Abstract
Omics and bioinformatics are essential to understanding the molecular systems that underlie various plant functions. Recent game-changing sequencing technologies have revitalized sequencing approaches in genomics and have produced opportunities for various emerging analytical applications. Driven by technological advances, several new omics layers such as the interactome, epigenome and hormonome have emerged. Furthermore, in several plant species, the development of omics resources has progressed to address particular biological properties of individual species. Integration of knowledge from omics-based research is an emerging issue as researchers seek to identify significance, gain biological insights and promote translational research. From these perspectives, we provide this review of the emerging aspects of plant systems research based on omics and bioinformatics analyses together with their associated resources and technological advances.
Keywords: Bioinformatics, Data integration, Genome-scale approach, Omics, Systems analysis
Introduction
In the first decade of the 21st century, genomic studies in model plants promoted gene discovery and provided a breadth of gene function knowledge. In the second decade, driven by recent technological advances, omics-based approaches have allowed us to address the complex global biological systems that underlie various plant functions. These technological advances have also accelerated the development of genome-scale resources in applied and emerging model plant species, and have promoted translational research by integrating knowledge across plant species (Mochida and Shinozaki 2010).
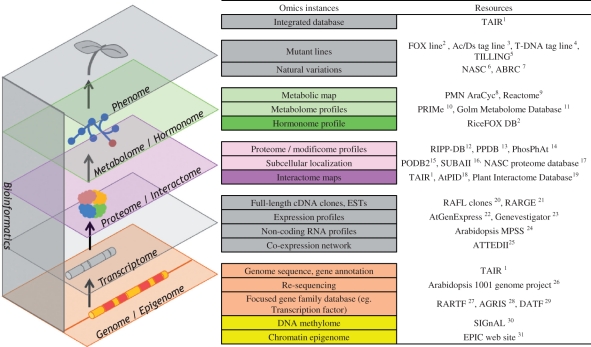
The recent game-changing advances in DNA sequencing technology have revolutionized the sciences field, featuring unprecedented innovations in sequencing scale and throughput, and implementations of various novel applications beyond genome sequencing. In particular, while accelerating genome projects, next-generation sequencing (NGS) technology provides feasible applications such as whole-genome re-sequencing for variation analysis, RNA sequencing (RNA-seq) for transcriptome and non-coding RNAome analysis, quantitative detection of epigenomic dynamics, and Chip-seq analysis for DNA–protein interactions (Lister et al. 2009). In addition to these approaches, which focus on transcriptional regulatory networks, other approaches have been developed, including interactome analysis for networks formed by protein–protein interactions (Arabidopsis Interactome Mapping Consortium 2011), hormonome analysis for phytohormone-mediated cellular signaling (Kojima et al. 2009), and metabolome analysis for metabolic systems (Saito and Matsuda 2010). These rapidly developing omics fields with associated genome-scale resources treat each molecular network as an interlocked component of the plant cellular system (Fig. 1). Bioinformatics has been crucial in every aspect of omics-based research to manage various types of genome-scale data sets effectively and extract valuable knowledge. Integration of accumulating omics outcomes will deepen and update our understanding and facilitate knowledge exchange with other model organisms (Shinozaki and Sakakibara 2009). Therefore, databases for omics outcomes derived from recent innovative analytical platforms comprise the essential infrastructure for systems analysis.
Fig. 1.
An updated omic space with emerging omics layers: epigenome, interactome and hormonome added to each of the closely related layers with illustrative resources for Arabidopsis available on the web. 1http://www.arabidopsis.org/, 2http://ricefox.psc.riken.jp/, 3http://rarge.gsc.riken.jp/dsmutant/index.pl, 4http://signal.salk.edu/tabout.html, 5http://tilling.fhcrc.org/, 6http://arabidopsis.org.uk/home.html, 7http://abrc.osu.edu/, 8http://plantcyc.org/, 9http://www.reactome.org/ReactomeGWT/entrypoint.html, 10http://prime.psc.riken.jp/, 11http://gmd.mpimp-golm.mpg.de/, 12http://phosphoproteome.psc.database.riken.jp/, 13http://ppdb.tc.cornell.edu/, 14http://phosphat.mpimp-golm.mpg.de/, 15http://podb.nibb.ac.jp/Organellome/, 16http://suba.plantenergy.uwa.edu.au/, 17http://proteomics.arabidopsis.info/, 18http://www.megabionet.org/atpid/webfile/, 19http://interactome.dfci.harvard.edu/A_thaliana/, 20http://www.brc.riken.jp/lab/epd/catalog/cdnaclone.html, 21http://rarge.psc.riken.jp/, 22http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp, 23https://www.genevestigator.com/gv/, 24http://mpss.udel.edu/at/, 25http://atted.jp/, 26http://1001genomes.org/index.html, 27http://rarge.psc.riken.jp/rartf/, 28http://arabidopsis.med.ohio-state.edu/, 29http://datf.cbi.pku.edu.cn/, 30http://signal.salk.edu/cgi-bin/methylome, 31https://www.plant-epigenome.org/links.
In this review, we provide an overview of several recently emerged resources derived from a systems approach to omics and bioinformatics analyses in plant science. We describe NGS-based applications and illustrative outcomes of plant genomics. We then review the status of emerging omics topics that have recently appeared in plant omics science, including the interactome, epigenome, hormonome and metabolome. We also highlight the status of genomic resource developments in the families Solanaceae, Gramineae and Fabaceae, each of which includes emerging plant species and/or important applied plant species. We also discuss the integration and visualization of omics data sets as key issues for effective analysis and better understanding of biological insights. Finally, we use omics-based systems analyses to understand plant functions. Throughout this review, we provide examples of recent outcomes in plant omics and bioinformatics, and present an overview of available resources.
Generational shifts in plant genomics by emerging sequencing technologies
Innovation in DNA sequencing technologies that rapidly produce huge amounts of sequence information has triggered a paradigm shift in genomics (Lister et al. 2009). A review on the historical changes in DNA sequencing was provided by Hutchison (2007). A number of so-called NGS are available (Gupta 2008) as widespread platforms, including the 454 FLX (Roche) (Margulies et al. 2005), the Genome Analyzer/Hiseq (Illumina Solexa) (Bennett 2004, Bennett et al. 2005) and the SOLiD (Life Technologies), and newer platforms such as Heliscope (Helicos) (Milos 2008) and PacBio RS (Pacific Biosciences) (Eid et al. 2009) for single molecular sequencing, and Ion Torrent (Life Technologies), based on a semi-conductor chip (Rothberg et al. 2011), are also available. For example, a long reader of NGS, the current version of the Roche 454 platform, is capable of generating ∼1 million reads in an ∼400 bp run, and, a short reader, the current version of the Illumina Hiseq2000, is capable of producing ∼600 Gb of sequence data in 100 bp × 2 paired end reads in a run. A number of reviews on NGS technologies including experimental applications and computational methods have recently been published (Lister et al. 2009, Varshney et al. 2009, Horner et al. 2010, Metzker 2010). We provide an overview of recent NGS-based approaches and outcomes in plants.
Genome and RNA sequencing by NGS
Several plant genomes have recently been sequenced using NGS technology (Table 1) (Huang et al. 2009, S. Sato et al. 2011, Shulaev et al. 2011, Wang et al. 2011, Xu et al. 2011). Since de novo assembly of complex plant genomes remains a challenge, combinatorial approaches using Sanger and/or Roche pyrosequencing methods with other NGS platforms provide more efficient methods of assembly than does a single NGS platform. Re-sequencing coupled with reference genome sequencing data is a pronounced application that fulfills the features of NGS technologies. Rapid acquisition of genome-scale variant data sets enables high-throughput identification of the candidate mutations and alleles associated with phenotypic diversity (DePristo et al. 2011). A methodological pipeline to identify ethyl methanesulfonate (EMS)-induced mutations in Arabidopsis rapidly was developed by taking advantage of the NGS technology (Uchida et al. 2011). DNA polymorphisms such as single nucleotide polymorphisms (SNPs) and insertion–deletion polymorphisms (InDels) have been identified using NGS-based re-sequencing, which enabled the identification of even those polymorphisms in closely related ecotypes and cultivars (Yamamoto et al. 2010, Arai-Kichise et al. 2011). High-throughput polymorphism analysis is an essential tool for facilitating any genetic map-based approach. Genome-wide association study (GWAS) is rapidly becoming an effective approach to dissect the genetic architecture of complex traits in plants (Atwell et al. 2010). The aim of the Arabidopsis 1001 Genomes Project, launched at the beginning of 2008, is to discover the whole-genome sequence variations in 1,001 Arabidopsis strains (accessions) using NGS technologies (http://1001genomes.org). In the first phase of the project, whole-genome re-sequencing of 80 Arabidopsis strains from eight geographic regions revealed 4,902,039 SNPs and 810,467 small InDels (Cao et al. 2011). According to the Arabidopsis 1001 genomes data center, nucleotide polymorphisms of >400 sequenced strains are available (http://www.1001genomes.org/datacenter/). Re-sequencing of plant genomes using NGS technologies has been made possible by decreased costs and increased throughput, improving the efficiency of our analyses of the genotype–phenotype relationship (Rounsley and Last 2010).
Table 1.
Outcomes from next-generation sequencing-based genome sequencing in plants
| Approach | Species | Analytical methods | Sequencing outcomes | Reference |
|---|---|---|---|---|
| Whole-genome shotgun sequencing | Cucumis sativus | De novo genome assembly | 26,682 protein-coding genes predicted | Huang et al. (2009) |
| Fragaria vesca | 34,809 protein-coding genes predicted | Shulaev et al. (2011) | ||
| Jatropha curcas | 40,929 protein-coding genes predicted | S. Sato et al. (2011) | ||
| Solanum tuberosum | 39,031 protein-coding genes predicted | Xu et al. (2011) | ||
| Brassica rapa | 41,174 protein-coding genes predicted | Wang et al. (2011) | ||
| Genome re-sequencing | Oryza sativa cv. Koshihikari | Mapping to reference genome | Identification of 67,051 SNPs | Yamamoto et al. (2010) |
| O. sativa cv. Omashi | Identification of 132,462 SNPs, 16,448 insertions and 19,318 deletions | Arai-Kichise et al. (2011) | ||
| O. sativa (517 rice landraces) | High-density haplotype map generation and GWAS for 14 agronomic traits | Huang et al. (2010) | ||
| Arabidopsis lyrata (pooled sample) | Identification of numerous mutations strongly associated with serpentine adaptation | Turner et al. (2010) | ||
| Arabidopsis thaliana (80 ecotypes) | Whole-genome re-sequencing of 80 Arabidopsis strains from 8 geographic regions. Identification of 4,902,039 SNPs and 810,467 small InDels | Cao et al. (2011) | ||
| Zea mays (6 elite inbred lines) | More than 1,000,000 SNPs, 30,000 InDels and 101 low-sequence-diversity chromosomal intervals | Lai et al. (2010) | ||
| Glycine max (17 wild and 14 cultivars) | Identification of 205,614 tag SNPs | Lam et al. (2010) | ||
NGS-based transcriptome analysis
NGS technologies in RNA-seq are a popular approach to collecting and quantifying the large-scale sequences of coding and non-coding RNAs rapidly (Wang et al. 2009c, Garber et al. 2011). RNA-seq has been used with NGS technologies to identify splicing variants accurately by mapping sequenced fragments onto a reference genome sequence such as in Arabidopsis and rice (Filichkin et al. 2010, Lu et al. 2010). These approaches have yielded information about alternative splicing events in transcriptomes at single base resolution as well as quantitative profiles of each isoform. Because it permits simultaneous acquisition of sequences, expression profiles and polymorphisms, NGS-based RNA-seq has been used for the rapid development of genomic resources in applied and emerging plant species (Table 2) (González-Ballester et al. 2010, Zenoni et al. 2010, Castruita et al. 2011, Gowik et al. 2011). Even when reference genome sequences are not available, de novo assembly from RNA-seq analysis enables rapid acquisition of preliminary genome-scale sequence data to provide early characterization of new plant species (Fu et al. 2011, Su et al. 2011, Wong et al. 2011).
Table 2.
Examples of recent outcomes from next-generation sequencing-based transcriptome sequencing in plants
| Approach | Species | Properties | Reference |
|---|---|---|---|
| RNA-seq with reference genome | A. thaliana | Confirmation of a majority of annotated introns and identification of thousands of novel, alternatively spliced mRNA isoforms | Filichkin et al. (2010) |
| O. sativa indica and japonicus subspecies | 15,708 novel transcriptionally active regions and ∼48% of the genes with alternative splicing patterns. Differential gene expression patterns and SNPs between indica and japonica rice | Lu et al. (2010) | |
| Chlamydomonas reinhardtii | Transcriptome analysis on acclimation to sulfur deprivation. RNA-seq showed larger dynamic range of expression than microarray hybridizations | González-Ballester et al. (2010) | |
| Transcriptome analysis of copper regulation | Castruita et al. (2011) | ||
| Vitis vinifera | Transcriptome analysis during berry development | Zenoni et al. (2010) | |
| RNA-seq with cross-species mapping | Flaveria species | Transcriptome analysis of species ranging from C3 to C3–C4 intermediate to C4. Transcriptome comparison between Flaveria bidentis (C4) and Flaveria pringlei (C3) | Gowik et al. (2011) |
| De novo assembly for RNA-seq data | Orchid species | A large collection of ESTs using NGS and in combination with Sanger sequencing | Fu et al. (2011) |
| Phalaenopsis aphrodite | A large collection of ESTs using Roche 454 pyrosequencing and Illumina Solexa | Su et al. (2011) | |
| Acacia species | Transcriptome data and polymorphism discovery in Acacia auriculiformis and Acacia mangium using NGS | Wong et al. (2011) | |
| Citrullus lanatus | A large collection of ESTs using Roche 454 pyrosequencing | Guo et al. (2011) | |
| Sesamum indicum | A large collection of ESTs using Illumina paired-end sequencing | Wei et al. (2011) |
NGS-based sequencing applications have rapidly expanded in plant genomics (Table 3). By browsing the Sequence Read Archive (SRA) in NCBI (http://www.ncbi.nlm.nih.gov/sra), European Nucleotide Archive (http://www.ebi.ac.uk/ena/home) and DDBJ Sequence Read Archive (http://trace.ddbj.nig.ac.jp/dra/index_e.shtml), all of which store raw sequencing data from NGS platforms, users can determine how thoroughly a given species has been sequenced and retrieve the publicly available sequencing data for further use.
Table 3.
Status of next-generation sequencing technology in plant species (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi, SRA Experiments, September 21, 2011)
| Taxonomic classification (common name) | No. of SRA experiments |
|---|---|
| Chlamydomonas reinhardtii | 131 |
| Physcomitrella patens | 8 |
| Solanum lycopersicum (tomato) | 44 |
| Solanum tuberosum (potato) | 48 |
| Glycine max (soybean) | 160 |
| Phaseolus vulgaris (kidney bean) | 83 |
| Medicago truncatula (barrel medic) | 118 |
| Quercus robur (truffle oak) | 11 |
| Manihot esculenta (cassava) | 20 |
| Populus (poplars) | 20 |
| Brassica napus (rape) | 60 |
| Brassica rapa (field mustard) | 10 |
| Arabidopsis lyrata (lyrate rockcress) | 10 |
| Arabidopsis thaliana (thale cress) | 578 |
| Vitis vinifera (wine grape) | 22 |
| Oryza sativa (rice) | 760 |
| Oryza sativa Japonica group (Japanese rice) | 23 |
| Hordeum vulgare (barley) | 38 |
| Triticum aestivum (bread wheat) | 34 |
| Triticum turgidum (poulard wheat) | 22 |
| Sorghum bicolor (sorghum) | 5 |
| Zea mays (maize) | 161 |
| Viridiplantae (green plants) | 3,745 |
Plant species with ≥5 entries of SRA experiments are basically listed.
Emerging layers in plant omics
Plant epigenome analysis
The epigenome, genome-scale properties of epigenetic modifications, has garnered attention as a new omics area that has been advanced by NGS technology-based solutions (Schmitz and Zhang 2011). Epigenetic modifications in plants can be directed and mediated by small RNAs (sRNAs) (Matzke et al. 2009). The epigenomic regulation of chromatin structure and genome stability is crucial to the interpretation of genetic information (Law and Jacobsen 2010, He et al. 2011). NGS technology-based sequencing of the cytosine methylome (methylC-seq), transcriptome (mRNA-seq) and Small RNA transcriptome (small RNA-seq) in Arabidopsis inflorescences revealed genome-scale methylation patterns and a direct relationship between the location of sRNAs and DNA methylation (Lister et al. 2008). At the whole plant level, a genome-scale map of methylated cytosine in Arabidopsis was generated by bisulfite sequencing using NGS technologies, the so-called BS-seq (Cokus et al. 2008). These NGS-based methylome analyses allowed for holistic understanding of genome-scale methylation patterns at a single base resolution. In addition to DNA methylation, histone N-terminal tail modifications such as acetylation, methylation are crucial in plant development (He et al. 2011) and defense mechanisms (Sokol et al. 2007, J.M. Kim et al. 2008). A genome-wide analysis of the nucleosomes positioning combined with profiles of DNA methylation revealed 10 base periodicities in the DNA methylation status of nucleosome-bound DNA (Chodavarapu et al. 2010).
Chromatin immune precipitation (ChIP) plus NGS technologies or tiling array, the so-called ChIP-seq or ChIP-chip, has been used to generate a genome-scale map of histone modifications. For example, the genome-scale epigenetic map for 8-histone modification—H3K4me2, H3K4me3, H3K27me1, H3K27me2, H3K36me3, H3K56ac, H4K20me1 and H2Bub—was recently generated in Arabidopsis (Roudier et al. 2011). The Epigenomics of Plants International Consortium web site (https://www.plant-epigenome.org/) provides hyperlinks to plant epigenome data resources. In fact, numerous efforts have been made to acquire epigenome information from plant species (Table 4) (Wang et al. 2009b. He et al. 2010).
Table 4.
Epigenome analysis and resources in plant
| Resource | Species | URL | Reference |
|---|---|---|---|
| SIGnAL | Arabidopsis | http://signal.salk.edu/cgi-bin/methylome | Zhang et al. (2006) |
| http://neomorph.salk.edu/epigenome/epigenome.html | Lister et al. (2008) | ||
| Jacobsen lab | Arabidopsis | http://www.mcdb.ucla.edu/research/jacobsen/ LabWebSite/P_EpigenomicsData.shtml | Zhang et al. (2006, 2007a, 2007b, 2009b); Cokus et al. (2008) |
| Yale Plant Genomics | Rice and maize | http://plantgenomics.biology.yale.edu/ | Li et al. (2008); Wang et al. (2009); He et al. 2010 |
| Epigara | Arabidopsis | http://epigara.biologie.ens.fr/cgi-bin/gbrowse/a2e/ | Bouyer et al. (2011); Moghaddam et al. (2011); Roudier et al. (2011) |
Plant interactome analysis
Protein–protein interactions are essential to almost all cellular processes. The interactome, a comprehensive set of all protein–protein interactions in an organism, is crucial to our understanding of the molecular networks underlying cellular systems (Cusick et al. 2005, Morsy et al. 2008). Interactome analysis has been used to characterize plant cellular functions such as the cell cycle, Ca2+/calmodulin-mediated signaling, auxin signaling and membrane protein–signaling protein interactions in Arabidopsis (Boruc et al. 2010, Lalonde et al. 2010, Reddy et al. 2011, Vernoux et al. 2011). The Arabidopsis Interactome Mapping Consortium recently presented a proteome-wide binary protein–protein interaction map of Arabidopsis containing about 6,200 highly reliable interactions between about 2,700 proteins (Arabidopsis Interactome Mapping Consortium 2011).
To generate the large-scale Arabidopsis interactome map, the consortium prepared about 8,000 open reading frames of Arabidopsis protein-coding genes, and then analyzed all pairwise combinations of the proteins encoded by these constructs using an improved high-throughput binary interactome mapping pipeline based on the yeast two-hybrid (Y2H) system. Using the Y2H pipeline method, a large-scale plant pathogen effector interactome network was generated (Mukhtar et al. 2011). In rice, a focused interactome analysis addressed biotic and abiotic stress responses (Seo et al. 2011). A number of databases for protein–protein interaction data sets have been available on the web (Table 5) (Stark et al. 2006, Swarbreck et al. 2008, Aranda et al. 2010). In addition to the curated data sets, predicted protein–protein interaction data sets are a valuable complement to experimental approaches (Cui et al. 2008, Lin et al. 2009, De Bodt et al. 2010, Li et al. 2011, Lin et al. 2011a, Lin et al. 2011b).
Table 5.
Interactome analysis and resources in plants
Plant hormonome analysis
Plant hormones play a crucial role as signaling molecules in the regulation of plant development and environmental responses. To date, a number of low molecular weight plant hormones have been identified, including auxin, ABA, cytokinin, gibberellins, ethylene, brassinosteroids, jasmonates and salicylic acid (Davies 2004). In addition, a novel plant hormone, strigolactone, was recently identified as a shoot branching inhibitor (Gomez-Roldan et al. 2008, Umehara et al. 2008). A special issue of Plant and Cell Physiology on strigolactone was published to collate current knowledge on the subject (Yamaguchi and Kyozuka 2010). Small peptides (peptide hormones) also function as signaling molecules in the regulation of plant growth and development (Matsubayashi and Sakagami 2006, Fukuda et al. 2007). A special issue on peptide hormones was also published to describe recent advances in the area of plant peptide research (Fukuda and Higashiyama 2011).
During the last decade, there have been many remarkable advances in our understanding of the molecular basis of plant hormones, including biosynthesis, transport, perception and response (Santner and Estelle 2009). Umezawa et al. in 2010 provided a review of recent exciting advances in understanding the molecular basis of regulatory networks in the ABA response. One recent remarkable advance was the discovery of the receptors for several important phytohormones, including auxin, gibberellins, ABA and jasmonates (Santner and Estelle 2009). Structural analysis of each complex revealed the structural basis of the interaction between each receptor and phytohormone and their signaling mechanisms (Tan et al. 2007, Shimada et al. 2008, Miyazono et al. 2009, Sheard et al. 2010). According to a number of recent mutant analyses, it is almost certain that all plant hormones cross-talk with one or more other hormones depending on the tissue, developmental stage and environmental changes (Santner and Estelle 2009, Depuydt and Hardtke 2011).
Because of the interplay between multiple plant hormones, a comprehensive analysis known as hormonome analysis, which is based on high-throughput, high-sensitivity and simultaneous profiling of plant hormones, is a key approach to a holistic understanding of the plant hormone network and its association with biological phenomena. A recently developed analytical platform for high-sensitivity, high-throughput measurements of plant hormones enables simultaneous measurement of 43 molecular species of cytokinin, auxin, ABA and gibberellin (Kojima et al. 2009). The platform was used to acquire hormonome profiles of the organ distribution patterns of plant hormones in rice. The hormonome analysis of endogenous levels of cytokinin, gibberellin, ABA and auxin in the wild type and gibberellin signaling mutants indicated that the metabolism of cytokinin, ABA and auxin cross-talk with the gibberellin signaling system. Comprehensive hormone profiling in Arabidopsis was used to analyze the accumulation of ABA, gibberellins, IAA, cytokinins, jasmonates and salicylic acid in the developing seeds of wild-type Arabidopsis and an ABA-deficient mutant. The results of the hormonome approach suggested that ABA interacts with other hormones to regulate seed development (Kanno et al. 2010).
Plant metabolome analysis
Plant metabolomics is now playing a significant role in systems approaches to plant functional analysis and applied plant biotechnology. Driven by advances in related technologies including instruments for metabolite measurement, analytical methodologies and information resources, there are many applications for functional genomics, systems biology and molecular breeding. A number of excellent metabolomics reviews have been published that describe emerging methodologies and attractive applications (Last et al. 2007, Saito and Matsuda 2010, Sumner 2010). Herein, we will briefly review recent advances in analytical platforms and then describe examples of practical applications to understanding plant metabolic systems.
Metabolome analysis deals with chemically diverse compounds. The plant metabolome consists of extremely large varieties of metabolites with various dynamic concentration ranges. Therefore, combined analytical techniques and data set integration from heterogeneous instruments have been key to a comprehensive understanding of diverse metabolites. Streamlined analytical platforms that integrate analytical steps such as sample preparation, data acquisition and data analysis enable us to address the complex plant metabolome (Saito and Matsuda 2010). Improved coverage and throughput for the simultaneous detection of large numbers of metabolites has significantly expanded the practical application of metabolome analysis. A widely targeted metabolomics platform that provides both coverage and throughput utilizes ultra-performance liquid chromatography–tandem quadrupole mass spectrometry (Sawada et al. 2009a). The approach enables us simultaneously to acquire accumulation patterns of hundreds or more metabolites for large numbers of samples. The platform enables us to address complex plant metabolic systems and develop practical approaches in genetics and breeding. The widely targeted metabolomics approach was used for metabolic profiling of the Arabidopsis knockout mutants for methionine chain elongation enzymes. The results suggest that these enzymes are involved in metabolism from methionine to primary and related secondary metabolites (Sawada et al. 2009b).
In addition to targeted metabolomics, non-targeted metabolome analysis with known and unknown metabolites has been important in the generation of comprehensive resources for the plant metabolome. Tandem mass spectrometry (MS/MS) spectral tag (MS2T) analysis, as a non-target metabolome analysis, was used to profile secondary plant metabolites and identify many previously unknown tissue-specific metabolites (Matsuda et al. 2009). The profile of accumulation of known and unknown metabolites was acquired in plant tissues throughout the Arabidopsis life cycle; the data set is available at AtMetExpress (Matsuda et al. 2010).
Metabolome profiling provides a snapshot of the accumulation patterns of metabolites in response to various kinds of biological conditions such as treatments, tissues and genotypes. For example, metabolome profiling approaches have been applied to monitor changing metabolite accumulation in response to stress conditions (Ishikawa et al. 2009, Kusano et al. 2011b). Metabolome profiling has also been applied to evaluate genetic resources, not only in the model plants Arabidopsis and rice but also in various crop species for metabolic phenotyping (Akihiro et al. 2008, Mochida et al. 2009a, Yin et al. 2010, Fujimura et al. 2011, Kusano et al. 2011a). Metabolome profiling has also been used to evaluate the metabolic phenotypes of natural and/or segregation populations. A number of approaches in metabolite quantitative trait locus (mQTL) analysis have been performed in various plant species in recent years (Rowe et al. 2008, Lisec et al. 2009, Do et al. 2010).
A number of information resources have become available in recent years to provide tools for metabolome analysis, such as PRIMe (http://prime.psc.riken.jp/), MeRy-B (http://www.cbib.u-bordeaux2.fr/MERYB/) and MetabolomeExpress (https://www.metabolome-express.org/) (Akiyama et al. 2008, Carroll et al. 2010, Ferry-Dumazet et al. 2011). The review by Saito and Matsuda (2010) provides a well summarized list of web resources for plant metabolomics. Synergistic application of comprehensive, high-throughput experimental procedures and computational approaches as well as genome-scale outcomes from other omics should enable the reconstruction of metabolic systems by metabolic modeling, simulation and theoretical network engineering (Stitt et al. 2010).
Omics resources in emerging plant species
The development of genomic resources has progressed in a number of plant species. As a typical example, genome sequence data sets of 25 plant species are available at Phytozome (ver. 7.0, http://www.phytozome.net/). Recently sequenced plant species were typically nominated for genomic resource development because: (i) they have particular systems not covered by ‘conventional’ model plants; (ii) they are evolutionarily important; or (iii) they provide a commodity resource such as food and energy. Here, we briefly review the current status of available genomic resources for emerging plant species with a focus on examples of the Solanaceae, Poaceae (Gramineae) and Fabaceae (Leguminosae) families.
Solanaceae species
The Solanaceae family includes a number of important agricultural crops, such as tomato, potato, pepper, paprika, petunia and tobacco. The tomato (Solanum lycopersicum) is a representative crop species for which there has been significant progress in genomic resources. The tomato is an important crop that is sold fresh and used in processed foods. In addition to its agricultural importance, the tomato is a model plant for studying Solanaceae species due to its small genome size and shared conserved synteny with other Solanaceae genomes. The tomato has also become a model plant for the study of fruit development, ripening, maturation and metabolic systems. The International Tomato Genome Sequencing Project was initiated in 2004. Following the initial BAC-by-BAC approach for the euchromatic regions, a whole-genome shotgun approach was initiated in 2009. The International Tomato Annotation Group provided the official annotation of the tomato genome assembly (http://solgenomics.net/organism/Solanum_lycopersicum/genome). A full-length cDNA resource from the tomato cultivar Micro-Tom was recently launched (Aoki et al. 2010; http://www.pgb.kazusa.or.jp/kaftom/). Transcriptome data from 296 samples of 16 series using the Affymetrix GeneChip tomato genome array can be found in NCBI GEO (September 8, 2011). The tomato GeneChip data deposited in NCBI GEO includes, for example, data sets acquired for co-expression analysis using cultivar Micro-Tom (Ozaki et al. 2010), for comparative transcriptome analysis between salt-tolerant and salt-sensitive wild tomato species (Sun et al. 2010) and for examining the transcriptome of the ripening process of an orange ripening mutant (Nashilevitz et al. 2010).
Significant progress has been made with metabolome analyses such as metabolome profiling and mQTL analysis (Schauer et al. 2008, Do et al. 2010, Enfissi et al. 2010, Schilmiller et al. 2010, Tieman et al. 2010). Information resources related to metabolome analysis are available and updated, and provide data archives for tomato metabolome data sets and analytical platforms such as Plant MetGenMAP (Joung et al. 2009), The Metabolome Tomato Database (MotoDB) (Moco et al. 2006), KaPPA-View4 SOL (Sakurai et al. 2011a) and KOMICS (Iijima et al. 2008). As an integrative information resource, the Tomato Functional Genomics Database provides data on gene expression, metabolites and microRNAs (miRNAs) through a web interface (Fei et al. 2011). As a tomato mutant resource, TOMATOMA was launched as a web-based database for a Micro-Tom EMS mutant collection with phenotypic classifications and a Targeting Induced Local Lesions IN Genomes (TILLING) resource was established (Okabe et al. 2011, Saito et al. 2011). The potato genome was recently sequenced using a homozygous doubled-monoploid potato clone, and assembly of 86% of the 844 Mb genome revealed 39,031 predicted protein-coding genes (Xu et al. 2011; http://www.potatogenome.net/index.php/Main_Page).
A large-scale expressed sequence tag (EST) analysis of the chili pepper yielded 116,412 ESTs with related annotations that are available at the pepper EST database (http://genepool.kribb.re.kr/pepper/) (Kim et al. 2008). To integrate collected ESTs from Solanaceae species, the SolEST database (D'Agostino et al. 2009) includes ESTs and related annotations from the tomato, potato and pepper (http://biosrv.cab.unina.it/solestdb/index.php). The Sol Genomics Network (SGN; http://solgenomics.net/) is an information portal for Solanaceae research that provides broad information on genomic resources for Solanaceae species and their close relatives (Bombarely et al. 2011).
Poaceae (Gramineae) species
The Poaceae family includes staple food crops such as rice, maize, wheat and barley, as well as grasses used for lignocellulose biomass production, such as switchgrass and Miscanthus (Somerville et al. 2010, Lobell et al. 2011). Since the completion of the japonica rice (Oryza sativa) genome project (International Rice Genome Sequencing Project 2005), whole-genome sequences have been completed in sorghum (Sorghum bicolor), maize (Zea mays) and Brachypodium (Brachypodium distachyon) (Paterson et al. 2009, Schnable et al. 2009, International Brachypodium Initiative 2010). Rice is a model species of monocot plants as well as one of the three major staple cereals in the world. So far, the genome sequence of japonica rice with high quality gene annotations has played a significant role in promoting the development of a number of genomic resources for the discovery and isolation of important genes for application in molecular breeding. The sorghum genome was sequenced as a representative species of the Saccharinae that includes biomass source plants for starch, sugar and cellulose. Maize is another of the major staple cereals for food and animal feed and is a model organism for fundamental research in complex inheritance and genomic properties such as domestication, epigenetics, evolution, chromosome structure and transposable elements (Walbot 2009). Accompanying the release of the maize B73 genome sequence, the ‘2009 Maize Genome Collection’ was edited (Walbot 2009).
Brachypodium is an emerging plant species of the Pooideae subfamily, a model plant for Triticeae crops such as wheat and barley, as well as for understanding systems for cellulose biomass production in grass species. Since the release of the Brachypodium Bd21 genome sequence, Brachypodium has garnered attention, and a number of genomic resource projects have been initiated at various institutions (Brkljacic et al. 2011). We thus have access to published reference genome sequences of four species from each of three important Poaceae subfamilies. Wheat and barley have been the subjects of ongoing genome sequencing attempts to regenerate their highly complex genomes. By incorporation of chromosome sorting, NGS, array hybridization and conserved synteny with the Brachypodium genome, a tentative linear order of 32,000 barley genes was recently regenerated (Mayer et al. 2011). A special issue on barley was recently published to introduce genetic and genomic resources and their applications (Saisho and Takeda 2011).
The large-scale collection of ESTs and cDNA clones has been performed in Poaceae species such as rice, maize, wheat and barley (Mochida and Shinozaki 2010). Those sequence and clone resources have facilitated gene discovery and structural annotation, large-scale expression analysis, genome-scale interspecific and intraspecific comparative analysis of expressed genes, and the design of expressed gene-oriented molecular markers and probes for microarrays (Mochida et al. 2003, Ogihara et al. 2003, Zhang et al. 2004, Kawaura et al. 2006, Mochida et al. 2006, Mochida et al. 2008, Sato et al. 2009a). Large-scale analyses of full-length cDNA sequences and clone resources have been performed in rice, maize, wheat and barley (Kikuchi et al. 2003, Liu et al. 2007, Kawaura et al. 2009, Sato et al. 2009b, Soderlund et al. 2009, Matsumoto et al. 2011). Full-length cDNA resources have been crucial for the annotation of the sequenced genome (Tanaka et al. 2008), large-scale gene discovery and functional analyses by creating transgenic plants (Kondou et al. 2009), and comparative analysis based on the entire sequence of transcripts (Mochida et al. 2009b) in grass species.
Data sets of transcriptome profiles are available for several Poaceae species. For example, the current version of Genevestigator provides processed transcriptome data from GeneChip hybridization data sets (1,626 for barley, 1275 for rice, 1000 for wheat and 458 for maize; https://www.genevestigator.com/gv/). The transcriptome throughout the reproductive process from primordia development through pollination/fertilization to zygote formation was analyzed in rice using an oligomicroarray as a rice expression atlas (Fujita et al. 2010). Accumulated transcriptome data sets have been made for co-expression analysis of the transcriptome in Poaceae species. The RiceArrayNet and OryzaExpress databases provide web-accessible co-expression data for rice (Lee et al. 2009, Hamada et al. 2011). The ATTED-II database also provides co-expression data sets for rice in addition to those for Arabidopsis (Obayashi et al. 2011). A co-expressed barley gene network was recently generated and then applied to comparative analysis to discover potential Triticeae-specific gene expression networks (Mochida et al. 2011a).
Microarray analyses coupled with laser microdissection have been applied to analyze transcriptomes of the male gametophyte and tapetum in rice (Hobo et al. 2008, Suwabe et al. 2008). Transcriptome data were recently collected from rice grown in field environments (Y. Sato et al. 2011). As a proteomics resource in Poaceae, for example, the plant proteome database (http://ppdb.tc.cornell.edu/) provides information on the maize and Arabidopsis proteomes. The RIKEN Plant Phosphoproteome Database (RIPP-DB, http://phosphoproteome.psc.database.riken.jp) was updated with a data set of large-scale identification of rice phosphorylated proteins (Nakagami et al. 2010, Nakagami et al. 2011). The OryzaPG-DB was launched as a rice proteome database based on shotgun proteomics (Helmy et al. 2011).
To date, various types of mutant resources have been developed and used to investigate gene functions in Poaceae species (Krishnan et al. 2009, Kuromori et al. 2009). In Brachypodium, a large-scale collection of T-DNA tagged lines have been generated and termed ‘the BrachyTAG program’ (Thole et al. 2010). As a recently developed novel gain-of-function system, the FOX (full-length cDNA overexpressor) gene hunting system combines overexpressor production with the large-scale resources of full-length cDNA clones (Kondou et al. 2009). The rice full-length cDNA overexpressed Arabidopsis mutant database (Rice FOX Database, http://ricefox.psc.riken.jp/) is a new information resource for the FOX line (Sakurai et al. 2011b). Driven by recent technological advances for genome-scale polymorphisms, mapping populations and natural variations have gained importance in identification of the genes associated with environmental adaptation through evolutionary history. Nested association mapping using recombinant inbred lines coupled with a high density haplotype map was used to identify genetic loci associated with quantitative traits (Kump et al. 2011, Poland et al. 2011, Tian et al. 2011). An analytical platform for genome-wide SNP genotyping is also available for barley and has been used to survey genomic variations among barley germplasms and to evaluate chromosomal distribution of introgressed segments of near-isogenic lines (Druka et al. 2011). A collection of natural barley variants was used to investigate the associations between nucleotide haplotypes and growth habits that were superimposed onto a geographical distribution (Saisho et al. 2011).
Fabaceae (Leguminosae) species
The large Fabaceae family includes economically important legume crops such as the soybean, common bean and alfalfa. The symbiotic nitrogen fixation that is produced by communication between plants and nitrogen-fixing bacteria is a biological phenomenon, particularly in legume species. Understanding this symbiosis is important for plant and microbial biology as well as for agriculture. Recent progress in symbiosis research between plants and microbes, including nitrogen-fixing symbiosis in legume plants, was presented in the reviews by Ikeda et al. (2010) and Kouchi et al. (2010) that appeared in a special issue on plant–microbe symbiosis (Kawaguchi and Minamisawa 2010). To investigate the symbiotic system and perform gene discovery in legume species, Lotus japonicus and Medicago truncatula served as models for molecular genetics and functional genomics studies (Stacey et al. 2006). In 2008, the genome sequence of L. japonicus was released with a 315.1 Mb sequence corresponding to 67% of the genome, covering 91.3% of the gene space (Sato et al. 2008). The TILLING resource for L. japonicus is used to identify allelic series for symbiosis genes (Perry et al. 2009). Proteome analyses on pod and seed development were performed in L. japonicus (Dam et al. 2009, Nautrup-Pedersen et al. 2010).
In M. truncatula, a number of genome resources have become available in recent years (Young and Udvardi 2009). For example, a gene expression atlas provides an information resource for the transcriptome (Benedito et al. 2008). Insertional mutagenesis by the Tnt1 transposon system and the flanking sequence data set have provided a reverse genetics resource (Tadege et al. 2008). The web site of the Medicago truncatula Genome Project in JCVI/TIGR (http://medicago.jcvi.org/cgi-bin/medicago/annotation.cgi) is an information resource that provides the current version of pseudomolecules (ver. 3.5) and an annotation of the M. truncatula genome. The web page of the Medicago truncatula HapMap Project (http://medicagohapmap.org/index.php) provides not only a reference genome sequence but also NGS re-sequencing data as a GWAS resource. sRNAs expressed in roots and nodules were analyzed using 454 pyrosequencing, and the MIRMED database (http://medicago.toulouse.inra.fr/Mt/RNA/MIRMED/LeARN/cgi-bin/learn.cgi) was constructed as an informative resource for M. truncatula miRNAs (Lelandais-Briere et al. 2009). Large-scale analysis of the phosphoproteome in M. truncatula roots was performed using immobilized metal affinity chromatography and MS/MS followed by launch of the Medicago PhosphoProtein Database (http://www.phospho.medicago.wisc.edu/db/index.php) (Grimsrud et al. 2010).
The soybean (Glycine max), the most important global legume crop, is widely grown for food and biofuel. A draft soybean genome assembly was released in 2010, and the first version of the genome annotation Glyma1.0 was built by homology and by ab initio-based gene predictions (Schmutz et al. 2010). A sequence data set of soybean full-length cDNAs was also favorably applied for the homology-based gene prediction (Umezawa et al. 2008). Several genomic resources are available for soybean genomics as well as for molecular breeding for improved productivity and stress tolerance (Manavalan et al. 2009). A recent review was published by Tran and Mochida (2010). By using the soybean genome sequence and annotated gene models, genome-scale exploration of gene families and those functional analyses have been performed to identify genes for molecular breeding (Mochida et al. 2009c, Mochida et al. 2010a, Mochida et al. 2010b, Le et al. 2011a, Le et al. 2011b). A transcriptome atlas of the soybean (http://digbio.missouri.edu/soybean_atlas/) has been developed using an NGS platform to perform RNA-seq of samples from 14 distinct conditions (Libault et al. 2010). NGS-based approaches were also applied to a genome-scale survey of sRNAs (Joshi et al. 2011, Song et al. 2011). As an information portal for soybean research, Soybase (http://soybase.org/) has played a significant role in integrating various resources and analytical platforms for soybean research (Grant et al. 2010).
Data integration to progress plant omics research
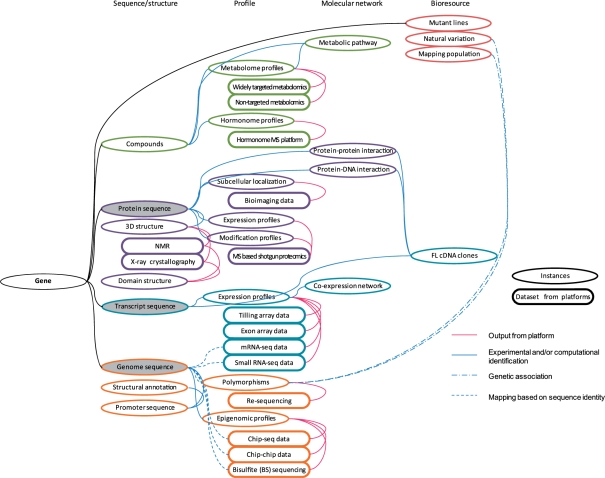
Recent remarkable innovations in omics platform research have produced a wealth of genome-scale data in the life sciences. The key problem in omics research now is developing ways to deal with such huge and heterogeneous data sets. A conceptual relationship of omics data sets inter-related with a gene is represented as an example for the integration of omics data sets (Fig. 2). Information resources such as databases and computational tools are becoming more and more important for effectively handling genome-scale data sets. Data storage for omics data sets must ensure persistence and retrieval functionalities for shared use. To integrate heterogeneous data sets effectively, the formation of procedures to generate inter-relationships between data sets and consolidate the data structure for each feature is another goal. User interface and visualization techniques to allow users to perform heuristic data mining and to be inspired by integrated genome-scale data sets are also important. Many information resources are available to facilitate plant science (Mochida and Shinozaki 2010). Here, we describe representative methods and visualizations of genome-scale data sets together with examples of currently available resources.
Fig. 2.
An example of the schematic relationship between biological instances gained from omics analysis that are associated with a gene. Each instance is roughly classified into four data set types: sequence/structure, profile, molecular network and bioresource. Shaded nodes are sequence-oriented instances in similarity searches. Each instance is inter-related based on experimental and/or computational identification, genetic association and sequence mapping.
Creating relationships using various kinds of data sets
With the completion of a number of plant genome sequences, genome-scale evolutionary and comparative analyses have allowed us to identify conserved and/or characteristic properties among plant species. Comparative genomics resources integrate genome sequences from multiple organisms and associated information with evolutionary insights. For example, PLAZA, an online platform for plant comparative genomics (http://bioinformatics.psb.ugent.be/plaza/), provides structural and functional annotations of published plant genomes and analytical tools (Proost et al. 2009). The schematic representation of the relationships between the data types and tools of the PLAZA platforms is illustrative of the current state of data integration in comparative genomics. As another example, the SALAD database (http://salad.dna.affrc.go.jp/salad/) provides comparative information on 10 sequenced plant species based on the organization of shared sequence motifs (Mihara et al. 2010). The SALAD database was used to discover candidate cis-regulatory elements by synergistic use of the phylogenetic relationships of genes with gene expression profiles and upstream flanking sequence features (Mihara et al. 2008).
Comparative analyses of plant species have also been performed on the transcriptome. PlaNet (http://aranet.mpimp-golm.mpg.de/), a database of co-expression networks for Arabidopsis and six plant crop species, uses a comparative network algorithm, NetworkComparer, to estimate similarities between network structures (Mutwil et al. 2011). The PlaNet platform integrates gene expression patterns, associated functional annotations and MapMan term-based ontology, and facilitates knowledge transfer from Arabidopsis to crop species for the discovery of conserved co-expressed gene networks.
The metabolic pathway map could be a knowledge foundation to superimpose profiles of each compound, proteome and transcriptome of the genes involved in each pathway. The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) is one of the most long established integrated information resources. The KEGG PLANT Resource provides information on biosynthetic pathways, sequenced plant genomes and phytochemicals, and it aims to integrate genomic information resources with the biosynthetic pathways of natural plant products (Masoudi-Nejad et al. 2007). Another information resource for biosynthetic pathways, PlantCyc, contains curated information from the literature and from computational analyses of the genes, enzymes, compounds, reactions and pathways involved in primary and secondary plant metabolism. PlantCyc recently added PoplarCyc for Populus trichocarpa in addition to AraCyc for Arabidopsis (Zhang et al. 2010), which is based on the BioCyc platform (Caspi et al. 2010). This platform has been used for a number of plant species. The pathways section in the Gramene databases provides RiceCyc, MaizeCyc, BrachyCyc and SorghumCyc, for rice, maize, Brachypodium and sorghum, respectively (http://www.gramene.org/pathway/). The web page also provides mirrors of species-specific pathway databases from Arabidopsis, tomato, potato, coffee, Medicago, Escherichia coli, and the MetaCyc and PlantCyc reference databases, enabling us to perform interspecific comparison between pathways (Youens-Clark et al. 2010).
Sequence identity- and/or similarity-based integration has been widely used to build cross-reference data sets between individual genes and their associated references, such as gene–genomic structure, gene–transcript/cDNA clones–protein, gene–transcript expression pattern and gene–polymorphism/mutants–phenotype. For example, an integrative database for the genes encoding transcription factors of the Gramineae species, GramineaeTFDB (http://gramineaetfdb.psc.riken.jp/) provides genomic sequence features, promoter regions, domain alignments, GO assignments, full-length cDNAs and gene expression profiles, and cross-references to various public databases and genetic resources (Mochida et al. 2011b). Genome sequence-oriented data integration is essential to the representation of genomic characteristics such as whole-genome transcription, genome-wide methylome patterns and genome-wide polymorphisms (Fig. 3). The ARabidopsis Tiling-Array-based Detection of Exons database 2 (http://artade.org) provides precisely predicted gene structures by tiling array-based transcriptome data sets (Iida et al. 2011). The database also infers gene function based on over-representation in gene model co-expression analyses. The SIGnAL Arabidopsis Methylome Mapping Tool (http://signal.salk.edu/cgi-bin/methylome) integrates existing Arabidopsis gene annotation and methylome information at single base pair resolution. The PhosPhAt database of Arabidopsis phosphorylation sites (http://phosphat.mpimp-golm.mpg.de/index.html) is built on mass spectrometry data (Durek et al. 2009). The database contains a collection of phosphopeptides and phosphorylation sites assigned to annotated Arabidopsis genes, and hyperlinks to external information resources such as The Arabidopsis Information Resource (TAIR). The RNA-editing site on protein 3D structure is a unique database of RNA-editing sites found in plant organelle genes with the results mapped onto amino acid sequences and 3D structures (Yura et al. 2009).
Fig. 3.
An example of integrated biological instances in a genome sequence. A gene annotation and related data, phosphorylation site, cDNA/EST, homology-based mapping, mutants and polymorphisms were retrieved from TAIR. Methylome data sets were retrieved from the Arabidopsis Epigenetics & Epigenomics group's web site. Each data set has been allocated onto the genomic region defined as a TAIR8 annotation.
Literature mining for gene associations is also an essential approach to knowledge integration in the life sciences (Krallinger et al. 2008, Winnenburg et al. 2008). PLAN2L (plant annotation to literature, http://zope.bioinfo.cnio.es/plan2l) is a web-based online application that integrates literature-derived bioentities and associated information in Arabidopsis (Krallinger et al. 2009). PosMed-plus (positional Medline for plant upgrading science, http://omicspace.riken.jp/PosMed-plus/) is a web-accessible tool that assists in candidate selection for positional cloning in plants (Makita et al. 2009). The accuracy of PosMed-plus is correlated with its ability to make correct associations between genes and documents that are based on direct searches, inference searches by co-citation and manual curation.
Visualization of plant omics data sets
Observation is the most basic approach in biology. Therefore, visualization of genome-scale data sets is an important task in bioinformatics. Gehlenborg et al. (2010) reviewed a number of practical tools for omics data visualization and integration. The goal of omics data visualization should be to create clear, meaningful and integrated resources without being overwhelmed by the intrinsic complexity of the data (Gehlenborg et al. 2010). We must remove scale dependency and implement seamless navigation between associated data to uncover their true value as a heuristic information resource.
The networks formed by associated biomolecules such as protein–protein interactions and co-expressed genes can be visualized to gain biological insights from genome-scale data sets of experimentally confirmed and/or computationally predicted molecular associations. For example, the node graph-based network visualization of co-expressed genes has been widely used in various plant species and has been implemented in web-accessible information resources such as ATTED-II, OryzaExpress and PlaNet (Hamada et al. 2011, Mutwil et al. 2011, Obayashi et al. 2011). SeedNet and SCoPNet, at The Virtual Seed Web Resource (http://vseed.nottingham.ac.uk/), are genome-wide network models of transcriptional interactions in dormant and germinating Arabidopsis seeds (Bassel et al. 2011a, Bassel et al. 2011b). AtCAST (Arabidopsis thaliana: DNA Microarray Correlation Analysis Tool, http://atpbsmd.yokohama-cu.ac.jp/cgi/network/home.cgi) is a unique information resource that combines multiple Arabidopsis microarray experiments into a single network (Sasaki et al. 2010).
Visualization of the spatio-temporal accumulation of biomolecules allows us to infer the biological significance of accumulated molecules and their function in different developmental stages, environmental conditions and tissues. The electronic fluorescent pictograph (eFP) browser (http://www.bar.utoronto.ca/) illustrates the visualization of spatio-temporal accumulation patterns of biomolecules (Winter et al. 2007). The current version of the Arabidopsis eFP browser consists of gene expression patterns of different developmental stages, tissues, stresses and natural variations. In addition to Arabidopsis, eFP browsers are available for poplar, M. truncatula, rice, barley, maize and soybean.
The genome browser is a key tool that integrates sequence-based information with genomic position. Several tools such as the genome browser are available in various web-accessible resources. For example, Gbrowse is a popular genome browser that is widely used on a number of web sites, such as TAIR, to visualize genome annotations (Podicheti and Dong 2011). With the increase in NGS-based sequencing data, the types of genomic features that can be visualized throughout genome sequences have been expanded and include, for example, transcript abundances based on RNA-seq, polymorphisms based on whole-genome re-sequencing, and quantitative Chip-seq data (Fig. 3). Circos (http://circos.ca/) is another genome browser that visualizes genome(s) in a circular layout. The circular layout of Circos facilitates visual annotations of the circular genomes of microbes as well as the structural relationships between chromosomal regions such as whole-genome duplications and syntenic relationships (Krzywinski et al. 2009).
Imaging provides immediate visualization of biological information. Along with technological advances in bioimaging, the informatics of bioimaging data sets is an emerging field (Moore et al. 2008). In plant cell biology, advanced imaging technologies allow us to visualize organelles and biomolecules and characterize cellular systems based on large-scale data sets of images and movies (Mano et al. 2009). Image databases in plant science were reviewed by Mano et al. in 2009. For example, the aim of the plant organelles database (http://podb.nibb.ac.jp/Organellome) is to promote the understanding of organelle dynamics such as organelle function, biogenesis, differentiation, movement and interactions with other organelles (Mano et al. 2008, Mano et al. 2011). Furthermore, comprehensive acquisitions of cellular images and movies are essential in the development of information resources to address computationally quantitative characterizations of plant cellular properties. Recently, various computational procedures have been developed to analyze cellular image data sets. For example, a computational algorithm based on machine learning and statistical modeling was applied to recognize subcellular localization of proteins in microscope images automatically (Hu et al. 2010). Multi-angle image acquisition, three-dimensional reconstruction and cell segmentation-automated lineage tracking was developed and applied to perform quantitative analysis of Arabidopsis flower development at cell resolution (Fernandez et al. 2010).
Systems analysis of plant functions
Systems analysis based on a combination of multiple omics analyses has been an efficient approach to determining the global picture of cellular systems. From the early period of plant metabolomics research, we have achieved a number of significant advances in our understanding of gene function in metabolic systems by integrating metabolome analysis with genome and transcriptome resources (Hirai et al. 2004, Tohge et al. 2005, Hirai et al. 2007, Hirai and Saito 2008, Saito et al. 2008, Watanabe et al. 2008, Yonekura-Sakakibara et al. 2008, Okazaki et al. 2009). Following these successes, multi-omics-based systems analyses have improved our understanding of plant cellular systems. For instance, integrated metabolome and transcriptome analyses were recently applied to analyze rice developing caryopses under high temperature conditions (Yamakawa and Hakata 2010), molecular events underlying pollination-induced and pollination-independent fruit sets (Wang et al. 2009a) and the effects of DE-ETIOLATED1 down-regulation in tomato fruits (Enfissi et al. 2010). Integrated metabolome and transcriptome analysis has also been applied to investigate changing metabolic systems in plants growing in field conditions, such as the rice Os-GIGANTEA (Os-GI) mutant and transgenic barley (Kogel et al. 2010, Izawa et al. 2011). Furthermore, a systems approach combined hormonome, metabolome and transcriptome analyses in Arabidopsis transgenic lines, displaying increased leaf growth to gain insight into the molecular mechanisms that control leaf size (Gonzalez et al. 2010).
An integrated proteome and metabolome analysis was applied to compare the differences in response to anoxia between rice and wheat coleoptiles (Shingaki-Wells et al. 2011). An integrated transcriptome, proteome and metabolome analysis was conducted to characterize the cascading changes in UV-B-mediated responses in maize (Casati et al. 2011). These illustrative examples demonstrate the power of multi-omics-based systems analysis for understanding the key components of cellular systems underlying various plant functions.
Thanks to recent technical advances of high-throughput and genome-scale genotyping platforms such as whole-genome re-sequencing, comprehensive exploration of the association between genomic diversity and quantitative instances in various omics aspects has facilitated the discovery of key genes involved in adaptive changes in various omics levels as another combinatorial approach (Kroymann 2011). Using a large set of Arabidopsis accessions and genome-scale variation data sets, GWAS identified genetic loci associated with enzyme activities, metabolome profiles and biomass (Chan et al. 2010, Sulpice et al. 2010). The hormonal responses of natural variations have been addressed to find relationships between physiological variations of hormonal response and other variations, such as in the genome and transcriptome (Delker et al. 2010). A combinatorial approach to population genomics using hormonome profiling would allow us to identify the association between genomic polymorphisms and plant hormone abundance as quantitative traits that might be closely related to environmental adaptation. Recently, relational instances of epigenomic modification, coding gene transcription and non-coding RNAs have been coupled with genome-scale nucleotide polymorphism data sets in human population genomics (Sigurdsson et al. 2009, Shoemaker et al. 2010). In a similar fashion, plant epigenome analysis can also be integrated with genome-scale variations to provide important clues to the epigenetic and genetic regulation associated with phenotypic diversity.
During the past few years, we have witnessed significant advances in plant omics, as shown in this review. Technological advances in analytical platforms are providing genome-scale outcomes, revitalizing our knowledge and opportunities to address long-standing basic questions in plant science. At the same time, we are also facing various global issues such as water, food and energy security; global warming; and climate changes. Understanding the specific plant functions that appear in particular plant species is also important to discover useful genes to improve plant functions. The integration of a wide spectrum of omics data sets from various plant species is then essential to promote translational research to engineer plant systems in response to the emerging demands of mankind.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan [Grant-in-Aid for Scientific Research on Innovative Areas (23119524) to K.M.].
Acknowledgments
The authors thank Tetsuya Sakurai, Takuhiro Yoshida, Lam-Son Phan Tran and Yuji Sawada of the RIKEN Plant Science Center, and Kei Iida of RIKEN BASE for their valuable suggestions and comments. The authors also thank Daisuke Saisho of Okayama University for critical reading of the manuscript.
Glossary
Abbreviations
- ChIP
chromatin immunoprecipitation
- EMS
ethyl methanesulfonate
- EST
expressed sequence tag
- FOX
full-length cDNA overexpressor
- GWAS
genome wide association study
- InDel
insertion–deletion polymorphism, miRNA, microRNA
- mQTL
metabolite quantitative trait locus
- NGS
next-generation sequencing, RNA-seq, RNA sequencing
- SNP
single nucleotide polymorphism
- SRA
Sequence Read Archive
- sRNA
small RNA
- TILLING
targeting induced local lesions in genomes
- Y2H
yeast two-hybrid.
References
- Akihiro T, Koike S, Tani R, Tominaga T, Watanabe S, Iijima Y, et al. Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol. 2008;49:1378–1389. doi: 10.1093/pcp/pcn113. [DOI] [PubMed] [Google Scholar]
- Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, et al. PRIMe: a Web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008;8:339–345. [PubMed] [Google Scholar]
- Aoki K, Yano K, Suzuki A, Kawamura S, Sakurai N, Sud K, et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics. 2010;11:210. doi: 10.1186/1471-2164-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai-Kichise Y, Shiwa Y, Nagasaki H, Ebana K, Yoshikawa H, Yano M, et al. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of Japonica rice by whole-genome sequencing. Plant Cell Physiol. 2011;52:274–282. doi: 10.1093/pcp/pcr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Glaab E, Marquez J, Holdsworth MJ, Bacardit J. Functional network construction in Arabidopsis using rule-based machine learning on large-scale data sets. Plant Cell. 2011a;23:3101–3116. doi: 10.1105/tpc.111.088153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, et al. Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl Acad. Sci. USA. 2011b;108:9709–9714. doi: 10.1073/pnas.1100958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, et al. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Mylle E, Duda M, De Clercq R, Rombauts S, Geelen D, et al. Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol. 2010;152:553–565. doi: 10.1104/pp.109.148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011;7:e1002014. doi: 10.1371/journal.pgen.1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, et al. Brachypodium as a model for the grasses: today and the future. Plant Physiol. 2011;157:3–13. doi: 10.1104/pp.111.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Badger MR, Harvey Millar A. The MetabolomeExpress Project: enabling web-based processing, analysis and transparent dissemination of GC/MS metabolomics datasets. BMC Bioinformatics. 2010;11:376. doi: 10.1186/1471-2105-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Campi M, Morrow DJ, Fernandes JF, Walbot V. Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics. 2011;12:321. doi: 10.1186/1471-2164-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, et al. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23:1273–1292. doi: 10.1105/tpc.111.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Kliebenstein DJ. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics. 2010;185:991–1007. doi: 10.1534/genetics.109.108522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li P, Li G, Xu F, Zhao C, Li Y, et al. AtPID: Arabidopsis thaliana protein interactome database—an integrative platform for plant systems biology. Nucleic Acids Res. 2008;36:D999–D1008. doi: 10.1093/nar/gkm844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14 Spec No. 2:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- D'Agostino N, Traini A, Frusciante L, Chiusano ML. SolEST database: a ‘one-stop shop’ approach to the study of Solanaceae transcriptomes. BMC Plant Biol. 2009;9:142. doi: 10.1186/1471-2229-9-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam S, Laursen BS, Ornfelt JH, Jochimsen B, Staerfeldt HH, Friis C, et al. The proteome of seed development in the model legume Lotus japonicus. Plant Physiol. 2009;149:1325–1340. doi: 10.1104/pp.108.133405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ, editor. Plant Hormones: Biosynthesis, Signal Transduction, Action. Dordrecht: Kluwer Academic Publishers; 2004. [Google Scholar]
- De Bodt S, Carvajal D, Hollunder J, Van den Cruyce J, Movahedi S, Inze D. CORNET: a user-friendly tool for data mining and integration. Plant Physiol. 2010;152:1167–1179. doi: 10.1104/pp.109.147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Poschl Y, Raschke A, Ullrich K, Ettingshausen S, Hauptmann V, et al. Natural variation of transcriptional auxin response networks in Arabidopsis thaliana. Plant Cell. 2010;22:2184–2200. doi: 10.1105/tpc.110.073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS. Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 2011;21:R365–R373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Ding X, Richter T, Chen M, Fujii H, Seo YS, Xie M, et al. A rice kinase–protein interaction map. Plant Physiol. 2009;149:1478–1492. doi: 10.1104/pp.108.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do PT, Prudent M, Sulpice R, Causse M, Fernie AR. The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol. 2010;154:1128–1142. doi: 10.1104/pp.110.163030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K, et al. Genetic dissection of barley morphology and development. Plant Physiol. 2011;155:617–627. doi: 10.1104/pp.110.166249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, et al. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2009;38:D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Enfissi EM, Barneche F, Ahmed I, Lichtle C, Gerrish C, McQuinn RP, et al. Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell. 2010;22:1190–1215. doi: 10.1105/tpc.110.073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z, Joung JG, Tang X, Zheng Y, Huang M, Lee JM, et al. Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 2011;39:D1156–D1163. doi: 10.1093/nar/gkq991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, et al. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat. Methods. 2010;7:547–553. doi: 10.1038/nmeth.1472. [DOI] [PubMed] [Google Scholar]
- Ferry-Dumazet H, Gil L, Deborde C, Moing A, Bernillon S, Rolin D, et al. MeRy-B: a web knowledgebase for the storage, visualization, analysis and annotation of plant NMR metabolomic profiles. BMC Plant Biol. 2011;11:104. doi: 10.1186/1471-2229-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CH, Chen YW, Hsiao YY, Pan ZJ, Liu ZJ, Huang YM, et al. OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol. 2011;52:238–243. doi: 10.1093/pcp/pcq201. [DOI] [PubMed] [Google Scholar]
- Fujimura Y, Kurihara K, Ida M, Kosaka R, Miura D, Wariishi H, et al. Metabolomics-driven nutraceutical evaluation of diverse green tea cultivars. PLoS One. 2011;6:e23426. doi: 10.1371/journal.pone.0023426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Horiuchi Y, Ueda Y, Mizuta Y, Kubo T, Yano K, et al. Rice expression atlas in reproductive development. Plant Cell Physiol. 2010;51:2060–2081. doi: 10.1093/pcp/pcq165. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Higashiyama T. Diverse functions of plant peptides: entering a new phase. Plant Cell Physiol. 2011;52:1–4. doi: 10.1093/pcp/pcq193. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Hirakawa Y, Sawa S. Peptide signaling in vascular development. Curr. Opin. Plant Biol. 2007;10:477–482. doi: 10.1016/j.pbi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- Gehlenborg N, O'Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, et al. Visualization of omics data for systems biology. Nat. Methods. 2010;7:S56–68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR. RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell. 2010;22:2058–2084. doi: 10.1105/tpc.109.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhont S, et al. Increased leaf size: different means to an end. Plant Physiol. 2010;153:1261–1279. doi: 10.1104/pp.110.156018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Brautigam A, Weber KL, Weber AP, Westhoff P. Evolution of C4 photosynthesis in the genus flaveria: how many and which genes does it take to make C4? Plant Cell. 2011;23:2087–2105. doi: 10.1105/tpc.111.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D, Nelson RT, Cannon SB, Shoemaker RC. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010;38:D843–D846. doi: 10.1093/nar/gkp798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsrud PA, den Os D, Wenger CD, Swaney DL, Schwartz D, Sussman MR, et al. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 2010;152:19–28. doi: 10.1104/pp.109.149625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zhu P, Jiao Y, Meng Y, Chen M. PRIN: a predicted rice interactome network. BMC Bioinformatics. 2011;12:161. doi: 10.1186/1471-2105-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Liu J, Zheng Y, Huang M, Zhang H, Gong G, et al. Characterization of transcriptome dynamics during watermelon fruit development: sequencing, assembly, annotation and gene expression profiles. BMC Genomics. 2011;12:454. doi: 10.1186/1471-2164-12-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK. Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 2008;26:602–611. doi: 10.1016/j.tibtech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Hamada K, Hongo K, Suwabe K, Shimizu A, Nagayama T, Abe R, et al. OryzaExpress: an integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 2011;52:220–229. doi: 10.1093/pcp/pcq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Elling AA, Deng XW. The epigenome and plant development. Annu. Rev. Plant Biol. 2011;62:411–435. doi: 10.1146/annurev-arplant-042110-103806. [DOI] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy M, Tomita M, Ishihama Y. OryzaPG-DB: rice proteome database based on shotgun proteogenomics. BMC Plant Biol. 2011;11:63. doi: 10.1186/1471-2229-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Saito K. Analysis of systemic sulfur metabolism in plants using integrated ‘-omics’ strategies. Mol. Biosyst. 2008;4:967–973. doi: 10.1039/b802911n. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Suwabe K, Aya K, Suzuki G, Yano K, Ishimizu T, et al. Various spatiotemporal expression profiles of anther-expressed genes in rice. Plant Cell Physiol. 2008;49:1417–1428. doi: 10.1093/pcp/pcn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner DS, Pavesi G, Castrignano T, De Meo PD, Liuni S, Sammeth M, et al. Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Brief. Bioinform. 2010;11:181–197. doi: 10.1093/bib/bbp046. [DOI] [PubMed] [Google Scholar]
- Hu Y, Osuna-Highley E, Hua J, Nowicki TS, Stolz R, McKayle C, et al. Automated analysis of protein subcellular location in time series images. Bioinformatics. 2010;26:1630–1636. doi: 10.1093/bioinformatics/btq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Hutchison CA., 3rd DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 2007;35:6227–6237. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Kawaguchi S, Kobayashi N, Yoshida Y, Ishii M, Harada E, et al. ARTADE2DB: improved statistical inferences for Arabidopsis gene functions and structure predictions by dynamic structure-based dynamic expression (DSDE) analyses. Plant Cell Physiol. 2011;52:254–264. doi: 10.1093/pcp/pcq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Nakamura Y, Ogata Y, Tanaka K, Sakurai N, Suda K, et al. Metabolite annotations based on the integration of mass spectral information. Plant J. 2008;54:949–962. doi: 10.1111/j.1365-313X.2008.03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Okubo T, Anda M, Nakashita H, Yasuda M, Sato S, et al. Community- and genome-based views of plant-associated bacteria: plant–bacterial interactions in soybean and rice. Plant Cell Physiol. 2010;51:1398–1410. doi: 10.1093/pcp/pcq119. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Takahara K, Hirabayashi T, Matsumura H, Fujisawa S, Terauchi R, et al. Metabolome analysis of response to oxidative stress in rice suspension cells overexpressing cell death suppressor Bax inhibitor-1. Plant Cell Physiol. 2009;51:9–20. doi: 10.1093/pcp/pcp162. [DOI] [PubMed] [Google Scholar]
- Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell. 2011;23:1741–1755. doi: 10.1105/tpc.111.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi T, Yan Z, Libault M, Jeong DH, Park S, Green PJ, et al. Prediction of novel miRNAs and associated target genes in Glycine max. BMC Bioinformatics. 2011;11(Suppl 1):S14. doi: 10.1186/1471-2105-11-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, et al. Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol. 2009;151:1758–1768. doi: 10.1104/pp.109.145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, et al. Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010;51:1988–2001. doi: 10.1093/pcp/pcq158. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Minamisawa K. Plant–microbe communications for symbiosis. Plant Cell Physiol. 2010;51:1377–1380. doi: 10.1093/pcp/pcq125. [DOI] [PubMed] [Google Scholar]
- Kawaura K, Mochida K, Enju A, Totoki Y, Toyoda A, Sakaki Y, et al. Assessment of adaptive evolution between wheat and rice as deduced from full-length common wheat cDNA sequence data and expression patterns. BMC Genomics. 2009;10:271. doi: 10.1186/1471-2164-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaura K, Mochida K, Yamazaki Y, Ogihara Y. Transcriptome analysis of salinity stress responses in common wheat using a 22k oligo-DNA microarray. Funct. Integr. Genomics. 2006;6:132–142. doi: 10.1007/s10142-005-0010-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Baek KH, Lee SW, Kim J, Lee BW, Cho HS, et al. Pepper EST database: comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. BMC Plant Biol. 2008;8:101. doi: 10.1186/1471-2229-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, et al. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1580–1588. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- Kogel KH, Voll LM, Schafer P, Jansen C, Wu Y, Langen G, et al. Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances. Proc. Natl Acad. Sci. USA. 2010;107:6198–6203. doi: 10.1073/pnas.1001945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, et al. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009;57:883–894. doi: 10.1111/j.1365-313X.2008.03733.x. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, et al. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–1397. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M, Morgan A, Smith L, Leitner F, Tanabe L, Wilbur J, et al. Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge. Genome Biol. 2008;9(Suppl 2):S1. doi: 10.1186/gb-2008-9-s2-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M, Rodriguez-Penagos C, Tendulkar A, Valencia A. PLAN2L: a web tool for integrated text mining and literature-based bioentity relation extraction. Nucleic Acids Res. 2009;37:W160–W165. doi: 10.1093/nar/gkp484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Guiderdoni E, An G, Hsing YI, Han CD, Lee MC, et al. Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 2009;149:165–170. doi: 10.1104/pp.108.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J. Natural diversity and adaptation in plant secondary metabolism. Curr. Opin. Plant Biol. 2011;14:246–251. doi: 10.1016/j.pbi.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 2011;43:163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Takahashi S, Kondou Y, Shinozaki K, Matsui M. Phenome analysis in plant species using loss-of-function and gain-of-function mutants. Plant Cell Physiol. 2009;50:1215–1231. doi: 10.1093/pcp/pcp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Redestig H, Hirai T, Oikawa A, Matsuda F, Fukushima A, et al. Covering chemical diversity of genetically-modified tomatoes using metabolomics for objective substantial equivalence assessment. PLoS One. 2011a;6:e16989. doi: 10.1371/journal.pone.0016989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Tohge T, Fukushima A, Kobayashi M, Hayashi N, Otsuki H, et al. Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 2011b;67:354–369. doi: 10.1111/j.1365-313X.2011.04599.x. [DOI] [PubMed] [Google Scholar]
- Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, et al. Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 2010;42:1027–1030. doi: 10.1038/ng.684. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Sero A, Pratelli R, Pilot G, Chen J, Sardi MI, et al. A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front. Physiol. 2010;1:24. doi: 10.3389/fphys.2010.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- Last RL, Jones AD, Shachar-Hill Y. Towards the plant metabolome and beyond. Nat. Rev. Mol. Cell. Biol. 2007;8:167–174. doi: 10.1038/nrm2098. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, et al. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011a;18:17–29. doi: 10.1093/dnares/dsq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011b;18:263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Kim YK, Pham TT, Song SI, Kim JK, Kang KY, et al. RiceArrayNet: a database for correlating gene expression from transcriptome profiling, and its application to the analysis of coexpressed genes in rice. Plant Physiol. 2009;151:16–33. doi: 10.1104/pp.109.139030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais-Briere C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, et al. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21:2780–2796. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zang W, Li Y, Xu F, Wang J, Shi T. AtPID: the overall hierarchical functional protein interaction network interface and analytic platform for Arabidopsis. Nucleic Acids Res. 2011;39:D1130–D1133. doi: 10.1093/nar/gkq959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, He K, Ma Y, Su N, He H, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, et al. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- Lin M, Hu B, Chen L, Sun P, Fan Y, Wu P, et al. Computational identification of potential molecular interactions in Arabidopsis. Plant Physiol. 2009;151:34–46. doi: 10.1104/pp.109.141317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Shen X, Chen X. PAIR: the predicted Arabidopsis interactome resource. Nucleic Acids Res. 2011a;39:D1134–D1140. doi: 10.1093/nar/gkq938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Zhou X, Shen X, Mao C, Chen X. The predicted Arabidopsis interactome resource and network topology-based systems biology analyses. Plant Cell. 2011b;23:911–922. doi: 10.1105/tpc.110.082529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Steinfath M, Meyer RC, Selbig J, Melchinger AE, Willmitzer L, et al. Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. Plant J. 2009;59:777–788. doi: 10.1111/j.1365-313X.2009.03910.x. [DOI] [PubMed] [Google Scholar]
- Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr. Opin. Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lu T, Yu S, Li Y, Huang Y, Huang T, et al. A collection of 10,096 indica rice full-length cDNAs reveals highly expressed sequence divergence between Oryza sativa indica and japonica subspecies. Plant Mol. Biol. 2007;65:403–415. doi: 10.1007/s11103-007-9174-7. [DOI] [PubMed] [Google Scholar]
- Lobell DB, Schlenker W, Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333:616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010;20:1238–1249. doi: 10.1101/gr.106120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita Y, Kobayashi N, Mochizuki Y, Yoshida Y, Asano S, Heida N, et al. PosMed-plus: an intelligent search engine that inferentially integrates cross-species information resources for molecular breeding of plants. Plant Cell Physiol. 2009;50:1249–1259. doi: 10.1093/pcp/pcp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan LP, Guttikonda SK, Tran LS, Nguyen HT. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009;50:1260–1276. doi: 10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- Mano S, Miwa T, Nishikawa S, Mimura T, Nishimura M. The plant organelles database (PODB): a collection of visualized plant organelles and protocols for plant organelle research. Nucleic Acids Res. 2008;36:D929–D937. doi: 10.1093/nar/gkm789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Miwa T, Nishikawa S, Mimura T, Nishimura M. Seeing is believing: on the use of image databases for visually exploring plant organelle dynamics. Plant Cell Physiol. 2009;50:2000–2014. doi: 10.1093/pcp/pcp128. [DOI] [PubMed] [Google Scholar]
- Mano S, Miwa T, Nishikawa S, Mimura T, Nishimura M. The Plant Organelles Database 2 (PODB2): an updated resource containing movie data of plant organelle dynamics. Plant Cell Physiol. 2011;52:244–253. doi: 10.1093/pcp/pcq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoudi-Nejad A, Goto S, Endo TR, Kanehisa M. KEGG bioinformatics resource for plant genomics research. Methods Mol. Biol. 2007;406:437–458. doi: 10.1007/978-1-59745-535-0_21. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Annu. Rev. Plant Biol. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Hirai MY, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart NJ, et al. AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol. 2010;152:566–578. doi: 10.1104/pp.109.148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 2009;57:555–577. doi: 10.1111/j.1365-313X.2008.03705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Tanaka T, Sakai H, Amano N, Kanamori H, Kurita K, et al. Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 2011;156:20–28. doi: 10.1104/pp.110.171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Martis M, Hedley PE, Simkova H, Liu H, Morris JA, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23:1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T. In silico identification of short nucleotide sequences associated with gene expression of pollen development in rice. Plant Cell Physiol. 2008;49:1451–1464. doi: 10.1093/pcp/pcn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T. SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 2010;38:D835–D842. doi: 10.1093/nar/gkp831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milos P. Helicos BioSciences. Pharmacogenomics. 2008;9:477–480. doi: 10.2217/14622416.9.4.477. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Mochida K, Furuta T, Ebana K, Shinozaki K, Kikuchi J. Correlation exploration of metabolic and genomic diversities in rice. BMC Genomics. 2009a;10:568. doi: 10.1186/1471-2164-10-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Kawaura K, Shimosaka E, Kawakami N, Shin IT, Kohara Y, et al. Tissue expression map of a large number of expressed sequence tags and its application to in silico screening of stress response genes in common wheat. Mol. Genet. Genomics. 2006;276:304–312. doi: 10.1007/s00438-006-0120-1. [DOI] [PubMed] [Google Scholar]
- Mochida K, Saisho D, Yoshida T, Sakurai T, Shinozaki K. TriMEDB: a database to integrate transcribed markers and facilitate genetic studies of the tribe Triticeae. BMC Plant Biol. 2008;8:72. doi: 10.1186/1471-2229-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol. 2010;51:497–523. doi: 10.1093/pcp/pcq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Uehara-Yamaguchi Y, Yoshida T, Sakurai T, Shinozaki K. Global landscape of a co-expressed gene network in barley and its application to gene discovery in Triticeae crops. Plant Cell Physiol. 2011a;52:785–803. doi: 10.1093/pcp/pcr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yamazaki Y, Ogihara Y. Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol. Genet. Genomics. 2003;270:371–377. doi: 10.1007/s00438-003-0939-7. [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Ogihara Y, Shinozaki K. TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol. 2009b;150:1135–1146. doi: 10.1104/pp.109.138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009c;16:353–369. doi: 10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010a;17:303–324. doi: 10.1093/dnares/dsq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. LegumeTFDB: an integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics. 2010b;26:290–291. doi: 10.1093/bioinformatics/btp645. [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. In silico analysis of transcription factor repertoires and prediction of stress-responsive transcription factors from six major Gramineae plants. DNA Res. 2011b doi: 10.1093/dnares/dsr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, et al. A liquid chromatography–mass spectrometry-based metabolome database for tomato. Plant Physiol. 2006;141:1205–1218. doi: 10.1104/pp.106.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam AM, Roudier F, Seifert M, Berard C, Magniette ML, Ashtiyani RK, et al. Additive inheritance of histone modifications in Arabidopsis thaliana intra-specific hybrids. Plant J. 2011;67:691–700. doi: 10.1111/j.1365-313X.2011.04628.x. [DOI] [PubMed] [Google Scholar]
- Moore J, Allan C, Burel JM, Loranger B, MacDonald D, Monk J, et al. Open tools for storage and management of quantitative image data. Methods Cell Biol. 2008;85:555–570. doi: 10.1016/S0091-679X(08)85024-8. [DOI] [PubMed] [Google Scholar]
- Morsy M, Gouthu S, Orchard S, Thorneycroft D, Harper JF, Mittler R, et al. Charting plant interactomes: possibilities and challenges. Trends Plant Sci. 2008;13:183–191. doi: 10.1016/j.tplants.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, et al. PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, et al. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010;153:1161–1174. doi: 10.1104/pp.110.157347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Ishihama Y, Shirasu K. Shotguns in the Front Line: Phosphoproteomics in Plants. Plant Cell Physiol. 2011 doi: 10.1093/pcp/pcr148. Oct 28 [Epub ahead of print] doi:10.1093/pcp/pcr148. [DOI] [PubMed] [Google Scholar]
- Nashilevitz S, Melamed-Bessudo C, Izkovich Y, Rogachev I, Osorio S, Itkin M, et al. An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant Cell. 2010;22:1977–1997. doi: 10.1105/tpc.110.074716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautrup-Pedersen G, Dam S, Laursen BS, Siegumfeldt AL, Nielsen K, Goffard N, et al. Proteome analysis of pod and seed development in the model legume Lotus japonicus. J. Proteome Res. 2010;9:5715–5726. doi: 10.1021/pr100511u. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K. ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 2011;52:213–219. doi: 10.1093/pcp/pcq203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y, Mochida K, Nemoto Y, Murai K, Yamazaki Y, Shin IT, et al. Correlated clustering and virtual display of gene expression patterns in the wheat life cycle by large-scale statistical analyses of expressed sequence tags. Plant J. 2003;33:1001–1011. doi: 10.1046/j.1365-313x.2003.01687.x. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Asamizu E, Saito T, Matsukura C, Ariizumi T, Mizoguchi T, et al. Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 2011 doi: 10.1093/pcp/pcr134. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Shimojima M, Sawada Y, Toyooka K, Narisawa T, Mochida K, et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell. 2009;21:892–909. doi: 10.1105/tpc.108.063925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S, Ogata Y, Suda K, Kurabayashi A, Suzuki T, Yamamoto N, et al. Coexpression analysis of tomato genes and experimental verification of coordinated expression of genes found in a functionally enriched coexpression module. DNA Res. 2010;17:105–116. doi: 10.1093/dnares/dsq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Perry J, Brachmann A, Welham T, Binder A, Charpentier M, Groth M, et al. TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiol. 2009;151:1281–1291. doi: 10.1104/pp.109.142190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podicheti R, Dong Q. Administering GBrowse sites with WebGBrowse. Curr. Protoc. Bioinformatics. 2011 doi: 10.1002/0471250953.bi0914s33. Chapter 9: Unit 9 14. [DOI] [PubMed] [Google Scholar]
- Poland JA, Bradbury PJ, Buckler ES, Nelson RJ. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl Acad. Sci. USA. 2011;108:6893–6898. doi: 10.1073/pnas.1010894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, et al. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Ben-Hur A, Day IS. Experimental and computational approaches for the study of calmodulin interactions. Phytochemistry. 2011;72:1007–1019. doi: 10.1016/j.phytochem.2010.12.022. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijoi S, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Last RL. Shotguns and SNPs: how fast and cheap sequencing is revolutionizing plant biology. Plant J. 2010;61:922–927. doi: 10.1111/j.1365-313X.2009.04030.x. [DOI] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell. 2008;20:1199–1216. doi: 10.1105/tpc.108.058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D, Ishii M, Hori K, Sato K. Natural variation of barley vernalization requirements: implication of quantitative variation of winter growth habit as an adaptive trait in East Asia. Plant Cell Physiol. 2011;52:775–784. doi: 10.1093/pcp/pcr046. [DOI] [PubMed] [Google Scholar]
- Saisho D, Takeda K. Barley: emergence as a new research material of crop science. Plant Cell Physiol. 2011;52:724–727. doi: 10.1093/pcp/pcr049. [DOI] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Saito K, Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 2010;61:463–489. doi: 10.1146/annurev.arplant.043008.092035. [DOI] [PubMed] [Google Scholar]
- Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, et al. TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 2011;52:283–296. doi: 10.1093/pcp/pcr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai N, Ara T, Ogata Y, Sano R, Ohno T, Sugiyama K, et al. KaPPA-View4: a metabolic pathway database for representation and analysis of correlation networks of gene co-expression and metabolite co-accumulation and omics data. Nucleic Acids Res. 2011a;39:D677–684. doi: 10.1093/nar/gkq989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Kondou Y, Akiyama K, Kurotani A, Higuchi M, Ichikawa T, et al. RiceFOX: a database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol. 2011b;52:265–273. doi: 10.1093/pcp/pcq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- Sasaki E, Takahashi C, Asami T, Shimada Y. AtCAST, a tool for exploring gene expression similarities among DNA microarray experiments using networks. Plant Cell Physiol. 2010;52:169–180. doi: 10.1093/pcp/pcq185. [DOI] [PubMed] [Google Scholar]
- Sato K, Nankaku N, Takeda K. A high-density transcript linkage map of barley derived from a single population. Heredity. 2009a;103:110–117. doi: 10.1038/hdy.2009.57. [DOI] [PubMed] [Google Scholar]
- Sato K, Shin IT, Seki M, Shinozaki K, Yoshida H, Takeda K, et al. Development of 5006 full-length CDNAs in barley: a tool for accessing cereal genomics resources. DNA Res. 2009b;16:81–89. doi: 10.1093/dnares/dsn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Hirakawa H, Isobe S, Fukai E, Watanabe A, Kato M, et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18:65–76. doi: 10.1093/dnares/dsq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, et al. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39:D1141–1148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Akiyama K, Sakata A, Kuwahara A, Otsuki H, Sakurai T, et al. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 2009a;50:37–47. doi: 10.1093/pcp/pcn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Kuwahara A, Nagano M, Narisawa T, Sakata A, Saito K, et al. Omics-based approaches to methionine side chain elongation in Arabidopsis: characterization of the genes encoding methylthioalkylmalate isomerase and methylthioalkylmalate dehydrogenase. Plant Cell Physiol. 2009b;50:1181–1190. doi: 10.1093/pcp/pcp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, et al. Mode of inheritance of primary metabolic traits in tomato. Plant Cell. 2008;20:509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Daniel Jones A, et al. Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 2010;62:391–403. doi: 10.1111/j.1365-313X.2010.04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Zhang X. High-throughput approaches for plant epigenomic studies. Curr. Opin. Plant Biol. 2011;14:130–136. doi: 10.1016/j.pbi.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Seo YS, Chern M, Bartley LE, Han M, Jung KH, Lee I, et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011;7:e1002020. doi: 10.1371/journal.pgen.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- Shingaki-Wells RN, Huang S, Taylor NL, Carroll AJ, Zhou W, Millar AH. Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol. 2011;156:1706–1724. doi: 10.1104/pp.111.175570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Sakakibara H. Omics and bioinformatics: an essential toolbox for systems analyses of plant functions beyond 2010. Plant Cell Physiol. 2009;50:1177–1180. doi: 10.1093/pcp/pcp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, et al. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ. HapMap methylation-associated SNPs, markers of germline DNA methylation, positively correlate with regional levels of human meiotic recombination. Genome Res. 2009;19:581–589. doi: 10.1101/gr.086181.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, Currie J, et al. Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 2009;5:e1000740. doi: 10.1371/journal.pgen.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol A, Kwiatkowska A, Jerzmanowski A, Prymakowska-Bosak M. Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta. 2007;227:245–254. doi: 10.1007/s00425-007-0612-1. [DOI] [PubMed] [Google Scholar]
- Somerville C, Youngs H, Taylor C, Davis SC, Long SP. Feedstocks for lignocellulosic biofuels. Science. 2010;329:790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011;11:5. doi: 10.1186/1471-2229-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G, Libault M, Brechenmacher L, Wan J, May GD. Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 2006;9:110–121. doi: 10.1016/j.pbi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Sulpice R, Keurentjes J. Metabolic networks: how to identify key components in the regulation of metabolism and growth. Plant Physiol. 2010;152:428–444. doi: 10.1104/pp.109.150821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CL, Chao YT, Alex Chang YC, Chen WC, Chen CY, Lee AY, et al. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011;52:1501–1514. doi: 10.1093/pcp/pcr097. [DOI] [PubMed] [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, Usadel B, Gibon Y, Witucka-Wall H, et al. Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. Plant Cell. 2010;22:2872–2893. doi: 10.1105/tpc.110.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW. Recent advances in plant metabolomics and greener pastures. F1000 Biol. Rep. 2010;2 doi: 10.3410/B2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Xu X, Zhu H, Liu A, Liu L, Li J, et al. Comparative transcriptomic profiling of a salt-tolerant wild tomato species and a salt-sensitive tomato cultivar. Plant Cell Physiol. 2010;51:997–1006. doi: 10.1093/pcp/pcq056. [DOI] [PubMed] [Google Scholar]
- Suwabe K, Suzuki G, Takahashi H, Shiono K, Endo M, Yano K, et al. Separated transcriptomes of male gametophyte and tapetum in rice: validity of a laser microdissection (LM) microarray. Plant Cell Physiol. 2008;49:1407–1416. doi: 10.1093/pcp/pcn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole V, Worland B, Wright J, Bevan MW, Vain P. Distribution and characterization of more than 1000 T-DNA tags in the genome of Brachypodium distachyon community standard line Bd21. Plant Biotechnol. J. 2010;8:734–747. doi: 10.1111/j.1467-7652.2010.00518.x. [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- Tieman D, Zeigler M, Schmelz E, Taylor MG, Rushing S, Jones JB, et al. Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 2010;62:113–123. doi: 10.1111/j.1365-313X.2010.04128.x. [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tran LS, Mochida K. Functional genomics of soybean for improvement of productivity in adverse conditions. Funct. Integr. Genomics. 2010;10:447–462. doi: 10.1007/s10142-010-0178-z. [DOI] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Uchida N, Sakamoto T, Kurata T, Tasaka M. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 2011;52:716–722. doi: 10.1093/pcp/pcr029. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, et al. Sequencing and analysis of approximately 40,000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 2008;15:333–346. doi: 10.1093/dnares/dsn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Nayak SN, May GD, Jackson SA. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009;27:522–530. doi: 10.1016/j.tibtech.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V. 10 reasons to be tantalized by the B73 maize genome. PLoS Genet. 2009;5:e1000723. doi: 10.1371/journal.pgen.1000723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, et al. Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell. 2009a;21:1428–1452. doi: 10.1105/tpc.108.060830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, et al. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009b;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009c;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Mochida K, Kato T, Tabata S, Yoshimoto N, Noji M, et al. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. Plant Cell. 2008;20:2484–2496. doi: 10.1105/tpc.108.060335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, et al. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics. 2011;12:451. doi: 10.1186/1471-2164-12-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnenburg R, Wachter T, Plake C, Doms A, Schroeder M. Facts from text: can text mining help to scale-up high-quality manual curation of gene products with ontologies? Brief. Bioinform. 2008;9:466–478. doi: 10.1093/bib/bbn043. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MM, Cannon CH, Wickneswari R. Identification of lignin genes and regulatory sequences involved in secondary cell wall formation in Acacia auriculiformis and Acacia mangium via de novo transcriptome sequencing. BMC Genomics. 2011;12:342. doi: 10.1186/1471-2164-12-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kyozuka J. Branching hormone is busy both underground and overground. Plant Cell Physiol. 2010;51:1091–1094. doi: 10.1093/pcp/pcq088. [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Hakata M. Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010;51:795–809. doi: 10.1093/pcp/pcq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Nagasaki H, Yonemaru J, Ebana K, Nakajima M, Shibaya T, et al. Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics. 2010;11:267. doi: 10.1186/1471-2164-11-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YG, Tominaga T, Iijima Y, Aoki K, Shibata D, Ashihara H, et al. Metabolic alterations in organic acids and gamma-aminobutyric acid in developing tomato (Solanum lycopersicum L.) fruits. Plant Cell Physiol. 2010;51:1300–1314. doi: 10.1093/pcp/pcq090. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, et al. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. Plant Cell. 2008;20:2160–2176. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youens-Clark K, Buckler E, Casstevens T, Chen C, Declerck G, Derwent P, et al. Gramene database in 2010: updates and extensions. Nucleic Acids Res. 2010;39:D1085–D1094. doi: 10.1093/nar/gkq1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Udvardi M. Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 2009;12:193–201. doi: 10.1016/j.pbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Yura K, Sulaiman S, Hatta Y, Shionyu M, Go M. RESOPS: a database for analyzing the correspondence of RNA editing sites to protein three-dimensional structures. Plant Cell Physiol. 2009;50:1865–1873. doi: 10.1093/pcp/pcp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, et al. Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 2010;152:1787–1795. doi: 10.1104/pp.109.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sreenivasulu N, Weschke W, Stein N, Rudd S, Radchuk V, et al. Large-scale analysis of the barley transcriptome based on expressed sequence tags. Plant J. 2004;40:276–290. doi: 10.1111/j.1365-313X.2004.02209.x. [DOI] [PubMed] [Google Scholar]
- Zhang P, Dreher K, Karthikeyan A, Chi A, Pujar A, Caspi R, et al. Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010;153:1479–1491. doi: 10.1104/pp.110.157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007a;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007b;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]