin is required for viral morphogenesis, syncytium formation, and cell-specific stress fiber induction by respiratory syncytial virus. BMC Microbiol. 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bracci L., Lozzi L., Rustici M., Neri P. 1992. Binding of HIV-1 gp120 to the nicotinic receptor. FEBS Lett. 311:115–118 [DOI] [PubMed] [Google Scholar]
- 9. Brass A. L., et al. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926 [DOI] [PubMed] [Google Scholar]
- 10. Brinton M. A. 2001. Host factors involved in West Nile virus replication. Ann. N. Y. Acad. Sci. 951:207–219 [DOI] [PubMed] [Google Scholar]
- 11. Broome D. 2005. The development of the horse. In Peplow E.(ed.), Encyclopedia of the horse. Barnes & Noble, New York, NY [Google Scholar]
- 12. Bryans J. T., Crowe M. E., Doll E. R., McCollum W. H. 1957. Isolation of a filterable agent causing arteritis of horses and abortion by mares; its differentiation from the equine abortion (influenza) virus. Cornell Vet. 47:3–41 [PubMed] [Google Scholar]
- 13. Bryans J. T., Doll E. R., Knappenberger R. E. 1957. An outbreak of abortion caused by the equine arteritis virus. Cornell Vet. 47:69–75 [PubMed] [Google Scholar]
- 14. Burgner D., et al. 2009. A genome-wide association study identifies novel and functionally related susceptibility loci for Kawasaki disease. PLoS Genet. 5:e1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burki F. 1965. Properties of the equine arteritis virus. Pathol. Microbiol. 28:939–949 (In German.) [PubMed] [Google Scholar]
- 16. Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629–633 [PubMed] [Google Scholar]
- 17. Chen W., et al. 2003. Beta-arrestin 2 mediates endocytosis of type III TGF-beta receptor and down-regulation of its signaling. Science 301:1394–1397 [DOI] [PubMed] [Google Scholar]
- 18. Chen W., et al. 2003. Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science 301:1391–1394 [DOI] [PubMed] [Google Scholar]
- 19. Coleman S. J., et al. 2010. Structural annotation of equine protein-coding genes determined by mRNA sequencing. Anim. Genet. 41(Suppl. 2):121–130 [DOI] [PubMed] [Google Scholar]
- 20. Davila S., Hibberd M. L. 2009. Genome-wide association studies are coming for human infectious diseases. Genome Med. 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeFea K. A., et al. 2000. The proliferative and antiapoptotic effects of substance P are facilitated by formation of a beta-arrestin-dependent scaffolding complex. Proc. Natl. Acad. Sci. U. S. A. 97:11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DeFea K. A., et al. 2000. Beta-arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 148:1267–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan H., et al. 2007. Beta-arrestins 1 and 2 differentially regulate LPS-induced signaling and pro-inflammatory gene expression. Mol. Immunol. 44:3092–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan J. B., Chee M. S., Gunderson K. L. 2006. Highly parallel genomic assays. Nat. Rev. Genet. 7:632–644 [DOI] [PubMed] [Google Scholar]
- 25. Fong A. M., et al. 2002. Defective lymphocyte chemotaxis in beta-arrestin2- and GRK6-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 99:7478–7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao H., et al. 2004. Identification of beta-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-kappaB pathways. Mol. Cell 14:303–317 [DOI] [PubMed] [Google Scholar]
- 27. Georgel P., et al. 2010. Virus-host interactions in hepatitis C virus infection: implications for molecular pathogenesis and antiviral strategies. Trends Mol. Med. 16:277–286 [DOI] [PubMed] [Google Scholar]
- 28. Go Y. Y., et al. 2010. Complex interactions between the major and minor envelope proteins of equine arteritis virus determine its tropism for equine CD3+ T lymphocytes and CD14+ monocytes. J. Virol. 84:4898–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gomez-Cambronero J., Di Fulvio M., Knapek K. 2007. Understanding phospholipase D (PLD) using leukocytes: PLD involvement in cell adhesion and chemotaxis. J. Leukoc. Biol. 82:272–281 [DOI] [PubMed] [Google Scholar]
- 30. Gregory S. M., et al. 2011. Discovery of a viral NLR homolog that inhibits the inflammasome. Science 331:330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hill A. V. 2006. Aspects of genetic susceptibility to human infectious diseases. Annu. Rev. Genet. 40:469–486 [DOI] [PubMed] [Google Scholar]
- 32. Hullinger P. J., Gardner I. A., Hietala S. K., Ferraro G. L., MacLachlan N. J. 2001. Seroprevalence of antibodies against equine arteritis virus in horses residing in the United States and imported horses. J. Am. Vet. Med. Assoc. 219:946–949 [DOI] [PubMed] [Google Scholar]
- 33. Izumi T., et al. 2010. HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication. Proc. Natl. Acad. Sci. U. S. A. 107:20798–20803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kerr J. R. 2005. Pathogenesis of parvovirus B19 infection: host gene variability, and possible means and effects of virus persistence. J. Vet. Med. B Infect. Dis. Vet. Public Health 52:335–339 [DOI] [PubMed] [Google Scholar]
- 35. Kerr J. R., et al. 2005. Single-nucleotide polymorphisms associated with symptomatic infection and differential human gene expression in healthy seropositive persons each implicate the cytoskeleton, integrin signaling, and oncosuppression in the pathogenesis of human parvovirus B19 infection. J. Infect. Dis. 192:276–286 [DOI] [PubMed] [Google Scholar]
- 36. Knoess M., et al. 2006. Nucleoporin 88 expression in hepatitis B and C virus-related liver diseases. World J. Gastroenterol. 12:5870–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Konishi S., Akashi H., Sentsui H., Ogata M. 1975. Studies on equine viral arteritis. I. Characterization of the virus and trial survey on antibody with Vero cell cultures. Nihon Juigaku Zasshi 37:259–267 [DOI] [PubMed] [Google Scholar]
- 38. Landro L., et al. 2009. CXCL16 in HIV infection—a link between inflammation and viral replication. Eur. J. Clin. Invest. 39:1017–1024 [DOI] [PubMed] [Google Scholar]
- 39. Lefkowitz R. J., Shenoy S. K. 2005. Transduction of receptor signals by beta-arrestins. Science 308:512–517 [DOI] [PubMed] [Google Scholar]
- 40. Lever A. M., Jeang K. T. 2011. Insights into cellular factors that regulate HIV-1 replication in human cells. Biochemistry 50:920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lilley B. N., Gilbert J. M., Ploegh H. L., Benjamin T. L. 2006. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J. Virol. 80:8739–8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Little T. V., Holyoak G. R., McCollum W. H., Timoney P. J. 1992. Output of equine arteritis virus from persistently infected stallions is testosterone dependent, p. 225–229 In Plowright W., Rossdale P. D., Wade J. F. (ed.), Proceedings of the 6th International Conference on Equine Infectious Diseases. R&W Publications, Newmarket, England [Google Scholar]
- 43. Ludwig A., Weber C. 2007. Transmembrane chemokines: versatile ‘special agents’ in vascular inflammation. Thromb. Haemost. 97:694–703 [PubMed] [Google Scholar]
- 44. Luttrell L. M., Daaka Y., Lefkowitz R. J. 1999. Regulation of tyrosine kinase cascades by G-protein-coupled receptors. Curr. Opin. Cell Biol. 11:177–183 [DOI] [PubMed] [Google Scholar]
- 45. Luttrell L. M., et al. 1999. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science 283:655–661 [DOI] [PubMed] [Google Scholar]
- 46. Luttrell L. M., et al. 2001. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc. Natl. Acad. Sci. U. S. A. 98:2449–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martin-Serrano J., Neil S. J. 2011. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 9:519–531 [DOI] [PubMed] [Google Scholar]
- 48. Matloubian M., David A., Engel S., Ryan J. E., Cyster J. G. 2000. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat. Immunol. 1:298–304 [DOI] [PubMed] [Google Scholar]
- 49. McCollum W. H., Bryans J. T. 1973. Serological identification of infection by equine arteritis virus in horses of several countries, p. 256–263 In Bryans J. T., Gerber H.(ed.), Proceedings of the 3rd International Conference on Equine Infectious Diseases. S. Karger AG, Basel, Switzerland [Google Scholar]
- 50. McCollum W. H., Doll E. R., Wilson J. C., Johnson C. B. 1961. Propagation of equine arteritis virus in monolayer cultures of equine kidney. Am. J. Vet. Res. 22:731–735 [Google Scholar]
- 51. McCollum W. H., Doll E. R., Wilson J. C., Cheatham J. 1962. Isolation and propagation of equine arteritis virus in monolayer cell cultures of rabbit kidney. Cornell Vet. 52:452–458 [PubMed] [Google Scholar]
- 52. McCollum W. H., Little T. V., Timoney P. J., Swerczek T. W. 1994. Resistance of castrated male horses to attempted establishment of the carrier state with equine arteritis virus. J. Comp. Pathol. 111:383–388 [DOI] [PubMed] [Google Scholar]
- 53. McDonald P. H., et al. 2000. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science 290:1574–1577 [DOI] [PubMed] [Google Scholar]
- 54. Moorman J., et al. 2009. HIV-1 gp120 primes lymphocytes for opioid-induced, beta-arrestin 2-dependent apoptosis. Biochim. Biophys. Acta 1793:1366–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Neu S. M., Timoney P. J., McCollum W. H. 1987. Persistent infection of the reproductive tract in stallions experimentally infected with equine arteritis virus, p. 149–154 In Powell D. G.(ed.), Proceedings of the 5th International Conference on Equine Infectious Diseases. The University Press of Kentucky, Lexington, KY [Google Scholar]
- 56. Newton J. R., Wood J. L., Castillo-Olivares F. J., Mumford J. A. 1999. Serological surveillance of equine viral arteritis in the United Kingdom since the outbreak in 1993. Vet. Rec. 145:511–516 [DOI] [PubMed] [Google Scholar]
- 57. Nitschke M., et al. 2008. Equine arteritis virus is delivered to an acidic compartment of host cells via clathrin-dependent endocytosis. Virology 377:248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nuesch J. P., Lachmann S., Rommelaere J. 2005. Selective alterations of the host cell architecture upon infection with parvovirus minute virus of mice. Virology 331:159–174 [DOI] [PubMed] [Google Scholar]
- 59. Paruch S., et al. 2007. CCR5 signaling through phospholipase D involves p44/42 MAP-kinases and promotes HIV-1 LTR-directed gene expression. FASEB J. 21:4038–4046 [DOI] [PubMed] [Google Scholar]
- 60. Perry S. J., Lefkowitz R. J. 2002. Arresting developments in heptahelical receptor signaling and regulation. Trends Cell Biol. 12:130–138 [DOI] [PubMed] [Google Scholar]
- 61. Pontillo A., et al. 2010. A 3′UTR SNP in NLRP3 gene is associated with susceptibility to HIV-1 infection. J. Acquir. Immune Defic. Syndr. 54:236–240 [DOI] [PubMed] [Google Scholar]
- 62. Revankar C. M., Vines C. M., Cimino D. F., Prossnitz E. R. 2004. Arrestins block G protein-coupled receptor-mediated apoptosis. J. Biol. Chem. 279:24578–24584 [DOI] [PubMed] [Google Scholar]
- 63. Schneider-Schaulies S., ter Meulen V. 2002. Modulation of immune functions by measles virus. Springer Semin. Immunopathol. 24:127–148 [DOI] [PubMed] [Google Scholar]
- 64. Snijder E. J., Meulenberg J. J. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961–979 [DOI] [PubMed] [Google Scholar]
- 65. Spiess M. 1990. The asialoglycoprotein receptor: a model for endocytic transport receptors. Biochemistry 29:10009–10018 [DOI] [PubMed] [Google Scholar]
- 66. Sun Y., Cheng Z., Ma L., Pei G. 2002. Beta-arrestin2 is critically involved in CXCR4-mediated chemotaxis, and this is mediated by its enhancement of p38 MAPK activation. J. Biol. Chem. 277:49212–49219 [DOI] [PubMed] [Google Scholar]
- 67. Swinburne J. 2009. Inherited disease in the horse: mapping complex disease variants is on the horizon. Vet. J. 179:317–318 [DOI] [PubMed] [Google Scholar]
- 68. Timoney P. J., McCollum W. H. 1993. Equine viral arteritis. Vet. Clin. North Am. Equine Pract. 9:295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Valsdottir R., et al. 2001. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 508:201–209 [DOI] [PubMed] [Google Scholar]
- 70. Wade C. M., et al. 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wagner H. M., Balasuriya U. B., James MacLachlan N. 2003. The serologic response of horses to equine arteritis virus as determined by competitive enzyme-linked immunosorbent assays (c-ELISAs) to structural and non-structural viral proteins. Comp. Immunol. Microbiol. Infect. Dis. 26:251–260 [DOI] [PubMed] [Google Scholar]
- 72. Wang Y., et al. 2006. Association of beta-arrestin and TRAF6 negatively regulates Toll-like receptor-interleukin 1 receptor signaling. Nat. Immunol. 7:139–147 [DOI] [PubMed] [Google Scholar]
- 73. Ward B. M. 2011. The taking of the cytoskeleton one two three: how viruses utilize the cytoskeleton during egress. Virology 411:244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wiermer M., et al. 2010. Nucleoporin MOS7/Nup88 contributes to plant immunity and nuclear accumulation of defense regulators. Nucleus 1:332–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Witherow D. S., Garrison T. R., Miller W. E., Lefkowitz R. J. 2004. Beta-arrestin inhibits NF-kappaB activity by means of its interaction with the NF-kappaB inhibitor IkappaBalpha. Proc. Natl. Acad. Sci. U. S. A. 101:8603–8607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xu Y., Ganem D. 2007. Induction of chemokine production by latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells. J. Gen. Virol. 88:46–50 [DOI] [PubMed] [Google Scholar]
- 77. Zhang Y. P., Zhang R. W., Chang W. S., Wang Y. Y. 2010. Cxcl16 interact with SARS-CoV N protein in and out of cell. Virol. Sin. 25:369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]