Abstract
The mutation of the axial ligand of the type I copper protein amicyanin from Met to Lys results in a protein that is spectroscopically invisible and redox inactive. M98K amicyanin acts as a competitive inhibitor in the reaction of native amicyanin with methylamine dehydrogenase indicating that the M98K mutation has not affected the affinity for its natural electron donor. The crystal structure of M98K amicyanin reveals that its overall structure is very similar to native amicyanin but that the type I binding site is occupied by zinc. Anomalous difference Fourier maps calculated using the data collected around the absorption edges of copper and zinc confirm the presence of Zn2+ at the type I site. The Lys98 NZ donates a hydrogen bond to a well-ordered water molecule at the type I site which enhances the ability of Lys98 to provide a ligand for Zn2+. Attempts to reconstitute M98K apoamicyanin with copper resulted in precipitation of the protein. The fact that the M98K mutation generated such a selective zinc-binding protein was surprising as ligation of zinc by Lys is rare and this ligand set is unique for zinc.
Keywords: cupredoxin, metalloprotein, protein folding, X-ray structure
1. Introduction
Type 1 copper sites are found in a wide range of redox proteins in bacteria, plants and animals, and function as electron transfer (ET) mediators [1]. A single copper is bound in the site and is always coordinated with three strong equatorial ligands; a Cys and two His residues, and with one weak axial ligand, usually a Met. Cupredoxins are small soluble type 1 copper proteins with a single copper site which are characterized by an intense blue color and absorption centered near 600 nm that results from a S(Cys)π→Cu(II)dx2-y2 ligand-to-metal charge transfer transition. The physical properties and X-ray crystal structures of several cupredoxins have been characterized [2, 3].
Amicyanin is a type 1 copper protein which serves as an electron acceptor for methylamine dehydrogenase (MADH) in many methylotrophic and autotrophic bacteria. Amicyanin from Paracoccus denitrificans mediates ET from MADH to cytochrome c-551i in the periplasmic space and allows the host bacteria to use methylamine as a sole source of carbon and energy [4]. The complex of MADH, amicyanin, and cytochrome c-551i is one of the best characterized physiological protein ET systems. High resolution crystal structures are available for the binary complex of amicyanin with MADH and a ternary complex of MADH, amicyanin and cytochrome c-551i [5, 6]. Structure-function relationships have been extensively studied by site-directed mutagenesis of amicyanin to identify roles of specific amino acid residues in determining its spectroscopic, redox, and ET properties [3, 7]. A high resolution neutron diffraction model for amicyanin has also been reported which revealed a dynamic nature of residues around the copper site and along the ET path [8, 9].
The type I copper site of the amicyanin is coordinated by three equatorial ligands provided by nitrogens of His53 and His95 and a sulfur of Cys92, with a fourth axial ligand provided by the sulfur of Met98, thereby forming a distorted tetrahedral geometry [10]. Met98 of P. denitrificans amicyanin was previously mutated to Ala, Gln, and Leu [11–14]. These studies indicated that the position and rigidity of the axial ligand of the type 1 copper site influences the overall protein stability, the active site ligation geometry, the spectroscopic properties, and ET properties. The axial ligand was also shown to exert a profound influence on the uptake specificity of the metal ion which occupies the type 1 site.
The role of the metal in the molecular mechanisms of metalloprotein biosynthesis and assembly is not well understood [15]. A comparative study of the specificity of the type 1 site of native and M98Q amicyanins for either Cu2+ or Zn2+ revealed that the influence of the axial ligand on metal specificity is strongest prior to the completion of protein folding and adoption of the final type I site geometry [13]. In this study, the role of the axial ligand in determining the metal specificity during and after protein folding was further examined by mutation of Met98 of amicyanin to Lys. This resulted in the isolation of a stable but inactive protein which contained Zn2+ in the metal site. In contrast to all other mutants of amicyanin which have been characterized, it was not possible to remove the zinc and then reconstitute M98K amicyanin with copper. The structure of this unusual zinc-specific amicyanin is presented together with data concerning its ability to interact with MADH, and properties of the metal-free M98K apoamicyanin.
2. Materials and Methods
2.1. Protein purification
Previously described procedures were used to purify native amicyanin [4] and MADH [4] from P. denitrificans. M98K amicyanin was expressed in Escherichia coli and purified from the periplasmic fraction as described previously for recombinant wild-type amicyanin [16].
2.2. Site-directed mutagenesis of the amicyanin gene
Site-directed mutagenesis was performed on double-stranded pMEG201[16] using two mutagenic primers with the QuickChange Site-Directed Mutagenesis Kit (Stratagene). The oligonucleotide sequences used to construct M98K were 5′-GACTATCACTGTACCCCGCATCCCTTCAAGCGCGGCAAGGTCG-3′ and its complementary DNA. The underlined bases are those that were changed to create the desired mutation. The entire 555-base mauC-containing fragment was sequenced to ensure that no second site mutations were present, and none were found.
2.3. Preparation of completely unfolded, partially folded, and fully folded apoamicyanin
M98K amicyanin was subjected to a procedure which was previously developed to remove Zn2+ from other amicyanin mutants and then to reconstitute those proteins with Cu2+ [14, 17]. To completely unfold the protein and remove any metal ions, M98K amicyanin was incubated in 10 mM HEPES buffer, pH 8.0, containing 6 M guandinine-HCl, 50 mM EDTA and 2 mM dithiothreitol. The completely unfolded apoprotein was then diluted 50-fold into 10 mM HEPES buffer, pH 8.0, containing 5 mM dithiothreitol. The protein was then dialyzed against 100 mM ammonium acetate, pH 8.0, at 4° C for 4 hr, followed by dialysis overnight against 250 mM ammonium acetate, pH 8.0, to generate the partially folded protein. The fully folded apoprotein was prepared by dialysis of the holoprotein for 20 h at 4 °C against 0.1 M Tris-HCl, pH 8.0, containing 0.1 M KCN, followed by dialysis against 0.05 M potassium phosphate, pH 7.5, to remove KCN and unbound metal, as previously described [18]. This native apo-amicyanin was shown by x-ray crystallography to have a structure essentially identical to that of holoamicyanin except for the absence of copper [10]. Reconstitution experiments with the unfolded, partially folded and fully folded M98K amicyanin apoproteins were performed at room temperature by incubation with CuSO4.
2.4. Test for reactivity of the single Cys sulfhydral group of amicyanin
The reactivity of the free sulfhydral of the sole Cys of amicyanin that provides one of copper ligands was probed with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). The reaction with DTNB was quantitated from the increase in absorbance at A412 nm with an extinction coefficient of 13600 M−1cm−1 as described previously [18].
2.5. Steady-state kinetic assays
Steady-state kinetic studies of the MADH-catalyzed reduction of native amicyanin by methylamine were performed as described previously [19]. Assay mixtures contained 18 nM MADH and varying concentrations of amicyanin in 10 mM potassium phosphate buffer at pH 7.5 at 30 °C. The reaction was initiated by the addition of 0.1 mM methylamine. As M98K amicyanin showed no activity in this reaction, it was tested as an inhibitor of the reaction with native amicyanin. In those studies the reactions with native amicyanin were repeated in the presence of different fixed concentrations of M98K amicyanin. Activity was monitored by the change in A595 nm caused by the reduction of amicyanin. Initial rates were measured from the linear portion of the progress curve and fit according to the standard Michaelis-Menten equation to extract kcat and Km values. The data for the inhibition of the reaction by M98K were best fit to eq 1, where Ki is the simple dissociation constant for the M98K amicyanin-MADH complex.
| (1) |
2.6. Crystallization
Prior to crystallographic trials, the M98K mutant of amicyanin was dialyzed against 5 mM sodium monobasic/potassium dibasic phosphate buffer, pH 6.6. As this mutant failed to crystallize using previously used protocols which were successful with native amicyanin and other mutant amicyanins [20, 21], a wide range of crystallization conditions was screened. The M98K amicyanin crystals were grown by the sitting drop vapor diffusion method, by mixing equal volumes of protein (13.8 mg/ml) and reservoir solution. The reservoir solution contained 0.01 M zinc sulphate heptahydrate, 0.1 M MES pH 6.5 and 25% w/v PEG monomethylether 550. Crystals suitable for x-ray diffraction analysis were cryoprotected with frombin oil (purchased from Sigma).
2.7. Data collection
X-ray diffraction data for M98K amicyanin was collected to 1.7 Å resolution at the NE-CAT beamline 24ID-C at the Advanced Photon Source, Argonne National Laboratory, Argonne equipped with Microdiffractometer-MD2 and ADSC Quantum 315 CCD detector. A fluorescence scan was carried out at the Zn and Cu absorption energy to verify the presence of Zn or Cu ion and the Zn scan showed clear peak indicating the presence of Zinc. In order to verify the presence of Cu or Zn ion at the type I site and to differentiate the contribution of zinc and copper ions from other elements including sulfur which is present naturally in the protein, data were collected around the copper and zinc absorption edges. The data collection energy for Zn was chosen from Zinc scan while copper foil fluorescence scan was used to choose data collection energy for Cu, as the Cu scan does not show any clear peak. For copper, data were collected at 1.3869 Å (below-peak), 1.3776 Å (peak) and 1.3700 Å (above-peak) and for zinc at 1.2861 Å (below-peak), 1.2819 Å (peak) and 1.2749 Å (above-peak). The data were processed, scaled and merged in HKL2000 [22]. The M98K amicyanin crystallized in the triclinic space group (P1) which is different than that was observed for previously crystallized native and mutant amicyanins. The unit-cell parameters and diffraction statistics are listed in Table 1.
Table 1.
Data Collection and Structure Determination and Refinementa
| Crystal | M98K Amicyanin |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.98 |
| Space group | P1 |
| Unit cell dimensions | |
| a , b, c (Å) | 37.4, 51.7,56.5 |
| α, β, γ (°) | 106, 96.5, 108.4 |
| Res. limit (Å) | 50–1.7 (1.76–1.7) |
| I/Sigma(I) | 9.8 (1.8) |
| Rmerge (%) | 7.0 (31.3) |
| Completeness (%) | 96.7 (95.5) |
| Redundancy | 2.1 |
| Refinement | |
| resolution range (Å) | 30.3–1.7 |
| R-work (%) | 19.0 |
| Rfree (%) | 21.5 |
| R (working + test) (%) | 19.2 |
| No. of Reflections | 39490 |
| Model | |
| No. of amino acids | 420 |
| No. of water molecules | 362 |
| No. of Zinc | 6 |
| Residues in generously allowed region | 0 |
| Residues in disallowed regions | 0 |
| No. of atoms with zero occupancy | 0 |
| Stereochemical ideality | |
| bonds (Å) | 0.018 |
| angles (deg) | 1.6 |
| dihedral angles (deg) | 11.9 |
| Planarity (Å) | 0.010 |
Values in parentheses are for the outer shell
2.8. Structure solution and refinement of M98K amicyanin
The M98K amicyanin structure was solved by molecular replacement method using PHASER [23] of PHENIX [24] using the coordinates for native amicyanin (PDB id 1AAC) with Met98 mutated to Ala. There were four molecules in the asymmetric unit. Inspection of the resulting difference electron density maps using COOT [25] revealed density for Lys98 in all the four molecules. To monitor the refinement, a random subset of all reflections (5%; 1992 reflections) was set aside for Rfree calculation [26]. The refinement of M98K amicyanin was carried out using PHENIX by subjecting the model to alternative positional and B-factor refinement. No restraints were applied to the metal, ligand distances or bond angles. A simulated annealing omit map was calculated around the copper region to remove the model bias using PHENIX. A total of 362 water molecules were added to the model. The final R and Rfree values of the model are 19.0 and 21.6, respectively. The final model contains six zinc ions, four of which were present in each of the type I sites that normally occupied by copper in native amicyanin. The average temperature factor is 18.8 Å2 for all protein atoms and 27.7 Å2 for water molecules and also 15.4 Å2 for zinc ions. The Ramachandran map calculated using PROCHECK [27] shows that all the non-glycine residues are either in most favored or in additional allowed regions. The rms deviation calculation and structure analysis were carried out using COOT [25], CCP4MG [28] and CCP4 [29].
3. Results
3.1. Spectroscopic and physical properties of M98K amicyanin
M98K amicyanin which was pure as judged by SDS-PAGE exhibited no visible absorption spectrum (Figure 1). Addition of Cu2+ caused no increase in A595 nm suggesting that the lack of visible absorbance was not because an apo-protein had been isolated that lacked metal. Addition of ferricyanide or H2O2 caused no increase in A595 indicating that the lack of visible absorbance was not because the copper in the protein was present in the invisible Cu1+ state.
Figure 1.
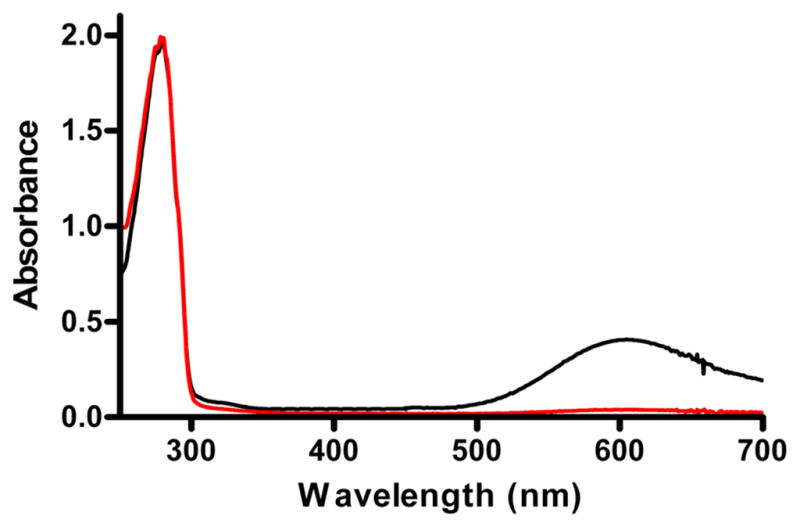
UV-visible absorption spectra of M98K and native amicyanin. Absorption spectra of native (black) and M98K (red) amicyanin were recorded in 10 mM potassium phosphate, pH 7.5.
It was previously observed that the as-isolated M98A, M98Q and M98L amicyanins from P. denitrificans did not have full occupancy of copper in the type 1 site [13, 14]. Instead there was partial occupancy of the metal site by zinc rather than copper. A procedure was developed to unfold the protein, remove any metal ions, and then incorporate Cu2+ exclusively into the partially folded protein [14, 17]. M98K amicyanin was subjected to the same procedure but it was not possible to reconstitute the protein with copper. Instead the procedure resulted in denaturation of the protein, as evidenced by the formation of a white precipitate. An alternative approach to attempt to incorporate copper into M98K amicyanin was to generate the fully folded apo form of M98K amicyanin. Native apoamicyanin has a very high affinity for Cu2+ [18]. However, addition of Cu2+ to M98K apoamicyanin again resulted in precipitation of the protein. In contrast, re-addition of Zn2+ to M98K apoamicyanin did not lead to precipitation.
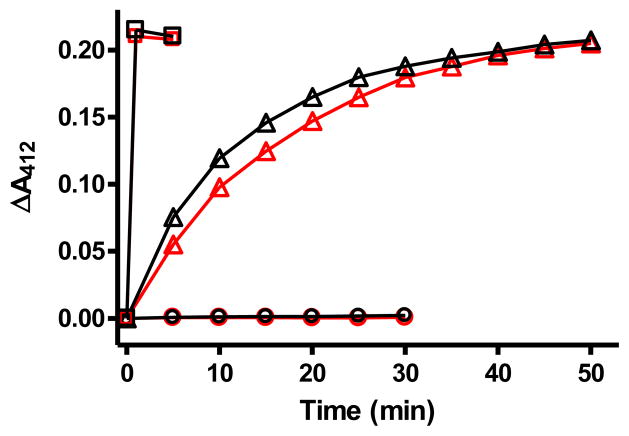
Given these results, it was important to verify that the accessibility and reactivity of the type1 site in M98K apoamicyanin was similar to that of native apoamicyanin, and that the tendency to denature during the reconstitution process with Cu2+ was not due to enhanced instability of the M98K apoamicyanin. To achieve this aim, the different folded forms of native and M98K amicyanins were probed with DTNB. DTNB reacts with free thiol groups and give rise to a color change which allows quantitation of the number of reactive thiols. Amicyanin contains only one Cys residue and it is a strong ligand for the bound copper. Thus, DTNB has been used to assess whether or not this Cys is ligating a metal or accessible to solvent. The as-isolated M98K amicyanin did not react with DTNB, suggesting that the Cys was ligating a metal and not exposed to solvent as is true for native amicyanin. Addition of DTNB to the completely unfolded metal-free native amicyanin or M98K amicyanin results in an immediate color change corresponding reaction of one Cys in each protein. Incubation of the folded M98K apoamicyanin with DTNB resulted in a slow increase in A412 nm corresponding to the slow reaction of 1 mol of Cys (Figure 2). The rate of the reaction of M98K apoamicyanin was very similar to that observed with native amicyanin suggesting that the extent of accessibility of the Cys to solvent is similar in the two apoproteins. These results indicate (i) that the as-isolated M98K amicyanin contains a tightly bound metal other than copper, (ii) that the M98K mutation does not significantly affect the accessibility of the metal to the type 1 site in either the unfolded or fully folded apoprotein, and (iii) that it is the introduction of copper that causes the denaturation of M98K apoamicyanin.
Figure 2.
Reactions of different forms of native amicyanins and M98K amicyanin with DTNB. Each reaction was performed with 15 μM protein at 25 °C in 50 mM potassium phosphate buffer, pH 7.5, and each reaction was initiated by the addition of 1 mM DTNB. Changes in absorbance were monitored at A412 nm. The reactions of native amicyanin are indicated in black and those for M98K amicyanin are shown in red. Circles describe the reactions of native amicyanin and pure as-isolated M98K amicyanin. Triangles describe the reactions of the folded apoproteins. Squares describe the reactions of the apoproteins which were fully unfolded by incubation in 6 M Guanidine-HCl.
3.2. X-ray structure of M98K amicyanin
The crystal structure of M98K amicyanin was determined at 1.7 Å resolution. The M98K amicyanin was crystallized in the triclinic space group (P1) which is different than that was observed for previously crystallized native and mutant amicyanins [14, 20, 21]. Inspection of difference electron-density map clearly revealed the mutated Lys98 residue in each of the four molecules in the asymmetric unit. Zinc rather than copper was present in each of the molecules in the type 1 site which is occupied by copper in native amicyanin. A fluorescent scan at the zinc and copper absorption edges and computed anomalous difference Fourier maps clearly confirmed the presence of zinc rather than copper in this mutant. Out of three data collected for copper, there should not be any contribution of copper in its below-peak data. There was no clear peak observed for copper in both peak and above-peak data when compared with data of below-peak. In the case of zinc’s below-peak data, there will not be any contribution from zinc, but contribution from copper could present. However, there were no peaks in the below-peak data of zinc. Both in peak and above-peak data, there are very strong peaks observed for both type I site and the zinc at monomer interface, with respect to below-peak data. The zinc peaks strength at type I site is in the range of 8–13σ for all the four molecules in the peak data and approximately half of their strength in the above-peak data. The total scattering factor f at the absorption edge of an element can be defined as f=fo+f′+f″ where fo is a normal scattering factor while f′ and f″ are real and imaginary anomalous components [30]. The strength of zinc peaks at the peak and above-peak data indicate the contribution of anomalous components, especially the f″ of the zinc ion. The zinc fluroresence scan indicates that the strength of the f″ is half at the energy of above-peak with respect to peak energy, which clearly reflected at the anomalous difference Fourier electron density map. Also, it clearly shows that the contribution to the anomalous density is solely by the zinc ion. The anomalous difference Fourier map based on the data collected at Zn-peak data is shown in Fig. 3. Two additional zinc ions were observed at the monomer interface of M98K amicyanin, with each bridging two different molecules of the asymmetric unit. Each of these monomer interface zinc ions is coordinated by nitrogen of His 36 and both oxygens of Glu105 of two different asymmetric unit molecules. The final model contains four molecules of M98K amicyanin, 362 water molecules and six zinc ions. The average b-factors for all the four molecules vary between 18.1 to 19.5 Å2. The thermal parameters of the zinc ion (15.2 Å2) at type I site is similar to its ligands/protein atoms (average thermal parameters 18.8 Å2 for protein atoms) which clearly indicate that site is fully occupied by the zinc.
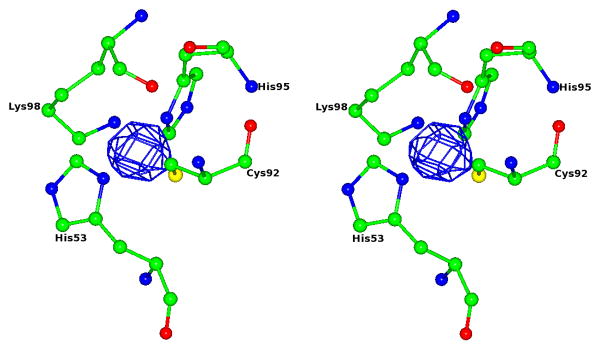
Figure 3.
Evidence for the presence of zinc in the type 1 site of M98K amicyanin. The stereoview of the anomalous difference Fourier map based on the data collected at zinc absorption energy is shown at 6σ level in blue.
3.2.1. Comparison of the structure of M98K with native and other mutant amicyanin structures
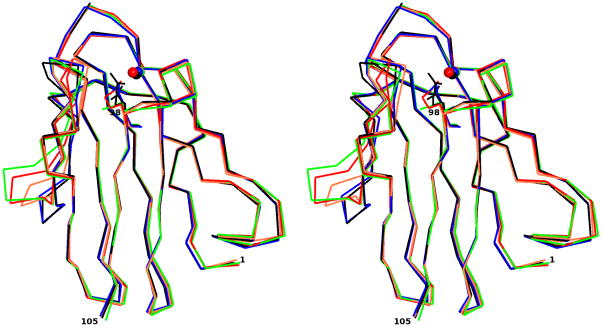
The rms deviation between native amicyanin (PDB entry 2OV0) and M98K amicyanin is 0.87 Å with 105 matched Cα atoms (Figure 4) based on the secondary structure matching [31] calculated with COOT [25]. As reported in earlier studies much of this deviation is attributed to the flexible N-terminal residues 1–21 [32]. When these are omitted the rms deviation between native amicyanin and M98K amicyanin is decreased to 0.39 Å. The rms deviation between the molecules in the asymmetric unit varies between 0.18 to 0.34 Å. For simplicity, molecule A is chosen for comparison with native and other mutant amicyanins. Superposition of the Cα trace of M98K amicyanin with those of native amicyanin, M98L, M98A and M98Q amicyanins from P. denitrificans is shown in Figure 4. The overall structures are very similar and indicate that neither the nature of the axial ligand nor the presence of zinc rather than copper has any significant effect on the overall protein structure. The zinc ion in the type I site is coordinated by the same four amino acid residues that provide ligands for copper in native amicyanin His53, His95, Cys92 and Met/Lys98. The copper/zinc coordination distances of M98K and native amicyanins are shown in Table 2. The superposition of the copper/zinc binding region of M98K and native amicyanin is shown in Figure 4.
Figure 4.
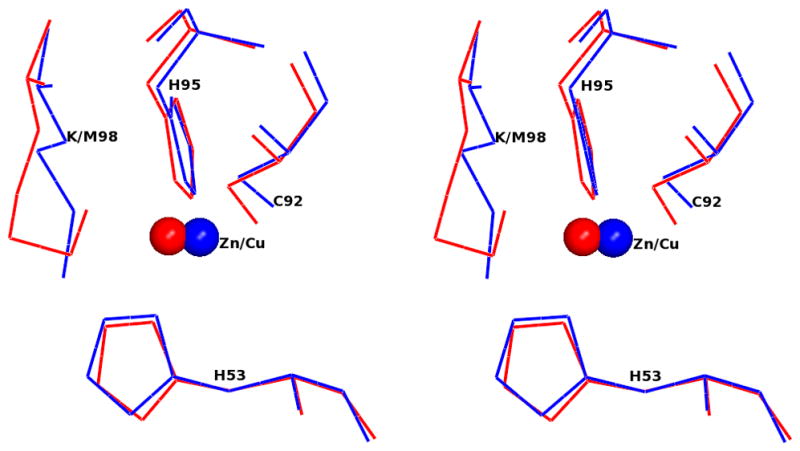
Stereoview of superposition of M98K amicyanin (red) with native amicyanin (PDB code 2OV0; blue) and M98L (PDB code 3IE9; green), M98A (PDB code 2IDQ; coral) and M98Q (PDB code 2IDT; black) amicyanin mutants. The Cu/Zn is shown as a sphere. The top panel shows the Cα trace of entire molecule. Residue 98 of each protein (Met/Leu/Ala/Gln) is shown as a stick. The Zn is colored red and Cu is the color of the respective protein molecules. In the bottom panel, the type I copper site of M98K amicyanin (red) is superimposed with native oxidized amicyanin (blue). Both Zn and Cu are shown as spheres and in colored red and blue, respectively.
Table 2.
Copper/Zinc coordination distances in M98K and native amicyanin.
| Ligands | M98K amicyanin (Å) | Native amicyanina (Å) |
|---|---|---|
| Cu-SG/Cys92 | 2.08 | 2.17 |
| Cu-ND1/His95 | 2.25 | 2.05 |
| Cu-ND1/His53 | 2.10 | 1.99 |
| Cu-NZ/Lys98 or Cu-SD/Met98 | 2.19 | 3.07 |
Taken from PDB entry 2OV0
A significant feature of the crystal structure of M98K amicyanin is the presence of a water molecule that is positioned such that it participates in hydrogen bonds with Lys98 NZ and the backbone O of Lys27 (Figure 5). In other words, this water molecule forms a hydrogen bond bridge connecting the axial Lys98 ligand with the protein backbone. Hydrogen bond donation by Lys98 will diminish the positive charge on Lys98 which in turn allows it to provide a strong ligand to Zn2+.
Figure 5.
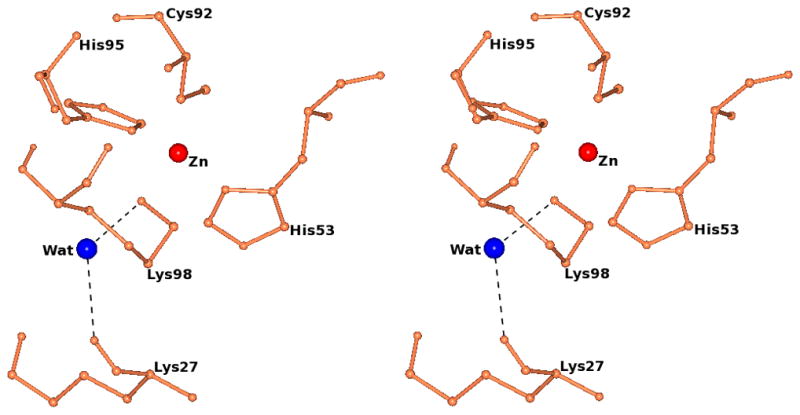
Stereview of the zinc site of M98K amicyanin. The water molecule that forms hydrogen bonds with NZ of Lys98 (NZ…..O distance 2.96 Å) and the carbonyl oxygen of Lys27 (O……O distance 2.83 Å) is shown as a blue sphere. The zinc ion is shown as a red sphere. The hydrogen bonds are shown as black dotted lines.
3.3. Characterization of M98K amicyanin as a non-reactive competitive inhibitor of native amicyanin
Since M98K amicyanin has no visible absorbance, it is not possible to use spectroscopic techniques to examine its interaction with MADH. There is no evidence for a redox reaction between M98K amicyanin and MADH in solution, as judged by monitoring the absorption spectrum of reduced MADH. To determine whether M98K amicyanin interacts with MADH in solution and to compare its affinity for MADH with that of native amicyanin, steady state kinetic studies were performed with the non-reactive M98K amicyanin tested as an inhibitor of native amicyanin. Methylamine-dependent amicyanin reduction by MADH was examined in the presence of different fixed concentrations of M98K amicyanin. Analysis of the data (Figure 6) reveals that M98K amicyanin is behaving as a competitive inhibitor of native amicyanin. From these data a Ki value of 7.2 ± 0.6 μM was determined for M98K amicyanin. This dissociation constant for the M98K amicyanin-MADH complex is very similar to the previously determined value of Kd = 5 μM for the native amicyanin-MADH complex. This result indicates that the M98K mutation does not affect the overall protein structure or the protein’s ability to specifically interact with MADH.
Figure 6.
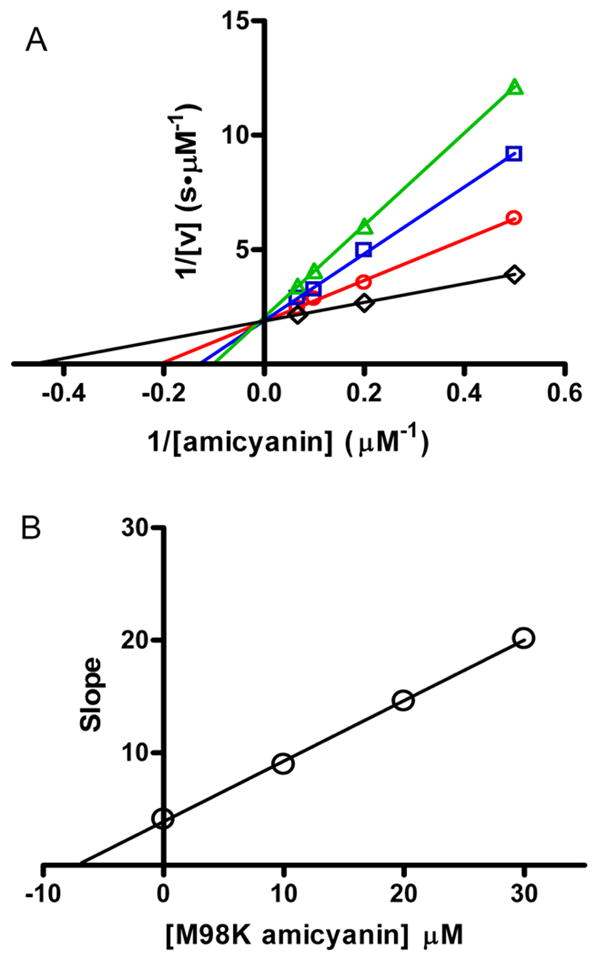
Competitive inhibition by M98K amicyanin of the reaction of methylamine-dependent reduction of amicyanin by MADH. A. Lineweaver-Burk plots of the initial velocity as a function of the concentration of amicyanin with different fixed concentrations of M98K amicyanin (black=0, μM, red=10 μM, blue=20 μM, green=30 μM). B. Secondary plot of slope versus [M98K amicyanin] from the data shown in A. The y-axis is the slope of the line for the data set for each concentration of inhibitor [M98K amicyanin]. The point of interaction of the line with the x-axis (y-intercept/slope) equals –Ki.
4. Discussion
The crystal structure of M98K amicyanin clearly shows that the metal-binding site is occupied by zinc rather than copper. Anomalous difference Fourier maps calculated using the data collected around the absorption edges of copper and zinc confirmed the presence of zinc (Figure 3) and the absence of copper, which have only one electron difference between them. The similar thermal parameters for zinc compared to its ligand and the other protein atoms indicated that type I is fully occupied by zinc ion. Lys98 provides one of ligands for zinc in addition to His53, Cys92 and His95. The axial ligand, Lys98 N-Zn coordination distance is significantly shorter than that of the Met98 S-Cu coordination distance in native amicyanin (Table 2). The distance between the other three ligands and Zn2+ in M98K amicyanin is comparable to the values observed in native amicyanin. The overall crystal structure of M98K amicyanin is also very similar to that of native amicyanin (Figure 4). Consistent with that finding is the demonstration that M98K amicyanin is a competitive inhibitor for reduction of amicyanin by MADH and exhibits a Ki value of 7.2 μM which is similar to the Kd value 5 μM for the interaction of native amicyanin with MADH. Thus, while the presence of zinc rather than copper renders the protein redox inactive, it does not affect its binding to MADH.
Recombinant type 1 copper proteins are sometimes isolated with zinc bound rather than copper when they are expressed in the cytoplasm of E. coli. That is because there is virtually no free copper in the cytoplasm. However, our recombinant amicyanin is expressed in the periplasmic space where the metal content is expected to be similar to the external medium which includes 120 μM CuSO4 [16]. We have previously expressed and purified recombinant native amicyanin and several amicyanin mutants which contained exclusively copper in the type 1 site. However, previously generated Met98 mutants of amicyanin were isolated with the site at least partially occupied by zinc. It was demonstrated that the influence of the axial ligand of the type 1 copper site on metal specificity for incorporation of copper or zinc into amicyanin is strongest prior to the completion of protein folding and adoption of the final type 1 site geometry [13]. However, in all other Met98 amicyanin mutants it was possible to remove the bound zinc and reconstitute the protein with copper. Removal of zinc from M98K amicyanin generated an apoprotein which was structurally similar to native apoamicyanin, as judged by reactivity towards DTNB. However, it could not be reconstituted with copper and addition of copper resulted in denaturation of the protein. Thus, this mutation has converted amicyanin to a zinc-binding protein rather than a copper-binding protein.
The crystal structure reveals that the type I site of M98K amicyanin is more positive than that of native and other Met98 mutant amicyanins. This is not only due to introduction of basic Lys residue at position 98, but also due of movement of Lys27 towards the type I site concomitant with movement of Met28 away from the site. Both the Lys27 and Met28 are part of type III’ reverse turn. This movement creates a more polar environment and there are three well-ordered water molecules forming triad in the vicinity of the type I site. Such a change in the environment of the metal binding site could be one of the reason why the introduction of Cu2+ into the site of M98K amicyanin results in precipitation while this was not the case with other Met98 mutants of amicyanin. Analogous Met to Lys mutations were made in azurin [33] and rusticyanin [34]. It was possible to incorporate Cu2+ into those mutant proteins. However, in each of those cases the spectral properties were significantly altered from those characteristic of the type 1 site. This suggests that there was at least some perturbation of structure. The reason why only M98K amicyanin precipitates when Cu2+ is reintroduced may relate to that fact that amicyanin has the shortest ligand loop of any type 1 protein like azurin and rusticyanin [2, 3]. The ligand loop is located between two β strands and contains three of the metal ligands Met/Lys98, His95 and Cys92. As the loops of the other two copper proteins contain more residues, those proteins may have enough flexibility to adapt their structure to accommodate any structural changes at the type 1 site that are caused by the M98K mutation, whereas amicyanin does not and instead precipitates.
Generally, Zn2+ plays a role in enzymatic catalysis or in maintaining protein structure [35]. A search of the PDB for a zinc-binding site with the similar ligands as those present in M98K amicyanin yielded no other protein and suggests that this “type 1” zinc-binding site is unique. An analysis of zinc binding sites of protein crystal structures deposited in the PDB indicated that the most common amino acids ligands for zinc are His, Cys, Asp and Glu [35]. His has highest preference with 48% and the Lys comes in seventh with 0.4%. So, while Lys has been observed to provide a ligand for zinc, it is rare. One such example is the Lys ligand for Zn2+ in leucine amino peptidase [36]. In the present structure, the Zn-ND of His and Zn-S of Cys inter-atomic distances are comparable to the values observed in the literature. The pKa of lysine in proteins is typically about 10.0 [37]. The crystals of M98K amicyanin were grown at pH 6.6 and so it is expected that Lys98 will be protonated. The ability of Lys to provide a ligand for Zn2+ is explained by the fact that it donates a hydrogen bond to a water in the metal-binding site thereby reducing the repulsive positive charge. The introduction of Lys as the fourth ligand endows such selectivity for zinc binding to the site rather than copper is surprising and not predicted from the existing structures of copper and zinc binding proteins.
Supplementary Material
Acknowledgments
This work and the 24ID-C beamline used to collect data were supported by RR-15301 (NE-CAT facility at the APS) from NCRR of NIH. This work was supported by NIH grant GM-41574 (V.L.D.). Use of the APS is supported by the U.S. DOE, Office of Science, Office of Basic Energy Science, Contract No. DE-AC02-06CH11357.
5. Abbreviations
- MADH
methylamine dehydrogenase
- ET
electron transfer
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- rms
root mean square
Footnotes
Data deposition: The x-ray coordinates of the M98K amicyanin have been deposited in the PDB under code 3RYM.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adman ET. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- 2.Dennison C. Coord Chem Rev. 2005;249:3025–3054. [Google Scholar]
- 3.Choi M, Davidson VL. Metallomics. 2011;3:140–151. doi: 10.1039/c0mt00061b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Husain M, Davidson VL. J Biol Chem. 1985;260:14626–14629. [PubMed] [Google Scholar]
- 5.Chen L, Durley R, Poliks BJ, Hamada K, Chen Z, Mathews FS, Davidson VL, Satow Y, Huizinga E, Vellieux FM, Hol WGJ. Biochemistry. 1992;31:4959–4964. doi: 10.1021/bi00136a006. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Durley RC, Mathews FS, Davidson VL. Science. 1994;264:86–90. doi: 10.1126/science.8140419. [DOI] [PubMed] [Google Scholar]
- 7.Davidson VL. Acc Chem Res. 2008;41:730–738. doi: 10.1021/ar700252c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sukumar N, Mathews FS, Langan P, Davidson VL. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0912672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sukumar N, Langan P, Mathews FS, Jones LH, Thiyagarajan P, Schoenborn BP, Davidson VL. Acta Crystallogr D Biol Crystallogr. 2005;61:640–642. doi: 10.1107/S090744490500541X. [DOI] [PubMed] [Google Scholar]
- 10.Durley R, Chen L, Lim LW, Mathews FS, Davidson VL. Protein Sci. 1993;2:739–752. doi: 10.1002/pro.5560020506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma JK, Mathews FS, Davidson VL. Biochemistry. 2007;46:8561–8568. doi: 10.1021/bi700303e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma JK, Wang Y, Carrell CJ, Mathews FS, Davidson VL. Biochemistry. 2007;46:11137–11146. doi: 10.1021/bi7012307. [DOI] [PubMed] [Google Scholar]
- 13.Ma JK, Lee S, Choi M, Bishop GR, Hosler JP, Davidson VL. J Inorg Biochem. 2008;102:342–346. doi: 10.1016/j.jinorgbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi M, Sukumar N, Liu A, Davidson VL. Biochemistry. 2009;48:9174–9184. doi: 10.1021/bi900836h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson CJ, Apiyo D, Wittung-Stafshede P. Quarterly reviews of biophysics. 2004;37:285–314. doi: 10.1017/S003358350500404X. [DOI] [PubMed] [Google Scholar]
- 16.Davidson VL, Jones LH, Graichen ME, Mathews FS, Hosler JP. Biochemistry. 1997;36:12733–12738. doi: 10.1021/bi971353m. [DOI] [PubMed] [Google Scholar]
- 17.Diederix RE, Canters GW, Dennison C. Biochemistry. 2000;39:9551–9560. doi: 10.1021/bi000648o. [DOI] [PubMed] [Google Scholar]
- 18.Husain M, Davidson VL, Smith AJ. Biochemistry. 1986;25:2431–2436. doi: 10.1021/bi00357a020. [DOI] [PubMed] [Google Scholar]
- 19.Brooks HB, Jones LH, Davidson VL. Biochemistry. 1993;32:2725–2729. doi: 10.1021/bi00061a034. [DOI] [PubMed] [Google Scholar]
- 20.Cunane LM, Chen ZW, Durley RC, Mathews FS. Acta Crystallogr D Biol Crystallogr. 1996;52:676–686. doi: 10.1107/S0907444996001072. [DOI] [PubMed] [Google Scholar]
- 21.Carrell CJ, Ma JK, Antholine WE, Hosler JP, Mathews FS, Davidson VL. Biochemistry. 2007;46:1900–1912. doi: 10.1021/bi0619674. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R, Thornton J, Moss D, MacArthur M. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 28.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 29.CCP4, Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Smith JL, Hendrickson WA, Terwilliger TC, Berendaen J. In: International Table of Crystallography. Rossmann MG, Arnold E, MAD F, MIR, editors. Kluwer Academic Publishers; Boston: 2001. pp. 299–309. [Google Scholar]
- 31.Krissinel E, Henrick K. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 32.Carrell CJ, Sun D, Jiang S, Davidson VL, Mathews FS. Biochemistry. 2004;43:9372–9380. doi: 10.1021/bi049634z. [DOI] [PubMed] [Google Scholar]
- 33.Pascher T, Karlsson BG, Nordling M, Malmstrom BG, Vanngard T. Eur J Biochem. 1993;212:289–296. doi: 10.1111/j.1432-1033.1993.tb17661.x. [DOI] [PubMed] [Google Scholar]
- 34.Hall JF, Kanbi LD, Strange RW, Hasnain SS. Biochemistry. 1999;38:12675–12680. doi: 10.1021/bi990983g. [DOI] [PubMed] [Google Scholar]
- 35.Tamames B, Sousa SF, Tamames J, Fernandes PA, Ramos MJ. Proteins. 2007;69:466–475. doi: 10.1002/prot.21536. [DOI] [PubMed] [Google Scholar]
- 36.Alberts IL, Nadassy K, Wodak SJ. Protein Sci. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canor C, Schimmel P. Biophysical Chemistry Part I. Freeman; San Francisco: 1980. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.