Abstract
Objective:
To use the Unified Batten Disease Rating Scale (UBDRS) to measure the rate of decline in physical and functional capability domains in patients with juvenile neuronal ceroid lipofuscinosis (JNCL) or Batten disease, a neurodegenerative lysosomal storage disorder. We have evaluated the UBDRS in subjects with JNCL since 2002; during that time, the scale has been refined to improve reliability and validity. Now that therapies are being proposed to prevent, slow, or reverse the course of JNCL, the UBDRS will play an important role in quantitatively assessing clinical outcomes in research trials.
Methods:
We administered the UBDRS to 82 subjects with JNCL genetically confirmed by CLN3 mutational analysis. Forty-four subjects were seen for more than one annual visit. From these data, the rate of physical impairment over time was quantified using multivariate linear regression and repeated-measures analysis.
Results:
The UBDRS Physical Impairment subscale shows worsening over time that proceeds at a quantifiable linear rate in the years following initial onset of clinical symptoms. This deterioration correlates with functional capability and is not influenced by CLN3 genotype.
Conclusion:
The UBDRS is a reliable and valid instrument that measures clinical progression in JNCL. Our data support the use of the UBDRS to quantify the rate of progression of physical impairment in subjects with JNCL in clinical trials.
The neuronal ceroid lipofuscinoses (NCLs) are degenerative, autosomal recessive storage diseases with common clinical and pathologic features, including blindness, seizures, dementia, motor decline, and lysosomal accumulation of autofluorescent material (ceroid and lipofuscin).1 The NCLs are categorized by genetic etiology. The juvenile form of neuronal ceroid lipofuscinosis (JNCL), also called Batten disease, is the most prevalent NCL. Clinically, JNCL begins with progressive visual loss between 4 and 8 years of age, followed by seizures, then loss of motor coordination and dementia between 10 and 12 years. Death occurs by the third or fourth decade.1,2
JNCL is due to mutations of the CLN33 gene that encodes a ubiquitously expressed protein of unknown function localized to the lysosomal membrane4,5; modeling studies suggest a role in substrate trafficking.5,6 Most cases of JNCL are caused by an approximately 1-kb deletion in the CLN33,7 gene, encompassing exons 7 and 8. Approximately 74% of patients with JNCL are homozygous for this common deletion, and 22% are compound heterozygotes for this deletion and another CLN3 mutation.8 There are no mutations consistently associated with better prognosis.8–10
With biological advances, there is hope for rational therapies to prevent, slow, or reverse the course of JNCL. We developed the Unified Batten Disease Rating Scale (UBDRS) to measure disease progression in JNCL.11 We used the UBDRS to evaluate and quantify disease burden in 82 subjects with JNCL from 2002 through 2010, the largest cohort of patients with JNCL reported to date.
METHODS
Protocol approval and subject consents.
The UBDRS and genetic samples were obtained at annual meetings of the Batten Disease Support & Research Association (BDSRA) and at the Batten Disease Diagnostic & Clinical Research Center at the University of Rochester, using a research protocol approved by the University of Rochester's Institutional Review Board. The parents of all subjects provided written informed consent for their child's participation.
CLN3 genotyping.
All subjects reported here have been confirmed to have CLN3 mutations; 80 of the 82 subjects were confirmed in the University of Rochester Molecular Diagnostics Laboratory. Two were tested in outside commercial laboratories and we reviewed the results. DNA samples were obtained via collection of blood before 2005; subsequently, we instituted a noninvasive method to derive the genotype from buccal epithelial cell samples. DNA was prepared from the specimen using standard methods. Analysis of the common deletion in the CLN3 gene was done as previously described.12 If the subject was not homozygous for the common deletion, additional sequencing of the CLN3 gene was performed.
Unified Batten Disease Rating Scale.
The Unified Batten Disease Rating Scale (UBDRS) is a reliable clinical rating scale developed to measure physical impairments and the severity of disease-associated symptoms over time.11 The UBDRS has measures to quantify physical impairment, daily function, seizure severity, behavioral symptoms, symptom onset, and clinical global impressions. In this study, we focused on 2 UBDRS measures: physical and motor dysfunction (Physical Impairment) and competence in activities of daily life (Capability). The Physical Impairment domain consists of 27 items evaluating vision, speech, tone, bulbar and motor function, and presence of abnormal movements. The items are scored by examiners trained in using the UBDRS. For each item, a score from 0 to 4 (normal to abnormal) is based on the degree of deviation from normal. In this domain, higher scores indicated greater impairment. The Capability domain consists of 10 items (2 items were based on a 3-point scale and the rest used a 4-point scale) using parent assessment of their child's performance of typical age-appropriate self-care and play tasks. In this domain, lower scores indicated poorer or more limited functioning. For our analyses, the scores of individual items were summed to give the domain totals, e.g., the Physical Impairment domain score was the sum of each of the 27 items queried under Physical Impairment. Some subjects were rated on the UBDRS by more than one examiner for concurrent interrater reliability studies; for these subjects, we utilized the median UBDRS scores across raters. Parents were asked to recall age when initial symptoms occurred in order to determine age at onset. For those who had multiple visits, the average age at onset was calculated based on these responses and this value was used as age at onset for these subjects. Disease duration was calculated as the difference between age at time of testing and age at onset.
The UBDRS was administered from 2002 through 2010 to 82 subjects with JNCL whose diagnosis was confirmed by CLN3 mutation analysis. Patients were evaluated either at the annual BDSRA meetings or at the University of Rochester. Subjects were invited to participate in annual serial research evaluations and over half of the participants came for multiple yearly evaluations. Until 2007, all subjects were rated on the UBDRS by more than one examiner for all evaluations. By 2007, reliability had been established for the original UBDRS raters (L.S.D., J.M.K., F.J.M., J.W.M.).11 After 2007, new evaluators (D.R.-M., E.F.A.) were expected to perform concurrent evaluations of 10 subjects and, for their data to be included in the sample, to have an interrater reliability of 0.85.
Statistical analysis.
Genotypic and demographic data were summarized with descriptive statistics. The change in the UBDRS domain scores over time was evaluated using both cross-sectional and longitudinal data analyses. Data from each subject's most recent evaluation were used to estimate pairwise Pearson correlation coefficients. These cross-sectional data were also used in linear regression analyses to estimate the rate of decline over time in the entire cohort, in CLN3 deletion homozygotes, and in those with other CLN3 genotypes separately. Pointwise 95% confidence intervals (CI) for these regression curves were constructed based on these models. To evaluate data from subjects who participated in multiple evaluations as well as those who only were tested once, we performed linear mixed model analyses to estimate the rate of impairment over time and to determine the effects of gender and CLN3 genotype on disease progression. The effects of age, genotype, and gender were quantified under various models using the likelihood ratio test and the corresponding χ2 test statistics. All analyses were performed using StataSE version 9 (2005; College Station, TX).
RESULTS
Subjects.
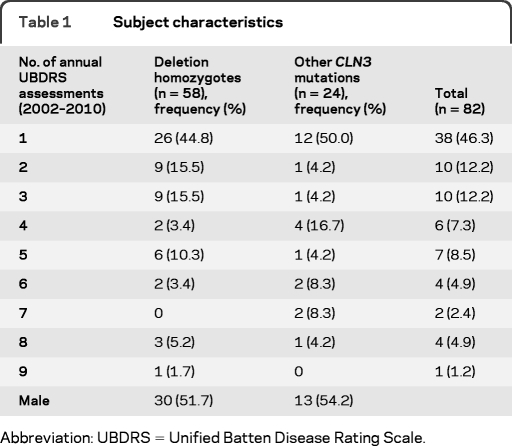
The diagnosis of JNCL was confirmed genetically in the 82 subjects evaluated using the UBDRS (table 1). Most subjects were of European (though generally not of Finnish or Scandinavian) ancestry. Forty-four subjects (53.6%) were seen by our group for UBDRS evaluations for more than one annual visit. The 82 subjects came for 226 total visits and of these visits 115 (50.9%) were rated by more than one rater. Forty-three subjects (52.4%) were male; 58 (70.7%) were homozygous for the common deletion, 22 (26.8%) were heterozygous for the common deletion and another CLN3 mutation. There were 2 subjects (2.4%) whose genotypes did not include the common deletion—one was homozygous for the R334H mutation and the other was a compound heterozygote for 2 previously reported exon 12 mutations (see subject 10 in table 2).
Table 1.
Subject characteristics
Abbreviation: UBDRS = Unified Batten Disease Rating Scale.
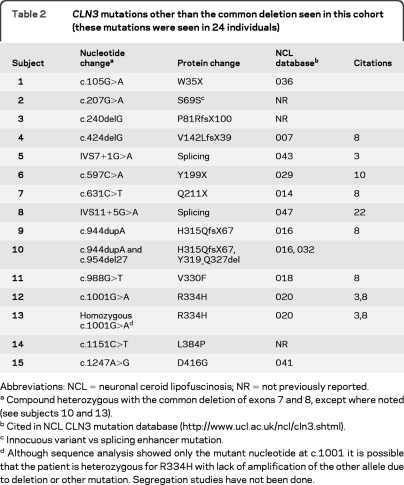
Table 2.
CLN3 mutations other than the common deletion seen in this cohort (these mutations were seen in 24 individuals)
Abbreviations: NCL = neuronal ceroid lipofuscinosis; NR = not previously reported.
Compound heterozygous with the common deletion of exons 7 and 8, except where noted (see subjects 10 and 13).
Cited in NCL CLN3 mutation database (http://www.ucl.ac.uk/ncl/cln3.shtml).
Innocuous variant vs splicing enhancer mutation.
Although sequence analysis showed only the mutant nucleotide at c.1001 it is possible that the patient is heterozygous for R334H with lack of amplification of the other allele due to deletion or other mutation. Segregation studies have not been done.
The mutations identified in our cohort included known mutations in the NCL mutation database (http://www.ucl.ac.uk/ncl/cln3.shtml) as well as 3 novel CLN3 variants (table 2). One variant results in the deletion of nucleotide G in position 240 of the cDNA (NCBI, NM_000086); this leads to a frameshift after amino acid 81 (Pro81). Another is a G to A substitution in Ser69 that does not alter the amino acid. The third variant is a missense mutation, Leu384Pro.
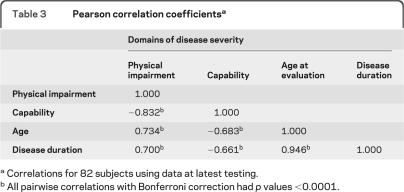
The mean ± SD age at onset of disease was 5.17 ± 1.70 years. Visual loss was the most common presenting symptom, though in some cases there were cognitive or behavioral changes (such as colic or unusually rigid temperament) that seemed unusual and were thought to mark the onset of a child's JNCL. To minimize the effects of bias in parental recall of age at onset, we evaluated whether the age at time of testing could be a valid surrogate for disease duration. Calculated disease duration was highly correlated with age at time of testing (table 3), and we chose to use this more reliable measure in subsequent analyses.
Table 3.
Pearson correlation coefficientsa
Correlations for 82 subjects using data at latest testing.
All pairwise correlations with Bonferroni correction had p values <0.0001.
Correlation of UBDRS domain scores and age.
The correlations between the domain scores, age, and disease duration (table 3) were notable for the high correlation between 1) Physical Impairment and Capability domain scores and 2) calculated disease duration and age at testing.
Physical impairment worsens over time.
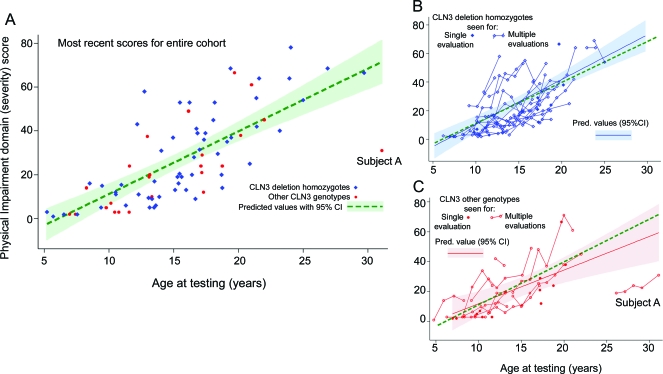
The figure, A–C, shows variation in the Physical Impairment scores over time. Greater disability is associated with higher Physical Impairment scores. The colored labels indicate the subject's CLN3 genotype (CLN3 deletion homozygotes, filled blue diamonds, and other CLN3 genotypes, filled red circles). In the figure, A, the most recent Physical Impairment scores for all subjects with genetic confirmation of JNCL are shown, and the fitted line of predicted values is based on ordinary linear regression analysis for the entire cohort with age as the only independent variable. The model predicts an average increase of 2.86 points per year (95% CI 2.27–3.45, p < 0.0001). Capability scores showed significant worsening over time (decline of 1.06 points per year, 95% CI 0.80–1.31, p < 0.0001; data not shown). The figure, B and C, shows the Physical Impairment scores of subjects by their underlying CLN3 genotype (the figure, B, shows deletion homozygotes and the figure, C, shows subjects with other CLN3 genotypes). In these figures, the scores of subjects who participated for multiple annual evaluations are connected by a solid line. To examine the effects of genotype on impairment, we fit similar regression curves for those who were homozygous for the common deletion (worsening of 3.12 points per year, 95% CI 2.41–3.84 p < 0.0001) and those with other CLN3 genotypes (worsening of 2.25 points per year, 95% CI 1.15–3.35, p < 0.0001). The figure, C, also shows the presence of an outlier (subject A) who alone accounts for much of the difference seen between the rate of progression in the 2 groups. When subject A's scores are not included in the analysis of those with other CLN3 genotypes, the rate of change of Physical Impairment scores is 3.31 points per year (95% CI 2.11–4.51, p < 0.0001). While a linear rate of progression is well supported by the data, extrapolation of these rates to subjects outside the age range of this cohort is speculative, especially when applied to younger subjects due to potential floor effects on Physical Impairment scores.
Figure. Physical Impairment domain scores worsen over time.
(A) Physical Impairment scores at the most recent time of testing for all subjects are shown. The fitted line of predicted values (green dashed line) is based on ordinary linear regression analysis for the entire cohort using the most recent Physical Impairment score with age as the only independent variable. The colored labels indicate the subject's CLN3 genotype (CLN3 deletion homozygotes, filled blue diamond, and other CLN3 genotypes, filled red circle). Subject A is described in the text. (B) Physical Impairment scores of the subset who are homozygous for the common CLN3 deletion (the most common genotype seen). When subjects participated for multiple evaluations, their scores are marked by connected hollow blue diamonds. Those seen for a single visit are marked by a filled blue diamond. The fitted regression line (blue) with 95% confidence interval (CI) reflects the regression estimates for this subset only. The dashed green line is included to show the fitted regression for the entire cohort, discussed in A. (C) Physical Impairment scores of those with other CLN3 genotypes (noncommon deletion homozygotes.) When subjects participated for multiple evaluations, their scores are marked by connected hollow red circles, and those seen for a single visit are marked with a filled red circle. The fitted regression line with 95% CI reflects the regression estimates for this subset only. Subject A is described in the text.
Genotype (comparing deletion homozygotes with other CLN3 genotypes) and gender did not contribute significantly to the association between Physical Impairment and age in multivariate regression analysis.
Physical impairment within and across subjects.
Linear mixed models (LMM) were used to fully incorporate the longitudinal data within subjects and again, there was a significant association between Physical Impairment scores and age. In the LMM, we used an unstructured covariance matrix in a model permitting random intercepts (with fixed slopes across subjects) and in a model allowing both random intercepts and slopes and found similar results. For the entire cohort, the random intercept and slopes model predicts an average increase in the Physical Impairment score of 3.14 points per year (95% CI 2.38–3.90, p < 0.0001). Addition of covariates for gender and genotype did not significantly improve these models (in all cases, p > 0.8).
The validity of estimates obtained from these longitudinal analyses in the presence of missing data relies on the missing at random (MAR) assumption13 that requires that the missing data not depend on the unobserved data. While this assumption is difficult to verify, we note that participation did not seem to depend on level of disability. In addition, estimates of progression rates from cross-sectional and longitudinal analyses are fairly consistent, suggesting that any bias induced by violating the MAR assumption is minimal.
These findings combined with the cross-sectional data provide a graphical and quantifiable picture of how measurable clinical signs and function steadily worsen over time. Given the size of our cohort, it is likely that the majority of patients with JNCL will follow a very similar course.
Using the UBDRS to highlight atypical JNCL course.
Subject A (figure, A and C) had a course that was unusual. The subject lost vision at age 7 years and the decline in physical disability began later than for the rest of the cohort. Subject A had a compound heterozygous genotype—the common deletion and the R334H missense mutation. This genotype has previously been reported in cases with a “protracted” disease course, but has also been seen in many individuals with fairly “classic” disease progression.8 In our sample, there were 24 individuals (29.3%) who were not homozygous for the CLN3 common deletion, including one who was compound heterozygous for the common mutation and R334H, and one who was homozygous for R334H. So far, we have not convincingly found an individual with as protracted a course as subject A. However, it should be noted that he declined quickly after his last UBDRS assessment and died 2 years later.
DISCUSSION
We have evaluated over 80 individuals with genetically confirmed JNCL using the UBDRS, and more than half have been evaluated on multiple occasions. This is the largest cohort of patients with JNCL followed clinically over time using a disease-specific rating scale. However, our cohort may diverge in some ways from the greater population of patients with JNCL. Anecdotal evidence and our own clinical experience suggest that the function of the subjects who participated in our study was representative of patients with JNCL as a whole.
Despite limitations in controlling for ascertainment bias, our data show the value of the UBDRS in quantifying JNCL progression. Correlation analyses show that Physical Impairment and Capability show quantifiable changes that appear to meaningfully reflect clinical morbidity. In our analyses, we focused on the Physical Impairment scores though we note its high correlation with Capability scores and that both domains demonstrate clinical worsening over time (table 3).
Physical Impairment is the one domain in the UBDRS that requires an experienced examiner to collect data by performing a physical examination. Other domains are based on caregiver reports in face-to-face interviews with these same examiners. The advantage of the Physical Impairment domain is that it does not rely on parents or guardians to provide history and information. The high correlation with Capability demonstrates convergent validity with this domain in evaluating patients with JNCL.
Statistical analyses allow some generalizations about the rate of worsening physical impairment over time. Based on this cohort, we predict that an individual's Physical Impairment scores would increase approximately 3 points each year, suggesting that changes could be detected in clinical assessments performed every 6–12 months. However, there is much variability between individuals that is not well explained. The Physical Impairment domain is still influenced by the emotional and behavioral state of the subject at the time of testing, for example. Some of these qualities are assessed in other UBDRS domains not presented here.
We used fairly simple models to evaluate the variation in the Physical Impairment scores with age. Yet these models are robust in supporting our conclusions that UBDRS Physical Impairment deteriorates over time. Some subjects decline more rapidly than others, but this effect is not clearly dependent on genotype or gender. Although we highlight a single exception (subject A), patients who are homozygous for the common deletion are generally not distinguishable from compound heterozygotes based on their clinical histories and the UBDRS.
The idea that genotype might explain some of the clinical variability in JNCL has been suggested since the first description of the CLN3 gene.3,8,9,14 Yet some patients homozygous for the common deletion can have very slow progression, mimicking a protracted course.8 The R334H mutation, often seen with a common deletion mutation, has been noted in some individuals with slower disease progression. This is similar to our subject A, who had later onset of physical impairment. In our cohort we follow 2 other individuals, one a compound heterozygote for the R334H mutation, and another homozygous for this mutation; their Physical Impairment scores do not show a protective effect and it is known that R334H is not consistently associated with a protracted clinical course.8
In the course of genetically characterizing our subjects, we have identified 3 novel variants (table 2). The c.207G>A variant does not change the amino acid (Ser69) and is not predicted to create or destroy a splice site. Segregation studies show that it is in trans with the common deletion. We have been unable to obtain a fresh specimen to fully evaluate potential splicing effects such as disruption of a splicing enhancer binding site.15 The c.240delG mutation causes a frameshift starting at Pro81. The Leu384Pro mutation is in a position conserved in all reported CLN3 sequences in GenBank, except in the yeast Schizosaccharomyces pombe (Ile instead of Leu). Thus, the evidence supports a pathogenic role for c.240delG and L384P.
These analyses demonstrate the utility of the UBDRS Physical Impairment domain in quantifying disease progression in patients with JNCL. Further modeling and incorporation of other UBDRS domains may help account for the intersubject variability seen in UBDRS Physical Impairment and Capability domains. Our ability to quantify disease progression despite this variability compares favorably with scales used in other neurodegenerative conditions, such as in Niemann-Pick type C,16 Friedreich ataxia,17 Parkinson disease,18,19 and Huntington disease.20,21
Results from this study suggest that the UBDRS is a valid, reliable, and sensitive tool for assessment of clinical change over time and will have utility in the evaluation of therapies for JNCL.
ACKNOWLEDGMENT
The authors thank the parents and children for their participation; Melissa Wang, MD, Tiffani McDonough, MD, and Danielle deCampo for assistance with data collection; and the Batten Disease Support and Research Association (BDSRA) for their assistance with subject recruitment.
GLOSSARY
- BDSRA
Batten Disease Support & Research Association
- CI
confidence interval
- JNCL
juvenile neuronal ceroid lipofuscinosis
- LMM
linear mixed model
- MAR
missing at random
- NCL
neuronal ceroid lipofuscinosis
- UBDRS
Unified Batten Disease Rating Scale
Footnotes
AUTHOR CONTRIBUTIONS
Dr. Kwon: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data; acquisition of data; statistical analysis. Dr. Adams: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data. Dr. Rothberg: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data; contribution of vital reagents/tools/patents. Dr. Augustine: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data; acquisition of data. Dr. Marshall: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data; acquisition of data. E.A. deBlieck: drafting/revising the manuscript for content, including medical writing for content; acquisition of data; study supervision or coordination. A. Vierhile: drafting/revising the manuscript for content, including medical writing for content; acquisition of data; study supervision or coordination. Dr. Beck: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data; statistical analysis. N.J. Newhouse: drafting/revising the manuscript for content, including medical writing for content; acquisition of data; study supervision or coordination. J. Cialone: drafting/revising the manuscript for content, including medical writing for content; acquisition of data. Dr. Levy: drafting/revising the manuscript for content, including medical writing for content; acquisition of data. Dr. Ramirez- Montealegre: drafting/revising the manuscript for content, including medical writing for content; acquisition of data. Dr. Dure: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data; acquisition of data. K.R. Rose: drafting/revising the manuscript for content, including medical writing for content; acquisition of data. Dr. Mink: drafting/revising the manuscript for content, including medical writing for content; study concept or design; analysis or interpretation of data; acquisition of data; study supervision or coordination; obtaining funding.
DISCLOSURE
Dr. Kwon receives research support from the NIH. Dr. Adams serves on a data safety monitoring board for the NIH/NINDS; receives research support from the NIH; and has reviewed records and provided written summary of record review in a medico-legal case. Dr. Rothberg has served on a scientific advisory board for Bio-Reference Laboratories, Inc.; and serves on the editorial boards of Leukemia Research, the Journal of Molecular Diagnostics, and Genetic Testing. Dr. Augustine has received funding from the NIH/NINDS, the FDA, and the Tourette Syndrome Association. Dr. Marshall serves on a data safety monitoring board (DSMB) for and has received funding for travel from Toyama Chemical Co., Ltd.; serves on the editorial board of the European Journal of Neurology; and receives research support from Medivation, Inc., Teva Pharmaceutical Industries Ltd., St. Jude Medical, Inc./Advanced Neuromodulation Systems, Inc., the NIH/NINDS, the US Veterans Administration (DSMB), the BDSRA, the High-Q Foundation, and the Michael J Fox Foundation for Parkinson's Research. E.A. deBlieck receives research support from the NIH. A. Vierhile reports no disclosures. Dr. Beck serves as a biostatistical reviewer for Neurology® and has received research support from Amarin Corporation, Guidant Corporation, Boston Scientific, CHDI Foundation, Inc., the National Parkinson Foundation, and the NIH. N.J. Newhouse reports no disclosures. J. Cialone has received research support from the NIH/NCRR. Dr. Levy and Dr. Ramirez-Montealegre report no disclosures. Dr. Dure serves on the editorial board of the Journal of Child Neurology. K.R. Rose reports no disclosures. Dr. Mink serves as an Associate Editor of Neurology and on the editorial boards of Journal of Child Neurology and Pediatric Neurology; and receives research support from the NIH, the CDC, and the Batten Disease Support and Research Association.
REFERENCES
- 1. Goebel HH, Wisniewski KE. Current state of clinical and morphological features in human NCL. Brain Pathol 2004; 14: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boustany RM. Neurology of the neuronal ceroid- lipofuscinoses: late infantile and juvenile types. Am J Med Genet 1992; 42: 533–535 [DOI] [PubMed] [Google Scholar]
- 3. Consortium TIBD. Isolation of a novel gene underlying Batten disease, CLN3. Cell 1995; 82: 949–957 [DOI] [PubMed] [Google Scholar]
- 4. Mitchison HM, Munroe PB, O'Rawe AM, et al. Genomic structure and complete nucleotide sequence of the Batten disease gene, CLN3. Genomics 1997; 40: 346–350 [DOI] [PubMed] [Google Scholar]
- 5. Phillips SN, Benedict JW, Weimer JM, Pearce DA. CLN3, the protein associated with Batten disease: structure, function and localization. J Neurosci Res 2005; 79: 573–583 [DOI] [PubMed] [Google Scholar]
- 6. Nugent T, Mole SE, Jones DT. The transmembrane topology of Batten disease protein CLN3 determined by consensus computational prediction constrained by experimental data. FEBS Lett 2008; 582: 1019–1024 [DOI] [PubMed] [Google Scholar]
- 7. Kitzmuller C, Haines RL, Codlin S, Cutler DF, Mole SE. A function retained by the common mutant CLN3 protein is responsible for the late onset of juvenile neuronal ceroid lipofuscinosis. Hum Mol Genet 2008; 17: 303–312 [DOI] [PubMed] [Google Scholar]
- 8. Munroe PB, Mitchison HM, O'Rawe AM, et al. Spectrum of mutations in the Batten disease gene, CLN3. Am J Hum Genet 1997; 61: 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lauronen L, Munroe PB, Jarvela I, et al. Delayed classic and protracted phenotypes of compound heterozygous juvenile neuronal ceroid lipofuscinosis. Neurology 1999; 52: 360–365 [DOI] [PubMed] [Google Scholar]
- 10. Sarpong A, Schottmann G, Ruther K, et al. Protracted course of juvenile ceroid lipofuscinosis associated with a novel CLN3 mutation (p. Y199X). Clin Genet 2009; 76: 38–45 [DOI] [PubMed] [Google Scholar]
- 11. Marshall FJ, de Blieck EA, Mink JW, et al. A clinical rating scale for Batten disease: reliable and relevant for clinical trials. Neurology 2005; 65: 275–279 [DOI] [PubMed] [Google Scholar]
- 12. Rothberg PG, Ramirez-Montealegre D, Frazier SD, Pearce DA. Homogeneous polymerase chain reaction nucleobase quenching assay to detect the 1-kbp deletion in CLN3 that causes Batten disease. J Mol Diagn 2004; 6: 260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Little RJA, Rubin DB. Statistical Analysis with Missing Data, Second Edition. Hoboken: John Wiley and Sons; 2002 [Google Scholar]
- 14. Wisniewski KE, Zhong N, Kaczmarski W, et al. Compound heterozygous genotype is associated with protracted juvenile neuronal ceroid lipofuscinosis. Ann Neurol 1998; 43: 106–110 [DOI] [PubMed] [Google Scholar]
- 15. Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 2007; 8: 749–761 [DOI] [PubMed] [Google Scholar]
- 16. Yanjanin NM, Velez JI, Gropman A, et al. Linear clinical progression, independent of age of onset, in Niemann-Pick disease, type C. Am J Med Genet B Neuropsychiatr Genet 2010; 153B 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedman LS, Farmer JM, Perlman S, et al. Measuring the rate of progression in Friedreich ataxia: implications for clinical trial design. Mov Disord 2010; 25: 426–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sampaio C. Can focusing on UPDRS Part II make assessments of Parkinson disease progression more efficient? Nat Clin Pract Neurol 2009; 5: 130–131 [DOI] [PubMed] [Google Scholar]
- 19. Shulman LM, Gruber-Baldini AL, Anderson KE, et al. The evolution of disability in Parkinson disease. Mov Disord 2008; 23: 790–796 [DOI] [PubMed] [Google Scholar]
- 20. Marder K, Zhao H, Myers RH, et al. Rate of functional decline in Huntington's disease. Huntington Study Group. Neurology 2000; 54: 452–458 [DOI] [PubMed] [Google Scholar]
- 21. Ruocco HH, Bonilha L, Li LM, Lopes-Cendes I, Cendes F. Longitudinal analysis of regional grey matter loss in Huntington disease: effects of the length of the expanded CAG repeat. J Neurol Neurosurg Psychiatry 2008; 79: 130–135 [DOI] [PubMed] [Google Scholar]
- 22. Zhong N. Molecular genetic testing for neuronal ceroid lipofuscinoses. Adv Genetics 2001; 45: 141–158 [DOI] [PubMed] [Google Scholar]