Abstract
Viral proteins and nucleic acids stimulate Toll-like receptors (TLRs) to elicit production of cytokines, chemokines, and interferons. Due to their immunostimulatory activity, several TLR agonists are being developed as vaccine adjuvants and cancer immunotherapeutics. However, TLR signaling is modified by disease state, which could enhance or impair therapeutic efficacy. For example, in the skin of psoriasis patients, the human cationic antimicrobial peptide LL37 is highly expressed and binds to host DNA. Association with LL37 enhances DNA uptake into intracellular compartments where it stimulates TLR9-dependent overproduction of interferons. Poly(I:C), an analogue of viral double-stranded (ds) RNA is recognized by TLR3, and is currently in preclinical trials as an inducer of type I interferon. If LL37 similarly enhanced interferon production, use of poly(I:C) might be contraindicated in certain conditions where LL37 is elevated. Here we show that TLR3 signaling was not enhanced, but was dramatically inhibited, by LL37 or mouse cathelicidin-related antimicrobial peptide (mCRAMP) in macrophages, microglial cells, and dendritic cells. Inhibition correlated with formation of a strong complex between antimicrobial peptides and poly(I:C), which partially inhibited poly(I:C) binding to TLR3. Therefore, after injury or during existing acute or chronic inflammation, when LL37 levels are elevated, the therapeutic activity of poly(I:C) will be compromised. Our findings highlight the importance of using caution when therapeutically delivering nucleic acids as immunomodulators.
Keywords: poly(I:C), antimicrobial peptide, Toll-like receptor 3, innate immunity, proinflammatory cytokines
Introduction
The innate immune system recognizes molecular structures from infectious agents through pattern recognition receptors including toll-like receptors (TLRs). Stimulation of the immune system through TLRs activates innate immune cells and facilitates inflammatory responses, which are necessary for host defense against invading pathogens (1–3). Due to their ability to activate the immune system, TLR agonists, especially those based on nucleic acids, are in current preclinical and clinical trials as immunomodulators.
Most TLR agonists are unique to the microbe and not present in the host; however, some detect shared nucleic acid structures such as DNA (TLR9), single stranded RNA (TLR7, TLR8) and double stranded RNA (TLR3). To limit response to host nucleic acids in the extracellular milieu, nucleic acid sensing TLRs are localized within the cell, traffic to endosomes, and are proteolytically processed (4–11). This permits detection of pathogen DNA released in phagosomes/endosomes. Host DNA is typically degraded extracellularly, and does not enter the endocytic compartment. However, in certain disease states host-DNA is chaperoned to TLR containing endosomes where it elicits TLR-dependent inflammatory responses. High Mobility Group Box proteins (12), the human antimicrobial peptide LL37 (13), and anti-DNA antibodies (14) bind directly to host-DNA and enhance uptake into the endosomal compartment. Enhanced uptake results in dramatically increased production of inflammatory cytokines and interferons (13, 15–19). Given the potential of these proteins to modulate responses to TLR ligands, and the development of TLR ligands as therapeutics, it is important to understand the impact of preexisting, or coexisting disease states on the therapeutic activity of TLR agonists.
Macrophages and dendritic cells are resident in tissues such as skin and are two of the cell types to first respond to topical therapeutic application of TLR ligands such as polyinosinic-polycytidylic acid (poly(I:C)). The antimicrobial peptide LL37 is locally elevated in diseases such as psoriasis (micromolar concentrations), but is not a significant product of either of these cell types (13). Therefore, it is important to test what effect elevated LL37 will have on macrophage and DC response to poly(I:C). If LL37 enhances response to poly(I:C), as it does for CpG DNA, use of poly(I:C) as an immunotherapeutic would be potentially harmful for individuals with diseases such as psoriasis.
In the present study, we show that TLR3 dependent responses were blocked by LL37 or the mouse equivalent peptide, mCRAMP. LL37 inhibited poly(I:C)-induced activation of macrophages, dendritic cells and microglial cells. TLR3-dependent, poly(I:C)-induced, nuclear factor-κB (NF-κB) and interferon regulatory factor 3 (IRF3) signaling pathways were inhibited by LL37. The inhibitory effect was due to the formation of a stable complex between antimicrobial peptide and poly(I:C), which partially blocked poly(I:C) binding to TLR3. Therefore, during infections, or in certain disease states, where LL37 is elevated, the therapeutic activity of poly(I:C) will be compromised. The interaction between immunomodulators and other immune components must be considered prior to widespread use of TLR ligands as therapeutic immunomodulators.
Materials and methods
Reagents
The following reagents and antibodies were used: poly(I:C) (EMD biosciences, San Diego, CA), LL37 (a kind gift from Dr. Dawn Bowdish, McMaster University), Scrambled peptide and mCRAMP (AnaSpec, Fremont, CA), CpG oligonucleotides, ODN-10104 (5’-TCGTCGTTTCGTCGTTTTGTCGTT-3’) (Eurofins MWG Operon, Huntsville, AL), Pam3CSK4 (Invivogen, San Diego, CA), DOTAP (Roche, Indianapolis, IN), Antibodies; phospho-IκBα (Ser32), phosphor-JNK (Thr183/Tyr185), phosphor-p38 (Thr180/Tyr182), phospho-IRF3 (Ser396) (Cell Signaling), α-Tubulin (eBioscience) and secondary antibodies (Southern Biotech).
Cell lines
RAW264.7 macrophages (American Type Culture Collection, Rockville, MD) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (v/v) heat-inactivated FCS, 2 mM L-glutamine, 10 mM HEPES and 1 mM sodium pyruvate (complete DMEM) with the addition of 100 U/ml penicillin, 100 mg/ml streptomycin. Wild type and TLR3 deficient microglial cell lines (BEI resources) were cultured in complete DMEM and 10 µg/ml ciprofloxacin. All cell lines were cultured at 37°C with 5% CO2 and routinely tested negative for mycoplasma by PCR.
Electrophoresis of RNA-antimicrobial peptide complexes
Cationic peptides were incubated with poly(I:C), or 540 bp dsRNA, for 1 hour at room temperature in 10 mM HEPES buffer at pH 7.2. The mixtures were then resolved by electrophoresis on an 0.8% agarose gel (containing ethidium bromide) in TAE buffer. Gels were imaged using UV light and images were analyzed in Photoshop.
RNA isolation and RT-PCR analysis
Total RNA was isolated using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA quantity and quality were confirmed using a NanoDrop® ND-1000 Spectrophotometer. cDNA was synthesized from 1 µg total RNA using SuperScript® III First-Strand Synthesis (Invitrogen, Carlsbad, CA). Real-time PCR was performed using the Power SYBR Green PCR master mix (Applied Biosystems, Foster city, CA). The GAPDH gene was used for RNA normalization. All samples were run in triplicate. Analysis was performed on an Applied Biosystems 7500 (Applied Biosystems, Carlsbad, CA). The 2−ΔΔCT method (20) was used to calculate relative changes in the gene expression determined from raw fluorescence data.
The following primers were used: IL-1β forward: 5’-CGCAGCAGCACATCAACAAGAGC-3’, reverse: 5’-TGTCCTCATCCTGGAAGGTCCACG-3’; IL-6 forward: 5’-CACAAGTCCGGAGAGGAGAC-3’, reverse: 5’-CAGAATTGCCATTGCACAAC-3’; GAPDH forward: 5’-ACTCCACTCACGGCAAATTCAACGG-3’, reverse: 5’-AGGGGCGGAGATGATGACCC-3’
Cytokine detection assay
TNF-α in the supernatants was measured by ELISA according to the manufacturer’s recommendations (Biolegend, San Diego, CA).
Nitrite assay
Accumulation of NO2− was measured after 18 hours of stimulation using the Griess assay system as described previously (21).
TLR3 binding assay
TLR3 binding assay was performed as previously described (22). Briefly, microplate wells were coated with goat anti-mouse IgG followed sequentially by mouse anti-GFP mAb and cell lysate (lysis buffer; 1% Triton X-100, 10 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, and protease inhibitors (Roche Applied Science)) containing TLR3-YFP. After blocking and washing, a mixture of peptides and biotinylated-dsRNA was added to the microplate wells and incubated at room temperature (RT) for 2 hours. Plates were then washed and incubated with streptavidin-HRP (Thermo Scientific) for 1 hour at RT. Using HRP substrate reagent (R&D Systems) and a FLUOstar OPTIMA plate reader (BMG Labtech) absorbance was measured at 450nm.
Circular dichroism spectroscopy
Circular dichroism was performed with 10 µg of poly(I:C) and 50 µM of LL37 either alone or in combinations. Spectra were recorded with a model 400 AVIV circular dichroism spectrometer (Lakewood, NJ) in a 1-cm-path-length cuvette at room temperature (RT). Structural analysis was performed using the Delta epsilon (DE) calculation method using K2D circular dichroism spectra Deconvolution software.
Western blotting
Cells were stimulated as indicated, washed with ice cold HBSS, and lysed with 300 µl of 1X SDS-PAGE reduced sample buffer (62.5 mM Tris pH 6.8, 12.5% glycerol, 1% SDS, 0.005% bromophenol blue, 1.7% β-mercaptoethanol). Lysates were incubated at 95°C for 5 min prior to resolving by 10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes and immunoblotted with the indicated antibodies. Membranes were incubated with Supersignal West Pico Chemiluminescence western blotting detection system (Thermo Scientific, Rockford, IL) and exposed to X-ray film. Films were scanned and images were assembled in Photoshop.
Preparation of DOTAP-poly(I:C) complexes
5 µg of poly(I:C) was prepared in HEPES buffered saline at pH 7.4 in a sterile reaction tube (final volume 50 µL). In a separate reaction tube 30 µL of DOTAP was mixed with HEPES buffered saline at pH 7.4 to a final volume of 100 µL. Then poly(I:C) containing solution was transferred to the tube containing DOTAP in HEPES buffered saline. The two solutions were mixed by gentle pipetting several times and incubated at room temperature for 15 min prior to adding to cells.
Generation of bone marrow derived macrophages and dendritic cells
Femurs and tibias were harvested from 8–10 week old C57BL/6 mice (Jackson Laboratory). For macrophages, bone marrow was flushed from the bones with cold DMEM supplemented with 20% L-929 cell-conditioned medium, 10% (v/v) heat-inactivated FCS, 2 mM L-glutamine, 10 mM HEPES and 1 mM sodium pyruvate and 100 U/ml penicillin, 100 mg/ml streptomycin. Bone marrow cells were cultured in 10 cm petri dishes (10 ml volume) at 37°C, 5% CO2 for 7 days. At day 3 and 6, fresh medium was added to the cultured cells. For Dendritic cells, bone marrow was flushed from the bones with cold RPMI containing 10 ng/ml mouse granulocyte-macrophage colony stimulating factor (PeproTech, Rocky Hill, NJ), 10% fetal bovine serum, 50 µM 2-mercaptoethanol, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 100 U/ml penicillin and 100 mg/ml streptomycin). Bone marrow cells were differentiated for 9 days in 10 cm petri dish in 20 ml complete RPMI 1640.
Results
Cationic antimicrobial peptides inhibit poly(I:C) induced signaling
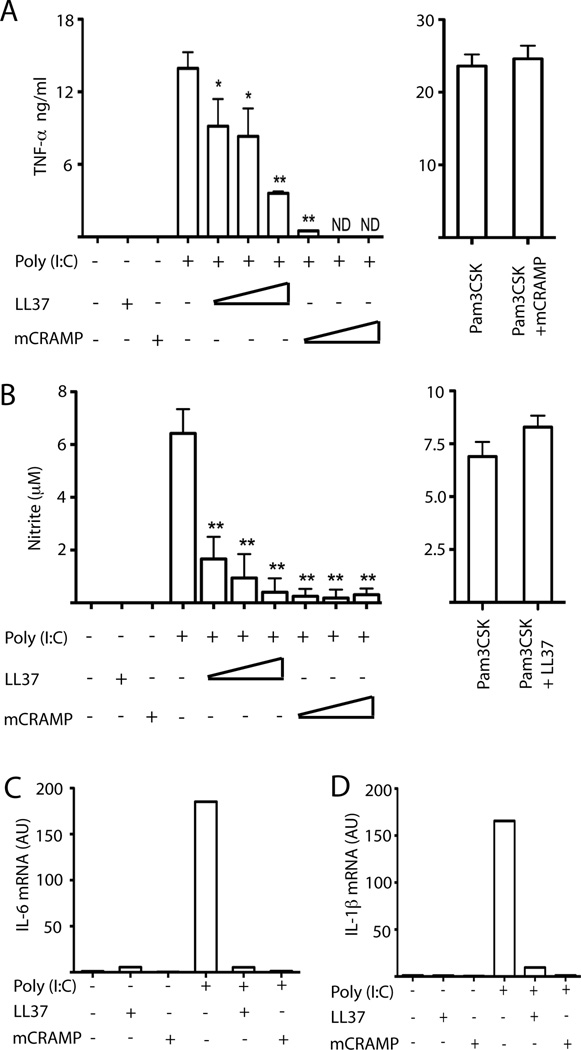
Since skin of psoriasis patients can have up to micromolar concentrations of LL37 that synergizes with DNA to induce inflammation (13). Therefore, we first asked whether high concentrations of LL37 enhanced poly(I:C) mediated signaling. RAW 264.7 macrophages produced TNF-α after poly(I:C) stimulation, which was dose-dependently inhibited by mCRAMP or LL37 (Fig. 1A). The mouse antimicrobial peptide was more effective suggesting some species specificity. Importantly, TLR2 response to Pam3CSK4 was not inhibited by mCRAMP or LL37 (Fig. 1A, and data not shown). Mouse, or human, antimicrobial peptide also inhibited nitrite production, a marker for nitric oxide (Fig. 1B). However, nitrite production induced by Pam3CSK4 was unaffected (Fig. 1B). Other pro-inflammatory cytokine messenger RNAs, such as IL-6 and IL-1β, were also induced by Poly(I:C) (Fig. 1C, D). These responses were inhibited by LL37 and mCRAMP (Fig. 1 C, D).
Figure 1.
Antimicrobial peptide inhibits poly(I:C) induced TNF-α and nitrite production from RAW 264.7 macrophages. (A, B) RAW 264.7 cells were treated with 5 µM LL37 alone, 1.5 µM mCRAMP alone, 27 pM (5 µg/ml) poly(I:C) alone, 27 pM poly(I:C) plus 1, 2 or 5 µM LL37, 27 pM poly(I:C) plus 0.5, 1 or 1.5 µM mCRAMP, or 1 µg/ml Pam3CSK4. After 24 hours, supernatants were collected and (A) TNF-α and (B) nitrite production were determined. Error bars indicate standard deviation (n=3). **, P< 0.01; *, P< 0.05 compared to poly(I:C) alone; ND, not detected. (C, D) RAW 264.7 cells were treated with 5 µM LL37, 1µM mCRAMP or 27 pM poly(I:C), either alone or in combination. After 24 hours, total mRNA was extracted and assayed by quantitative RT-PCR for (C) IL-6 and (D) IL-1β. Results are representative of three independent experiments.
Early interaction of poly(I:C) and antimicrobial peptide is required for inhibitory signaling
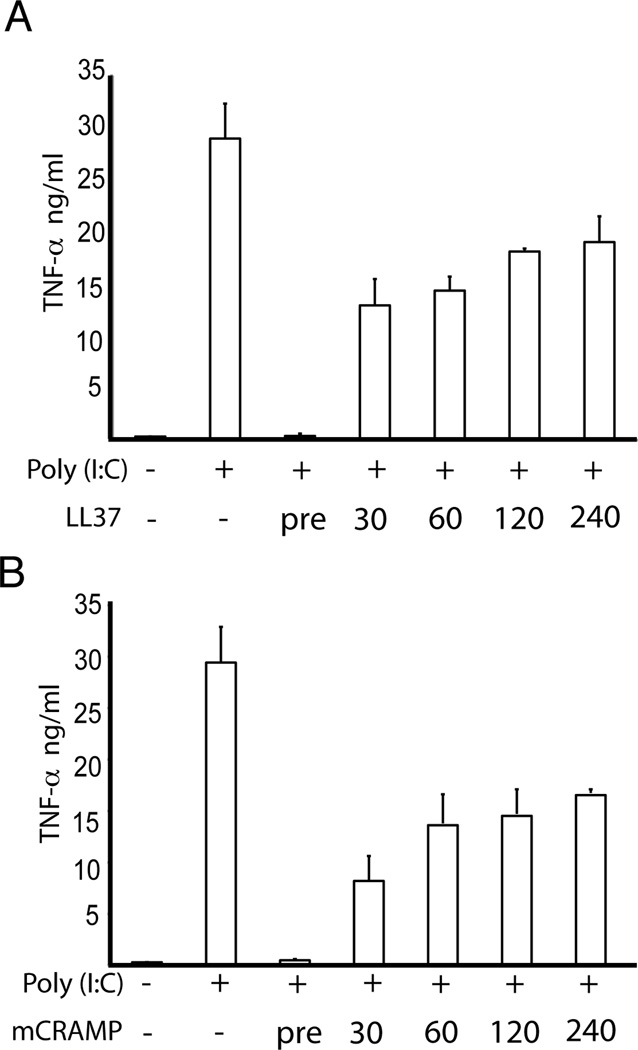
To determine whether delayed addition of antimicrobial peptide has any effect on poly(I:C) induced TNF-α production, we stimulated RAW cells either with premixed poly(I:C) and LL37 or LL37 was added later time points as indicated. With a delayed addition of LL37 as short as 30 minutes, the inhibitory effect of LL37 is dramatically reduced (Fig. 2). With longer delays, up to 4 hours, there is still some inhibitory effect on poly(I:C) induced signaling (Fig. 2). Similar results were obtained using mouse antimicrobial peptide, mCRAMP (unpublished observation, MH and CL). Therefore, these data provide the evidence that antimicrobial peptide is much more efficient as an inhibitor when it is preincubated with poly(I:C).
Figure 2.
Delayed addition of LL37, or mCRAMP, has less inhibitory effect on poly(I:C) induced TNF-α production. RAW 264.7 cells were treated with 27 pM (5 µg/ml) poly(I:C) alone, 27 pM poly(I:C) plus 5 µM LL37 or 1.5 µM mCRAMP, where LL37 or mCRAMP were added as a preformed complex with poly(I:C) or 30 minutes, 1, 2 or 4 hours after the addition of poly(I:C). Supernatants were collected after 24 hours and TNF-α production was determined. Error bars indicate standard deviation (n=4). Results are representative of two experiments.
Antimicrobial peptides inhibit poly(I:C) signaling in primary macrophages and dendritic cells
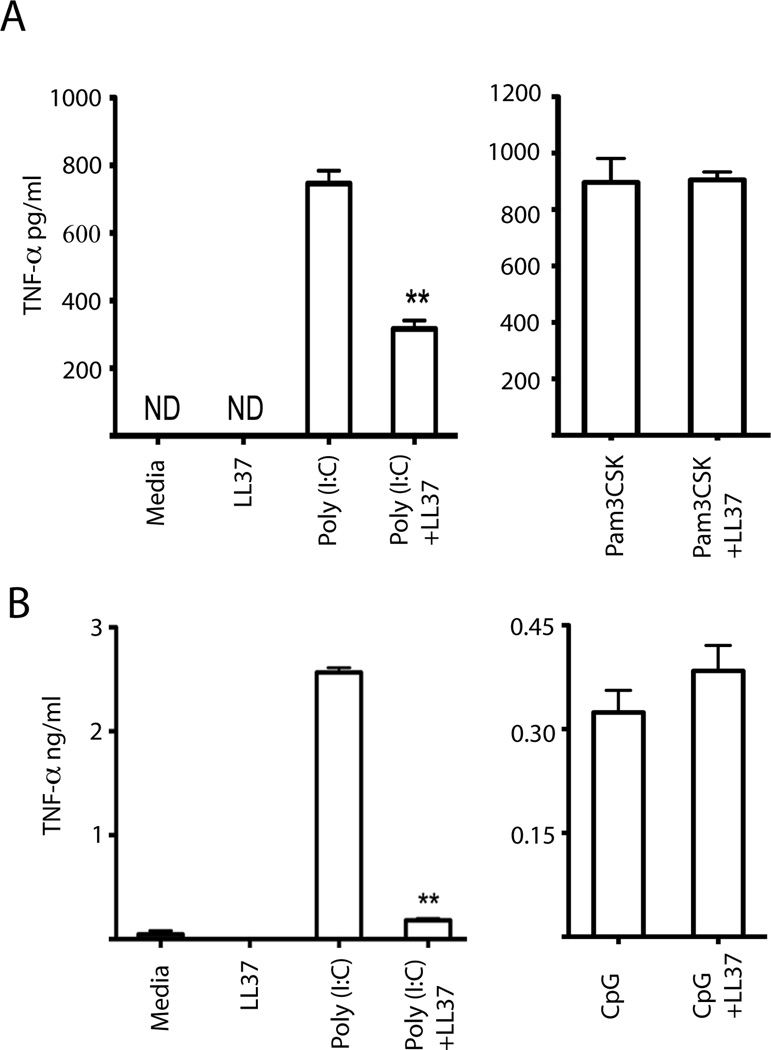
We next examined whether LL37 and mCRAMP inhibited poly(I:C) induced signaling in primary macrophages and dendritic cells. As expected, LL37 had no effect on Pam3CSK4-mediated, TLR2-dependent, TNF-α production by bone marrow-derived macrophages (BMM) (Fig. 3A). However, response to poly(I:C) was dramatically inhibited (Fig. 3A). LL37 had similar inhibitory activity on poly(I:C)-induced bone marrow-derived dendritic cell (BMDC) production of TNF-α (Fig 3B). In BMDC, LL37 slightly increased CpG DNA-induced TNF-α; however, the this effect was not as dramatic as that previously observed in plasmacytoid dendritic cells (13) (Fig. 3B). Importantly, LL37 alone did not have any stimulatory activity in either cell type (Fig. 3 A, B). Similar results were obtained using mouse antimicrobial peptide, mCRAMP (unpublished observation, MH and CL). Therefore, cationic antimicrobial peptide significantly inhibited poly(I:C)-induced TNF-α production.
Figure 3.
Antimicrobial peptides inhibit poly(I:C)-induced TNF-α production. (A) Bone marrow-derived macrophages were exposed to 5 µM LL37, 27 pM poly(I:C), or 1 µg/ml Pam3CSK4, either alone or in combination. Supernatants were collected after 24 hours and assayed for TNF-α production by ELISA. Error bars indicate standard deviation (n=3). (B) As in (A) except bone marrow-derived dendritic cells were stimulated with 5 µM LL37, 27 pM poly(I:C), or 1 µM CpG DNA, either alone or in combination. Error bars indicate standard deviation (n=3). **, P<0.01 compared to poly(I:C) alone; ND, not detected. Results are representative of three independent experiments.
Antimicrobial peptide suppresses poly(I:C)-induced NFκB and MAP kinase activation
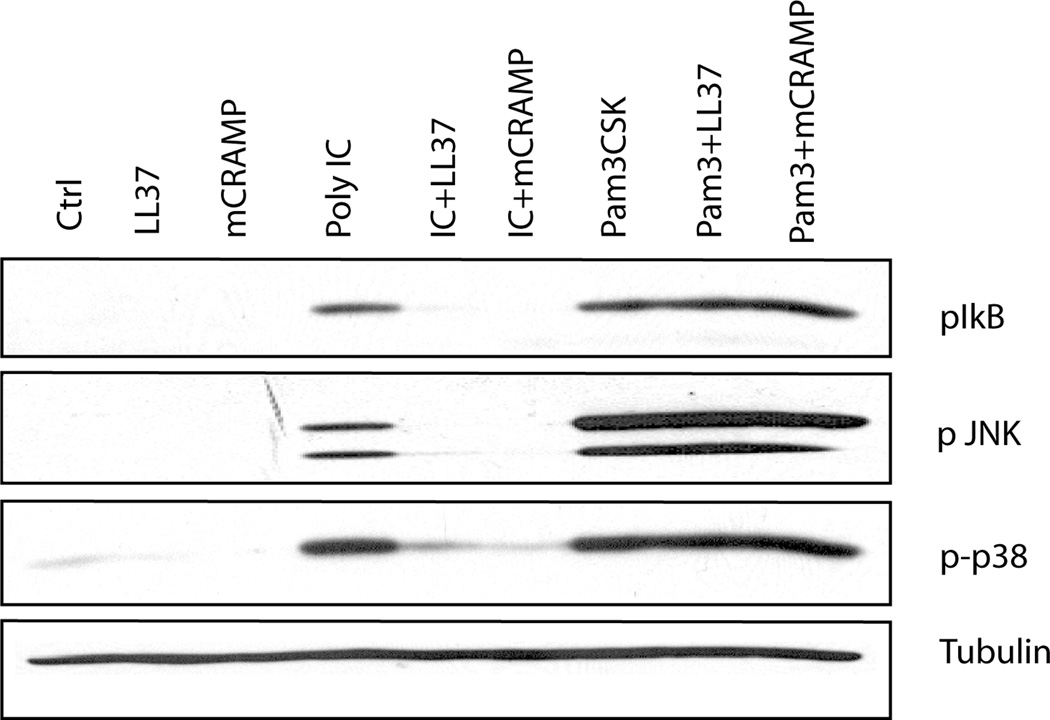
The induction of proinflammatory cytokines depends on activation of mitogen-activated protein (MAP) kinases and the transcription factor NF-κB. Phosphorylation and subsequent degradation of the inhibitor protein IκB relieves repression of NF-κB, which translocates to the nucleus and initiates transcription of target genes (23, 24). In macrophages, treatment with poly(I:C), but not LL37 or mCRAMP, alone induced phosphorylation of IkB, and the MAP kinases p38 and JNK (Fig. 4). Both mCRAMP, and LL37, inhibited poly(I:C)-induced IκB, p38, and JNK phosphorylation (Fig. 4). However, Pam3CSK4-induced phosphorylation of these signaling mediators was not inhibited by mCRAMP or LL37 (Fig. 4). Therefore, antimicrobial peptides selectively inhibit poly(I:C) signaling pathways.
Figure 4.
Antimicrobial peptide inhibits poly(I:C) induced phosphorylation of IκB-α, JNK and p38. RAW 264.7 cells were stimulated for 30 minutes with media (Ctrl), 5 µM LL37, 1 µM mCRAMP, 27 pM poly(I:C), or 1 µg/ml Pam3CSK4 either alone or in combination. Whole cell lysates were resolved by SDS-PAGE and immunoblotted for phosphorylated IκB-α (Ser32), JNK (Thr183/Tyr185), p38 (Thr180/Tyr182) or α-tubulin. Results are representative of three independent experiments.
The inhibitory effect of cationic antimicrobial peptide is TLR3 dependent
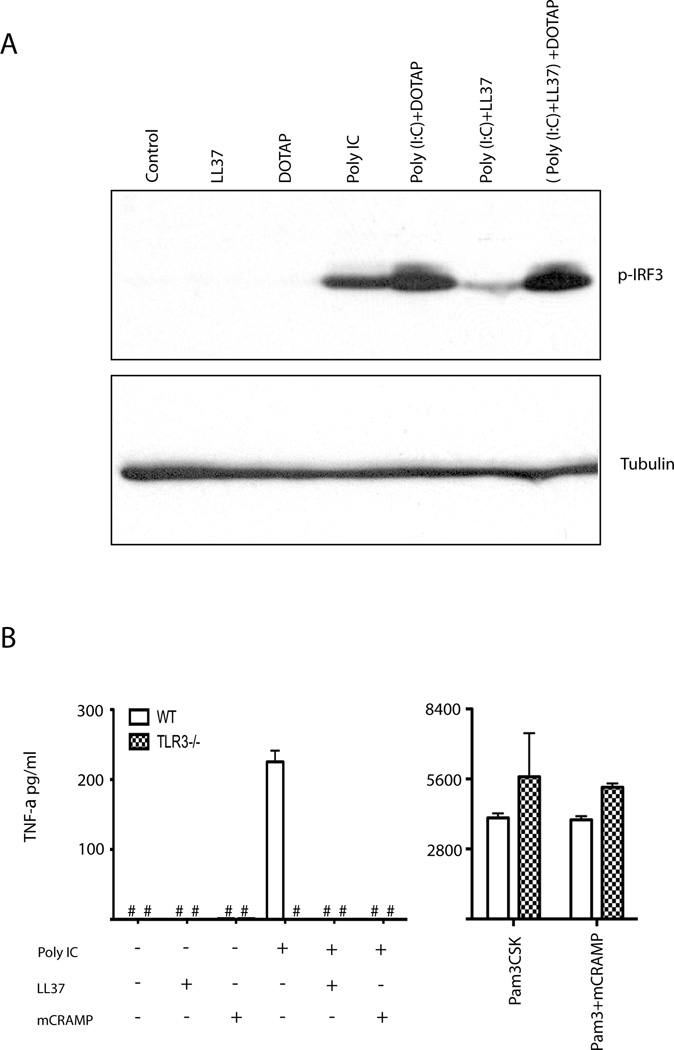
Double stranded RNA is detected by both TLR3 and by the cytoplasmic receptor melanoma differentiation-associated gene 5 (MDA5) (25). While poly(I:C) alone stimulates TLR3, in complex with the lipid DOTAP poly(I:C) also activates MDA5 (26, 27). Therefore, we compared the ability of LL37 to inhibit phosphorylation of IRF3 in macrophages treated with poly(I:C) alone, or in complex with the lipid DOTAP. Poly(I:C) stimulation induced phosphorylation of IRF3, which was enhanced by lipid encapsulation with DOTAP, indicating both the TLR3 and the MDA5 pathways were functional in macrophages (Fig. 5A). LL37 inhibited poly(I:C) alone-induced, but not poly(I:C) in complex with DOTAP-induced, IRF3 phosphorylation (Fig. 5A). Neither LL37 alone nor DOTAP alone, induced phosphorylation of IRF3 (Fig. 5A). These data supported the conclusion that LL37 inhibited TLR3-dependent poly(I:C) signaling.
Figure 5.
Antimicrobial peptide selectively inhibits TLR3-dependent, but not MDA5-dependent, signaling. (A) RAW 264.7 cells were stimulated for 90 minutes with media (Control), 5 µM LL37, 27 pM poly(I:C), either alone or in combination. In some samples, poly(I:C) or poly(I:C)-LL37 complexes were incubated with DOTAP prior to adding to cells, as described in the methods. Whole cell lysates were resolved by SDS-PAGE and immunoblotted for phosphorylated IRF3 (Ser396) and α-tubulin. Representative of two independent experiments. (B) Wild type and TLR3−/− microglial cells were stimulated with 5 µM LL37, 1 µM mCRAMP, 27 pM poly(I:C), or 1 µg/ml Pam3CSK, either alone or in combination. At 24 hours supernatants were assayed for TNF-α production by ELISA. Error bars indicate standard deviation (n=3). #, not detected. Representative of two independent experiments.
To confirm the TLR3 specificity of antimicrobial peptide inhibition, we compared poly(I:C) induced signaling in wild-type and TLR3 deficient cells. Microglial cells respond to poly(I:C) through TLR3 (28). While wild type microglial cells secreted TNF-α in response to poly(I:C), TLR3 deficient microglial cells did not (Fig. 5B). These data confirm that in the microglial cells, poly(I:C)-induced TNF-α production was TLR3-dependent but MDA5-independent (Fig. 5B). Neither LL37 nor mCRAMP alone induced TNF-α (Fig. 5B). The poly(I:C)-induced TNF-α response was completely abolished when cells were treated with LL37 or mCRAMP (Fig. 5B). However, Pam3CSK4-mediated TNF-α production was unaffected by mCRAMP or lack of TLR3 expression (Fig. 5B). Therefore, antimicrobial peptides selectively inhibit TLR3-dependent poly(I:C)-induced responses.
Antimicrobial peptides bind directly to synthetic dsRNA and form a stable complex
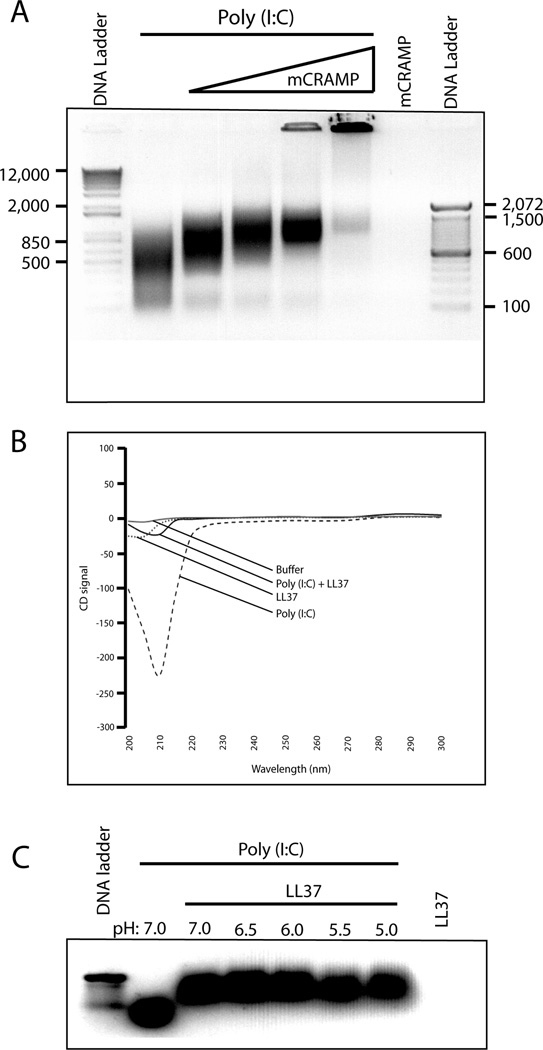
We next asked whether the antimicrobial peptide and poly(I:C) directly associate to form a complex. Poly(I:C) alone migrated as a smear by agarose gel electrophoresis (Fig. 6A). Increasing concentrations of mCRAMP reduced the migration of poly(I:C) demonstrating that they form a stable complex (Fig. 6A). In fact, with the highest concentration of mCRAMP, the majority of poly(I:C) never migrated into the gel, but remained in the well (Fig. 6A). These data suggest that bulky, high molecular weight complexes are formed, or that LL37 neutralizes the charge of poly(I:C) preventing it from entering the gel. Circular dichroism analysis confirmed a strong interaction between LL37 and poly(I:C). The circular dichroism signal for poly(I:C) alone was between −200 and −250, but in the presence of LL37, the signal was dramatically reduced to less than −50 (Fig. 6B).
Figure 6.
Antimicrobial peptides bind to synthetic poly(I:C) and form a pH resistant complex. (A) Poly(I:C) (10.8 picomoles) was incubated for one hour with 131, 263, 526, or 1000 picomoles of mCRAMP, representing approximately a 10 to 100 fold molar excess. Complexes were electrophoresed on a 0.8% agarose gel and imaged on a Syngene ChemiGenius Bio Imaging System. Representative of three independent experiments. (B) Poly(I:C) (50 pM), or 50 µM LL37 alone, or in combination, were incubated for 1 hour and circular dichroism spectra were measured at room temperature (25 °C) in a 1-cm path length quartz cell with a 2 ml volume. Spectra were recorded for each condition from 200 to 300 nm in 5-nm increments with 30-s temperature equilibrations, followed by 30-s data averaging. Representative of two independent experiments. (C) Poly(I:C) (10 pM) was incubated with 5 µM LL37 for 1 hour in neutral buffer. The buffer was then adjusted to pH 7.0, 6.5, 6.0, 5.5 or 5.0 for 3 hours. Complexes were electrophoresed on a 0.8% agarose gel and imaged on a Syngene ChemiGenius Bio Imaging System. Results are representative of three independent experiments.
Since TLR3 signaling initiates in endosomes (29), we next asked whether the poly(I:C)-LL37 complex was stable at low pH. Complexes were formed in neutral pH buffer and transferred to reduced pH buffer (from pH 7.0 to 5.0). The complexes were then subjected to electrophoretic mobility shift assay. LL37-poly(I:C) complexes were not dissociated, even at a pH as low as 5.0 (Fig. 6C). Therefore, the cationic peptide-poly(I:C) complexes are stable and pH resistant.
Antimicrobial peptide partially inhibits dsRNA binding to TLR3
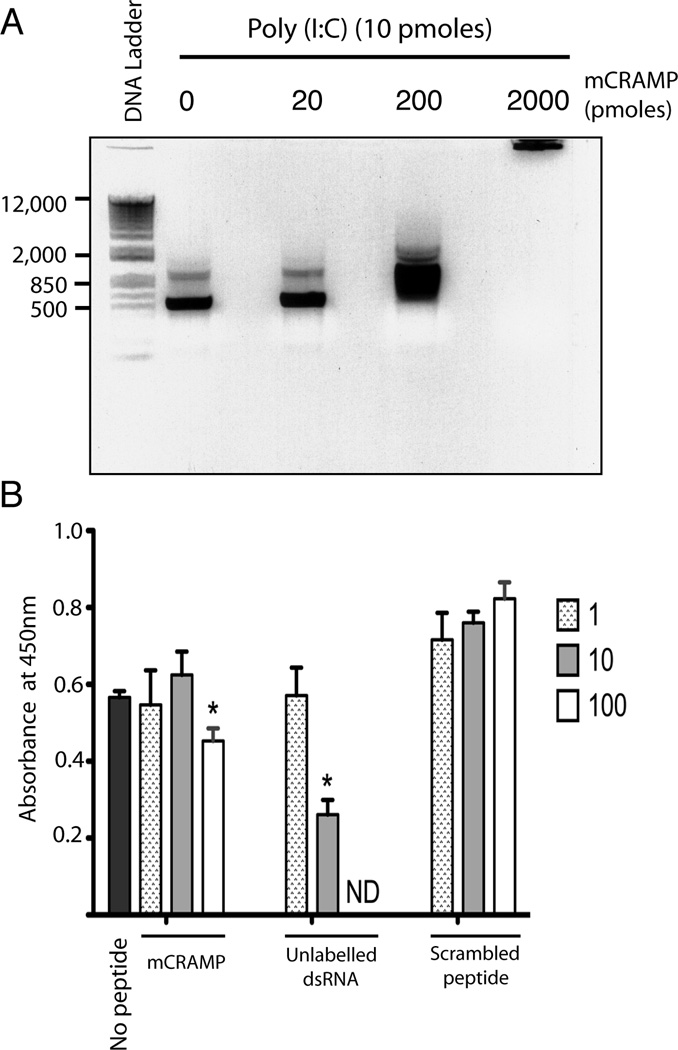
We next asked whether antimicrobial peptides inhibited dsRNA binding to TLR3. Similar to poly(I:C), a synthetic 540 bp dsRNA bound to mCRAMP (Fig. 7A). Using our previously described assay, TLR3 was immobilized on a plate and incubated with a defined biotinylated dsRNA, (22). Without any peptide, the biotinylated dsRNA bound to TLR3 (Fig. 7). This binding was strongly, and dose dependently, inhibited by increasing molar ratios of unlabled dsRNA (Fig. 7). A 100 fold molar excess of mCRAMP, but not its scrambled peptide control, inhibited dsRNA binding to TLR3 (Fig. 7). However, inhibition by mCRAMP was much less efficient than that observed with unlabeled dsRNA. Therefore, although mCRAMP partially inhibited poly(I:C) binding to TLR3, other mechanisms likely account for the dramatic inhibition of signaling.
Figure 7.
Antimicrobial peptide partially inhibits dsRNA binding to TLR3. (A) 540 bp dsRNA (10 picomoles) was incubated for 1 hr at 37°C with 20 to 2000 fold molar excess of mCRAMP and examined by gel shift analysis as in Figure 6. Representative of two experiments. (B) Biotinylated dsRNA (29 pmol/ml) was incubated alone (no peptide), or with equivalent, 10 or 100 molar excess of mCRAMP, scrambled peptide, or unlabeled 540 bp dsRNA as previously described (22). Binding to TLR3 (absorbance at 450 nM) was determined as described in methods. Error bars indicate standard deviation (n=3). * P<0.02. Representative of two independent experiments.
Discussion
Recognition of microbial products is critical for innate immunity, but increasing evidence suggests pathogenic roles for TLR signaling in autoimmune and inflammatory disorders (13, 30, 31). The pathogenic role of TLRs in disease has been linked to facilitated uptake of self nucleic acids, which are normally poorly endocytosed and do not stimulate inflammation. The antimicrobial peptide LL37 is one protein that stabilizes and enhances uptake of self nucleic acids (13, 16). Given their therapeutic activity as immunomodulators (32), and their current evaluation in clinical trials (33, 34), it is important to understand how the immunomodulatory activity of TLR agonists is altered by host factors, such as LL37. Here we showed that LL37 inhibits the inflammatory activity of poly(I:C), a model double stranded RNA. The inhibitory effect was more pronounced when poly(I:C) was pre-mixed with antimicrobial peptides. If the addition of LL37 was delayed, the inhibitory effect of LL37 was reduced significantly.
LL37 inhibited poly(I:C)-induced proinflammatory responses in mouse macrophages (Fig. 1), yet it enhanced CpG DNA-induced type I interferon responses in human plasmacytoid dendritic cells (pDCs) (13). Interestingly, in human myeloid dendritic cells and in HEK293 cells, both poly(I:C) and CpG DNA are endocytosed by a shared clathrin-dependent pathway (35). Recent studies have implicated CD14 and scavenger receptor in endocytosis of poly(I:C) (36, 37), and CpG DNA (38, 39). Therefore, it is unclear why LL37 has opposite effects on CpG and poly(I:C) signaling. Further studies are needed to determine if uptake is dependent on as yet unidentified receptors in different cell types.
Although LL37 and mCRAMP dramatically inhibit poly(I:C) induced responses (Fig. 1–3), they minimally inhibit binding of poly(I:C) to TLR3 (Fig. 7). Due to the presence of several positive charges, LL37, and mCRAMP, can condense short, single-stranded, CpG DNAs. In contrast, poly(I:C) length is variable (from 200 base pairs to 8 kilobases) and forms concatemer-like structures in solution. Therefore, unlike complete coverage that might be achieved by LL37 association with CpG DNA, only part of the poly(I:C) might be covered by LL37, or higher concentrations of LL37 would be required to saturate binding. This model would explain our data that show LL37 forms a complex with poly(I:C), but high concentrations are required to inhibit poly(I:C) binding to TLR3. Since LL37 does not efficiently block binding to TLR3, but TLR3 requires dimerization to signal (40), it is possible that LL37 inhibits signaling by preventing receptor dimerization. However, dsRNA does not stably bind to TLR3 without receptor dimerization (22) and if LL37 blocked dimerization, it should have inhibited poly(I:C) binding to TLR3. Since it did not inhibit binding (Fig. 6), it is unlikely that LL37 blocks ligand-induced receptor dimerization. Therefore, inhibition of poly(I:C) signaling by LL37 likely occurs by other mechanisms such as inhibition of poly(I:C) uptake into endosomes.
Together, our studies show that antimicrobial peptides form a stable complex with poly(I:C) and block TLR3 signaling. This is due, at least in part, to inhibition of poly(I:C) binding to TLR3. Since poly(I:C), and other nucleic acids, are currently in clinical trials, it is important to understand complex interactions between these nucleic acids and host proteins that modify their activity. Our data suggest that during acute infection, or chronic inflammatory conditions, where LL37 is elevated, that the therapeutic activity of poly(I:C) will be compromised. Therefore, more information is needed on how disease conditions and host proteins modify the therapeutic potential of dsRNA.
Acknowledgements
We thank Dr. D. Bowdish for LL37, and members of the lab for discussions and assistance with the manuscript. The following reagent was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: TLR3 deficient microglial cells.
Abbreviations
- poly(I:C)
polyinosinic-polycytidylic acid
- ds
double stranded
- MDA5
melanoma differentiation-associated gene 5
- IRF
interferon regulatory factor
- mCRAMP
cathelicidin-related antimicrobial peptide
Footnotes
This work was supported by RO1AI076588 and RO1AI076588-S1 (CAL).
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.O'Neill LA, Fitzgerald KA, Bowie AG. The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol. 2003;24:286–290. doi: 10.1016/s1471-4906(03)00115-7. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 4.Chockalingam A, Brooks JC, Cameron JL, Blum LK, Leifer CA. TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol Cell Biol. 2009;87:209–217. doi: 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 6.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 7.Leifer CA, Brooks JC, Hoelzer K, Lopez JL, Kennedy MN, Mazzoni A, Segal DM. Cytoplasmic targeting motifs control localization of toll-like receptor 9. J Biol Chem. 2006;281:35585–35592. doi: 10.1074/jbc.M607511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173:1179–1183. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chockalingam A, Cameron JL, Brooks JC, Leifer CA. Negative Regulation of Signaling by a Soluble Form of Toll-Like Receptor 9. Eur J Immunol. 2011 doi: 10.1002/eji.201041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, Savitsky D, Ronfani L, Akira S, Bianchi ME, Honda K, Tamura T, Kodama T, Taniguchi T. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 13.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, Zal T, Mellman I, Schroder JM, Liu YJ, Gilliet M. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 14.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 15.Hurtado P, Peh CA. LL-37 promotes rapid sensing of CpG oligodeoxynucleotides by B lymphocytes and plasmacytoid dendritic cells. J Immunol. 2010;184:1425–1435. doi: 10.4049/jimmunol.0902305. [DOI] [PubMed] [Google Scholar]
- 16.Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, Homey B, Barrat FJ, Zal T, Gilliet M. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. 2009;206:1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustafsson A, Sigel S, Ljunggren L. The antimicrobial peptide LL37 and its truncated derivatives potentiates proinflammatory cytokine induction by lipoteichoic acid in whole blood. Scand J Clin Lab Invest. 2010;70:512–518. doi: 10.3109/00365513.2010.521255. [DOI] [PubMed] [Google Scholar]
- 18.Kandler K, Shaykhiev R, Kleemann P, Klescz F, Lohoff M, Vogelmeier C, Bals R. The anti-microbial peptide LL-37 inhibits the activation of dendritic cells by TLR ligands. Int Immunol. 2006;18:1729–1736. doi: 10.1093/intimm/dxl107. [DOI] [PubMed] [Google Scholar]
- 19.Nagaoka I, Hirota S, Niyonsaba F, Hirata M, Adachi Y, Tamura H, Heumann D. Cathelicidin family of antibacterial peptides CAP18 and CAP11 inhibit the expression of TNF-alpha by blocking the binding of LPS to CD14(+) cells. J Immunol. 2001;167:3329–3338. doi: 10.4049/jimmunol.167.6.3329. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Ding AH, Nathan CF, Stuehr DJ. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J Immunol. 1988;141:2407–2412. [PubMed] [Google Scholar]
- 22.Wang Y, Liu L, Davies DR, Segal DM. Dimerization of Toll-like receptor 3 (TLR3) is required for ligand binding. J Biol Chem. 2010;285:36836–36841. doi: 10.1074/jbc.M110.167973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 25.Tamassia N, Le Moigne V, Rossato M, Donini M, McCartney S, Calzetti F, Colonna M, Bazzoni F, Cassatella MA. Activation of an immunoregulatory and antiviral gene expression program in poly(I:C)-transfected human neutrophils. J Immunol. 2008;181:6563–6573. doi: 10.4049/jimmunol.181.9.6563. [DOI] [PubMed] [Google Scholar]
- 26.Reimer T, Brcic M, Schweizer M, Jungi TW. poly(I:C) and LPS induce distinct IRF3 and NF-kappaB signaling during type-I IFN and TNF responses in human macrophages. J Leukoc Biol. 2008;83:1249–1257. doi: 10.1189/jlb.0607412. [DOI] [PubMed] [Google Scholar]
- 27.De Miranda J, Yaddanapudi K, Hornig M, Lipkin WI. Astrocytes recognize intracellular polyinosinic-polycytidylic acid via MDA-5. FASEB J. 2009;23:1064–1071. doi: 10.1096/fj.08-121434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Town T, Jeng D, Alexopoulou L, Tan J, Flavell RA. Microglia recognize double-stranded RNA via TLR3. J Immunol. 2006;176:3804–3812. doi: 10.4049/jimmunol.176.6.3804. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, Seya T. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 30.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, Meller S, Chamilos G, Sebasigari R, Riccieri V, Bassett R, Amuro H, Fukuhara S, Ito T, Liu YJ, Gilliet M. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicodemus CF, Berek JS. TLR3 agonists as immunotherapeutic agents. Immunotherapy. 2010;2:137–140. doi: 10.2217/imt.10.8. [DOI] [PubMed] [Google Scholar]
- 33.Hendrix CW, Margolick JB, Petty BG, Markham RB, Nerhood L, Farzadegan H, Ts'o PO, Lietman PS. Biologic effects after a single dose of poly(I):poly(C12U) in healthy volunteers. Antimicrob Agents Chemother. 1993;37:429–435. doi: 10.1128/aac.37.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson KA, Strayer DR, Salvato PD, Thompson CE, Klimas N, Molavi A, Hamill AK, Zheng Z, Ventura D, Carter WA. Results of a double-blind placebo-controlled study of the double-stranded RNA drug polyI:polyC12U in the treatment of HIV infection. Eur J Clin Microbiol Infect Dis. 1996;15:580–587. doi: 10.1007/BF01709367. [DOI] [PubMed] [Google Scholar]
- 35.Itoh K, Watanabe A, Funami K, Seya T, Matsumoto M. The clathrin-mediated endocytic pathway participates in dsRNA-induced IFN-beta production. J Immunol. 2008;181:5522–5529. doi: 10.4049/jimmunol.181.8.5522. [DOI] [PubMed] [Google Scholar]
- 36.Lee HK, Dunzendorfer S, Soldau K, Tobias PS. Double-stranded RNA-mediated TLR3 activation is enhanced by CD14. Immunity. 2006;24:153–163. doi: 10.1016/j.immuni.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Limmon GV, Arredouani M, McCann KL, Corn Minor RA, Kobzik L, Imani F. Scavenger receptor class-A is a novel cell surface receptor for double-stranded RNA. FASEB J. 2008;22:159–167. doi: 10.1096/fj.07-8348com. [DOI] [PubMed] [Google Scholar]
- 38.Gursel M, Gursel I, Mostowski HS, Klinman DM. CXCL16 Influences the Nature and Specificity of CpG-Induced Immune Activation. J Immunol. 2006;177:1575–1580. doi: 10.4049/jimmunol.177.3.1575. [DOI] [PubMed] [Google Scholar]
- 39.Jozefowski S, Sulahian TH, Arredouani M, Kobzik L. Role of scavenger receptor MARCO in macrophage responses to CpG oligodeoxynucleotides. J Leukoc Biol. 2006;80:870–879. doi: 10.1189/jlb.0705357. [DOI] [PubMed] [Google Scholar]
- 40.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]