Abstract
Type 1 diabetes (T1D) is one of the most widely studied complex genetic disorders, and the genes in HLA are reported to account for approximately 40% to 50% of the familial aggregation of T1D. The major genetic determinants of this disease are polymorphisms of class II HLA genes encoding DQ and DR. The DR-DQ haplotypes conferring the highest risk are DRB1*03:01-DQA1*05:01-DQB1*02:01 (abbreviated “DR3”) and DRB1*04:01/02/04/05/08-DQA1*03:01-DQB1*03:02/04 (or DQB1*02; abbreviated “DR4”). The risk is much higher for the heterozygote formed by these two haplotypes (OR = 16.59; 95% CI, 13.7–20.1) than for either of the homozygotes (DR3/DR3, OR = 6.32; 95% CI, 5.12–7.80; DR4/DR4, OR = 5.68; 95% CI, 3.91). In addition, some haplotypes confer strong protection from disease, such as DRB1*15:01-DQA1*01:02-DQB1*06:02 (abbreviated “DR2”; OR = 0.03; 95% CI, 0.01–0.07). After adjusting for the genetic correlation with DR and DQ, significant associations can be seen for HLA class II DPB1 alleles, in particular, DPB1*04:02, DPB1*03:01, and DPB1*02:02. Outside of the class II region, the strongest susceptibility is conferred by allele B*39:06 (OR =10.31; 95% CI, 4.21–25.1) and other HLA-B alleles. In addition, several loci in the class III region are reported to be associated with T1D, as are some loci telomeric to class I. Not surprisingly, current approaches for the prediction of T1D in screening studies take advantage of genotyping HLA-DR and HLA-DQ loci, which is then combined with family history and screening for autoantibodies directed against islet-cell antigens. Inclusion of additional moderate HLA risk haplotypes may help identify the majority of children with T1D before the onset of the disease.
Keywords: Type 1 diabetes, Genetic risk, HLA class II, HLA class I, HLA class III, Risk prediction
Introduction
Type 1 diabetes (T1D) is an autoimmune disease in which insulin is functionally absent because of the destruction of the β cells in the pancreas by the immune system [1•].The triggers for the autoimmune attack are not fully understood, but it is now widely accepted that both environmental and genetic factors contribute to it. T1D most commonly occurs in children, and its usual clinical symptoms include polyuria, polydipsia, and weight loss. A total of 15% to 67% of new T1D patients present in diabetic ketoacidosis, a potentially life-threatening complication [2].
T1D is estimated to represent 5% to 10% of cases of diabetes [3]. Its incidence varies dramatically in different countries, ranging from 0.1/100,000 per year in China to greater than 20/100,000 per year in Sardinia, Finland, Sweden, Norway, Portugal, the United Kingdom, Canada, and New Zealand [4]. Even within the United States, the incidence varies among different ethnic groups, with the highest incidence in non-Hispanic whites and the lowest in Native Americans [5]. T1D was originally described as “juvenile diabetes” and, later, “insulin-dependent diabetes mellitus” (IDDM) and is characterized by the loss of endogenous insulin production due to destruction of pancreatic β cells. T1D occurs most commonly in juveniles but can occur in adults, especially those in their late 30s and early 40s [1•]. The incidence of T1D has risen over the past 20 years [5], highlighting the importance of environmental triggers in its etiology.
Genetics of T1D
T1D is one of the most frequently studied complex genetic disorders [6]. The strong genetic contribution to T1D is illustrated by the fact that by age 60, 65% of identical twins of T1D probands will develop T1D themselves [7]. Furthermore, children born to a family with an affected family member have a 5% risk of T1D by age 20, compared with a 0.3% risk for children without affected family members [8], yet the great majority of T1D cases do not have a first-degree relative with diabetes [1•]. The major histocompatibility complex (MHC) is reported to account for approximately 40% to 50% of the familial aggregation of T1D [9, 10]. The gene products from this region were originally discovered on the surface of white blood cells, so they became known as leukocyte antigens; thus, the human MHC is also referred to as the HLA complex. HLA molecules were originally studied for their ability to confer tolerance (histocompatibility) following tissue grafts [11]. Later studies revealed that their primary function is to provide protection against pathogens. HLA class I molecules present endogenous antigens, and class II molecules present exogenous antigens to T cells, creating the “tri-molecular complex” (HLA-peptide-TCR) that initiates the immune response.
Although T1D susceptible HLA alleles are very common in the general population, the HLA genotype, which is the combination of HLA alleles inherited from both parents, is key for the development of T1D [12]. The major genetic determinants of T1D are polymorphisms of class II HLA genes encoding DQ, DR, and, to a lesser extent, DP. Alleles of the class I HLA-B gene are strongly associated as well.
In this review, we discuss the contribution of variation at genes within the MHC to the risk of T1D, the prediction models that have been used to predict risk, and the application of HLA genes for screening infants at risk of T1D.
HLA Function
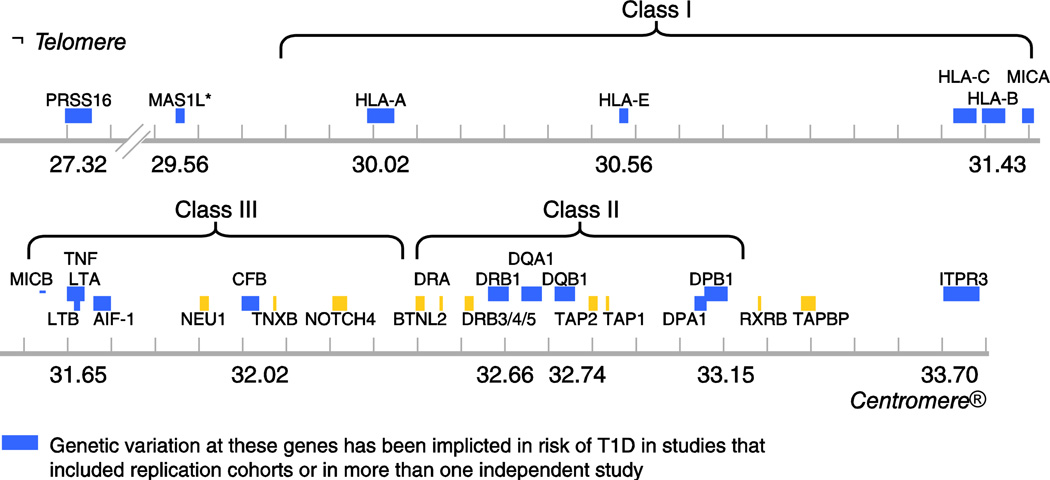
The MHC is the most important region in the vertebrate genome with respect to infection and autoimmunity, and is crucial in adaptive and innate immunity [11]. In humans, the HLA region maps to chromosome 6p21.31. The classical HLA loci are encoded in a region of DNA approximately 4 Mb long, with the class II loci at the centromeric end of the region and the class I loci at the telomeric end. The region contains more than 200 identified genes, over half of which are predicted to be expressed. Only some of the HLA region genes are involved in the immune response; in particular, the genes that encode the classical class I (A, B, and C) and class II (DR, DQ, and DP) antigens. Genes encoding classical HLA class I and class II antigens flank a chromosomal region that is sometimes referred to as the “class III region,” which contains some immunologically relevant genes (eg, TNFA) but no classical HLA genes (Fig. 1). Products of loci encoding the six classical class I (A, B, and C) and II (DR, DQ, and DP) antigens are structurally homologous, cell-surface proteins that bind antigenic peptides and present them to T cells. Nearly all of the polymorphism in the HLA genes is found in the regions encoding the amino acid residues that form the peptide-binding groove. Thus, the polymorphic residues affect the shape of the groove and, therefore, determine the repertoire of peptides that can bind to a given allele.
Figure 1.
Schematic map of the HLA region showing genes reported to be associated with type 1 diabetes (T1D). Map positions to chromosome 6 are from HapMap Rel 28 NCBI B36. Asterisk (*) indicates the signal maps to a 500-kb region between the UBD gene and the MAS1L gene also comprising several other genes [58].
To date, although the HLA class II DRB1-DQA1-DQB1 genotypes are widely recognized as the strongest genetic risk factors for T1D, several other genes within the HLA region and outside it contribute to disease risk. The most commonly used measure of effect size, describing the strength of association between a risk factor, such as a gene variant, and binary outcome such as disease status is the odds ratio (OR). An OR of 1.0 indicates no disease risk; OR values greater than 1 indicate disease susceptibility, and OR values less than 1 indicate protection from disease. Unlike many common diseases, the risk associated with HLA genes is extremely large and the highest risk genotype has an OR greater than 16 (see below and Fig. 2).
Figure 2.
Forest plot showing study-specific estimates for the association between the HLA class II DR3/DR4 genotype and type 1 diabetes (T1D). DR3 = DRB1*03:01-DQA1*05:01-DQB1*02. DR4 = DRB1*04:01/02/04/05/08-DQA1*03:01-DQB1*03:02/04 (or DQB1*02). Total sample size = 5130 T1D cases and 6366 controls from 14 studies. P = 6 × 10[−293] (fixed effects meta-analysis). No significant interstudy heterogeneity was observed: I2 = 13% p(Cochran’s Q) < 0.3 (see [55] for a description of the meta-analysis methods). Case and control genotype counts were derived from the following published studies: Sardinia [65] T1D = 1052, controls = 1917; Italy [66] T1D = 134, controls = 128; USA-HBDI collection [9] T1D = 283, controls = 199; USA-DAISY collection [62,63] T1D = 837, controls = 115; United Kingdom [10] T1D = 753, controls = 753; Sweden [67] T1D = 628, controls = 501; Finland [68] T1D = 622, controls = 622; Hungary [69] T1D = 149, controls = 177; Slovenia [70] T1D = 171, controls = 117; Turkey [71] T1D = 178, controls = 248; Hong Kong [72] T1D = 76, controls = 250; Singapore [71, 73] T1D = 73, controls = 80; Philippines [74] T1D = 90, controls 191; USA Mexican American [75] T1D = 84, controls = 68; For the non-Caucasian studies the control genotype counts were zero, the frequency in controls was estimated as ½ × number of controls. See Thomson et al. [12] for details on study selection criteria. DAISY—Diabetes Autoimmunity in the Young; HBDI—Human Biological Data Interchange.
HLA Notation
The extremely polymorphic nature of the HLA-encoding loci can create challenges with maintaining a consistent nomenclature among laboratories. Before the advent of molecular genotyping technology, HLA variation was determined serologically, using antisera derived from women who have had multiple pregnancies, and had a simple nomenclature consisting of a letter to describe the locus and a number to specify the allele (eg, A1 or DR4). As serologic genotyping technology became more refined, and as more variation was discovered in HLA, some of the original designations were subdivided (eg, the DR2 group was subdivided into DR15 and DR16).
The emergence of DNA-based genotyping led to the creation of a nomenclature system that included the locus name, followed by an asterisk, followed by a numerical designation for the allele (eg, DRB1*04:05). In the previous example, the “04” refers to the serologic group of the DRB1 allele, and the “05” refers to the individual allele within the DRB1*04 group. DPB1 is an exception to the serology-based nomenclature because no adequate serologic typing system was available for DPB1. Nearly all DPB1 polymorphisms have been discovered since the advent of DNA-based genotyping, with alleles named in order of discovery, rather than by serologic group. Thus, with two exceptions (DPB1*02:02 and DPB1*04:02), the second section of DPB1 allele designations is written “01.”
Adding to the complexity of the nomenclature system are silent polymorphisms (ie, changes to the third nucleotide in a codon that do not change which amino acid it codes for). For example, a G to T change in the third position of the “TCG” codon that encodes serine would result in a “TCT” codon, which also encodes serine. Silent polymorphisms were originally designated as an additional digit (eg, A*68011), but later changed to an additional two digits (eg, A*010101). In 2010, the nomenclature system was again changed, this time to add a colon delimiter between the sections of an allele designation such that each section is no longer limited to two digits (eg, A*02:171:01). Allele designations can become even longer when intronic polymorphisms are reported, and alleles with known expression anomalies can be designated with “L” (low expression) or “N” (null) at the end. Reporting of HLA data is still a mixed bag, with some papers using the new nomenclature, other papers using the old, and others still using old, serologic nomenclature. For clarity throughout this text we will report alleles with four-digit resolution (the first two numeric sections of the allele designation), because this is sufficient to indicate the amino acid sequence of the encoded protein.
In addition to variation in the reporting of allele designations, published HLA and disease association reports also vary with respect to the extent of HLA information reported. In some cases, reported data are given only for alleles at a single genetic locus, such as DQB1. In other cases, haplotypes (ie, alleles at multiple genetic loci that lie on the same chromosome) are reported (eg, DRB1*03:01-DQA1*05:01-DQB1*02:01). Haplotypes can only be directly determined in family-based studies, where transmission of sets of alleles from parent to child can be documented; however, many allele combinations are commonly found together due to linkage disequilibrium (LD) in the HLA region, thus allowing haplotypes to be assumed and/or estimated with specialized software programs. Some studies report data for genotypes, which represent genetic information taken from both chromosomes. A genotype can be reported for alleles at a single locus, or for haplotypes. The most T1D predisposing genotype is comprised of haplotypes DRB1*03:01-DQA1*05:01-DQB1*02:01 and DRB1*04:01/02/04/05/08-DQA1*03:01-DQB1*03:02/04 (or DQB1*02) and is commonly abbreviated with the short serology notation “DR3/DR4.”
Polymerase chain reaction–based HLA typing has greatly refined and increased our understanding of these HLA associations with T1D, initially observed and reported based on serological typing, and has led to an explosion of newly discovered alleles in the past decade, with more than 6000 total alleles currently named. Alleles can differ by as few as a single nucleotide in the DNA sequence, or they can have multiple differences. Most of the extensive polymorphism in the HLA genes is found in the portions of the DNA sequence that encode the amino acid residues that form the peptide-binding groove, where these differences can change the shape or the charge within the groove, thus changing the repertoire of peptides that can bind to and be presented by HLA.
Genetic Association Studies of HLA and T1D
Association studies compare marker frequencies in unrelated cases and controls. If a particular variant increases susceptibility to a disease of interest, it will be more common among affected individuals than among random controls. Random markers in LD (the tendency for alleles at two genetic loci to be found together more often than expected) with a disease susceptibility mutation may also appear associated with the disease, which usually implies close physical linkage of the marker and disease-predisposing gene [13].
A strong HLA association with T1D was reported as early as 1973 [14]. By the early 1980s, several associations between juvenile diabetes and various HLA antigens in case-control studies had been reported [14], and by 1990 it had become clear that a heterodimer formed by class II molecules showed the strongest risk [15]. An initial study of the HLA component of T1D risk gave results compatible with a simple recessive model (with incomplete penetrance) [16]. In later studies, the extensive heterogeneity of the HLA component to T1D emerged. Genetic heterogeneity was definitively established by the demonstration that the HLA class II DR3 and DR4 serological associations at the DRB1 locus showed increased risk for DR3/DR4 heterozygotes [17]. Furthermore, a hierarchy of very susceptible, through intermediate, to very protective HLA T1D allele, haplotype, and genotype effects are seen, which no current molecular model fully explains (Table 1) [18••].
Table 1.
DRB1-DQA1-DQB1 haplotypes most strongly associated with T1D in European descent populations
|
Predisposing haplotypes | |||||||
|---|---|---|---|---|---|---|---|
| DRB1 | DQA1 | DQB1 | Controls (%) |
T1D (%) | OR | 95% CIa | |
| 04:05 | 03:01 | 03:02 | 0.2 | 2.5 | 11.37 | 2.71 | − 47.68 |
| 04:01 | 03:01 | 03:02 | 4.5 | 28.1 | 8.39 | 5.97 | − 11.80 |
| 03:01 | 05:01 | 02:01 | 12.5 | 34.1 | 3.64 | 2.89 | − 4.58 |
| 04:02 | 03:01 | 03:02 | 1.0 | 3.5 | 3.63 | 1.76 | − 7.49 |
|
Protective haplotypes | |||||||
| DRB1 | DQA1 | DQB1 |
Controls (%) |
T1D (%) | OR | 95% CIa | |
| 13:03 | 05:01 | 03:01 | 1.0 | 0.1 | 0.08 | 0.01 | − 0.64 |
| 11:04 | 05:01 | 03:01 | 2.3 | 0.2 | 0.07 | 0.02 | − 0.30 |
| 15:01 | 01:02 | 06:02 | 12.0 | 0.4 | 0.03 | 0.01 | − 0.07 |
| 07:01 | 02:01 | 03:03 | 4.3 | 0.1 | 0.02 | 0.00 | − 0.13 |
| 14:01 | 01:01 | 05:03 | 2.1 | 0.0 | 0.02 | 0.00 | − 0.32 |
95% confidence interval for the OR.
OR—odds ratio; T1D—type 1 diabetes.
(Data from Erlich et al. [18••].)
Determining the precise HLA components of disease is difficult due to the multiple genetic factors involved, the strong LD between HLA genes, the very high polymorphism of the HLA region loci, and the likely interaction effects among HLA and/or non-HLA loci. These characteristics necessitate a multi-strategy approach, in which complementary methods are used, including analysis of data from various ethnically or geographically defined groups, as differences in HLA polymorphism and disease prevalence across groups can point toward the precise genetic factors that are relevant to disease suceptibility. The methods that have been applied to HLA data to identify the actual disease-predisposing factors, and to determine all disease factors in the region, and the difficulties encountered, apply equally to non-HLA regions.
Although LD can help detect genetic regions involved in disease, it confounds attempts to identify the actual gene in the region involved in disease and to identify additional genes in the region contributing to disease.
The role of HLA region genes additional to HLA DR-DQ in T1D was first demonstrated using affected sib pairs with parents homozygous for the DR3 haplotype [19]. HLA class I B locus data were used to distinguish between the two DR3 haplotypes of the homozygous parent. Using the haplotype method, LD patterns, matching of case/control data for specific DR-DQ combinations, and transmission disequilibrium test analysis of heterozygote microsatellite data from parents homozygous for the DR-DQ genes, the role of additional HLA region genes in T1D has been clearly demonstrated in a number of additional studies. Other approaches used to detect additional loci within the HLA region include conditional logistic regression (eg, [20]), which is a robust and powerful method but requires modeling the susceptibility component of the loci in LD with the test locus. The large number of HLA-DQB1 and HLA-DRB1 alleles, and even more so, the large number of HLA class I-class II haplotypes, result in sparse data, thus the probability is high that the class I-class II risks could be inaccurately modeled. Allele and haplotype grouping strategies must be implemented for each analysis to model the effects of the HLA class II-class I loci; each grouping strategy makes a different set of assumptions. An alternative method, conditional haplotype analysis, is a model free approach that requires the estimation of haplotype phase. In this method, under the null hypothesis (H0), the expected allele frequencies at the test locus can be computed using the equation derived by Thomson [21].
where Dij denotes the pairwise LD coefficient between the ith DRB1-DQB1 (or HLA-B/HLA-A/DPB1) haplotype and the jth test locus allele in the control sample, q denotes the allele or haplotype frequency in patients, p denotes the frequency in the affected family-based controls, and qexp denotes the expected frequency in patients if it is not involved in disease risk. One advantage of this method is its ability to derive an expected frequency and an OR estimate after adjusting for LD (see [22] for further details).
These and other methods, discussed in Thomson et al. [17], have been used in the past decade to detect the role of additional HLA molecules such as HLA-DPB1 -A, -B, and -C, as well as genes in the “class III” region and in gene regions both telomeric and centromeric to the classical HLA (see section below and Table 1). The genes in or near the HLA region that have most convincingly been shown to associate with T1D are shown in Figure 1.
DRB1-DQA1-DQB1
The strongest associations between T1D and the HLA region are seen with haplotypes and genotypes formed by the DRB1-DQA1-DQB1 loci. An unusual feature of this association is that the highest risk is conferred by a specific heterozygous genotype, comprised of DRB1*03:01-DQA1*05:01-DQB1*02 on one chromosome and DRB1*04:01/02/04/05/08-DQA1*03:01-DQB1*03:02/04 (or DQB1*02) on the other. The common abbreviation for this highest-risk genotype is “DR3/DR4.” We have used published data from 14 studies (see legend to Fig. 2 for details), totaling 5196 T1D cases and 6359 controls, to compute a summary OR for this genotype. The results (Fig. 2) clearly indicate a very high risk from this genotype. A similar exercise resulted in a summary OR= 6.32 (95% CI, 5.12–7.80) for DR3/DR3 and OR= 5.68 (95% CI, 3.91–8.23) for DR4/DR4 genotypes (not shown). Thus, although these three genotypes are all highly predisposing, the composite OR of 16.59 (95% CI, 13.7–20.1) for DR3/DR4 illustrates the extremely high risk conferred by this genotype.
Early reports in the 1980s noted increased risk for DR3/DR4 heterozygotes relative to DR3/DR3 and DR4/DR4 homozygotes [23] and that the presence of the serologically defined DQ8 molecule (DQB1*03:02), rather than DQ7 (DQB1*03:01 ), on the DR4 haplotype, could distinguish between high- and low-risk DR4 serotypes [23, 24]. One explanation for the very high susceptibility of the DR3/DR4 heterozygous genotype is the presence of DQ molecules that can be formed from polypeptide chains encoded in trans in this genotype, in addition to those encoded in cis on each of the two haplotypes. The putative DQ molecule formed by DQA1*03:01 on the DR3 haplotype and DQB1*02:01 on the DR4 haplotype is found in cis on some African haplotypes; however, DQA1*05:01 on the DR3 haplotype and DQB1*03:02 on the DR4 haplotype are not found encoded in cis on any haplotype and may be highly diabetogenic.
Some haplotypes strongly contribute to protection from T1D. The most notable example is DRB1*15:01-DQA1*01:02-DQB1*06:02, which is the most common DR-DQ haplotype in Caucasians. Protection from T1D conferred by this haplotype, in a dominant fashion, is almost complete, although some studies suggest that the protection is modified by other loci within the HLA region [25]. Other haplotypes strongly associated with protection in Caucasians include DRB1*11:04-DQA1*05:01-DQB1*03:01, DRB1*07:01-DQA1*02:01-DQB1*03:03, and DRB1*14:01-DQA1*01:01-DQB1*05:03 (Table 1).
DPB1
The HLA DPA1 and DPB1 genes are the third set of classical HLA class II loci. They encode the DP antigen and have been associated with a lower immunostimulatory capacity and level of expression than other class II antigens [26], although differences at individual DPB1-encoded amino acids have been associated with an increased proliferative response in the mixed lymphocyte reaction [27, 28]. In addition to T1D (see below), other diseases are reproducibly associated with DPB1 alleles, such as pauciarticular juvenile rheumatoid arthritis and chronic beryllium disease [29, 30].
Association studies of HLA-DPB1 and T1D have shown multiple associations with conflicting results. The following have been reported as susceptibility alleles in populations of different ethnic backgrounds: DPB1*02:01, *02:02, *03:01, *04:01, *04:02, *17:01 and the following as protective alleles; DPB1*01:01, *02:02, *04:01, *04:02, *17:01 [31–38]. Some studies have also reported weak or no association with HLA DPB1 alleles [39–41], although the sample sizes of these were small. The consensus, based on analysis of high-resolution genotyping data from a large number of samples in the Type 1 Diabetes Genetics Consortium (T1DGC) [42•], is that DPB1*02:02 and DPB1*03:01 increase disease risk whereas DPB1*04:02 decreases risk after adjusting for LD with DR-DQ-encoding loci. The LD-adjusted ORs for these three alleles in the T1DGC data are shown in Table 2.
Table 2.
HLA DPB1, HLA-A, HLA-B, and HLA-C alleles significantly different from expected values when accounting for LD with DRB1-DQB1
| Allele | T1D (%) | Expected frequency given LD with DRB1- DQB1 (%) |
OR (95% CI) a |
|---|---|---|---|
| DPB1*02:02 | 2.8 | 1.4 | 2.0 (1.41–3.01) |
| DPB1*03:01 | 15.6 | 10.1 | 1.65 (1.42–1.98) |
| DPB1*04:02 | 5.3 | 10.9 | 0.49 (0.40–0.59) |
| A*02:01 | 30.6 | 24.7 | 1.35 (1.17–1.54) |
| A*11:01 | 3.4 | 6.2 | 0.53 (0.40–0.70) |
| A*24:02 | 11.4 | 7.0 | 1.71 (1.37–2.12) |
| A*32:01 | 2.1 | 3.8 | 0.54 (0.37–0.75) |
| A*66:01 | 0.1 | 0.6 | 0.16 (0.04–0.57) |
| B*07:02 | 7.0 | 11.4 | 0.58 (0.47–0.70) |
| B*18:01 | 10.2 | 5.2 | 2.05 (1.59–2.61) |
| B*35:02 | 0.3 | 1.2 | 0.29 (0.14–0.60) |
| B*39:06 | 3.2 | 0.3 | 10.31 (4.21–25.1) |
| B*44:03 | 1.9 | 4.5 | 0.42 (0.30–0.59) |
| B*57:01 | 0.5 | 2.8 | 0.19 (0.11–0.32) |
| C*03:03 | 5.5 | 3.8 | 1.48 (1.10–1.99) |
HLA Class I Classical Loci
Products of the HLA class I genes bind and present peptide antigens to CD8+ T cells. They function both in shaping the T-cell repertoire in the thymus and in initiating antigen-specific T-cell–mediated cytotoxicity, providing a plausible immunological rationale to explain the genetic association with T1D. Evidence for a role of MHC class I alleles in diabetes susceptibility comes from studies of animal models. Some common MHC class I variants, when coexpressed with other susceptibility genes, aberrantly mediate autoreactive CD8 T-cell responses essential to T1D development [43]. Evidence from nonobese diabetic mice reveals that MHC class I and class II variants interactively regulate not only the development of diabetogenic T cells, but also the T-cell receptor promiscuity of autoreactive effectors; further evidence indicates an effect on disease susceptibility of allelic variation at the class I K locus [44]. In humans, the presence of HLA-A*24 has been demonstrated to correlate with little or no residual β-cell activity in patients with T1D [45].
Some well-powered studies have investigated the role of class I alleles in susceptibility to T1D adjusting for LD with class II loci. Significant T1D associations have been observed with class I HLA-A, -B, and -C loci. The most significantly T1D–associated alleles are B*57:01 (protective) and B*39:06 (predisposing) [20, 46••, 47, 48]. Other significantly T1D associated alleles include A*24:02, A*02:01, B*18:01, and C*05:01 (predisposing); A*11:01, A*32:01, A*66:01, B*07:02, B*44;03, B*35:02, C*16:01, and C*04:01 (protective) (Table 2).
HLA allele and haplotype frequencies differ among populations. In some cases, these differences may, at least partially, explain differences in disease prevalence. For example, in general, a North-South gradient in T1D prevalence is seen among European countries, with Scandinavians having the highest T1D risk. The Mediterranean island of Sardinia, however, is an exception, with very high T1D risk. This high risk may be partially accounted for by the high frequencies of the T1D–predisposing DR3 haplotype and the DR4 haplotype that carries DRB1*04:05, the highest-risk of the DRB1*04 alleles. In addition, the frequency of the highly protective DR2 haplotype, DRB1*15:01-DQA1*01:02-DQB1*06:02, is lower in Sardinia than in other Caucasian populations [49, 50].
Genes in the “Class III” Region
Although no classical HLA loci are encoded in the region, a 700-kb sequence located between the centromeric class II (HLA-DRA) and the telomeric class I regions (MICB) is commonly referred to as the HLA “class III” region. This region includes more than 60 genes and is one of the most gene-dense regions of the human genome [11]. Class III region genes include those encoding components of the activation cascades of the complement system (C2, factor B [BF], C4), hormonal synthesis (steroid 21-hydroxylase, CYP21), inflammation and cell stress (eg, tumor necrosis factor [TNF-α]), lymphotoxins α and β, 70-kD heat shock proteins (HSPA1A, HSPA1B, HSPA1L), extracellular matrix organization (Tenascin [TNX]), as well as immunoglobulin superfamily (Ig-SF) members (1C7, G6f and G6b). The remainder (the majority) of loci are primarily involved in more “core” biological functions with no immediate implication for the immune system [51]. The class III region has been implicated in risk for T1D, and the associations observed are not due to LD with the DR-DQ genes [25, 52], including the TNFA gene [53]. A recent study identified polymorphisms in the AIF-1 gene, mapping to class III, to be significantly associated with T1D risk after conditioning on DRB1, DQB1, HLA-A, and HLA-B variation [54]. Furthermore, two other loci, rs4151659 (mapping to CFB) and rs7762619 (mapping 5’ of LTA) were found to be strongly associated with T1D on the most predisposing DR3 and DR4 haplotypes and remained significantly associated after stratifying individuals in analyses for HLA-B, HLA-A, and DPB1 [55].
Other HLA Region Loci Implicated in T1D Susceptibility
A fine mapping study of the HLA extended region in a Swedish case-control population sample reported a novel association from the inositol 1-, 4-, 5-trisphosphate receptor type 3 gene (ITPR3), reportedly independent of the HLA class II effect, although this was assessed simply by showing the pairwise LD with DR-DQ [56]. A Canadian group attempted to replicate this association and found that association of the ITPR3 marker identified was in a direction opposite to that previously reported and that the marker identified was in significant LD with alleles at the HLA DQB1 gene [57]. Genes telomeric to the classical HLA region have also been implicated in T1D risk after adjusting for LD with the main susceptibility loci. Among these are the genes mapping to the UBD/MAS1L region, immediately telomeric to the classical class I genes [58] and the PRSS16 gene [59].
Risk Prediction
The predictive power of a given diagnostic is usually summarized by a receiver operating characteristic (ROC) curve. In this type of analysis, subjects are ranked in descending order of their predicted risk and the cumulative proportion of subjects who develop disease (cases) is plotted against the corresponding cumulative proportion of the population (ie, the sensitivity [true positive fraction] is plotted in the y-axis vs 1-specificity [the false-negative fraction] in the x-axis). [60]. A perfect diagnostic would be represented by a line that starts at the origin, travels up the y axis to 1 and then across the origin to an x-axis value of 1, thus having a total area under the curve (AUC) of 1. Greater AUC indicates a better diagnostic test. Clayton [61] derived equations that can generate ROC curves where, instead of focusing on the AUC, the measure was the sib recurrence risk λs, a measure of familial aggregation, which indicates the additional T1D risk for an individual who has an affected sibling, compared with the general population. In this model, therefore, the higher the λs, the closer that the ROC curve will be to an ideal diagnostic. He then attempted an ROC curve for HLA prediction based on six tag single nucleotide polymorphisms (SNPs), rather than the actual HLA DRB1-DQA1-DQB1 genotypes known to be involved in T1D risk, in a large collection of T1D cases and controls. That report further assessed the predictive value of 40 SNPs outside the HLA region typed in this collection. Although the actual values of AUC for HLA versus HLA and other SNPs were not included in that analysis, it is clear that the largest prediction is achieved by HLA. Some improvement would be achieved by including other loci as well, specifically a λs = 3.13 for HLA alone versus λs = 4.75 for a multiplicative model including the 40 other loci. Importantly, the association of many of the 40 non-HLA SNPs had been discovered originally in the samples used in the analysis. A more reliable estimate of the effect of HLA loci (using actual HLA high-resolution genotyping) and non-HLA SNPs might be expected from independent samples.
Current approaches for the prediction of T1D in screening studies take advantage of the major genetic risk factors, genotyping for HLA-DR and HLA-DQ loci (which is then combined with family history), and screening for autoantibodies directed against islet-cell antigens [6]. The TrialNet study, for example, targets first-degree relatives of T1D patients and includes autoantibody testing and a limited genetic screen. Antibody-positive individuals who do not test positive for the DQB1*06:02 allele, found on the highly T1D–protective DR2 haplotype, are invited to participate in the trial and can then be randomized for various interventions and followed for development of multiple antibody positivity and progression to T1D.
Children with the highest-risk HLA genotype (DR3/DR4-DQB1*03:02) have a risk of approximately 1 in 20 for a diagnosis of T1D by the age of 15 years [62]. If the child has the high-risk genotype and has a sibling who has T1D, the risk is even higher (~ 55%) [63]. However, the DR3/DR4 genotype is only found in approximately 2% to 3% of the US population. Inclusion of additional moderate HLA risk haplotypes and screening for autoantibodies would add cost and complexity to a population-screening approach, but this would have the potential to identify the majority of all children with diabetes before the onset of the disease. The discovery of an efficacious intervention might justify the additional screening cost. Screening for both high- and moderate-risk genotypes, followed by autoantibody screening, may have clinical relevance; suggestive evidence exists that for some therapies, earlier intervention, at a stage at which an increased β-cell mass remains, may improve outcomes [64].
Conclusions
After decades of research and thousands of reports, HLA remains, by far, the strongest predictor of T1D risk, consistent with the idea that the human genome is prone to develop T1D when mutations in genes controlling tolerance override the normal polygenic prevention of autoimmunity [1•]. However, the complexity of the genetics of T1D is greater than might have been predicted by the early reports. “HLA” does not refer to a single genetic locus, but to a region of the genome that includes genes encoding three classical HLA class II and three classical class I antigens as well as a number of other genes whose products influence susceptibility. Although the exact biological mechanism of HLA-conferred T1D susceptibility remains elusive, any efforts aimed at risk prediction for early intervention in children must necessarily be based, first and foremost, on HLA genotyping that is more comprehensive than just screening for DR3/DR4. Thus, cost-effective improvements in this area, leading toward a goal of complete and inexpensive HLA genotyping, are highly desirable.
Acknowledgment
This work was supported by National Institutes of Health R01 DK61722.
Footnotes
Disclosure
No potential conflicts of interest relevant to this article were reported.
Contributor Information
Janelle A. Noble, Children's Hospital Oakland Research Institute, 5700 Martin Luther King, Jr. Way, Oakland, CA 94609, USA, jnoble@chori.org
Ana M. Valdes, Department of Twin Research King’s College London, London, UK
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1. Eisenbarth GS. Banting Lecture2009: An unfinished journey: molecular pathogenesis to prevention of type 1A diabetes. Diabetes. 2010;59:759–774. doi: 10.2337/db09-1855. This lecture gives an excellent overview of T1D pathogenesis important to the understanding of the genetic susceptibility.
- 2.Dunger DB, Sperling MA, Acerini CL, Bohn DJ, Daneman D, Danne TP, Glaser NS, Hanas R, Hintz RL, Levitsky LL, Savage MO, Tasker RC, Wolfsdorf JI. European Society for Paediatric Endocrinology/Lawson Wilkins Pediatric Endocrine Society consensus statement on diabetic ketoacidosis in children and adolescents. Pediatrics. 2004;113:e133–e140. doi: 10.1542/peds.113.2.e133. [DOI] [PubMed] [Google Scholar]
- 3.Maahs DM, West NA, Lawrence JM, Mayer-Davis EJ. Epidemiology of type 1 diabetes. Endocrinol Metab Clin North Am. 2010;39:481–497. doi: 10.1016/j.ecl.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karvonen M, Viik-Kajander M, Moltchanova E, Libman I, LaPorte R, Tuomilehto J. Incidence of childhood type 1 diabetes worldwide. Diabetes Mondiale (DiaMond) Project Group. Diabetes Care. 2000;23:1516–1526. doi: 10.2337/diacare.23.10.1516. [DOI] [PubMed] [Google Scholar]
- 5.Dabelea D, Bell RA, D'Agostino RB, Jr, Imperatore G, Johansen JM, Linder B, Liu LL, Loots B, Marcovina S, Mayer-Davis EJ, Pettitt DJ, Waitzfelder B. Incidence of diabetes in youth in the United States. Jama. 2007;297:2716–2724. doi: 10.1001/jama.297.24.2716. [DOI] [PubMed] [Google Scholar]
- 6.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 7.Redondo MJ, Jeffrey J, Fain PR, Eisenbarth GS, Orban T. Concordance for islet autoimmunity among monozygotic twins. N Engl J Med. 2008;359:2849–2850. doi: 10.1056/NEJMc0805398. [DOI] [PubMed] [Google Scholar]
- 8.Bonifacio E, Ziegler AG. Advances in the prediction and natural history of type 1 diabetes. Endocrinol Metab Clin North Am. 2010;39:513–525. doi: 10.1016/j.ecl.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert AP, Gillespie KM, Thomson G, Cordell HJ, Todd JA, Gale EA, Bingley PJ. Absolute risk of childhood-onset type 1 diabetes defined by human leukocyte antigen class II genotype: a population-based study in the United Kingdom. J Clin Endocrinol Metab. 2004;89:4037–4043. doi: 10.1210/jc.2003-032084. [DOI] [PubMed] [Google Scholar]
- 11.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 12.Thomson G, Valdes AM, Noble JA, Kockum I, Grote MN, Najman J, Erlich HA, Cucca F, Pugliese A, Steenkiste A, Dorman JS, Caillat-Zucman S, Hermann R, Ilonen J, Lambert AP, Bingley PJ, Gillespie KM, Lernmark A, Sanjeevi CB, Ronningen KS, Undlien DE, Thorsby E, Petrone A, Buzzetti R, Koeleman BP, Roep BO, Saruhan-Direskeneli G, Uyar FA, Gunoz H, Gorodezky C, Alaez C, Boehm BO, Mlynarski W, Ikegami H, Berrino M, Fasano ME, Dametto E, Israel S, Brautbar C, Santiago-Cortes A, Frazer de Llado T, She JX, Bugawan TL, Rotter JI, Raffel L, Zeidler A, Leyva-Cobian F, Hawkins BR, Chan SH, Castano L, Pociot F, Nerup J. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens. 2007;70:110–127. doi: 10.1111/j.1399-0039.2007.00867.x. [DOI] [PubMed] [Google Scholar]
- 13.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 14.Singal DP, Blajchman MA. Histocompatibility (HL-A) antigens, lymphocytotoxic antibodies and tissue antibodies in patients with diabetes mellitus. Diabetes. 1973;22:429–432. doi: 10.2337/diab.22.6.429. [DOI] [PubMed] [Google Scholar]
- 15.Tait BD. Genetic susceptibility to type I diabetes: a review. J Autoimmun. 1990;3 Suppl 1:3–11. doi: 10.1016/s0896-8411(09)90003-8. [DOI] [PubMed] [Google Scholar]
- 16.Payami H, Thomson G, Motro U, Louis EJ, Hudes E. The affected sib method. IV. Sib trios. Ann Hum Genet. 1985;49:303–314. doi: 10.1111/j.1469-1809.1985.tb01706.x. [DOI] [PubMed] [Google Scholar]
- 17.Thomson G, Robinson WP, Kuhner MK, Joe S, MacDonald MJ, Gottschall JL, Barbosa J, Rich SS, Bertrams J, Baur MP, et al. Genetic heterogeneity, modes of inheritance, and risk estimates for a joint study of Caucasians with insulin-dependent diabetes mellitus. Am J Hum Genet. 1988;43:799–816. [PMC free article] [PubMed] [Google Scholar]
- 18. Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, Mychaleckyj JC, Todd JA, Bonella P, Fear AL, Lavant E, Louey A, Moonsamy P. HLAR-DQ Haplotypes Genotypes Type 1 Diabetes Risk: Analysis of the Type 1 Diabetes Genetics Consortium Families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. This paper summarizes the analyses of HLA class II DR- and DQ-encoding genes on T1D risk in the T1DGC samples.
- 19.Robinson WP, Barbosa J, Rich SS, Thomson G. Homozygous parent affected sib pair method for detecting disease predisposing variants: application to insulin dependent diabetes mellitus. Genet Epidemiol. 1993;10:273–288. doi: 10.1002/gepi.1370100502. [DOI] [PubMed] [Google Scholar]
- 20.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, Reynolds P, Hardy M, King E, Masters J, Hulme J, Maier LM, Smyth D, Bailey R, Cooper JD, Ribas G, Campbell RD, Clayton DG, Todd JA. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson G. HLA DR antigens and susceptibility to insulin-dependent diabetes mellitus. Am J Hum Genet. 1984;36:1309–1317. [PMC free article] [PubMed] [Google Scholar]
- 22.Noble JA, Valdes AM, Bugawan TL, Apple RJ, Thomson G, Erlich HA. The HLA class I A locus affects susceptibility to type 1 diabetes. Hum Immunol. 2002;63:657–664. doi: 10.1016/s0198-8859(02)00421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svejgaard A, Ryder LP. HLA and insulin-dependent diabetes: an overview. Genet Epidemiol. 1989;6:1–14. doi: 10.1002/gepi.1370060104. [DOI] [PubMed] [Google Scholar]
- 24.Ronningen KS, Markussen G, Iwe T, Thorsby E. An increased risk of insulin-dependent diabetes mellitus (IDDM) among HLA-DR4,DQw8/DRw8,DQw4 heterozygotes. Hum Immunol. 1989;24:165–173. doi: 10.1016/0198-8859(89)90057-8. [DOI] [PubMed] [Google Scholar]
- 25.Valdes AM, Thomson G, Graham J, Zarghami M, McNeney B, Kockum I, Smith A, Lathrop M, Steenkiste AR, Dorman JS, Noble JA, Hansen JA, Pugliese A, Lernmark A. D6S265*15 marks a DRB1*15, DQB1*0602 haplotype associated with attenuated protection from type 1 diabetes mellitus. Diabetologia. 2005;48:2540–2543. doi: 10.1007/s00125-005-0011-8. [DOI] [PubMed] [Google Scholar]
- 26.Pawelec G, Buhring HJ. Expression of MHC class II epitopes on human T lymphocyte clones. Cell Immunol. 1990;127:520–526. doi: 10.1016/0008-8749(90)90152-h. [DOI] [PubMed] [Google Scholar]
- 27.Cesbron A, Moreau P, Milpied N, Harousseau JL, Muller JY, Bignon JD. Crucial role of the third and fourth hypervariable regions of HLA-DPB1 allelic sequences in the mixed lymphocyte reaction. Hum Immunol. 1992;33:202–207. doi: 10.1016/0198-8859(92)90072-u. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson I, Varney M, Kanaan C, Grigg A, Szer J, Tiedemann K, Tait BD. Alloresponses to HLA-DP detected in the primary MLR: correlation with a single amino acid difference. Hum Immunol. 1997;55:163–169. doi: 10.1016/s0198-8859(97)00091-8. [DOI] [PubMed] [Google Scholar]
- 29.Begovich AB, Bugawan TL, Nepom BS, Klitz W, Nepom GT, Erlich HA. A specific HLA-DP beta allele is associated with pauciarticular juvenile rheumatoid arthritis but not adult rheumatoid arthritis. Proc Natl Acad Sci U S A. 1989;86:9489–9493. doi: 10.1073/pnas.86.23.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenman KD, Rossman M, Hertzberg V, Reilly MJ, Rice C, Kanterakis E, Monos D. HLA class II DPB1 and DRB1 polymorphisms associated with genetic susceptibility to beryllium toxicity. Occup Environ Med. 2011;68:487–493. doi: 10.1136/oem.2010.055046. [DOI] [PubMed] [Google Scholar]
- 31.Cruz TD, Valdes AM, Santiago A, Frazer de Llado T, Raffel LJ, Zeidler A, Rotter JI, Erlich HA, Rewers M, Bugawan T, Noble JA. DPB1 alleles are associated with type 1 diabetes susceptibility in multiple ethnic groups. Diabetes. 2004;53:2158–2163. doi: 10.2337/diabetes.53.8.2158. [DOI] [PubMed] [Google Scholar]
- 32.Al-Hussein KA, Rama NR, Ahmad M, Rozemuller E, Tilanus MG. HLA-DPB1*0401 is associated with dominant protection against type 1 diabetes in the general Saudi population and in subjects with a high-risk DR/DQ haplotype. Eur J Immunogenet. 2003;30:115–119. doi: 10.1046/j.1365-2370.2003.00369.x. [DOI] [PubMed] [Google Scholar]
- 33.Cucca F, Dudbridge F, Loddo M, Mulargia AP, Lampis R, Angius E, De Virgiliis S, Koeleman BP, Bain SC, Barnett AH, Gilchrist F, Cordell H, Welsh K, Todd JA. The HLA-DPB1--associated component of the IDDM1 and its relationship to the major loci HLA-DQB1, -DQA1, and -DRB1. Diabetes. 2001;50:1200–1205. doi: 10.2337/diabetes.50.5.1200. [DOI] [PubMed] [Google Scholar]
- 34.Noble JA, Valdes AM, Thomson G, Erlich HA. The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes. 2000;49:121–125. doi: 10.2337/diabetes.49.1.121. [DOI] [PubMed] [Google Scholar]
- 35.Nishimaki K, Kawamura T, Inada H, Yagawa K, Nose Y, Nabeya N, Isshiki G, Tatsumi N, Niihira S. HLA DPB1*0201 gene confers disease susceptibility in Japanese with childhood onset Type I diabetes, independent of HLA-DR and DQ genotypes. Elsevier-Diabetes Research and Clinical Practice. 2000;47:49–55. doi: 10.1016/s0168-8227(99)00103-5. [DOI] [PubMed] [Google Scholar]
- 36.Erlich HA, Rotter JI, Chang JD, Shaw SD, Raffel LJ, Klitz W, Bugawan TL, Zeidler A. Association of HLA-DPB1*0301 with insulin dependent diabetes mellitus in Mexican-Americans. Diabetes. 1996;45:610–614. doi: 10.2337/diab.45.5.610. [DOI] [PubMed] [Google Scholar]
- 37.Balducci-Silano PL, Layrisse ZE. HLA-DP and susceptibility to insulin-dependent diabetes mellitus in an ethnically mixed population. Associations with other HLA-alleles. J Autoimmun. 1995;8:425–437. doi: 10.1006/jaut.1995.0034. [DOI] [PubMed] [Google Scholar]
- 38.Baisch JM, Capra JD. Analysis o HLA genotypes susceptibility to insulin-dependent diabetes mellitus: association maps telomeric to HLA-DP. Scand J Immunol. 1992;36:331–340. doi: 10.1111/j.1365-3083.1992.tb03106.x. [DOI] [PubMed] [Google Scholar]
- 39.Johansson S, Lie BA, Pociot F, Nerup J, Cambon-Thomsen A, Kockum I, Thorsby E, Undlien DE. HLA associations in type 1 diabetes: DPB1 alleles may act as markers of other HLA-complex susceptibility genes. Tissue Antigens. 2003;61:344–351. doi: 10.1034/j.1399-0039.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- 40.Lie BA, Akselsen HE, Joner G, Dahl-Jorgensen K, Ronningen KS, Thorsby E, Undlien DE. HLA associations in insulin-dependent diabetes mellitus: no independent association to particular DP genes. Hum Immunol. 1997;55:170–175. doi: 10.1016/s0198-8859(97)00095-5. [DOI] [PubMed] [Google Scholar]
- 41.Mbanya JC, Sobngwi E, Mbanya DN. HLA-DRB1, -DQA1, -DQB1 and DPB1 susceptibility alleles in Cameroonian type 1 diabetes patients and controls. Eur J Immunogenet. 2001;28:459–462. doi: 10.1046/j.0960-7420.2001.00247.x. [DOI] [PubMed] [Google Scholar]
- 42. Varney MD, Valdes AM, Carlson JA, Noble JA, Tait BD, Bonella P, Lavant E, Fear AL, Louey A, Moonsamy P, Mychaleckyj JC, Erlich H. HLA DPA1 DPB1 alleles haplotypes contribute to the risk associated with type 1 diabetes: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2010;59:2055–2062. doi: 10.2337/db09-0680. This paper summarizes the results of T1D association analyses of the HLA class II genes encoding the DP molecule in the T1DGC samples.
- 43.Serreze DV, Holl TM, Marron MP, Graser RT, Johnson EA, Choisy-Rossi C, Slattery RM, Lieberman SM, DiLorenzo TP. MHC class II molecules play a role in the selection of autoreactive class I-restricted CD8 T cells that are essential contributors to type 1 diabetes development in nonobese diabetic mice. J Immunol. 2004;172:871–879. doi: 10.4049/jimmunol.172.2.871. [DOI] [PubMed] [Google Scholar]
- 44.Inoue K, Ikegami H, Fujisawa T, Noso S, Nojima K, Babaya N, Itoi-Babaya M, Makimo S, Ogihara T. Allelic variation in class I K gene as candidate for a second component of MHC-linked susceptibility to type 1 diabetes in non-obese diabetic mice. Diabetologia. 2004;47:739–747. doi: 10.1007/s00125-004-1370-2. [DOI] [PubMed] [Google Scholar]
- 45.Nakanishi K, Kobayashi T, Murase T, Naruse T, Nose Y, Inoko H. Human leukocyte antigen-A24-DQA1*0301 in Japanese insulin-dependent diabetes mellitus: independent contributions to susceptibility to the disease and additive contributions to acceleration of beta-cell destruction. J Clin Endocrinol Metab. 1999;84:3721–3725. doi: 10.1210/jcem.84.10.6045. [DOI] [PubMed] [Google Scholar]
- 46. Noble JA, Valdes AM, Varney MD, Carlson JA, Moonsamy P, Fear AL, Lane JA, Lavant E, Rappner R, Louey A, Concannon P, Mychaleckyj JC, Erlich HA. HLA class I genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 2010;59:2972–2979. doi: 10.2337/db10-0699. This paper describes the results of T1D association for HLA class I A, B, and C genes from the T1DGC samples.
- 47.Valdes AM, Erlich HA, Noble JA. Human leukocyte antigen class I B and C loci contribute to Type 1 Diabetes (T1D) susceptibility and age at T1D onset. Hum Immunol. 2005;66:301–313. doi: 10.1016/j.humimm.2004.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baschal EE, Baker PR, Eyring KR, Siebert JC, Jasinski JM, Eisenbarth GS. The HLA-B*3906 allele imparts a high risk of diabetes only on specific HLA-DR/DQ haplotypes. Diabetologia. 2011;54:1702–1709. doi: 10.1007/s00125-011-2161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cucca F, Lampis R, Frau F, Macis D, Angius E, Masile P, Chessa M, Frongia P, Silvetti M, Cao A, et al. The distribution of DR4 haplotypes in Sardinia suggests a primary association of type I diabetes with DRB1 and DQB1 loci. Hum Immunol. 1995;43:301–308. doi: 10.1016/0198-8859(95)00042-3. [DOI] [PubMed] [Google Scholar]
- 50.Cucca F, Muntoni F, Lampis R, Frau F, Argiolas L, Silvetti M, Angius E, Cao A, De Virgiliis S, Congia M. Combinations of specific DRB1, DQA1, DQB1 haplotypes are associated with insulin-dependent diabetes mellitus in Sardinia. Hum Immunol. 1993;37:85–94. doi: 10.1016/0198-8859(93)90146-r. [DOI] [PubMed] [Google Scholar]
- 51.Hauptmann G, Bahram S. Genetics of the central MHC. Curr Opin Immunol. 2004;16:668–672. doi: 10.1016/j.coi.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 52.Johansson S, Lie BA, Todd JA, Pociot F, Nerup J, Cambon-Thomsen A, Kockum I, Akselsen HE, Thorsby E, Undlien DE. Evidence of at least two type 1 diabetes susceptibility genes in the HLA complex distinct from HLA-DQB1, -DQA1 and -DRB1. Genes Immun. 2003;4:46–53. doi: 10.1038/sj.gene.6363917. [DOI] [PubMed] [Google Scholar]
- 53.Noble JA, Valdes AM, Lane JA, Green AE, Erlich HA. Linkage disequilibrium with predisposing DR3 haplotypes accounts for apparent effects of tumor necrosis factor and lymphotoxin-alpha polymorphisms on type 1 diabetes susceptibility. Hum Immunol. 2006;67:999–1004. doi: 10.1016/j.humimm.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eike MC, Olsson M, Undlien DE, Dahl-Jorgensen K, Joner G, Ronningen KS, Thorsby E, Lie BA. Genetic variants of the HLA-A, HLA-B and AIF1 loci show independent associations with type 1 diabetes in Norwegian families. Genes Immun. 2009;10:141–150. doi: 10.1038/gene.2008.88. [DOI] [PubMed] [Google Scholar]
- 55.Valdes AM, Arden NK, Tamm A, Kisand K, Doherty S, Pola E, Cooper C, Tamm A, Muir KR, Kerna I, Hart D, O'Neil F, Zhang W, Spector TD, Maciewicz RA, Doherty M. A meta-analysis of interleukin-6 promoter polymorphisms on risk of hip and knee osteoarthritis. Osteoarthritis Cartilage. 2010;18:699–704. doi: 10.1016/j.joca.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Roach JC, Deutsch K, Li S, Siegel AF, Bekris LM, Einhaus DC, Sheridan CM, Glusman G, Hood L, Lernmark A, Janer M. Genetic mapping at 3-kilobase resolution reveals inositol 1,4,5-triphosphate receptor 3 as a risk factor for type 1 diabetes in Sweden. Am J Hum Genet. 2006;79:614–627. doi: 10.1086/507876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu HQ, Marchand L, Szymborski A, Grabs R, Polychronakos C. The association between type 1 diabetes and the ITPR3 gene polymorphism due to linkage disequilibrium with HLA class II. Genes Immun. 2008;9:264–266. doi: 10.1038/gene.2008.12. [DOI] [PubMed] [Google Scholar]
- 58.Aly TA, Baschal EE, Jahromi MM, Fernando MS, Babu SR, Fingerlin TE, Kretowski A, Erlich HA, Fain PR, Rewers MJ, Eisenbarth GS. Analysis of single nucleotide polymorphisms identifies major type 1A diabetes locus telomeric of the major histocompatibility complex. Diabetes. 2008;57:770–776. doi: 10.2337/db07-0900. [DOI] [PubMed] [Google Scholar]
- 59.Viken MK, Blomhoff A, Olsson M, Akselsen HE, Pociot F, Nerup J, Kockum I, Cambon-Thomsen A, Thorsby E, Undlien DE, Lie BA. Reproducible association with type 1 diabetes in the extended class I region of the major histocompatibility complex. Genes Immun. 2009;10:323–333. doi: 10.1038/gene.2009.13. [DOI] [PubMed] [Google Scholar]
- 60.Bossuyt P. Evidence-Based Laboratory Medicine: Principles, Practice, and Outcomes. Washington, DC, USA: AACC Press; 2007. pp. 67–81. [Google Scholar]
- 61.Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet. 2009;5:e1000540. doi: 10.1371/journal.pgen.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aly TA, Ide A, Humphrey K, Barker JM, Steck A, Erlich HA, Yu L, Miao D, Redondo MJ, McFann K, Roberts CM, Babu SR, Norris JM, Eisenbarth GS, Rewers MJ. Genetic prediction of autoimmunity: initial oligogenic prediction of anti-islet autoimmunity amongst DR3/DR4-DQ8 relatives of patients with type 1A diabetes. J Autoimmun. 2005;25 Suppl:40–45. doi: 10.1016/j.jaut.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 63.Aly TA, Ide A, Jahromi MM, Barker JM, Fernando MS, Babu SR, Yu L, Miao D, Erlich HA, Fain PR, Barriga KJ, Norris JM, Rewers MJ, Eisenbarth GS. Extreme genetic risk for type 1A diabetes. Proc Natl Acad Sci U S A. 2006;103:14074–14079. doi: 10.1073/pnas.0606349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keymeulen B, Vandemeulebroucke E, Ziegler AG, Mathieu C, Kaufman L, Hale G, Gorus F, Goldman M, Walter M, Candon S, Schandene L, Crenier L, De Block C, Seigneurin JM, De Pauw P, Pierard D, Weets I, Rebello P, Bird P, Berrie E, Frewin M, Waldmann H, Bach JF, Pipeleers D, Chatenoud L. Insulin needs after CD3-antibody therapy in new-onset type 1 diabetes. N Engl J Med. 2005;352:2598–2608. doi: 10.1056/NEJMoa043980. [DOI] [PubMed] [Google Scholar]
- 65.Cucca F, Lampis R, Congia M, Angius E, Nutland S, Bain SC, Barnett AH, Todd JA. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum Mol Genet. 2001;10:2025–2037. doi: 10.1093/hmg/10.19.2025. [DOI] [PubMed] [Google Scholar]
- 66.Petrone A, Bugawan TL, Mesturino CA, Nistico L, Galgani A, Giorgi G, Cascino I, Erlich HA, Di Mario U, Buzzetti R. The distribution of HLA class II susceptible/protective haplotypes could partially explain the low incidence of type 1 diabetes in continental Italy (Lazio region) Tissue Antigens. 2001;58:385–394. doi: 10.1034/j.1399-0039.2001.580607.x. [DOI] [PubMed] [Google Scholar]
- 67.Pugliese A, Dorman JS, Steenkiste A. The 13th International Histocompatibility Working Group for Type 1 Diabetes (T1D) Joint Report. In: Hanson JA, editor. 13th International Histocompatibility Workshop and Conference. Seattle, WA USA: IHWG Press; 2007. pp. 788–796. [Google Scholar]
- 68.Hermann R, Turpeinen H, Laine AP, Veijola R, Knip M, Simell O, Sipila I, Akerblom HK, Ilonen J. HLA DR-DQ-encoded genetic determinants of childhood-onset type 1 diabetes in Finland: an analysis of 622 nuclear families. Tissue Antigens. 2003;62:162–169. doi: 10.1034/j.1399-0039.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 69.Hermann R, Mijovic CH, Rayner M, Croft N, Kelly MA, Jenkins D, Soltesz G, Barnett AH. HLA alleles IDD Min children in Hungary: a comparison with Finland. Hum Immunol. 2001;62:391–398. doi: 10.1016/s0198-8859(01)00224-5. [DOI] [PubMed] [Google Scholar]
- 70.Petrone A, Battelino T, Krzisnik C, Bugawan T, Erlich H, Di Mario U, Pozzilli P, Buzzetti R. Similar incidence of type 1 diabetes in two ethnically different populations (Italy and Slovenia) is sustained by similar HLA susceptible/protective haplotype frequencies. Tissue Antigens. 2002;60:244–253. doi: 10.1034/j.1399-0039.2002.600306.x. [DOI] [PubMed] [Google Scholar]
- 71.Saruhan-Direskeneli G, Uyar FA, Bas F, Gunoz H, Bundak R, Saka N, Darendeliler F. HLA-DR and -DQ associations with insulin-dependent diabetes mellitus in a population of Turkey. Hum Immunol. 2000;61:296–302. doi: 10.1016/s0198-8859(99)00182-2. [DOI] [PubMed] [Google Scholar]
- 72.Chang YW, Lam KS, Hawkins BR. Strong association between DQA1/DQB1 genotype early-onset IDDM in Chinese: the association is with alleles rather than specific residues. Eur J Immunogenet. 1998;25:273–280. doi: 10.1046/j.1365-2370.1998.00109.x. [DOI] [PubMed] [Google Scholar]
- 73.Chan SH, Thai AC, Lin YN, Liu KF, Wee GB. Influence of gender and age at onset on the HLA associations in Chinese with insulin-dependent diabetes mellitus. Hum Immunol. 1995;44:175–180. doi: 10.1016/0198-8859(95)00085-2. [DOI] [PubMed] [Google Scholar]
- 74.Bugawan TL, Klitz W, Alejandrino M, Ching J, Panelo A, Solfelix CM, Petrone A, Buzzetti R, Pozzilli P, Erlich HA. The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens. 2002;59:452–469. doi: 10.1034/j.1399-0039.2002.590602.x. [DOI] [PubMed] [Google Scholar]
- 75.Erlich HA, Zeidler A, Chang J, Shaw S, Raffel LJ, Klitz W, Beshkov Y, Costin G, Pressman S, Bugawan T, et al. HLA class II alleles and susceptibility and resistance to insulin dependent diabetes mellitus in Mexican-American families. Nat Genet. 1993;3:358–364. doi: 10.1038/ng0493-358. [DOI] [PubMed] [Google Scholar]