Abstract
Human infections of H5N1 highly pathogenic avian influenza virus have continued to occur in China without corresponding outbreaks in poultry, and there is little conclusive evidence of the source of these infections. Seeking to identify the source of the human infections, we sequenced 31 H5N1 viruses isolated from humans in China (2005 to 2010). We found a number of viral genotypes, not all of which have similar known avian virus counterparts. Guided by patient questionnaire data, we also obtained environmental samples from live poultry markets and dwellings frequented by six individuals prior to disease onset (2008 and 2009). H5N1 viruses were isolated from 4 of the 6 live poultry markets sampled. In each case, the genetic sequences of the environmental and corresponding human isolates were highly similar, demonstrating a link between human infection and live poultry markets. Therefore, infection control measures in live poultry markets are likely to reduce human H5N1 infection in China.
INTRODUCTION
The first confirmed human case of H5N1 highly pathogenic avian influenza (HPAI) was detected in Hong Kong in 1997. Of the 18 patients confirmed to be infected, 6 succumbed (5, 19, 33). Massive slaughter of poultry, which were the primary source of the viruses, stopped the human outbreak (16). However, in November 2003, two further human cases were identified in Hong Kong, confirming that the viruses continued to circulate in the region (14). H5N1 viruses have now spread to avian species in many countries (9), all the while undergoing mutation and frequent reassortment. As of 12 February 2011, there had been 520 laboratory-documented human cases, 307 of which were fatal (27). Although no sustained human-to-human transmission has been confirmed, several family clusters have been reported.
More than 40 genotypes of H5N1 viruses were identified in China between 1996 and 2006 (6, 34). After poultry outbreaks in 2004, massive H5N1 vaccination campaigns were initiated to prevent both further outbreaks and subsequent spread to humans. From January to September 2006 alone, approximately 8.2 billion poultry were vaccinated in China (12). The vaccination campaign rapidly reduced the number of H5N1 outbreaks in poultry from 50 in 2004 to 32 in 2005 and then to only 12 in 2006. There have been very few subsequent outbreaks detected on domestic poultry farms, but sporadic human H5N1 infections continued to occur in China. The most recent case was identified in China on 4 June 2010 after the introduction of the 2009 H1N1 influenza pandemic. With the threat of a new pandemic strain caused by reassortment between H5N1 HPAIV and 2009 H1N1 pandemic viruses, it is important to identify the direct sources of the sporadic human H5N1 cases so that effective prevention and control can be developed.
Live poultry markets have been termed potential “hotbeds” of influenza A viruses, but their role in human H5N1 infection has not been firmly established. Live poultry markets have been speculated to be a major risk factor for human infection. Based on a retrospective epidemiology study, it was recently reported that most patients in urban areas in China during 2005 and 2006 visited live bird markets (32). However, there has been a lack of direct evidence showing an association between human isolates and those from live poultry markets. On the other hand, various subtypes, including H5N1 HPAIV, have been isolated from poultry or the environment in Asian markets (8, 9, 13, 18; H. L. Yen et al., presented at the World Congress on Options for the Control of Influenza IV, Excerpta Medica, Amsterdam, The Netherlands, 2001). Genotypic analyses suggest that the genotypes present in the live poultry markets periodically change as these viruses evolve (10; H. L. Yen et al., presented at the World Congress on Options for the Control of Influenza IV, Excerpta Medica, Amsterdam, The Netherlands, 2001).
In this study, we sequenced the genomes of 31 H5N1 HPAIVs identified by the Chinese National Influenza Center (CNIC) as having been isolated from humans between November 2005 and June 2010. We found that these isolates comprised multiple genotypes. To identify the direct sources of recent human infections, we collected environmental samples associated with six of the seven 2009 cases. Phylogenetic analyses and sequence comparisons strongly suggested that live poultry markets were the direct source of infection. These findings show that the live bird markets are likely to be a major factor in human H5N1 infections in China.
MATERIALS AND METHODS
Patients, sample collection, and virus isolation.
A confirmed case of H5N1 infection was defined as a case of pneumonia- or influenza-like illness with laboratory evidence of H5N1 infection based on multiple methods, including viral culture, reverse transcriptase PCR (RT-PCR), real-time PCR, and/or a serology test (30, 36). Briefly, each sample was tested by real-time PCR using matrix gene-specific primers, as well as H5 hemagglutinin (HA) and N1 neuraminidase (NA) gene-specific primers. If any real-time RT-PCR was positive, the samples were inoculated into three specific-pathogen-free (SPF) embryonated chicken eggs for viral isolation. The HA and NA subtypes of the viral isolates were determined through both antigenic characterization using hemagglutination inhibition (HI) and neuraminidase inhibition assays and genetic characterization through sequences.
For six patients who presented during the 2008–2009 season, environmental samples were collected from patient households and from live poultry markets visited within 2 weeks before the onset of illness. The environmental samples comprised swabs of feces or bird droplets on the floors of cages in the live poultry markets and water on the floors or in ditches of the live poultry markets or the households. The swabs were placed in Hank's or Eagle's medium with 0.5% bovine serum albumin (BSA) and antibiotics and shipped to Beijing at 4°C within 24 h. Samples not shipped immediately were kept frozen at or below −70°C until shipment. The environmental and clinical samples were processed using the same procedures. Viruses were isolated in SPF embryonated chicken eggs in enhanced biosafety level 3 (BSL3) facilities at the CNIC (28).
RNA extraction, RT-PCR, real-time PCR, and genomic sequencing.
RNA extraction was performed using the QIAamp Virus RNA Mini Kit (Qiagen, Shanghai, China). Real-time PCR assays were performed using a QuantiTect Probe RT-PCR Kit (Qiagen, Shanghai, China). The genomes of all H5N1 isolates were fully sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit (AB) and AB 3730 analyzer.
Identification of precursor candidates.
We applied the complete composition vector (CCV)-minimum spanning tree (MST) method (1, 23) (available at http://sysbio.cvm.msstate.edu/IPMiner). Briefly, we constructed a genetic distance matrix using CCV and then applied MST to group sequences separated by small evolutionary distances. This method efficiently analyzes a large number of sequences and identifies potential precursors as viruses showing the smallest genetic distance from an isolate. Phylogenetic tree construction, statistical validation by bootstrap analysis, and selection of potential precursors have been described elsewhere (22).
Phylogenetic analysis.
Phylogenetic trees were generated by using the maximum likelihood method in Garli version 0.95 (37), and the bootstrap values were generated by using neighbor-joining methods implemented in PAUP* version 4.0 beta (20). Clades were assigned on the basis of the WHO H5N1 nomenclature (29).
Antigenic cartography construction.
Influenza antigenic cartography is analogous to geographic cartography, and it projects influenza antigens (viruses) onto a two- or three-dimensional map, through which we can visualize and measure the antigenic distances between influenza antigens (viruses) as we visualize and measure geographic distances between cities in geographic cartography. The antigenic cartography was constructed using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap) (2, 3) based on the HI assays, which were performed using horse red blood cell (HRBC) and chicken reference antisera. As a comparison, the HI assays were also performed using HRBC and three ferret reference antisera, including those against A/Indonesia/1/05(H5N1) (clade 2.1), A/Turkey/1/05(H5N1) (clade 2.2), and A/Anhui/1/05(H5N1) (clade 2.3.4) (see Table S3 in the supplemental material).
RESULTS
Human H5N1 cases in China.
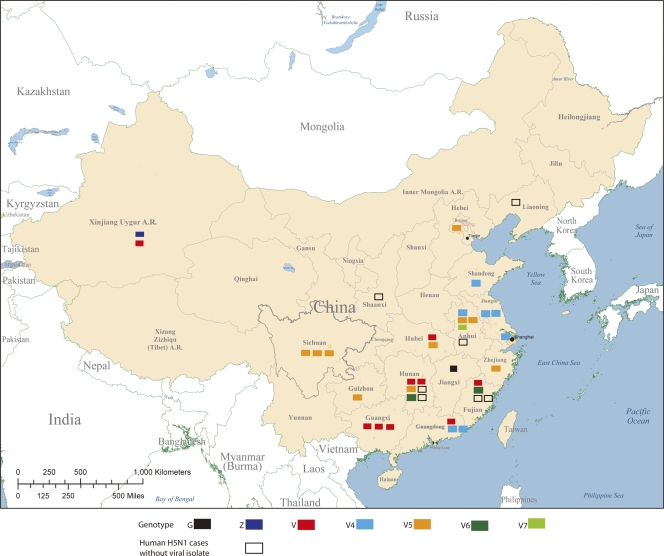
Between October 2005 and June 2010, there were 38 confirmed human cases of H5N1 infection in China, with a mortality rate of 65.7% (see Table S1 in the supplemental material). These cases were widely distributed across 17 districts or provinces (Fig. 1) and occurred in individuals aged 2 to 62 years (median, 32 years). No significant geographic clusters were found, although one possible case of human-to-human transmission was detected (24). As part of the national enhanced human avian influenza surveillance program, patients who were confirmed to have H5N1 infection were asked to complete a questionnaire and epidemiological investigation (http://www.moh.gov.cn/uploadfile/200671294330162.doc) that included their activities within 2 weeks before the onset of the clinical disease, such as the avian influenza cases/outbreaks nearby; the history of bird and other animal exposure; the history of bird and other animal-slaughtering and meat-processing work; the history of avian influenza exposure in a laboratory environment; the details of travels, including live bird market visits; and the patients' living environments, including backyard and wild bird and other animal activity status. The questionnaire data showed that only 3 cases were associated with known H5N1 outbreaks in poultry, whereas 20 cases were associated with contact with sick or dead birds and 11 cases were associated with visits to live poultry markets (Table 1 ). These findings were consistent with previous reports suggesting that direct contact with sick or dead birds and visits to live poultry markets are major risk factors for human H5N1 infection in China (15, 35). Interestingly, five of nine recent cases were associated with neither avian outbreaks nor dead birds (see Table S1 in the supplemental material).
Fig. 1.
Geographic distribution of 38 confirmed human cases of H5N1 avian influenza in China, November 2005 to May 2010; 24 of these cases were fatal. The virus genotypes are color coded (see Fig. S1 and Table S1 in the supplemental material).
Table 1.
Detection of H5N1 avian influenza viruses in environmental samples associated with six cases of human H5N1 infection in China during the 2008–2009 season
| Patient identifier | Date of clinical disease onset (mo/day/yr) | Patient city or province | Patient isolate | Environmental sample |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Sample site | Date of sample collection (mo/day/yr) | Total no. of samples | No. influenza A virus positive | No. H5N1 positive | H5N1 isolate (genotype) | ||||
| 30 | 12/4/2008 | Beijing | A/Beijing/1/2009(H5N1) | Sanheqingong, Hebei; market | 1/06/2009 | 6 | 6 | 6 | A/water/Hebei/1/2009(H5N1) (V5) |
| A/water/Hebei/2/2009(H5N1) (V5) | |||||||||
| A/water/Hebei/3/2009(H5N1) (V5) | |||||||||
| A/duck feces/Hebei/5/2009(H5N1) (V5) | |||||||||
| 32 | 1/07/2009 | Shanxi | None | Changsha, Hunan; market | 1/16/2009 | 8 | 8 | 4 | A/water/Hunan/3/2009(H5N1) (V4) |
| A/water/Hunan/7/2009(H5N1) (V) | |||||||||
| 33 | 1/08/2009 | Hunan | A/Hunan/1/2009(H5N1) | Guizhou, Guiyang; household | 1/17/2009 | 18 | 1 | 0 | 0 |
| 34 | 1/10/2009 | Xinjiang | A/Xinjiang/1/2009(H5N1) | Changjie, Xinjiang; market | 1/22/2009 | 17 | 6 | 2 | A/water/Xinjiang/3/2009(H5N1) (V) |
| A/environment/Xinjiang/6/2009(H5N1) (V) | |||||||||
| 35 | 1/15/2009 | Guizhou | A/Guizhou/1/2009(H5N1) | Guiyang, Guizhou; market | 01/24/2009 | 9 | 6 | 4 | A/environment/Guizhou/2/2009(H5N1) (V5) |
| A/environment/Guizhou/4/2009(H5N1) (V5) | |||||||||
| A/environment/Guizhou/7/2009(H5N1) (V5) | |||||||||
| A/environment/Guizhou/9/2009(H5N1) (V5) | |||||||||
| 37 | 1/23/2009 | Hunan | A/Hunan/2/09(H5N1) | Xupu, Hunan; household | 1/29/2009 | 11 | 4 | 1 | 0 |
Genetic and antigenic diversity of H5N1 isolates.
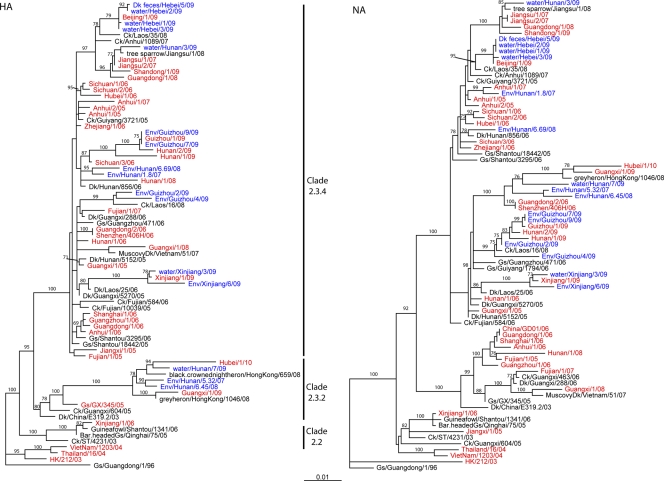
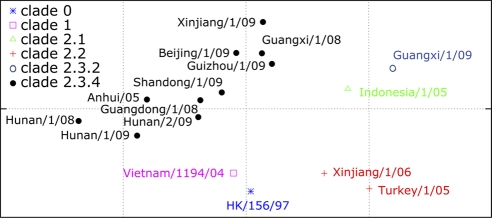
To identify the genetic composition of the human isolates and to identify their closest avian equivalents, we fully sequenced the 31 available isolates from the 38 confirmed H5N1 cases (see Table S2 in the supplemental material). Phylogenetic analysis of the HA genes showed that 28 isolates belonged to clade 2.3.4, 1 (Xinjiang, 2006) belonged to clade 2.2, and 2 (Guangxi, 2009, and Hubei, 2010) belonged to clade 2.3.2 (Fig. 2; see Table S1 in the supplemental material) (31). Antigenic characterization was consistent with these findings (see Table S3 in the supplemental material). As shown in antigenic cartography (Fig. 3), the viruses in clade 2.3.4 were antigenically closer to each other. There was quite a large antigenic divergence in these clade 2.3.4 viruses isolated from the 2008–2009 season. For instance, the antigenic distance between A/Hunan/1/2008(H5N1) (clade 2.3.4) and A/Guangxi/1/2008(H5N1) (clade 2.3.4) was more than 1 unit, with each unit corresponding to a 2-fold change in HI assay results. This is similar to the results of another study (7).
Fig. 2.
Phylogenetic analysis of the HA and NA genes of 31 H5N1 highly pathogenic avian influenza viruses isolated from patients in China. Viruses of human origin are shown in red, and those of environmental origin are in blue. Ck, chicken; Dk, duck; Env, environment; Gs, goose.
Fig. 3.
The antigenic cartography of highly pathogenic H5N1 avian influenza virus made by using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap) (2, 3). The cartography is based on the results from hemagglutination inhibition assays in horse red blood cells. One unit (grid) represents a 2-fold change in HI assay results. The cartography includes 16 influenza viruses (see Table S3 in the supplemental material), which includes four reference viruses, HK/156/97 (clade 0), Vietnam/1194/04 (clade 1), Indonesia/1/05 (clade 2.1), and Turkey/1/05 (clade 2.2) (26, 29).
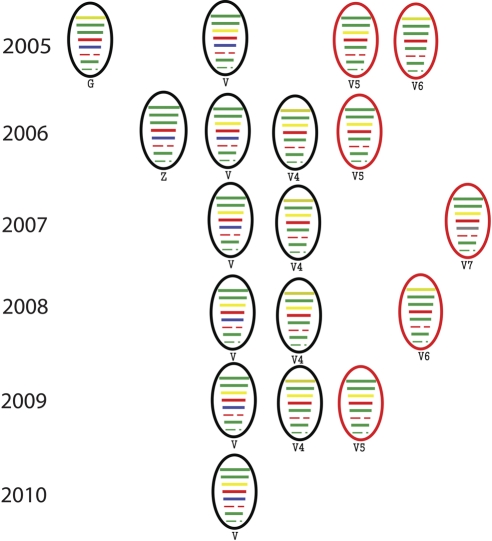
Seven different genotypes were detected among the 31 H5N1 isolates, including three not previously detected in humans (Fig. 2 and 4; see Fig. S1 in the supplemental material). Consistent with a previous report showing approximately 8.9% of H5N1 viruses circulating in China to be adamantane resistant (4), 3 of the 31 viruses (A/Fujian/1/07, A/Shandong/1/09, and A/Beijing/1/09) had the M2 S31N mutation, which is linked to adamantane resistance.
Fig. 4.
Temporal distribution of the genotypes of highly pathogenic H5N1 influenza viruses isolated in China. Genotypes V5, V6, and V7 are new genotypes identified in this study.
To identify potential avian virus gene precursors of each human isolate, we applied the CCV-MST method (1, 22, 23). One or more gene segments for most of the human H5N1 viruses isolated before 2007 had a corresponding gene segment in the public influenza database that was isolated from birds. These genes from avian-origin isolates and those from human H5N1 isolates had high sequence similarity (e.g., more than 95%) and were also located in the same cluster in the phylogenetic tree. However, we were unable to identify such genes for those isolated during or after 2007, probably because of the reduced number of avian H5N1 HPAIV isolates available for comparison (due to markedly reduced outbreaks in domestic poultry). In fact, no reported avian outbreaks were temporally or geographically related to any human case identified after 11 February 2006 (see Table S1 in the supplemental material). However, many patients who presented after February 2006 had visited live poultry markets shortly before the onset of clinical disease (15, 36).
Phylogenetic analyses demonstrated that all gene segments of the human H5N1 isolates were of avian origin. The lineages of their NA and internal gene segments were distinct from those of the 1997 H5N1 human clinical isolates in Hong Kong. Nine of the 31 human isolates belonged to genotype V, 7 to V4, 1 to G, and 1 to Z (6) (see Fig. S1 in the supplemental material). The genotypes V and V4 were widely distributed among 10 districts or provinces in China (Fig. 1) throughout the study period (Fig. 4). Genotype V was predominant in the bird population of China during 2005 and 2006 (6). Thus, not surprisingly, the H5N1 genotype distribution in the human population is related to the H5N1 genotype prevalence in poultry outbreaks and in live poultry markets.
We identified three new genotypes of H5N1 virus among the human isolates; we termed these genotypes V5, V6, and V7, according to the nomenclature of H5N1 HPAIVs (6) (see Fig. S1 in the supplemental material). Viruses of the V5 genotype have gene segment origins similar to those of the V genotype with the exception of the nucleoprotein (NP) gene, which is phylogenetically close to that of genotype G. The genomic configuration of genotype V6 is similar to that of V4, with the exception of the polymerase PA subunit gene (PA gene), which is phylogenetically close to that of genotype Z. Genotype V7 has an NP gene that appears to have originated in the H9N2 avian influenza viruses (see Fig. S1 in the supplemental material). Because two of the environmental H5N1 isolates (2007 and 2009) were also of the V7 genotype, it is possible that this genotype was circulating in the avian population during or before 2007.
H5N1 HPAIV isolated from the environment.
To investigate the possible sources of human infection, we collected environmental samples associated with six cases that occurred during the 2008–2009 season (Table 1), using the CNIC's influenza surveillance environmental-sampling protocol. All samples were collected within 5 weeks of the onset of illness. Multiple samples were collected for each patient.
Thirty-one of the 69 samples (44.9%) were positive for influenza A virus, and 17 of these 31 samples (54.8%) were positive for H5N1 virus by RT-PCR. This high detection rate demonstrated that large quantities of H5N1 viruses are shed into the environment. H5N1 viruses were isolated from 12 environmental samples associated with four of the human cases, and all of these samples were obtained from live poultry markets. The markets were located in a suburb of Beijing (4 isolates) and in the provinces of Guizhou (4 isolates), Hunan (2 isolates), and Xinjiang (2 isolates). An additional sample from the household of patient 37 was PCR positive for H5 hemagglutinin, but no viral isolate was recovered.
All 12 environmental isolates were completely sequenced (see Table S2 in the supplemental material), and the sequence analysis identified all 12 H5N1 viruses as HPAIVs. The genotype analyses showed that these viruses belonged to genotypes V, V3, and V5, confirming the genotypic diversity of the H5N1 viruses in the live poultry markets in China.
Genetic comparison of the human and environmental H5N1 HPAIV isolates.
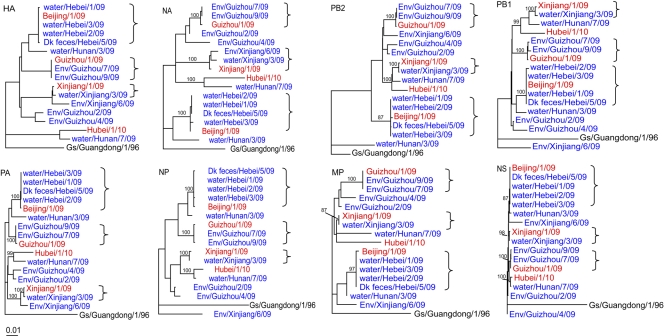
The viruses isolated from each market were phylogenetically similar to those isolated from the corresponding patient who had visited that market (Fig. 5), and these paired clinical and environmental isolates had a sequence identity greater than 99% (see Table S4 in the supplemental material). These results suggest that the live poultry markets were the source of the human infections. H5N1 HPAI viruses were isolated from a live poultry market visited by patient 32, although no virus was isolated from the patient.
Fig. 5.
Phylogenetic relationships of H5N1 influenza viruses isolated from patients in China and from their environments during the 2008-2009 influenza season. Although no direct environmental samples are associated with it, A/Hubei/1/2010(H5N1), the most recent human clinical isolate, is also included in these phylogenetic trees. The human clinical isolates are shown in red. The environmental samples isolated from this study are shown in blue. The similarities between the clinical isolates and the environmental isolates are shown in Table S4 in the supplemental material.
It is interesting that four phylogenetically distinct H5N1 viruses were isolated from a single market in Guizhou and that two of these isolates were similar to the corresponding human isolate; two distinct viruses were also isolated from the market in Hunan. These findings demonstrate that multiple H5N1 viruses are cocirculating within the same market environments.
DISCUSSION
Our findings suggest that the live poultry markets of China are an important source of human infection with H5N1 HPAI viruses. Enhanced infection control measures are warranted in these markets, not only to reduce human H5N1 infection, but also to minimize the likelihood of coinfection with H5N1 and 2009 H1N1 viruses. The sporadic cases of human H5N1 HPAI infection, the H5N1 outbreaks in birds, and the simultaneous circulation of the 2009 H1N1 pandemic virus in China raise concern that a deadly reassortant virus may emerge. The 2009 H1N1 pandemic virus has been introduced into multiple animal species, especially swine and poultry (11, 17, 21), and a subsequent reassortant of the virus has also been reported recently in swine in Hong Kong (21).
Prior to this report, another study (25) tried to establish a correlation between a human H5N1 isolate and those from the live bird market the patient visited. The authors amplified three genes (HA, NA, and M) from influenza virus RT-PCR-positive swabs collected from cages or bird feces in the wet poultry markets in Guangzhou. Although the authors claimed that these viruses are associated with each other, phylogenetic analysis indicates the environmental strain A/goose/Guangzhou/471/06(H5N1) they isolated from the wet poultry market was not directly related to the H5N1 isolate causing human disease. As shown in Fig. 2, the NA gene of A/goose/Guangzhou/471/06(H5N1) belongs to a different lineage from that of the human isolate A/Guangzhou/1/06(H5N1). Thus, this previous study was still not able to provide direct links between these environmental isolates and human isolates. Thus, until this study, there was still a lack of direct evidence supporting the linkage between the widespread presence of influenza A viruses in the wet poultry market and the viruses causing human infections.
Our genetic analysis of the 31 H5N1 HPAIVs isolated from the 38 human H5N1 cases identified in China during the past 5 years revealed diverse genotypes (G, V, V4, V5, V6, V7, and Z) that were consistent with those identified in poultry outbreaks or in live poultry markets. Live poultry markets, backyard poultry breeding, and domestic poultry farms are all potential sources of human H5N1 infection, and visiting live poultry markets has been identified as an important factor in such infections in China (15, 36).
Most of the six 2008–2009 patients whose environments were investigated were reported to have visited the live bird markets at least once. Our evolutionary analyses showed that the H5N1 isolates from these patients were highly genetically related (sequence identity > 99%) to the viruses isolated from their local live poultry markets. These results strongly suggest that the live poultry markets are the source of recent human H5N1 infections in China. These live poultry markets continue to act as reservoirs of H5N1 viruses despite fewer reported outbreaks on poultry farms. Therefore, control measures are needed, not only in the domestic bird population, but also in the live poultry markets to reduce human H5N1 infection.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the provincial CDCs in China for sample collection and field investigation, and especially to those in Beijing, Hunan, Guizhou, and Xinjiang for live poultry market investigations. We thank John Harkness for critical comments and Sharon Naron for editorial support.
L.-P.L., Z.C., and X.-F.W. were supported by RC1AI086830 from the U.S. National Institutes of Health (NIH) and NSF-EPS-0903787 to X.-F.W. Y.S. received financial support from the Ministry of Health of China (Major National Earmark Project for Infectious Diseases 2008ZX10004-013), the Ministry of Science and Technology (National Science and Technology Pillar Program 2006BAD06A15 and 2006BAD06A02), a U.S. CDC-China CDC collaboration program (U51/IP000334-02), and grant U19 AI051915-05s1 from the U.S. NIH. R.J.W. was partially supported by contract no. HHSN266200700005C with the U.S. National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Cai Z., et al. 2011. IPMiner: a progenitor gene identifier for influenza A virus. Influenza Other Respir. Viruses 5(Suppl. 1):413–415 [PMC free article] [PubMed] [Google Scholar]
- 2. Cai Z., Zhang T., Wan X.-F. 2011. Concepts and applications for influenza antigenic cartography. Influenza Other Respir. Viruses 5:204–207 [PMC free article] [PubMed] [Google Scholar]
- 3. Cai Z., Zhang T., Wan X. F. 2010. A computational framework for influenza antigenic cartography. PLoS Comput. Biol. 6:e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheung C. L., et al. 2006. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J. Infect. Dis. 193:1626–1629 [DOI] [PubMed] [Google Scholar]
- 5. Claas E. C., et al. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477 [DOI] [PubMed] [Google Scholar]
- 6. Duan L., et al. 2008. The development and genetic diversity of H5N1 influenza virus in China, 1996-2006. Virology 380:243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ducatez M. F., et al. 2011. Extent of antigenic cross-reactivity amongst highly pathogenic H5N1 influenza viruses. J. Clin. Microbiol. 49:3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jadhao S. J., et al. 2009. Genetic analysis of avian influenza A viruses isolated from domestic waterfowl in live-bird markets of Hanoi, Vietnam, preceding fatal H5N1 human infections in 2004. Arch. Virol. 154:1249–1261 [DOI] [PubMed] [Google Scholar]
- 9. Li K. S., et al. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209–213 [DOI] [PubMed] [Google Scholar]
- 10. Liu M., et al. 2003. The influenza virus gene pool in a poultry market in south central china. Virology 305:267–275 [DOI] [PubMed] [Google Scholar]
- 11. Mathieu C., et al. 2010. Pandemic (H1N1) 2009 in breeding turkeys, Valparaiso, Chile. Emerg. Infect. Dis. 16:709–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ministry of Agriculture, People's Republic of China 23 October 2006, posting date. Poultry avian influenza vaccination in China. [Google Scholar]
- 13. Nguyen D. C., et al. 2005. Isolation and characterization of avian influenza viruses, including highly pathogenic H5N1, from poultry in live bird markets in Hanoi, Vietnam, in 2001. J. Virol. 79:4201–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peiris J. S., et al. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shu Y., Yu H., Li D. 2006. Lethal avian influenza A (H5N1) infection in a pregnant woman in Anhui Province, China. N. Engl. J. Med. 354:1421–1422 [DOI] [PubMed] [Google Scholar]
- 16. Sims L. D., et al. 2003. Avian influenza in Hong Kong 1997-2002. Avian Dis. 47:832–838 [DOI] [PubMed] [Google Scholar]
- 17. Song M. S., et al. 2010. Evidence of human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. J. Clin. Microbiol. 48:3204–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sturm-Ramirez K. M., et al. 2005. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J. Virol. 79:11269–11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Subbarao K., et al. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396 [DOI] [PubMed] [Google Scholar]
- 20.Swofford D. L. PAUP*: Phylogenic Analysis Using Parsimony. Sinauer, Sunderland, MA: 1998. [Google Scholar]
- 21. Vijaykrishna D., et al. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wan X. F., et al. 2008. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One 3:e3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wan X. F., Ozden M., Lin G. 2008. Ubiquitous reassortments in influenza A viruses. J. Bioinform. Comput. Biol. 6:981–999 [DOI] [PubMed] [Google Scholar]
- 24. Wang H., et al. 2008. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet 371:1427–1434 [DOI] [PubMed] [Google Scholar]
- 25. Wang M., et al. 2006. Food markets with live birds as source of avian influenza. Emerg. Infect. Dis. 12:1773–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. WHO 2009. Continuing progress towards a unified nomenclature for the highly pathogenic H5N1 avian influenza viruses: divergence of clade 2.2 viruses. Influenza Other Respir. Viruses 3:59–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. WHO 18 October 2010, posting date. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. [Google Scholar]
- 28. WHO 2002. Manual on animal influenza diagnosis and surveillance., vol. 2008 WHO, Geneva, Switzerland [Google Scholar]
- 29. WHO 2008. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. WHO 29 August 2006. WHO case definitions for human infections with influenza A (H5N1) virus. WHO, Geneva, Switzerland [Google Scholar]
- 31. Xu C., et al. 2009. Human avian influenza A (H5N1) virus infection in China. Sci. China C Life Sci. 52:407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu H., et al. 2007. Human influenza A (H5N1) cases, urban areas of People's Republic of China, 2005–2006. Emerg. Infect. Dis. 13:1061–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuen K. Y., et al. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471 [DOI] [PubMed] [Google Scholar]
- 34. Zhao Z. M., Shortridge K. F., Garcia M., Guan Y., Wan X. F. 2008. Genotypic diversity of H5N1 highly pathogenic avian influenza viruses. J. Gen. Virol. 89:2182–2193 [DOI] [PubMed] [Google Scholar]
- 35. Zhou L., et al. 2009. Risk factors for human illness with avian influenza A (H5N1) virus infection in China. J. Infect. Dis. 199:1726–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zou S., et al. 2007. Human influenza A virus (H5N1) detection by a novel multiplex PCR typing method. J. Clin. Microbiol. 45:1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. University of Texas, Austin, TX [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.