Summary
The oxidative burst is an early response to pathogen attack leading to the production of reactive oxygen species (ROS) including hydrogen peroxide. Two major mechanisms involving either NADPH oxidases or peroxidases that may exist singly or in combination in different plant species have been proposed for the generation of ROS. We identified an Arabidopsis thaliana azide-sensitive but diphenylene iodonium-insensitive apoplastic oxidative burst that generates H2O2 in response to a Fusarium oxysporum cell-wall preparation. Transgenic Arabidopsis plants expressing an anti-sense cDNA encoding a type III peroxidase, French bean peroxidase type 1 (FBP1) exhibited an impaired oxidative burst and were more susceptible than wild-type plants to both fungal and bacterial pathogens. Transcriptional profiling and RT-PCR analysis showed that the anti-sense (FBP1) transgenic plants had reduced levels of specific peroxidase-encoding mRNAs, including mRNAs corresponding to Arabidopsis genes At3g49120 (AtPCb) and At3g49110 (AtPCa) that encode two class III peroxidases with a high degree of homology to FBP1. These data indicate that peroxidases play a significant role in generating H2O2 during the Arabidopsis defense response and in conferring resistance to a wide range of pathogens.
Keywords: apoplastic peroxidase, Arabidopsis, oxidative burst, reactive oxygen species
Introduction
One of the earliest events in the plant-defense response against pathogen attack is the production of reactive oxygen species (ROS) including hydrogen peroxide and superoxide (Bolwell and Wojtaszek, 1997; Lamb and Dixon, 1997; Wojtaszek, 1997). Two major mechanisms have been described for generating ROS during this so-called oxidative burst. Depending on the host and pathogen species involved, these ROS-generating mechanisms involve plasma membrane NADPH oxidases or cell-wall peroxidases (Bolwell et al., 1998; Grant et al., 2000a,b; Martinez et al., 1998; Torres et al., 2002).
NADPH oxidases, also referred to as respiratory burst oxidases, have been implicated in biotic interactions, abiotic stress responses and development (Torres and Dangl, 2005). In Arabidopsis thaliana, a 10-gene family of Atrboh genes encodes homologues of the mammalian NADPH oxidase gp91phox (Keller et al., 1998; Torres et al., 1998). The highly expressed AtrbohD and AtrbohF genes are required for the production of a full oxidative burst in response to avirulent strains of the bacterial and oomycete pathogens Pseudomonas syringae and Hyaloperonospora parasitica, respectively (Torres et al., 2002). Surprisingly, however, neither the atrbohD nor atrbohF mutants, either singly or doubly, was more susceptible to either P. syringae or H. parasitica.
In addition to NADPH oxidases, heme-containing perox-idases are also able to generate hydrogen peroxide (Bolwell and Wojtaszek, 1997). Unlike the NADPH oxidase reaction, the peroxidase-mediated oxidative burst is sensitive to azide and cyanide, but relatively insensitive to the NADPH oxidase inhibitor diphenylene iodonium (DPI) (Bolwell et al., 1998). Previously, we described an oxidative burst that could be elicited in Phaseolus vulgaris (French bean) tissue culture cells that is characterized by a cell wall-associated type III peroxidase, French bean peroxidase type 1 (FBP1), that is azide-sensitive but DPI-insensitive (Blee et al., 2001; Bolwell, 1999; Bolwell et al., 2002). In the experiments described here, we have turned to Arabidopsis to elucidate further the role of peroxidases in the pathogen-elicited oxidative burst. Although the large size of the type III peroxidase gene family in Arabidopsis (at least 73 type III members) and presumed functional redundancy precluded a systematic study of Arabidopsis peroxidase mutants, we expressed the heterologous anti-sense French bean FBP1 cDNA in transgenic Arabidopsis plants. The FBP1 transgenic lines showed normal morphology, but proved enormously susceptible to spontaneous infections, with the result that we could recover only two heterozygous lines and a homozygous line derived from one of these. However, these lines showed an impaired oxidative burst and enhanced susceptibility to both bacterial and fungal pathogens.
Results
Development of an elicitor-mediated oxidative burst assay for Arabidopsis suspension cultures
As described in detail in Experimental procedures, an oxidative burst assay was developed for an Arabidopsis cell-suspension culture treated with an elicitor derived from a preparation of cell walls from Fusarium oxysporum. f.sp. matthioli race 1. In contrast to a previously developed assay for monitoring the oxidative burst in French bean cells that utilized luminol to detect hydrogen peroxide (Bindschedler et al., 2001), the Arabidopsis assay utilized xylenol orange. This was necessary because the Arabidopsis suspension cultures were green and quenched luminol fluorescence. As shown in Figure 1(a), the F. oxysporum elicitor at 100 µg ml−1 glucose equivalents, the optimal concentration, induced a robust and highly reproducible oxidative burst. Addition of catalase completely abolished the xylenol orange signal, indicating it was primarily due to H2O2 rather than other reactive species (Figure 1a). The response was elicitor-specific as the Arabidopsis cells did not respond to Colletotrichum lindemuthianum elicitor, which was shown previously to elicit an oxidative burst in French bean suspension cultures (Bindschedler et al., 2001). The equivalence of the luminol and xylenol orange assays was demonstrated previously in the French bean system (Bindschedler et al., 2001).
Figure 1. Elicitation of an oxidative burst in Arabidopsis tissue cultures.
(a) Comparative level of the oxidative burst in Arabidopsis cell-suspension cultures treated with 100 µg ml −1 glucose equivalents of Fusarium oxysporum elicitor in the absence (●) or presence (○) of 10 U catalase. Also shown is the burst in response to 25 µg ml−1 Colletotrichum lindemuthianum elicitor (■), the optimum for reactive oxygen species (ROS) production in French bean cells. Production of ROS was measured by the xylenol orange assay as described in Experimental procedures.
(b) Dose-response curve for inhibition of ROS production in F. oxysporum elicitor-treated Arabidopsis cell-suspension cultures by sodium azide.
(c) Comparative dose-response curve for diphenylene iodonium (DPI) inhibition of the oxidative burst in Arabidopsis cell-suspension cultures treated with F. oxysporum elicitor (○) or French bean cells treated with C. lindemuthianum elicitor (□) using the xylenol orange method. Values are means ± SD from replicates from at least five independent experiments.
To distinguish whether the Arabidopsis oxidative burst is mediated by an NADPH/NADH oxidase or a peroxidase-dependent mechanism, inhibition studies were carried out using diphenylene iodonium (DPI) and sodium azide. Inhibition of an oxidative burst by DPI with an I50 of approximately 0.2 µm is indicative of the involvement of an NADPH/NADH oxidase, whereas inhibition by azide with an I50 > 50 µm suggests a peroxidase-dependent mechanism (Bolwell et al., 1998; Frahry and Schopfer, 1998). Figure 1(b) shows that sodium azide inhibited F. oxysporum-elicited hydrogen peroxide formation in Arabidopsis tissue culture cells with an I50 of approximately 500 µm and that 2 mm sodium azide considerably reduced hydrogen peroxide formation. Similar results were obtained with potassium cyanide (not shown). The relatively high I50 for sodium azide may be a consequence of detoxification of the inhibitor by the suspension cells, as observed previously for bean cells with potassium cyanide (Bolwell et al., 1998).
In contrast to sodium azide and potassium cyanide, the Arabidopsis oxidative burst was remarkably insensitive to DPI (Figure 1c). The insensitivity of the Arabidopsis oxidative burst to DPI was significantly greater than the insensitivity observed in the French bean system, where a peroxidase was previously shown to be active: The DPI I50 was about 2.5 µm for French bean, whereas in Arabidopsis 50 µm DPI decreased the production of hydrogen peroxide by only 20% (Figure 1c). The reason the I50 for DPI in French bean observed in these experiments is lower than the previously reported value of 25 µm (Bolwell et al., 1998) is probably a consequence of the different detection systems employed. The xylenol orange method used here directly measures ROS concentration, whereas the luminol assay used previously requires the activity of endogenous peroxidases that oxidize the luminol with H2O2. Because peroxidases are inhibited by DPI at higher concentrations (Frahry and Schopfer, 1998), the luminol assay is likely to overestimate the I50 of DPI. In any case, these data suggest that peroxidases contribute to the generation of a significant proportion of the H2O2 released in the Arabidopsis oxidative burst elicited by the Fusarium cell wall preparation.
Transgenic Arabidopsis plants expressing anti-sense FBP1 mRNA exhibit a diminished oxidative burst
A previously constructed full-length French bean cDNA encoding FBP1 peroxidase in an anti-sense orientation (Blee et al., 2001, 2003) was used to transform Arabidopsis eco-type Col-0 with the expectation that the transgenic plants might express lower levels of peroxidase(s) involved in generating the oxidative burst (a preliminary report of this experiment was published by Bolwell et al., 2002). Interestingly, almost all of more than 20 kanamycin-resistant transformants that were recovered succumbed to opportunistic fungal infections, and only two lines (referred to as 1.1 and 1.2) survived to maturity and set seed. Homozygous transgenic line H4 was derived from line 1.1 by repeated selection of seeds on kanamycin. Despite repeated attempts, we were not able to derive a homozygous derivative of line 1.2.
We developed a xylenol orange-based Arabidopsis leaf-disc assay for measuring an elicitor-induced oxidative burst, so that we could compare the oxidative burst in wild-type and FBP1 transgenic plants (see Experimental procedures). As shown in Figure 2(a), the two T1 FBP1 anti-sense lines (1.1 and 1.2) exhibited significantly less F. oxysporum-elicited release of H2O2 in the surrounding medium than wild-type Col-0 leaf disks. To confirm these results, we used an independent method that involves 3,3′-diaminobenzidine (DAB) staining of leaves (Thordal-Christensen et al., 1997) following infection with the avirulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) expressing the avirulence gene avrRpm1. Pto DC3000 (avrRpm1) was infiltrated at the relatively high dose of 107 CFU cm−2 leaf area into one half of a leaf, and infiltrated leaves were detached 2 h later and stained with DAB for 3 h. As shown in Figure 2(b), in wild-type Col-0 leaves a strong precipitate appeared in the inoculated leaf halves, whereas no precipitate was observed in the control non-inoculated leaf halves. In comparison with Col-0 wild type, the homozygous FBP1 anti-sense line H4 showed a reduced amount of DAB staining at both macroscopic and microscopic levels, comparable to that reported for NADPH oxidase (Atrboh) mutants similarly infiltrated with Pto DC3000(avrRpm1) (Torres et al., 2002). Leaf halves that were mock-inoculated with 10 mm MgSO4 did not show any DAB staining within the experimental time frame.
Figure 2. Elicitation of reactive oxygen species (ROS) in Arabidopsis leaves.
(a) Production of ROS in leaf discs of wild-type Col-0 (●) or FBP1 transgenic lines 1.1 (◇) or 1.2 (△) treated with 25 µg ml−1 Fusarium oxysporum elicitor.
(b) In situ detection of H2O2 by 3,3′-diaminobenzidine (DAB) staining. Leaves of non-transformed Col-0 plants or homozygous transgenic anti-sense FBP1 H4 plants were inoculated with Pto DC3000(avrRpm1) at OD600 = 2.0 (107 CFU cm−2), detached after 2 h and infiltrated under gentle vacuum with 1 mg ml−1 DAB containing 0.05% v/v Tween 20 and 10 mm sodium phosphate buffer pH 7.0. The reaction was terminated at 6–7 h post-inoculation when a brown precipitate began to be visible in Col-0 leaves. Leaves were examined after bleaching by light microscopy (100× magnification).
(c) In situ detection of H2O2 by cerium chloride staining. Leaves of non-transformed Col-0 plants or homozygous transgenic FBP1 H4 plants were infiltrated with Pto DC3000(avrRpm1) at 107 CFU cm−2 followed by CeCl3 staining and electron microscope detection. Scale bar, 0.5 µm. Ps, Pseudomonas syringae; CW, cell wall.
Finally, the production of hydrogen peroxide production at the site of infection in the FBP1 anti-sense plants was monitored using a third independent method that involves the reaction of cerium chloride with hydrogen peroxide to form cerium perhydroxides that are opaque in the electron microscope. By co-infiltrating catalase, Grant et al. (2000b) demonstrated that the electron-opaque deposits observed after cerium chloride staining of Pto DC3000(avrRpm1)-inoculated Arabidopsis leaves are due to reaction of cerium chloride with hydrogen peroxide. As shown in Figure 2(c), we observed dark cerium perhydroxides deposits in Pto DC3000(avrRpm1)-infected Col-0 leaves at the site of infection that were co-localized with the bacteria at the plant cell wall. In contrast, no deposits were observed in non-inoculated Col-0 plants (data not shown) or in infected transgenic H4 plants expressing anti-sense FBP1 cDNA (Figure 2c).
Transgenic Arabidopsis expressing anti-sense FBP1 mRNA exhibit enhanced susceptibility to both virulent and avirulent pathogens
The observation that the primary FBP1-transformed seedlings frequently succumbed to spontaneous opportunistic infections suggested that they would exhibit enhanced susceptibility to a variety of pathogens. This was confirmed under controlled conditions by testing the T1 transgenic lines 1.1 and 1.2 and the homozygous FBP1 transgenic line H4 for susceptibility to a variety of pathogens. Figure 3(a) shows that the homozygous FBP1 line H4 exhibits a significant level of enhanced susceptibility to two unrelated virulent P. syringae strains, Pto DC3000 and P. syringae pv. maculicola strain ES4326 (Psm ES4326). Similar results were obtained with the T1 transgenic lines 1.1 and 1.2, and with eight out of eight different T2 lines derived from T1 line 1.1 (data not shown). Surprisingly, the avirulent strain Psm ES4326 carrying avrRpt2 grew to at least the same titer as the virulent strain Psm ES4326 in FBP1 H4 transgenic plants, suggesting that avr-gene mediated resistance is severely compromised in the FBP1 anti-sense lines (Figure 3b). Normally, avirulent P. syringae strains expressing avrRpt2 grow very poorly in plants such as Col-0, which express the RPS2 resistance gene product that corresponds to the avrRtp2 avirulence gene protein (Figure 3b). The enhanced susceptibility of FBP1 transgenic lines to P. syringae growth was reflected in more severe disease symptoms when Arabidopsis leaves were infiltrated either with virulent strains of Pto DC3000 or Psm ES4326, or with isogenic avirulent strains expressing the avirulence gene avrRpt2 (Bolwell et al., 2002; data not shown).
Figure 3. Susceptibility of FBP1 transgenic plants to bacterial and fungal pathogens.
(a) Growth of Pto DC3000 (black bars) or Psm ES4326 (open bars) 3 days post-inoculation of wild-type Col-0 or homozygous FBP1 transgenic H4 leaves at a dose of 104 CFU cm−2 leaf area. Means ± SE of six determinations are shown.
(b) Growth of virulent (ES4326, black bars) or avirulent [ES4326(avrRpt2), open bars] Psm strains 3 days post-inoculation of wild-type Col-0 or homozygous FBP1 transgenic H4 leaves with 103 CFU cm−2 leaf area. Means ± SE of eight determinations are shown. Differences between Col and H4 were statistically significant according to a t-test, with P ≤ 0.001. The experiment was repeated twice with similar results.
(c) Leaf pairs of Col-0 (left) and FBP1 transgenic line H4 plants (right) either mock-inoculated or inoculated with virulent spores of Botrytis cinerea.
(d) Opportunistic infection of FBP1 transgenic line H4 by the powdery mildew pathogen Golovinomyces orontii. Wild-type Col-0 and FBP1 transgenic plants were grown side-by-side in a flat (left panel) in a glasshouse contaminated with G. orontii spores. The FBP1 transgenic plants, but not the wild-type plants, develop symptoms of G. orontii infection. The two right-hand panels show close-ups of wild-type and FBP1 plants.
The enhanced susceptibility phenotype of the transgenic FBP1-expressing plants was also observed when challenged with several different fungal pathogens. Figure 3(c) shows large lesions in transgenic lines compared with relatively small lesions in Col-0 in response to inoculation with the necrotrophic pathogen Botrytis cinerea. Figure 3(d) shows that FBP1 transgenic plants exhibited greatly enhanced susceptibility to opportunistic powdery mildew infections that were grown in the same flats side-by-side.
Arabidopsis transgenic plants expressing anti-sense FBP1 are more resistant to fumonisin FB1
Alvarez et al. (1998) demonstrated a requirement for ROS during activation of programmed cell death (PCD) associated with the hypersensitive response (HR). Reactive oxygen species may also be involved in signaling that induces PCD in response to fumonisin B1 (FB1), a phytotoxin produced by Fusarium moniliforme that elicits PCD (Gilchrist, 1998). As shown in Figure 4, the FBP1 anti-sense homozygous line H4 was strikingly insensitive to FB1, with greatly reduced cell death in both primary infiltrated leaves and secondary non-infiltrated leaves. These data suggest that peroxidases may play a significant role in elicitation of the Arabidopsis HR.
Figure 4. Susceptibility of FBP1 transgenic lines to the fungal toxin fumonisin B1.
Top row, Col-0 leaf pairs (left) compared with leaf pairs of FBP1 transgenic line H4 (right) mock-infiltrated with 0.14% methanol or infiltrated with 10 µM FB1. Bottom row, non-infiltrated leaves from plants treated with 0.14% methanol or 10 µm FB1, respectively.
Identification of the Arabidopsis peroxidase(s) affected by the FBP1 anti-sense transgene
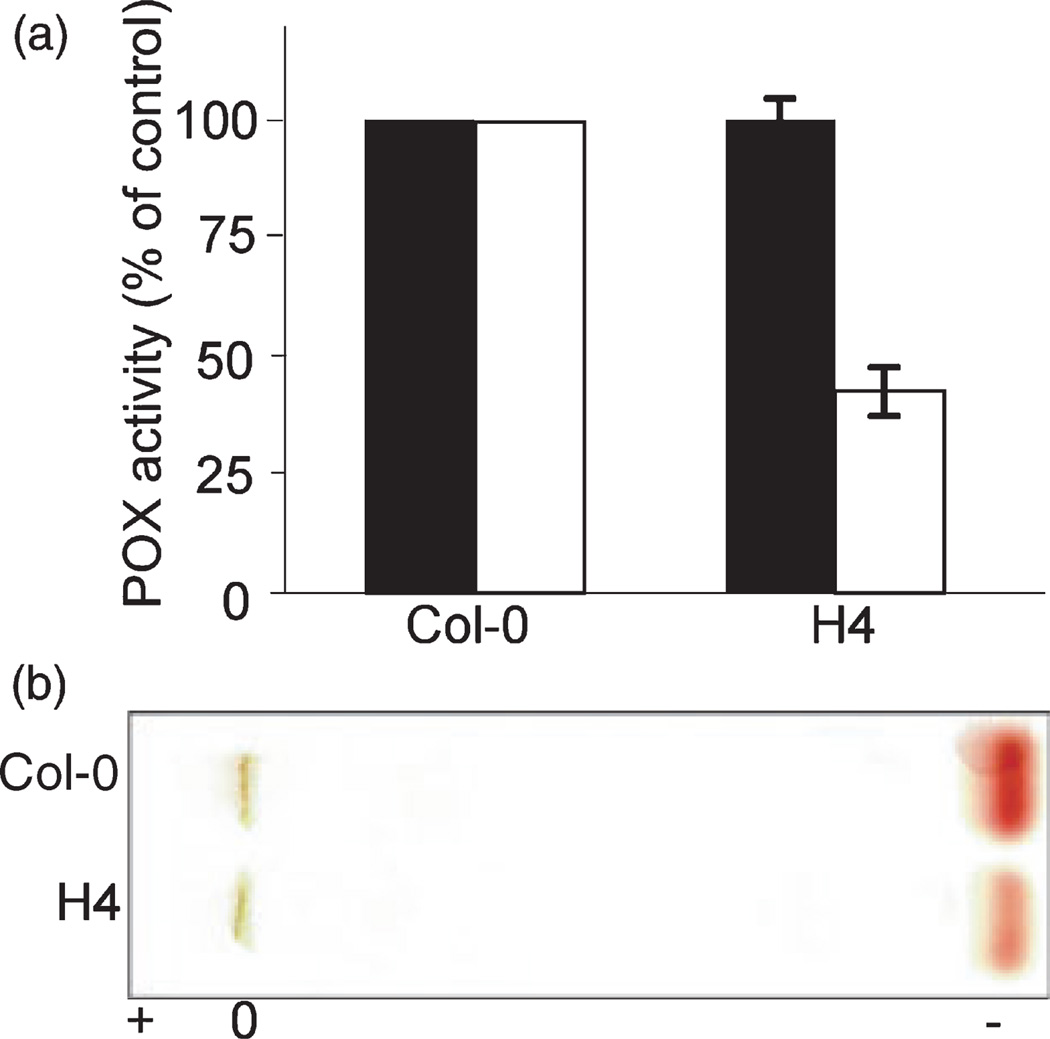
Based on the results obtained previously in French bean, the FBP1 transgenic lines would be expected to show lowered peroxidase specific activity in the ionically bound fraction, which should correspond to the cell wall-bound subset of peroxidases. Notwithstanding such a prediction, the actual target for FBP1 transgene-mediated silencing may represent a further sub-division of this subset of leaf peroxidases. Figure 5(a) shows that homozygous FBP1 anti-sense line H4 showed a significant reduction in bound peroxidase activity to approximately 40–65% of the Col-0-specific activity. These differences were confirmed by isoelectric profiling using equal loading of protein fractions and staining for peroxidase isoforms that gives a semi-quantitative indication of abundance. There were no differences in the profile of soluble peroxidases (data not shown), but a cationic isoform was reduced in the bound fraction in line H4 (Figure 5b).
Figure 5. Peroxidases in leaves of FBP1 transgenic lines.
(a) Relative specific activities of peroxidases in soluble (■) and bound (□) fractions were determined for Col-0 and FBP1 transgenic line H4 extracts using ABTS as substrate, as described in Experimental procedures. The experiment was repeated three times with similar results.
(b) Isoelectric focusing profiling of bound peroxidases from Col-0 and FBP1 transgenic line H4. Equal amounts (20 µg) of protein were loaded per track and the zymogram was developed using o-danisodine as described in Experimental procedures.
To identify specifically the peroxidase(s) that are down-regulated in the transgenic FBP-1 anti-sense lines, RNA-expression profiles of FBP-1 transgenic plants were compared with those of wild-type Col-0 using the Affymetrix full-genome ATH1 GeneChip (Affymetrix, Santa Clara, CA, USA). Of the 86 peroxidases (including 66 type III) annotated on the Affymetrix ATH1 GeneChip, three were significantly down-regulated (P ≤ 0.01) in transgenic line H4 (the entire data set is available in Appendix S1 and at http://ausubellab.mgh.harvard.edu/imds). However, two of the downregulated peroxidase genes (At1g63460, At3g63080) are annotated as encoding glutathione peroxidases (The Arabidopsis Information Resource TAIR; The Institute for Genomic Research, TIGR), which would not be expected to be present in the cell wall-associated peroxidase fraction that has reduced activity in the transgenic plants (Figure 5). The third Arabidopsis peroxidase gene, At3g49120, that was identified as downregulated by the GeneChip analysis encodes a predicted secreted type III cationic peroxidase, and is therefore a likely candidate for being ionically bound to the cell wall. This peroxidase is also the most likely target of the FBP1 anti-sense RNA, as it is the Arabidopsis peroxidase-encoding gene with the longest region of exact homology (25 bases) with the French bean FBP1 oxidative burst peroxidase gene. Only four other Arabidopsis peroxidases have sufficient identity with FBP1 (19–23 bases) to enable RNAi (Semizarov et al., 2003). Of these four, no difference in transcript level was detected by the GeneChip analysis for three; the fourth (At3g49110), which has strong homology to At3g49120 (75%, Valeério et al., 2004), is not represented on the ATH1 array.
To confirm the reduction in At3g49120 transcript levels and to determine whether At3g49110 is an additional target of the anti-sense FBP1, expression levels of these two genes in the FBP1 transgenic line H4 and wild-type plants were assayed by quantitative-real time RT-PCR analysis. As shown in Figure 6(a,b), transcript levels of both At3g49110 and At3g49120 are reduced in mock-inoculated H4 compared with similarly treated wild-type plants. After inoculation with Pto DC3000(avrRpm1), At3g49120 expression in H4 is around 37% of wild type (Figure 6b). Although At3g49110 mRNA levels do not appear to be decreased in the FBP1 anti-sense line after inoculation (Figure 6a), this may be a consequence of the strong upregulation of this gene in response to infection (Figure 6c). In any case, the fact that both genes are expressed at higher levels following inoculation (Figure 6c; Tao et al., 2003) supports the hypothesis that these perox-idases have a role in defense. Given the similarity in sequence between the two peroxidases, it is likely that they have similar activities, but as indicated by the RT-PCR assays and proteomics data (Charmont et al., 2005, discussed below), they are differentially regulated: At3g49120 is constitutively expressed whereas At3g49110 is highly induced on infection.
Figure 6. Transcript levels of the peroxidase-encoding genes AtPCa (At3g49110) and AtPCb (At3g49120) in Col-0 and FBP1 transgenic line H4.
(a) Ratio of At3g49110 expression in H4 versus wild-type Col-0 in plants that were either mock-inoculated or inoculated with Pto DC3000(avrRpm1). Leaves were harvested 5 h post-inoculation. Values are averages of three independent experiments.
(b) Ratio of At3g49120 expression in H4 versus wild-type Col-0 in plants that were either mock-inoculated or inoculated with Pto DC3000(avrRpm1).
(c) Change in transcript level of At3g49110 and At3g49120 in wild-type Col-0 in response to inoculation with Pto DC3000(avrRpm1). Fold-change is relative to mock-inoculated. Steady-state mRNA in 4-week old Col-0 and H4 was assayed by real-time RT-PCR and results from three independent biological replicates were averaged.
Discussion
Role of peroxidases in the Arabidopsis defense response
It is now recognized that various plants exhibit either a plasma membrane-localized NADPH/NADH oxidase-dependent, or a cell wall peroxidase-dependent oxidative burst, or both (for reviews see Apel and Hirt, 2004; Grant and Loake, 2000). It is therefore important to determine in which situations either or both enzymes are involved in the generation of ROS, and the respective roles of the two types of ROS-generation mechanisms in the plant-defense response. In two biochemically well characterized systems, French bean cells and rose cells, different mechanisms are involved in the generation of ROS (Bolwell et al., 1998). In French bean, the peroxidase appears to be the dominant mechanism; in rose cells, the oxidase with additional specificity towards NADH as well as NADPH is dominant (Bolwell et al., 1998). On the other hand, other plants, including cotton, exhibit both mechanisms of ROS generation, albeit temporally separated (Martinez et al., 1998).
Here we determined the extent to which cell wall-localized peroxidases contribute to the generation of ROS in Arabidopsis in response to pathogens and pathogen-derived elicitors. Using an Arabidopsis cell suspension-culture system and an elicitor from the fungal pathogen F. oxysporum, we showed that production of ROS is sensitive to azide and cyanide, inhibitors that in other species have been shown to target primarily cell wall-derived peroxidases. Consistent with these results, the F. oxysporum-elicited production of H2O2 was relatively insensitive to DPI, an inhibitor that is relatively specific for NADPH-oxidases. To confirm that peroxidases play a role in the Arabidopsis oxidative burst, we expressed a cDNA encoding the French bean peroxidase FBP1 in the anti-sense orientation in transgenic Arabidopsis plants. These FBP1 transgenic plants exhibited decreased production of H2O2 in response to the F. oxysporum elicitor and to infection by an avirulent strain of P. syringae; were more susceptible to both virulent and avirulent bacterial and fungal pathogens; had reduced levels of ionically cell wall-bound peroxidase activity; and exhibited lower levels of mRNAs corresponding to two close Arabidopsis homologs of FBP1, At3g49110 and At3g49120.
The reduced levels of the oxidative burst that we observed in transgenic FBP1 lines, as well as their enhanced susceptibility to both virulent and avirulent P. syringae strains, need to be reconciled with previous observations that indicate that NADPH oxidase(s) play a key role in the oxidative burst (Grant et al., 2000b; Torres et al., 2002). In the Grant et al. (2000b) study, H2O2 production following inoculation with Pto DC3000(avrRpm1) was sensitive to 7 µm DPI, and inhibition at this concentration was interpreted as demonstrating that an NADPH oxidase was most likely the origin of ROS detected with cerium chloride staining. Although the concentration of DPI used would favor inhibition of the NADPH oxidase, the peroxidase system is also sensitive to DPI, albeit with lower specificity, and 7 µm DPI would also inhibit peroxidase-generated ROS to some extent (Frahry and Schopfer, 1998). In another study utilizing a ROS-responsive GST-luciferase gene expression system, the oxidative burst proved to be sensitive to both 3 µm DPI and 1 µm azide when the inhibitor was co-inoculated with either avrB- or avrRpt2-expressing P. syringae strains (Grant et al., 2000a). This was interpreted as indicating that both oxidases and peroxidases could be engaged in generating ROS in Arabidopsis.
By generating a series of Arabidopsis NADPH oxidase mutants, Torres et al. (2002) showed that the AtrbohD and AtrbohF genes were required for production of a full oxidative burst. Because our results show that peroxidases are also required for H2O2 production in response to the same avirulent P. syringae strains, it seems likely that both membrane-associated NADPH oxidases and cell wall-bound peroxidases are required to generate H2O2. A major difference in our study, in comparison with the Torres et al. (2002) study, is that we observed that the FBP1 anti-sense transgenic plants are highly susceptible to pathogen infection, whereas the atrboh mutants in the Torres et al. (2002) study were not more susceptible. This raises a question as to the respective roles of peroxidases and oxidases in the oxidative burst.
A possible explanation of the role of peroxidases in the oxidative burst comes from recent work of Torres et al. (2005), in which they constructed an lsd1 atrbohD atrbohF triple mutant. The lsd1 (lesions simulating disease) mutant exhibits spreading lesions and, surprisingly, the triple mutant exhibited an enhanced spreading lesion phenotype compared with lsd1, contrary to what was expected if the NADPH oxidase-generated burst was responsible for triggering cell death. Torres et al. (2005) concluded, counter-intuitively, that the role of NADPH oxidases is to limit the spread of a salicylic acid-elicited cell-death program in cells surrounding an infection site. Moreover, they showed that the NADPH oxidases need to be activated by an independent source of ROS to generate their own oxidative burst.
A model, consistent with our observations as well as those of Torres et al. (2005), is that apoplastic peroxidases are an initial rapid source of ROS, and are essential for conferring at least partial resistance independently of any involvement of NADPH oxidases. Subsequently, the peroxidase-generated ROS activate NADPH oxidases, which, in turn, generate a plasma membrane-associated oxidative burst, the primary role of which is to limit the extent of cell death in neighboring cells. If this model is correct, the failure to detect an oxidative burst in Arabidopsis atrbohD/F mutants could be a consequence of the fact that the oxidative burst assays were carried out several hours after elicitation, long after a peroxidase-mediated burst may have occurred (Torres et al., 2002, 2005).
For some species, there is evidence that recognition of virulent and avirulent pathogens produces different oxidative burst profiles, with early ROS production observed in non-host, compatible and incompatible interactions, and a later, more extensive and sustained burst only in R genemediated resistance responses (Apel and Hirt, 2004; Grant and Loake, 2000). Data presented here support a role for cell wall-associated peroxidase(s) in both phases, as hydrogen peroxide production is reduced in the FBP1 transgenic line compared with wild-type plants following elicitation with an elicitor from cell walls of F. oxysporum within a 1-h time frame, or elicitation with an avirulent strain of bacteria within a 5-h time frame. One possibility is that peroxidase(s) catalyze ROS production during basal resistance triggered by recognition of pathogen-associated molecular patterns of virulent pathogens, and that this initial oxidative burst is essential for subsequent activation of NADPH oxidase during an R gene-mediated HR, as well as for activation of basal defenses. In this model, peroxidases play an essential role in basal resistance, independently of their role in activating NADPH oxidases, explaining why the FBP1 anti-sense line has a much more severe phenotype than an atrbohD/F mutant. Consistent with this interpretation is our observation that the FBP1 transgenic plants are more susceptible to highly virulent necrotrophic (B. cinerea) and biotrophic (powdery mildew, Golovinomyces orontii) fungal pathogens as well as virulent and avirulent P. syringae strains. Additional support for this model comes from recent work showing differential effects of DPI on H2O2 production following challenge of Arabidopsis leaves with hrpL mutants of P. syringae pv. phaseolicola, or with P. syringae pv. phaseolicola carrying an avirulence gene that triggers an HR. Interestingly, the oxidative burst elicited by the hrpL mutant, which induced papilla formation but not HR, was much less susceptible to DPI inhibition than the burst elicited by the avr gene, suggesting that ROS may be generated by different mechanisms, depending on the challenge (Soylu et al., 2005). Alternatively, overexpressing FBP1 might compromise resistance by preventing oxidative cell-wall cross-linking and the formation of barriers. However, the loss of resistance to avirulent P. syringae argues against an effect solely on physical barriers. Distinguishing between these possibilities requires further biochemical studies, such as those carried out in French bean (Bolwell et al., 2002), and assays of defense gene expression. Insertion mutants that target specifically either At3g49120 or At3g49110 would help clarify the involvement of these two peroxidases during defense. However, lines with insertions in regions of At3g49120 that would be likely to interfere with protein function are not available: although Arabidopsis stock collections list such mutants, it is not possible to obtain them; in at least one case the line could not be maintained, consistent with our own experience. It is likely that tests of the requirement of AtPCb and/or AtPCa for the production of H2O2 in response to potential pathogens will require an inducible system for targeted reduction of At3g49110 and At3g49120 expression.
Arabidopsis peroxidases
Although the cationic French bean FBP1 peroxidase shows reasonable similarity to a number of Arabidopsis type III peroxidases, including the anionic AtPA2, which has a putative role in lignification (Ostergaard et al., 2000), expression of anti-sense cDNA coding for FBP1 appears to target specifically At3g49120 and At3g49110 that encode AtPCb and AtPCa, respectively (Valeério et al., 2004). Both peroxidases have been grouped by a detailed phylogenetic analysis in a clade of seven out of the 73 type III peroxidases in the Arabidopsis genome that is closest to FBP1 (Duroux and Welinder, 2003). Comparison of FBP1 and AtPCb amino acid sequences indicates that they share 53.1% identity and 65.4% similarity, while FBP1 and AtPCa have 53.3% identity and 65.0% similarity. Furthermore, FBP1, AtPCa and AtPCb are all cationic, have a similar number of potential glycosylation sites, and have an unusual carboxy-terminal extension that is known to be cleaved in the case of FBP1 (Blee et al., 2001). All three have predicted amino-terminal secretion sequences leading to cell-wall localization. These properties of AtPCa and AtPCb are consistent with the data in Figure 5 that show a reduction of a wall-bound cationic peroxidase activity in the FBP1 transgenic plants. AtPCb and AtPCa have been characterized previously (Ostergaard et al., 1998; Tognolli et al., 2002; Welinder et al., 2002). AtPCb is expressed throughout the plant, whereas AtPCa has been detected only in aerial organs (Welinder et al., 2002). Like FBP1, AtPCb is induced in response to a variety of different pathogens and elicitors including virulent and avirulent H. parasitica, avirulent Pto DC3000, B. cinerea, and oligogalacturonides (Maleck et al., 2000; Scheideler et al., 2002; Tao et al., 2003; S. Ferrari, J.P., C.D., J.D. and F.M.A., unpublished data). Less is known about the regulation of AtPCa because it is not represented on the Affymetrix ATH1 GeneChip, although the data in Figure 6 show that it is strongly induced following P. syringae infection. At the protein level, AtPCb is one of only eight extracellular peroxidases detected in the culture medium of liquid-grown 2-week-old seedlings; AtPCa was not detected (Charmont et al., 2005). Data from the FBP1 anti-sense transgenic line do not allow us to determine the relative importance of AtPCb and AtPCa in limiting pathogen growth, as both peroxidases have reduced expression in this line. However, the difference in protein levels prior to infection, and the difficulty of maintaining lines with insertions in At2g49120, suggest that, of the two, AtPCb has the more critical role in defense.
Interestingly, two glutathione peroxidases were also downregulated in uninfected leaves of the FBP1 anti-sense plants. Presumably, these would function in scavenging ROS. It may well be that levels of ROS accumulating over time in these plants under normal light conditions are reduced compared with the wild type, resulting in lower levels of induction of protective peroxidases. It is unlikely that these two glutathione peroxidases are direct targets of the FBP1 anti-sense transgene, as they have both low overall similarity and regions of identity with FBP1 that are no longer than 10 nucleotides.
Susceptibility of transgenic FBP1 plants to pathogens
Despite all efforts, most of the Arabidopsis transgenic FBP1 lines succumbed to opportunistic fungal infections during culturing. However, two T1 lines survived and homozygous lines were derived from one of these. The plants that survived were normal phenotypically, although the homozygous lines formed somewhat larger rosette leaves and inflorescence initiation was delayed compared with Col-0. However, because of their susceptibility to opportunistic infections, even these surviving FBP1 transgenic lines are very difficult to maintain. The FBP1 transgenic lines demonstrated reduced capacity for ROS generation when challenged with the F. oxysporum elicitor in the leaf-disc assay, and exhibited a level of disease susceptibility comparable with previously isolated Arabidopsis enhanced disease susceptibility (eds) mutants including npr1 and pad4 (Glazebrook et al., 1996; Rogers and Ausubel, 1997) when challenged with the virulent bacterial pathogens Psm ES4326 or Pto DC3000.
In contrast to most other eds mutants, however, the transgenic FBP1 plants exhibited enhanced susceptibility to avirulent P. syringae strains. While Psm ES4326(avrRpt2) grew less than 10-fold in wild-type Col-0 plants over 3 days, the same strain multiplied 20 000-fold in line H4 and actually exceeded growth of a near-isogenic virulent strain of Psm by a small but reproducible amount. A number of studies have demonstrated that, in the absence of an R gene-mediated resistance response, bacterial avirulence proteins (including AvrRpt2) have effector or virulence functions (Abramovitch et al., 2003; Chen et al., 2004; Chisholm et al., 2005; Greenberg and Vinatzer, 2003; Hauck et al., 2003; Kim et al., 2005; Metz et al., 2005). Our data suggest that not only basal resistance, but also R gene-mediated resistance, are compromised by a reduction in AtPCb and/or AtPCa activity. Consistent with an increase in bacterial growth, tissue collapse 3 days after inoculation with either virulent or avirulent P. syringae strains was more extensive in the peroxidase anti-sense lines than in the wild type.
The peroxidase anti-sense lines are also unusual in having increased susceptibility to a broad range of virulent pathogens. Many of the mutants isolated on the basis of enhanced susceptibility to P. syringae do not show a similar eds phenotype with G. orontii or B. cinerea (Ferrari et al., 2003; Reuber et al., 1998) and, conversely, a majority of mutants isolated in a screen for enhanced susceptibility to G. orontii are not significantly more susceptible to P. syringae ES4326 (Dewdney et al., 2000). However, FBP1 anti-sense line H4 is strikingly more susceptible than the wild-type to all three of these pathogens, which represent bacterial, biotrophic fungal and necrotrophic fungal phytopathogens.
Although FBP1 anti-sense plants exhibited increased susceptibility to pathogens, they were more resistant to the fungal toxin fumonisin B1. In wild-type plants, fumonisin B1 triggers the generation of reactive oxygen intermediates (Stone et al., 2000), which may be essential for FB1 induction of HR-like cell death (Gilchrist, 1998). A number of studies (Alvarez et al., 1998; Bennett et al., 2005; Delledonne, 2005; Kliebenstein et al., 1999; Mach et al., 2001; Torres et al., 2005) indicate that cell-death programs are impacted by multiple ROS, including superoxide, hydrogen peroxide and nitric oxide, which may activate or repress cellular suicide. Although the precise mechanism of FB1-induced cell death in Arabidopsis is unclear, the attenuation of HR-like cell death in FB1-treated FBP1 anti-sense plants suggests that peroxidase-generated H2O2 is involved. It would be of interest to test whether the atrbohD/F double mutant is also resistant to fumonisin B1.
Experimental procedures
Plant material
Arabidopsis thaliana ecotype Columbia (Col-0) plants were germinated and grown in a glasshouse or climate-controlled chamber as described by Reuber et al. (1998). Arabidopsis suspension-culture cell lines were a kind gift of Professor A. R. Slabas, Durham University, UK, and were maintained in Murashige and Skoog basal salts with minimal organics (MSMO) medium (Sigma, Poole, Dorset, UK) containing sucrose (30 g l−1)aphthylene acetic acid (0.5 mg l−1) and kinetin (0.05 mg l−1) under low light intensity in a 16 h light/8 h dark regime.
Bacterial strains and growth conditions
The virulent bacterial strains Pseudomonas syringae pv. maculicola ES4326 (Dong et al., 1991) and P. syringae pv. tomato DC3000 (Cuppels, 1986) have been described previously. The corresponding isogenic avirulent strains carry avrRpt2 on the plasmid pLH12 (Whalen et al., 1991) or avrRPM1 on plasmid pVSP61 (Debener et al., 1991). Pseudomonas syringae strains were grown at 28°C in King’s B medium (10 mg ml−1 protease peptone, 2 mg ml−1 K2HPO4, 10 mg ml−1 glycerol, 6 mm MgSO4 pH 7.0) supplemented with 100 µg ml−1 streptomycin for strain ES4326 or 25 µg ml−1 rif-ampicin for strains DC3000. Strains containing pLH12 and pVSP61 were maintained on media containing 10 µg ml−1 tetracycline or 25 µg ml−1 kanamycin, respectively.
Fungal strains, toxins and elicitor preparation
Golovinomyces orontii (formerly Erysiphe orontii) isolate MGH (Plotnikova et al., 1998), Botrytis cinerea (Ferrari et al., 2003) and Fusarium oxysporum f.sp. matthioli race 1 (Kistler et al., 1991) have been described previously. An F. oxysporum elicitor preparation was prepared by culturing F. oxysporum as a yeast form in half-strength potato dextrose broth (Sigma P-6685), collecting yeast cells by centrifugation, resuspending in 100 ml 500 mm KH2PO4, and centrifuging at 5000 g in an SS-34 rotor (Sorvall, Asheville, NC, USA) for 30 min. The pellet was resuspended in 200 ml 50 mm KH2PO4 and centrifuged as above. The pellet was then sequentially washed and centrifuged in chloroform:methanol (1:1 v/v) and acetone. The pellet was resuspended in ddH2O (10 g pellet l−1) and autoclaved for 30 min at 121°C. The elicitor preparation was stored at −20°C until use. Fumonisin B1 was obtained from Sigma and kept as a stock solution of 7 mm in methanol.
Transformation of Arabidopsis
The isolation of a French bean oxidative burst peroxidase cDNA (FBP1 cDNA) has been described (Blee et al., 2001). Agrobacterium tumefaciens strain LB4044, harboring the binary vector pBin19 expressing FBP1 in the anti-sense direction under the control of the CaMV/35S promoter and the A. tumefaciens nopaline synthase terminator (Blee et al., 2003), was used to transform Col-0 plants as described (Clough and Bent, 1998). Transformed plants were placed in the dark with high humidity overnight, then returned to the glasshouse to allow seed setting. Putative transgenic seeds were collected, surface sterilized, resuspended in 0.1% agar, germinated on solid MS salts medium (0.8% phytoagar) containing 50 µg ml−1 kanamycin, transferred to soil, and screened for the presence of the transgene using the PCR as follows. DNA was isolated from leaf tissue using the fast DNA kit for plants (Bio 101, Carlsbad, CA, USA) and added to Ready to Go PCR beads (Amersham Pharmacia Biotech, Buckinghamshire, UK) with CaMV/35S promoter and nopaline synthase terminator primers 5′-CAATCCCACTATCCTTCGC-3′ and 5′-CATCGCAAGACCGGCAACAG-3′, respectively. Cycling proceeded after 7 min at 95°C with 40 cycles of 1 min at 95°C, 1 min at 58°C and 2 min at 72°C. Tn lines were derived by repeated selection on 50 µg ml−1 kanamycin leading to homozygous lines as determined by PCR analysis of T3 progeny.
Elicitation of suspension cultured cells and leaf discs: measurement of the oxidative burst
Suspension-cultured cells and leaf discs (0.13 cm2) were treated directly with the F. oxysporum elicitor preparation at a concentration of 25 µg ml−1 glucose equivalents. In the case of leaf discs, 30 discs excised using a cork borer were floated on 2 ml H2O containing elicitor in Petri dishes (90-mm diameter), vacuum-infiltrated three times for 10 sec each time, and gently swirled on a platform shaker. For both the suspension-cell and leaf-disc assays, 150 µl medium was withdrawn at various times, briefly centrifuged, and the amount of H2O2 in 100 µl was measured using xylenol orange, as described (Bindschedler et al., 2001).
Pathogen-susceptibility assays
For bacterial growth assays, plants grown in a climate-controlled chamber were infiltrated with the relevant bacteria at the doses indicated. Dose was determined by OD600, with approximately 103 CFU cm−2 leaf area being equivalent to an OD600 of 0.0002. Bacterial growth was assayed by excising a leaf sample consisting of one or two 0.13-cm2 disks from each infected leaf using a cork borer and grinding the sample in a microfuge tube in 200 µl 10 mm MgSO4 using a plastic pestle. Appropriate dilutions were plated on King’s B medium containing the appropriate antibiotic(s). Growth data are reported as the log of the bacterial CFU cm−2 leaf area (log CFU cm−2). For B. cinerea susceptibility assays, leaves of 4-week-old plants were inoculated with 5 µl drops of a 5 × 105 spore suspension and scored 4 days post-inoculation. Golovinomyces orontii infection occurred opportunistically.
Detection of reactive oxygen species
For in situ detection of H2O2, DAB staining was carried out using an adaptation of the method of Thordal-Christensen et al. (1997). Bacteria were inoculated at OD600 = 2.0 (107 CFU cm−2) into leaves, which were collected after 2 h and infiltrated under gentle vacuum with 1 mg ml−1 DAB containing 0.05% v/v Tween 20 and 10 mm sodium phosphate buffer pH 7.0. The reaction was terminated at 6–7 h post-inoculation, when a brown precipitate started to be visible in Col-0 leaves. Leaves were fixed and then boiled for 15 min in ethanol:acetic acid:glycerol 3:1:1. Bleaching solution was replaced and leaves were incubated until the chlorophyll was completely bleached. Leaves were observed by light microscopy under bright field at 100× magnification. Hydrogen peroxide was also detected in situ by electron microscopy with cerium chloride staining. Leaves were inoculated as described above; after 3 h, 1–2-mm2 pieces were incubated for 1 h in CeCl3. Leaf tissue was dehydrated and fixed as described by Bestwick et al. (1997). Samples were observed at 20 000- or 30 000-fold magnification using a Hitachi H600 transmission electron microscope.
Peroxidase assays
Peroxidases were extracted from leaves by grinding in 100 mm Tris-HCl buffer pH 7.2 containing 5 mm 2-mercaptoethanol and 0.25 m sucrose. Homogenates were centrifuged at 14 000 g for 5 min and the supernatant collected as the soluble fraction. The pellet was resuspended in 20 mm Tris-HCl pH 7.2, 1 m NaCl, 1 mm CaCl2, 1 mm MgCl2, 1 mm MnCl2, and extracted end-over-end for 30 min at 4°C. Following centrifugation at 14 000 g for 5 min, the supernatants were pooled and referred to as the ‘ionically-bound’ peroxidase fraction. Peroxidase activity was determined spectrophotometrically at 405 nm using 10 mm 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) as substrate in 1 ml 10 mm sodium acetate buffer pH 4.4 and 0.1 mm H2O2 to which 50 µl enzyme extract was added to start the reaction. Peroxidase fractions were also separated by isoelectric focusing (IEF). Equal amounts (20 µg) of protein were applied to precast IEF gels (pH range 3–10; Pharmacia, Sandwich, Kent, UK) and proteins separated using 0.4% (w/v) arginine and lysine, respectively, in 12% aqueous ethylene diamine as cathiodic buffer and 0.33% (w/v) aspartic acid and 0.37% (w/v) glutamic acid as aniodic buffers. Electrophoresis was performed at 6 W for 3 h. Plates were developed using 0.04% o-danisidine and 0.15% H2O2 in 10 mm sodium acetate buffer pH 4.5.
Genome-wide expression profiling
Two samples of Col-0 or FBP1 transgenic H4 plants, each sample consisting of leaves from six plants, were harvested from 4-week-old uninfected plants. Total RNA was extracted using the Qiagen RNeasy Plant RNA Miniprep kit (Qiagen, Valencia, CA, USA). Each sample was split into two before homogenization and re-pooled before loading on the RNA-binding column. RNA quality was assessed by determining the A260/280 ratio of RNA in Tris buffer and by checking the integrity of RNA on an Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, USA). Target labeling and hybridization to Affymetrix ATH1 GeneChips were performed according to the protocol given in the Affymetrix GeneChip Expression Analysis Technical Manual 701025, rev. 1 (for details see Appendix S1). Arrays were scanned using an Affymetrix GeneArray 2500 Scanner and Affymetrix microarray suite ver. 5.0 software. Data analysis was performed with rosetta resolverver. 3.2 gene expression data analysis system (Rosetta Inpharmatics, Kirkland, WA, USA), using Affymetrix CEL files of array feature intensities and standard deviations as input. Determination of absolute intensity values, propagation of error and P values, and normalization for comparing arrays in the resolver system have been described by Waring et al. (2001). To increase detection sensitivity, data from the two replicate samples of each line were combined after global normalization, and average intensity, error and P value were derived for each gene. For comparison of H4 with wild type, ratios of average normalized intensities were log10-transformed and P values of differential expression were determined by t-test. For gene annotation we used the TAIR and TIGR databases.
Real-time RT-PCR
RNA, isolated as described for expression profiling, was reverse-transcribed with iScript Reverse Transcriptase (Bio-Rad, Hercules, CA, USA) using the manufacturer’s protocol. At3g49120 and At3g49110 cDNAs were amplified using iQ SYBR green Supermix (Bio-Rad) with primers TATGCTCACCATTGCAGCTC and GGACG-ATCGAGACCAACATT for At3g49120 and primers TGTCCTCGC-AATGGTAATCA and GATTGTGTCAGTGGCATTGG for At3g49110. The amplification protocol included an initial 3′ incubation at 95° followed by 45 cycles of 95°10′, 55°30′, and 72°30′. Ubiquitin (At5g25760) was used as a standard, and amplified using primers TTACGAAGGCGGTGTTTTTCAG and TTCCCTGAGTCGCAGTTAA-GAG. Relative levels of transcripts were calculated using the method of Pfaffl (2001).
Acknowledgements
We thank the Harvard University Bauer Center for Genomics Research, especially J. Couget, P. Grosu, R. Gali and C. Bailey, for assistance with microarray analysis of expression profiles. We also thank M. Torres for helpful comments on the manuscript. This work was funded by NIH Grant R37 GM48707 and NSF 2010 Grant DBI-0114783 awarded to F.M.A.
Footnotes
Supplementary Material
The following supplementary material is available for this article online:
Appendix S1. Microarray hybridization.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 2003;22:60–69. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer P-J, Ishikawa A, Dixon RA, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;93:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Bennett M, Mehta M, Grant M. Biophoton imaging; a non-destructive method for assaying R gene responses. Mol. Plant Microb. Interact. 2005;18:95–102. doi: 10.1094/MPMI-18-0095. [DOI] [PubMed] [Google Scholar]
- Bestwick CS, Brown IR, Bennett MHR, Mansfield JW. Localisation of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv. phaseolicola. Plant Cell. 1997;9:209–221. doi: 10.1105/tpc.9.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, Minibayeva F, Gardber SL, Gerrish C, Davies DR, Bolwell GP. Early signalling events in the apoplastic oxidative burst in suspension cultured French bean cells involve cAMP and Ca2+ New Phytol. 2001;151:185–194. doi: 10.1046/j.1469-8137.2001.00170.x. [DOI] [PubMed] [Google Scholar]
- Blee KA, Jupe SC, Richard G, Zimmerlin A, Davies DR, Bolwell GP. Molecular identification and expression of the peroxidase responsible for the oxidative burst in French bean (Phaseolus vulgaris L.) and related members of the gene family. Plant Mol. Biol. 2001;47:607–620. doi: 10.1023/a:1012307324782. [DOI] [PubMed] [Google Scholar]
- Blee KA, Choi J-W, O’Connell AP, Schuch W, Lewis NG, Bolwell GP. Alignin-specific peroxidasein tobacco whose antisense suppression leads to vascular tissue modification. Phytochemistry. 2003;64:163–176. doi: 10.1016/s0031-9422(03)00212-7. [DOI] [PubMed] [Google Scholar]
- Bolwell GP. Role of active oxygen species and NO in plant defence responses. Curr. Opin. Plant Biol. 1999;2:287–294. doi: 10.1016/S1369-5266(99)80051-X. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defence - a broad perspective. Physiol. Mol. Plant Pathol. 1997;51:347–366. [Google Scholar]
- Bolwell GP, Davies DR, Gerrish C, Auh CK, Murphy TM. Comparative biochemistry of the oxidative burst produced by rose and French bean cells reveals two distinct mechanisms. Plant Physiol. 1998;116:1379–1385. doi: 10.1104/pp.116.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Bindschedler LV, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F. The apoplastic oxidative burst in response to biotic stress in plants: a three component system. J. Exp. Bot. 2002;53:1367–1376. [PubMed] [Google Scholar]
- Charmont S, Jamet E, Pont-Lezica R, Canut H. Proteomic analysis of secreted proteins from Arabidopsis thaliana seedlings: improved recovery following removal of phenolic compounds. Phytochemistry. 2005;66:453–461. doi: 10.1016/j.phytochem.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Cuzick A, Moeder W, Tang D, Innes RW, Klessig DF, McDowell JM, Kunkel BN. The Pseudomonas syringae type III effector AvrRpt2 functions down-stream or independently of SA to promote virulence on Arabidopsis thaliana. Plant J. 2004;37:494–504. doi: 10.1111/j.1365-313x.2003.01984.x. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc. Natl Acad. Sci.USA. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cuppels DA. Generation and characterisation of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 1986;52:323–327. doi: 10.1128/aem.51.2.323-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debener T, Lehnackers H, Arnold M, Dangl JL. Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1991;1:289–302. doi: 10.1046/j.1365-313X.1991.t01-7-00999.x. [DOI] [PubMed] [Google Scholar]
- Delledonne M. NO news is good news for plants. Curr. Opin. Plant Biol. 2005;8:390–396. doi: 10.1016/j.pbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000;24:205–218. doi: 10.1046/j.1365-313x.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroux L, Welinder KG. The peroxidase gene family in plants: a phylogenetic overview. J. Mol. Evol. 2003;57:397–407. doi: 10.1007/s00239-003-2489-3. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003;35:193–205. doi: 10.1046/j.1365-313x.2003.01794.x. [DOI] [PubMed] [Google Scholar]
- Frahry G, Schopfer P. Inhibition of O2-reducing activity of horseradish peroxidase by diphenyleneiodonium. Phytochemistry. 1998;48:223–227. doi: 10.1016/s0031-9422(98)00004-1. [DOI] [PubMed] [Google Scholar]
- Gilchrist DG. Programmed cell death in plant disease: the purpose and promise of cellular suicide. Annu. Rev. Phytopathol. 1998;36:393–414. doi: 10.1146/annurev.phyto.36.1.393. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JJ, Loake GJ. Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 2000;124:21–29. doi: 10.1104/pp.124.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JJ, Yun B-W, Loake GJ. Oxidative burst and cognate redox signalling reported by luciferase imaging: identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant J. 2000a;24:569–582. doi: 10.1046/j.1365-313x.2000.00902.x. [DOI] [PubMed] [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, Mansfield J. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 2000b;23:441–450. doi: 10.1046/j.1365-313x.2000.00804.x. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Vinatzer BA. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 2003;6:20–28. doi: 10.1016/s1369-5274(02)00004-8. [DOI] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl Acad. Sci. USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, Debroy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors Inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Kistler HC, Momol EA, Benny U. Repetitive genomic sequences for determining relatedness among strains of Fusarium-Oxysporum. Phytopathology. 1991;81:331–336. [Google Scholar]
- Kliebenstein DJ, Dietrich RA, Martin AC, Last RL, Dangl JL. LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 1999;12:1022–1026. doi: 10.1094/MPMI.1999.12.11.1022. [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl Acad. Sci. USA. 2001;98:771–776. doi: 10.1073/pnas.021465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Martinez C, Montillet JL, Bresson E, Agnel JP, Daı¨ GH, Daniel JF, Geiger JP, Nicole M. Apoplastic peroxidase generates superoxide anions in cells of cotton cotyledons undergoing the hypersensitive reaction to Xanthomonas campestris pv. malvacearum race 18. Mol. Plant Microbe Interact. 1998;11:1038–1047. [Google Scholar]
- Metz M, Dahlbeck D, Morales CQ, Al Sady B, Clark ET, Staskawicz BJ. The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 2005;41:801–814. doi: 10.1111/j.1365-313X.2005.02338.x. [DOI] [PubMed] [Google Scholar]
- Ostergaard L, Pedersen AG, Jespersen HM, Brunak S, Welinder KG. Computational analyses and annotations of the Arabidopsis peroxidase gene family. FEBS Lett. 1998;433:98–102. doi: 10.1016/s0014-5793(98)00849-7. [DOI] [PubMed] [Google Scholar]
- Ostergaard L, Teilum K, Mirza O, Petersen M, Welinder KG, Mundy J, Gajhede M, Hendriksen A. Arabidopsis ATPA2 peroxidase: expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 2000;44:231–243. doi: 10.1023/a:1006442618860. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova JM, Reuber TL, Ausubel FM, Pfister DH. Powdery mildew pathogenesis of Arabidopsis thaliana. Mycologia. 1998;90:1009–1016. [Google Scholar]
- Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausubel FM. Correlation of defence gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 1998;16:473–485. doi: 10.1046/j.1365-313x.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM. Arabidopsis enhanced disease susceptibility mutants exhibit enhances susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD. Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem. 2002;277:10555–10561. doi: 10.1074/jbc.M104863200. [DOI] [PubMed] [Google Scholar]
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl Acad. Sci. USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soylu S, Brown I, Mansfield JW. Cellular reactions in Arabidopsis following challenge by strains of Pseudomonas syringae: from basal resistance to compatibility. Physiol. Mol. Plant Pathol. 2005;66:232–243. [Google Scholar]
- Stone JM, Heard JE, Asai T, Ausubel FM. Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell. 2000;12:1811–1822. doi: 10.1105/tpc.12.10.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang H-S, Han B, Zhu T, Zou G, Katagiri F. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- Tognolli M, Penel C, Greppin H, Simon P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene. 2002;288:129–138. doi: 10.1016/s0378-1119(02)00465-1. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Torres MA, Onouchi H, Hamada S, Machida C, Hammond-Kosak KE, Jones JDG. Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox) Plant J. 1998;14:365–370. doi: 10.1046/j.1365-313x.1998.00136.x. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG. Arabidopsis gp91phox homologues, AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl Acad. Sci. USA. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL. Pathogen-induced NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005;37:1130–1134. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- Valeério L, de Meyer M, Penel C, Dunand C. Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry. 2004;65:1331–1342. doi: 10.1016/j.phytochem.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Waring JF, Jolly RA, Ciurlionis R, Lum PY, Praestgaard JT, Morfitt DC, Buratto B, Roberts C, Schadt E, Ulrich RG. Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol. Appl. Pharmacol. 2001;175:28–42. doi: 10.1006/taap.2001.9243. [DOI] [PubMed] [Google Scholar]
- Welinder KG, Justesen AF, Kjaersgard IV, Jensen RB, Rasmussen SK, Jespersen HM, Duroux L. Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 2002;269:6063–6081. doi: 10.1046/j.1432-1033.2002.03311.x. [DOI] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P. The oxidative burst: a plant’s early response against infection. Biochem. J. 1997;322:4158–4163. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]