Abstract
The cardiovascular restricted bHLH transcription factor CHF1/Hey2 has been reported to play an important role in regulation of vascular smooth muscle phenotype and gene expression, but the downstream target genes that mediate these effects have not been completely elucidated. We have previously found that loss of CHF1/Hey2 in vascular smooth muscle cells leads to dysregulated expression of the matrix metalloproteinase MMP10 after treatment with PDGF. Here we report that loss or knockdown of CHF1/Hey in vascular smooth muscle cells leads to increased expression and activity of MMP10 at baseline, suggesting a direct effect of CHF1/Hey2 on MMP10 promoter regulation. To test this hypothesis, we assessed the effects of CHF1/Hey2 on a 2.5kb MMP10 promoter region upstream of the transcriptional start site. We found that this region contains multiple elements including 12 E-boxes that mediate constitutive activity and repression by CHF1/Hey2 in 293T cells and A7r5 smooth muscle cells. Surprisingly, mutation of these E-boxes not only abolished CHF1/Hey2 repression, but also diminished constitutive expression. In addition, we observed that some of these mutations unmasked an activator function for CHF1/Hey2, which has not been previously described. These findings support the hypothesis that CHF1/Hey2 is an important regulator of MMP10 expression.
Keywords: transcription factor, Hey2, MMP10, matrix metalloproteinase, vascular smooth muscle cells
Introduction
Cardiovascular basic helix-loop-helix factor 1 (CHF1, also referred to as Hey2, Hesr, Hrt, HERP and gridlock) belongs to a family of hairy-related transcription factors that play important roles in cardiovascular development and adult cardiovascular biology. CHF1/Hey2 is expressed in the ventricle and vasculature during embryonic stages and at lower levels after birth. The importance of CHF1/Hey2 in the development of the vascular system has been shown in zebrafish [1,2] and in mice [3]. A mutation in gridlock, which encodes the ortholog of CHF1/Hey2, selectively disrupts assembly of the aorta and causes coarctation at the aortic bifurcation. In mice, global deletion of CHF1/Hey2 leads to abnormal patterning and maturation of the coronary arteries, characterized by abnormal smooth muscle development. CHF1/Hey2 has also been shown to function as a transcriptional repressor of myocardin-dependent smooth muscle marker genes, SM-MHC and SM22α, through the disrupting the binding between SRF and the CArG boxes in the promoter region of these smooth muscle marker genes [4] or through some other mechanism that does not involve protein-protein interaction [5]. It has also been shown to repress Notch-dependent activation of the smooth muscle alpha actin promoter [6]. Knockout mice also show decreased neointimal formation after femoral arterial wire injury and decreased proliferation in CHF1/Hey2-deficient smooth muscle cells (SMCs) compared with wild-type SMCs [7]. The downstream targets of CHF1/Hey2 that regulate smooth muscle phenotype have not been completely elucidated.
Our previous study identifying potential target genes downstream of CHF1/Hey2 has demonstrated that matrix metalloproteinase-10 (MMP-10) was upregulated in SMCs isolated from CHF1/Hey2 knockout mice and treated with platelet-derived growth factor (PDGF) [8]. MMP-10 (also called stromelysin-2) belongs to the matrix metalloproteinase (MMP) family, which contains at least 28 zinc-dependent endopeptidases [9] that are divided into six groups: collagenases (MMP-1, -8 and -13), gelatinases (MMP-2 and MMP-9), stromelysins (MMP-3, -10 and -11), matrilysins (MMP-7 and MMP-26), membrane-type (MT)-MMPs (MMP-14, -15, -16, -17, -24 and -25) and others (MMP-12, -19, -20, -21, -23, -27 and -28). MMPs are found as secreted enzymes, cell surface enzymes and intracellular enzymes [10] and function in the degradation of collagen, elastin, and other bioactive components of the extracellular matrix (ECM). MMPs are initially synthesized as inactive zymogens (pro-MMPs) and proteolytically converted to the active enzymes. MMP-10 is known to play an important role in cell proliferation, migration, differentiation and angiogenesis, as well as vascular integrity [11]. Highly expressed MMP-10 has been founded in several human tumors of epithelial origin and several stages of the tumorigenic process [12].
To verify that CHF1/Hey2 regulates MMP10 expression, we measured expression and enzyme activity in knockout cells and after siRNA knockdown. We also tested whether CHF1/Hey2 directly regulates the MMP10 promoter and performed mutational analysis to identify important elements that mediate this effect. Here we report that knockdown of CHF1/Hey2 in SMCs directly leads to increased expression of MMP10, that CHF1/Hey2 directly represses the MMP10 promoter in reporter assays and that its effects are mediated by multiple E boxes.
Materials and methods
Construction of Plasmids and Antibodies
The mouse MMP10 upstream promoter fragments 2.5kb, 2kb,1.5 kb,1kb and 0.5kb upstream of exon1 were amplified by PCR from C57BL/6 mouse genomic DNA and cloned into the commercially available firefly luciferase reporter vector, pGL3-basic (Promega) at the XhoI and HindIII (underlined) sites, using the following primers: forward: 2.5kb: 5’aattcaaactcgagccttcattagaatctttccttgtacc 3’; 2kb: 5’aattcaaactcgagtttctacatgccaccctgagtcc 3’;1.5kb: 5’aattcaaactcgagtccccttcccagggaaatttttgt 3’;1kb: 5’aattcaaactcgagtatgaggcatctcagaatgagtcg 3’and 0.5kb: 5’aattcaaactcgagtacctctgtacatttcctcctccg 3’; reverse, 5’aattcaaaaagcttctagcacctggtttccatatcttc 3’). The E-boxes (CANNTG) in the promoter were mutated using a QuikChange Site-Directed Mutagenesis Kit (Invitrogen). Primer sequences for each mutagenic oligonucleotide are available upon request. Site1, 3, 4 and site6 E-boxes are replaced with CGGTCG and the other 8 E-boxes were replaced with CGTTCG. The CHF1/Hey2 expression vector pCDNA3-hCHF1 has been described previously [13]. The MMP-10 mouse monoclonal antibody was purchased from Millipore (MAB13422).
Transient transfection and luciferase reporter assays
293T and A7r5 cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum. Cells were plated onto six-well trays and transfected when 50–70% confluency with the indicated plasmids and an internal control plasmid (pcDNA3.1/Hygro/LacZ, Invitrogen) encoding beta-galactosidase, mixed with FuGENE HD transfection reagent according to the manufacturer's instructions (Roche). Cells were harvested with passive lysis buffer after 24 h and luciferase assays were performed according to the manufacturer’s protocol (Promega). Beta-galactosidase activity was measured using a beta-galactosidase enzyme assay system (Promega) and was used to normalize the results for transfection efficiency. All experiments were repeated independently at least 3 times, and the results are shown as mean ± SD.
Culture of mouse aortic SMCs (MASMCs)
Primary MASMCs were isolated from mouse aortas from littermates at 2–3 months of age as described previously (Sakata et al. 2004). Cells were plated in complete culture medium DMEM supplemented with 20% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 mg/mL), and incubated at 37°C in a humidified atmosphere of 5% CO2. The purity of the cells was verified by immunostaining for both smooth muscle α-actin and calponin (Santa Cruz). MASMCs were used before passage 10 for each experiment.
Western blotting
Cells were lysed with RIPA buffer (50mM Tris pH 7.4, 150 mM NaCl, 0.1 % SDS, 0.5 % sodium Deoxycholate, 1%NP-40, 1 mM EDTA) containing protease inhibitors. The lysates were then separated on sodium dodecyl sulfate–polyacrylamide gels by electrophoresis and transferred onto a Hybond-P membrane. The membranes were blocked with 5% milk in phosphate buffered saline (PBS) with 0.1% Tween 20 for 1 hour at room temperature, and probed with MMP-10 antibody (Millipore) and GAPDH. The signal was visualized with ECL Western blotting detection reagents (Amersham Pharmacia Biotech, Inc., Piscataway, NJ).
Total RNA Preparation and Quantitative -Polymerase Chain Reaction (RT-PCR)
Quantitative RT-PCR was performed using an ABI Prism 7500 Fast Real-time PCR System (Applied Biosystems). Total RNA was extracted from cultured smooth muscle cells and A7r5 cells using Qiagen RNeasy kit according to the manufacturer's instructions (Qiagen). Reverse transcription was performed with 2µg of RNA using with SuperScript® III First-Strand Synthesis System (Invitrogen). Primers spanning exon-intron junctions were designed to avoid amplification of genomic DNA. PCR conditions were 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Fluorescence changes were monitored with SYBR Green PCR Supermix (Applied Biosystems)) after every cycle, and dissociation curve analysis was performed at the end of 40 cycles to verify PCR product (0.5 °C/s increase from 55–99 °C with continuous fluorescence reading). The GAPDH gene was used to normalize samples for comparison. To quantify changes in gene expression, the ΔΔCt method was used to calculate the relative fold changes as previously described [14]. Primer sets for qPCR were used as following: for GAPDH: 5’-ccttcattgacctcaactac-3’ and 5’-ggaaggccatgccagtgagc-3’; for CHF1: 5’-tggggagcgagaacaattac-3’ and 5’-ttttcaaaagctgttggcact-3’.
shRNA Knockdown of CHF1/Hey2
Clones of CHF1/Hey2 shRNA contained within pLKO.1 plasmid containing puromycin resistance were purchased from Open Biosystems (Huntsville, AL). HEK293T cells were transfected with the CHF1/Hey2 shRNA plasmid, a packaging plasmid psPAX2, and an envelope plasmid pMD2.G using the calcium phosphate precipitate method. Lentivirus particles were collected by centrifugation and tittered using an HIV-1 p24 Antigen assay (Advanced BioScience Laboratories, Kensington, MD). A7R5 cells were transduced with CHF1/Hey2 shRNA lentiviral particles at a MOI of 10, followed by selection with 2ug/ml puromycin.
MMP-10 activity Assay
MMP-10 activity was measured with a SensoLyte® 520 MMP-10 Assay Kit according to the manufacturer’s protocol (ANASPEC). Cultured smooth muscle cells were lysed in assay buffer containing 0.1% (v/v) TritonX-100. MMP-10 was activated by incubating the MMP-10 containing-samples with APMA (4-aminophenylmercuric acetate) for 24 hours at 37°C. Activated enzyme was then mixed with the MMP-10 substrate FAM/QXL™520 FRET peptide. Fluorescence signal was measured at Excitation/Emission=490 nm+20 /520+20 nm.
Bioinformatic analysis
Approximately 2.5bp of the region upstream of the transcription start site of the mouse MMP10 gene was screened for putative transcription factor binding sites using TFSEARCH (version 1.3, 1995, Kyoto University, Japan) and MotifMogul (version 1.0, November 2008, Institute of Systems Biology). The taxonomy matrix was entered as ‘vertebrate’ and the threshold score was set at 90.0 for the analysis.
Statistical analysis
Relative promoter activities were compared by a two-tailed Student’s T-test, and a p-value<0.05 was considered statistically significant in all tests.
Results
Decreased expression of CHF1/Hey2 results in elevated MMP10 expression and activity
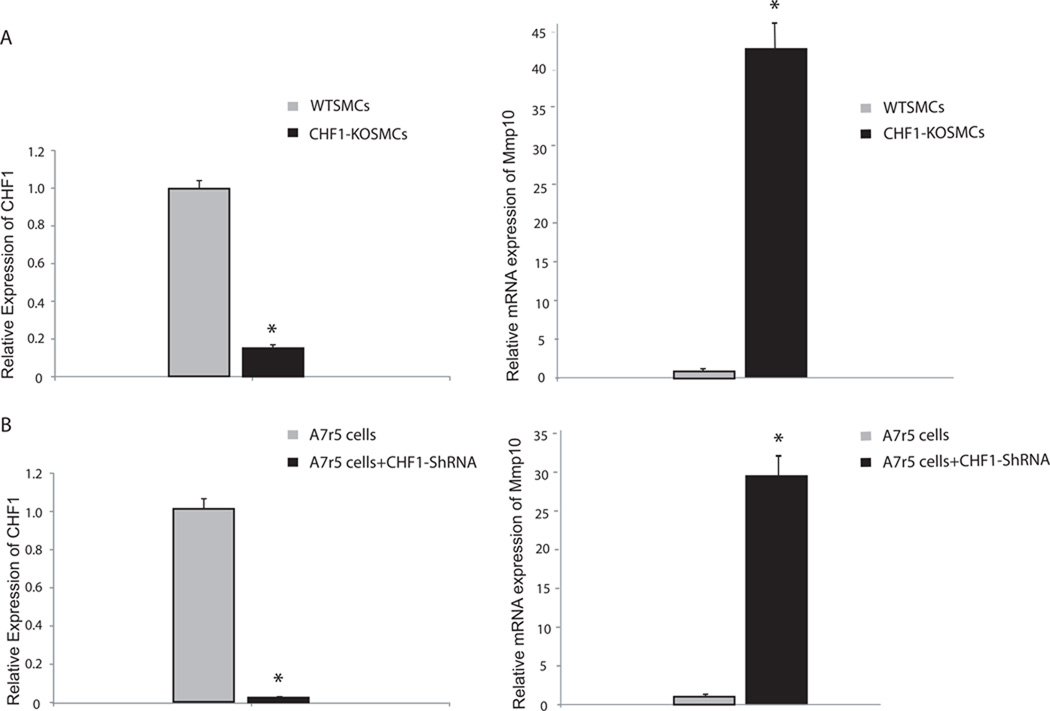
Our previous study demonstrated that the mRNA expression level of MMP10 was increased in vascular smooth muscle cells (VSMCs) isolated from CHF1/Hey2 global knockout mice, after treatment with PDGF, as measured by oligonucleotide microarrays [8]. To determine whether changes in CHF1/Hey2 expression lead to changes in MMP10 expression in the absence of PDGF stimulation, quantitative RT-PCR was performed on RNA isolated from WT and KO SMCs, as shown in Fig1a. Western blot analysis was also used to compare the expression level of the MMP-10 protein between WT and knockout SMCs, as shown in Supplemental Fig. 1a. The results demonstrate that the expression level of MMP10 mRNA and protein in knockout SMCs is higher than that in WT control cells, implying that CHF1/Hey2 represses MMP10 expression.
Fig. 1. Decreased expression of CHF1 promotes increased expression of MMP10 in SMCs and A7r5 cells.
(a) Quantitative RT-PCR analysis of mRNA expression of CHF1/Hey2 (left panel), and MMP10 RNA (right panel) isolated from WT and KO SMCs (b) Quantitative RT-PCR analysis of mRNA expression of CHF1/Hey2 and MMP10 RNA extracted from CHF1/Hey2 shRNA treated A7r5 cells and untreated cells. Data were normalized to GAPDH RNA expression in the same sample and reported as relative expression (mean ± SE) compared to levels in wild type control cells (*p = 0.017). The experiments were repeated three times and similar results were obtained.
To validate whether changes in MMP10 expression are direct effects of CHF1/ Hey2 in vascular smooth muscle cells, we further compared MMP10 expression level in untreated and CHF1/Hey2 shRNA treated A7r5 vascular smooth muscle cells. MMP10 mRNA expression was found to be highly increased when CHF1/Hey2 was knocked down by CHF1/Hey2 shRNA (Fig. 1b). This result is consistent with the observation we had in CHF1 knockout SMCs and implies that increased MMP10 expression is likely a direct consequence of CHF1/Hey2 loss of function.
Elevated MMP10 expression in CHF1/Hey2 KO SMCs results in elevated MMP-10 Activity
To determine the functional significance of elevated MMP10 mRNA and protein in KO SMCs, we tested MMP-10 activity in CHF1/Hey2 knockout mouse aortic SMCs. MMP-10 is normally expressed as an inactive zymogen. The results demonstrated that MMP-10 enzymatic activity was significantly upregulated in CHF1/Hey2 knockout SMCs (Supplemental Fig. 1b).
Identification of constitutive and CHF1/Hey2 responsive elements within the MMP10 promoter
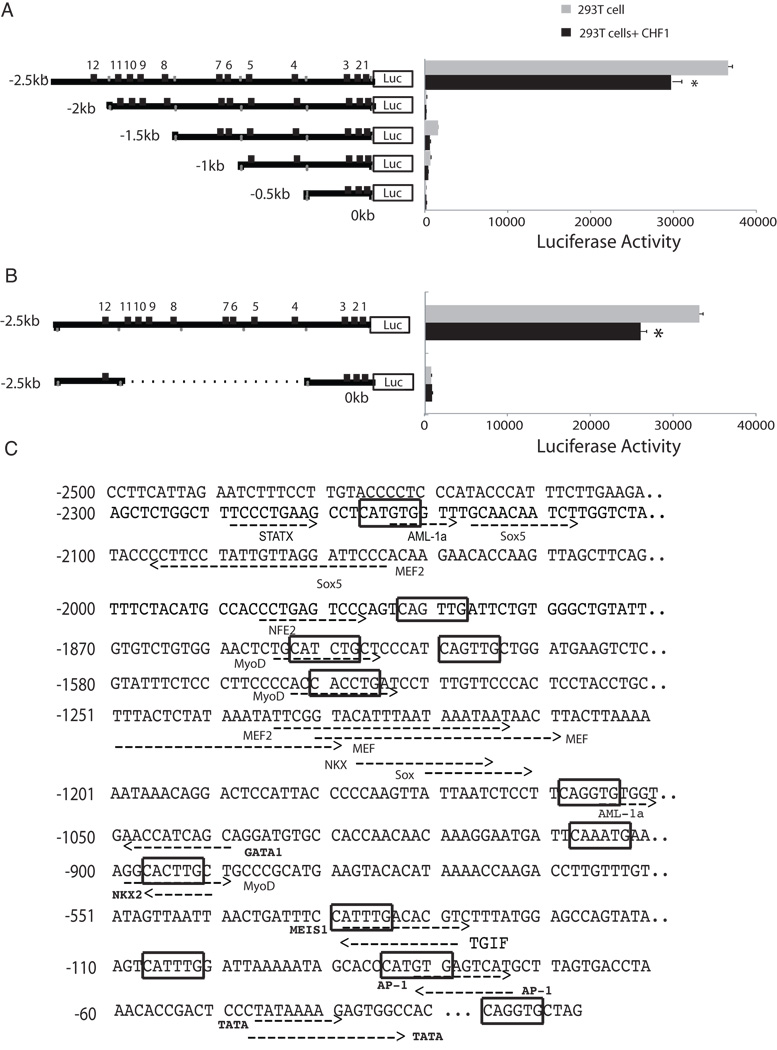
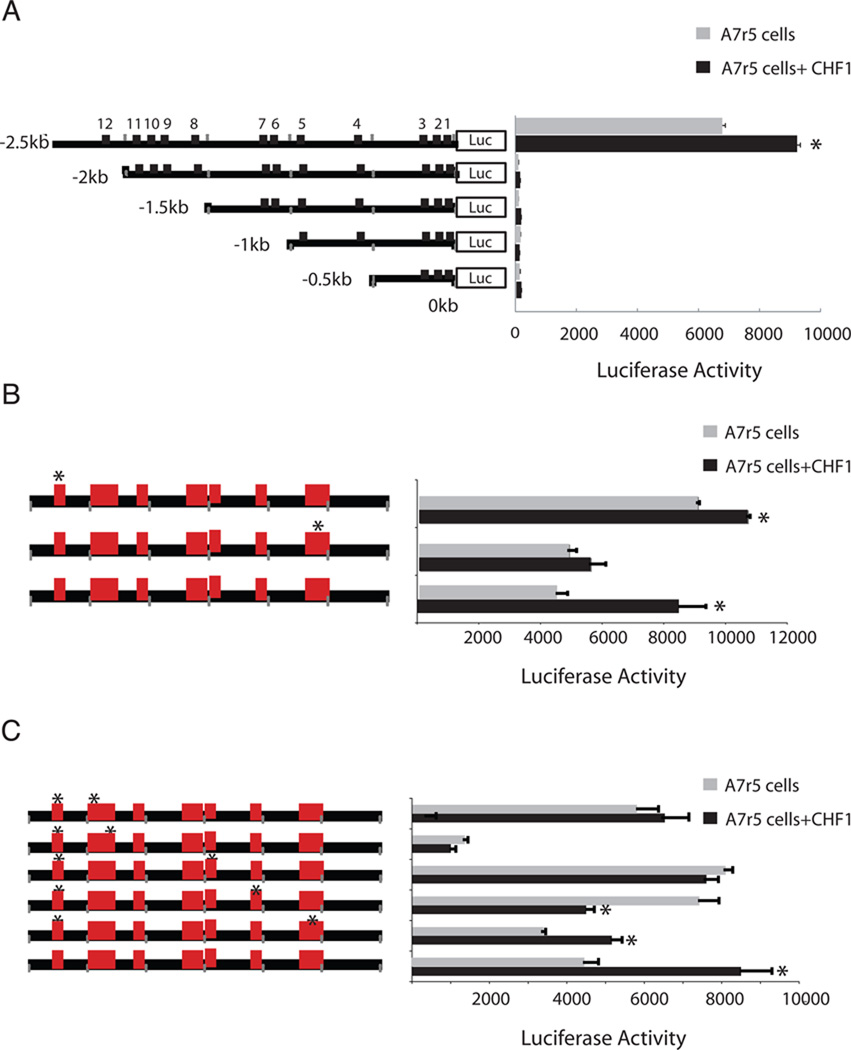
To study MMP10 promoter activity, promoter DNA fragments of different lengths (2.5kb, 2kb,1.5kb,1kb and 0.5 kb) located immediately upstream of the transcription start site of the mouse MMP10 gene were cloned from mouse genomic DNA by PCR into the pGL3-Basic luciferase reporter vector. The sequence of each construct was then confirmed by DNA sequencing. These five constructs were transfected into 293T and A7r5 cells, and luciferase activity was then determined. A comparison of luciferase activity revealed that the luciferase reporter plasmid containing the longest promoter sequence (2.5kb) demonstrated in the highest luciferase activity (Fig. 2a). Similar results were obtained in A7r5 smooth muscle cells (Fig. 4a). These results suggested that the MMP10 promoter segment between −2.5kb and −2kb contains elements that confer constitutive activation in both types of cells. Reconstitution of a minimal promoter containing the constitutive 500bp element linked to the 500bp proximal promoter did not restore promoter activity (Fig. 2b), however, indicating that this constitutive element is necessary but not sufficient for full promoter activity.
Fig. 2. Mutational analysis of the mouse MMP10 gene promoter region.
(a–b) The structures of a series of MMP10-promoter luciferase reporter plasmids are shown schematically (left) and their relative luciferase activities are shown to the right of each construct. Numbers indicate the location of E-boxes relative to the transcription start site. MMP10 reporter constructs and pCDNA3-CHF1 were transfected into 293T cells. Results are representative of experiments done in triplicate. (c) Sequence analysis of the 2.5 kb upstream promoter sequence of the mouse MMP10 gene for potential regulatory elements with TFSEARCH (version 1.3, 1995, Kyoto University, Japan) and MotifMogul (version 1.0, November 2008, Institute of Systems Biology). E-boxes are marked in boxes. Putative recognition sites for other transcription factors are underlined and labeled.
Fig. 4. Analysis of MMP10 promoter regulation in A7r5 cells.
(a) Deletion mutants of the MMP10 promoter were transfected into A7r5 smooth muscle cells. (b) Selected single E-box mutants were transfected into A7r5 cells in the absence or presence of CHF1/Hey2. (C) E-box mutations in the site 12 E-box were combined with selected mutations in other E-boxes and assessed for luciferase activity in A7r5 cells, in either the absence or presence of CHF1/Hey2. The assays were carried out in triplicate to confirm the reproducibility of the observations and the error bars indicate the standard error of the mean. *indicates significant changes in the presence of CHF1/Hey2, p<0.05.
To examine the possible direct influence of CHF1/Hey2 on MMP10 promoter activity, we contransfected a CHF1/Hey2 expression plasmid. As expected, the MMP10 promoter activity was repressed in 293T cells (Fig. 2a). Surprisingly, CHF1/Hey2 activated the MMP10 promoter in A7r5 cells (Fig. 4). These observations indicated that CHF1/Hey2 is involved in the transcriptional regulation of MMP10, but its precise role in vivo may be complex.
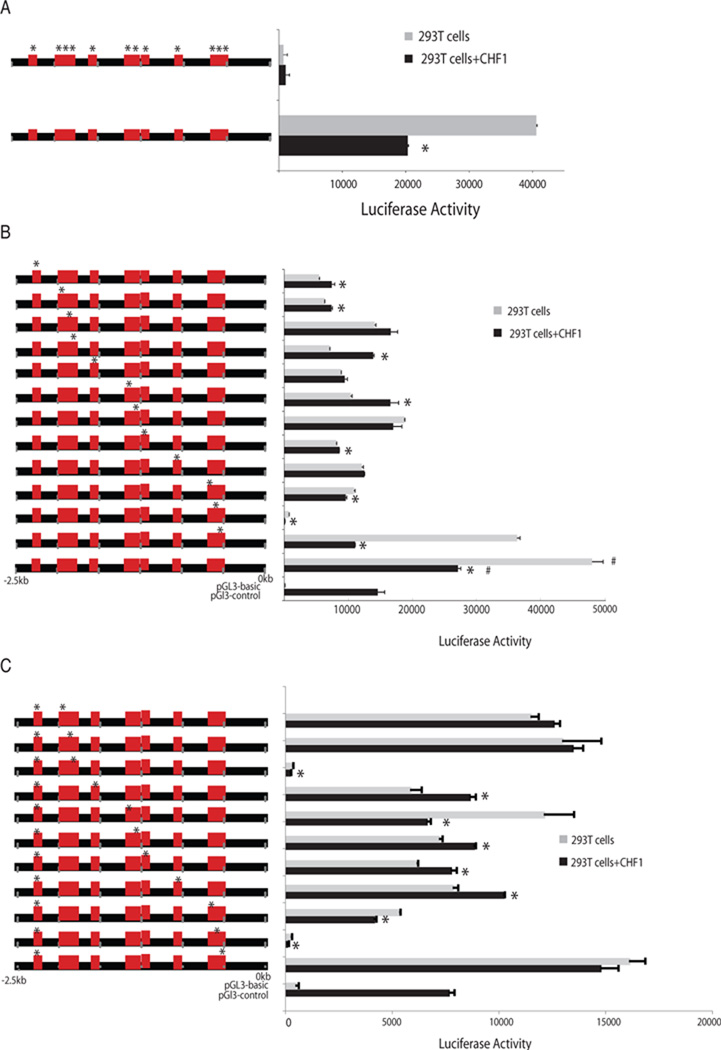
To identify precise nucleotide sequences in the MMP10 Promoter that confer responsiveness to CHF1/Hey2, we performed a search for regulatory elements for transcription factors in the promoter region of the MMP10 gene. CHF1/Hey2 has previously been reported to bind the E-box sequence (CANNTG) [15]. Within the 2.5kb proximal promoter, we identified 12 canonical E-boxes (Fig. 2c). To identify their contribution to MMP10 promoter activity, a mutated MMP10 reporter plasmid containing mutations in all twelve E-boxes was transfected into both 293T cells and A7r5 cells. Surprisingly, MMP10 reporter luciferase activity was totally abolished when all the E-boxes are mutated (Fig. 3a).
Fig. 3. E-boxes mediate both constitutive and CHF1/Hey2-dependent MMP10 promoter activity in 293 T cells.
(a) Single E-box mutations were introduced into the 2.5kb promoter fragment and assessed for luciferase activity in 293T cells, in the presence of either the expression vector pcDNA3 or pcDNA-hCHF1. (b) E-box mutations in the site 12 E-box were combined with mutations in each of the other E-boxes and assessed for luciferase activity in 293T cells, in either the absence or presence of CHF1/Hey2. The activity was normalized to β-galactosidase activity induced by cotransfection with the pcDNA/Hygro/LacZ plasmid. The assays were carried out in triplicate to confirm the reproducibility of the observations and the error bars indicate the standard error of the mean. *indicates significant changes in the presence of exogenous CHF1/Hey2, p<0.05. # indicates significant changes in the luciferase activities of all E-box mutated promoters in comparison to the wild type promoter, p<0.05.
To better define the importance of these E-boxes for MMP10 expression, the twelve E-boxes in the 2.5kb luciferase reporter plasmid were mutated individually. Our results showed that all the single E-box mutations in MMP10 promoter attenuated reporter luciferase activity in 293T cells (Fig. 3b). Mutation of the Site 2 E-box resulted in a significant drop in reporter activity to the basal level of luciferase activity generated by a promoterless pGL3-Basic construct (Fig. 3b). Moreover, mutations in sites 4, 8 and 10 E-boxes individually abolished repression of the MMP10 promoter by CHF1/Hey2. Furthermore, mutations in sites 5,7,9,11 and 12 led to CHF1/Hey2-dependent activation of luciferase reporter activity (Fig. 3b). In A7r5 cells, however, the individual site 2 and site 12 E-box mutations did not change the reporter luciferase activity significantly (Fig4b).
Base on the critical role of the 500bps between −2.5kb and −2kb for MMP10 constitutive promoter activity, the site 12 E-box located within this region was mutated in combination with each of the other 11 E-boxes. Double mutants combining site 12 and site 2 or site 12 and site 9 conferred the lowest luciferase activity comparable to the promoterless control plasmid (Fig. 3c). Double mutants of sites 12/1, 12/10 and 12/11 restored constitutive activity and abolished repression or activation by CHF1/Hey2. Double mutants in sites 12/3 and 12/5 were similar to mutation in sites 3 and 5 alone. Combined mutation in sites 12/4, 12/6 or 12/8 demonstrated activation by CHF1/Hey2, similar to mutation in site 12 alone. Consistently, the site 12/site 9 combined mutations also decreased reporter luciferase activity in A7r5 cells (Fig. 4c). In A7r5 cells, the combined site 12/ site 5 mutation had no effect on constitutive activity but completely abolished CHF1/Hey2 responsiveness. These results imply that specific E-boxes play a critical role for both constitutive and CHF1/Hey2-dependent MMP10 promoter activity.
Discussion
In this study, we extended our previously published finding that MMP10 expression is increased in CHF1/Hey2 knockout SMCs after PDGF treatment by demonstrating that this expression is also increased at the protein level under basal, untreated conditions. In addition, we found that shRNA knockdown of CHF1/Hey2 expression in a smooth muscle cell line A7r5 led to an increase in MMP10 expression. Furthermore, we demonstrated that MMP-10 enzymatic activity in CHF1 knockout aortic SMCs was increased which is consistent with the increased MMP10 expression level. To gain further insight into how CHF1/Hey2 regulates the MMP10 promoter, we have characterized the mouse MMP10 promoter in reporter assays. A constitutively active promoter element was mapped between −2.5kb and −2kb, and multiple E-boxes were found to be important for the regulation of expression by CHF1/Hey2. Previous studies have shown that a MEF2 site located within the −886 bp promoter is important for constitutive activity in endothelial cells [11] and that two AP-1 sites and two PEA3 sites are responsible for constitutive expression and induction by inflammation [16]. These sites do not appear to be active in smooth muscle cells. Interestingly, a MEF2 binding site is located within the constitutively active promoter element at −2096 to −2075 (Fig. 3c) and may be contributing to activity, but such a contribution would be surprising in the absence of activity of other MEF2 sites known to be active in other cell types.
To identify potential transactivators binding to the promoter region between −2.5kb and −2kb that may confer constitutive activity, we performed additional sequence analysis. The sequence analysis of this region revealed the presence of consensus binding sequences for several transcription factors including STAT1 and AML-1a. STAT1 is a well-known transactivator and is possibly the main activator for this promoter. AML-1a lacks the transactivation domain compared to another two alternative splice variants of the AML1 gene, AML-1b and AML-1c, and inhibits transcriptional activity [17]. The DNA binding domain for AML-1a overlaps with the site 12 E-box and the mutated site 12 E-box is expected to prevent the binding of AML-1a to the promoter. Our data showing that mutation of the site 12 E-box in −2.5kb to −2kb region enhanced the promoter activity (Fig. 4a) is consistent with a model of co-repression of MMP10 by CHF1/Hey2 and AML-1a, although further investigation is needed to explore this possibility.
A surprising finding is that CHF1/Hey2 functions as a transcriptional activator in reporter gene assays done in A7r5 cells (Fig. 4) instead of a repressor as is seen in 293T cells. ShRNA directed against CHF1/Hey2 in A7r5 cells leads to increased RNA expression, suggesting that endogenous CHF1/Hey2 functions as a transcriptional repressor. The possible reason for this discrepancy is that other cofactors such as HDACs, histones and other chromatin proteins influence endogenous transcription in ways that are not assessed in our reporter gene assays.
Another interesting result of this study is that single E-box mutations and combinations of E-box mutations in the MMP10 promoter resulted in CHF1/Hey2-dependent activation of the reporter gene. CHF1/Hey2 has previously been shown to be a transcriptional repressor [4,5,13,15,18,19,20,21]. Our data indicate the potential novel role of CHF1/Hey2 as a transcriptional activator, which has not been previously reported. A possible explanation may be that CHF1/Hey2 contains both activation and repression domains for transcriptional regulation, although no activation domains have been reported previously. Other possibilities include effects on recruitment of other coactivators in certain contexts, such as the activator STAT1 near the site 12 E-box or through inhibition of the binding of a repressor such as AML1a.
In conclusion, we have identified an important element in the mouse MMP10 promoter that drives expression in smooth muscle cells that has not been previously reported. In addition, we have determined that E-box sequences in the mouse MMP10 promoter play an important role in the regulation of MMP10 expression by CHF1/Hey2. These findings may have important implications for understanding the role of CHF1/Hey2 and MMP10 in vascular smooth muscle biology and may provide useful information for treatment and prevention of the vascular diseases.
Highlights.
Knockout or knockdown of CHF1/Hey2 in vascular smooth muscle cells leads to increased expression and activitiy of MMP10
CHF1/Hey2 directly represses the MMP10 promoter in reporter assays
Regulation of MMP10 by CHF1/Hey2 is mediated by multiple E-box elements
Supplementary Material
Increased MMP10 expression leads to increased MMP-10 enzymatic activity in CHF1/Hey2 knockout SMCs. (a) Western blot analysis of MMP-10 protein from mouse aortic SMCs. GAPDH was used as a loading control. (b) SMCs were lysed and total proteins were used for measuring MMP-10 activity. Fluorescence signal was recorded at Excitation/Emission=490nm/520nm. Experiments were done in triplicate. Results are shown as mean±SEM.*p<0.05.
Acknowledgements
This work was supported by NIH grant HL076232 (M.T.C.) and an American Heart Association postdoctoral fellowship (L.W.). M.E.H and F.M. were supported by NIH Training Grant T32 HL07828.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhong TP, Childs S, Leu JP, Fishman MC. Gridlock signalling pathway fashions the first embryonic artery. Nature. 2001;414:216–220. doi: 10.1038/35102599. [DOI] [PubMed] [Google Scholar]
- 2.Zhong TP, Rosenberg M, Mohideen M-APK, Weinstein B, Fishman MC. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe T, Koibuchi N, Chin MT. Transcription factor CHF1/Hey2 regulates coronary vascular maturation. Mech Dev. 2010;127:418–427. doi: 10.1016/j.mod.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doi H, Iso T, Yamazaki M, Akiyama H, Kanai H, Sato H, Kawai-Kowase K, Tanaka T, Maeno T, Okamoto E, Arai M, Kedes L, Kurabayashi M. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler Thromb Vasc Biol. 2005;25:2328–2334. doi: 10.1161/01.ATV.0000185829.47163.32. [DOI] [PubMed] [Google Scholar]
- 5.Proweller A, Pear WS, Parmacek MS. Notch signaling represses myocardin-induced smooth muscle cell differentiation. J Biol Chem. 2005;280:8994–9004. doi: 10.1074/jbc.M413316200. [DOI] [PubMed] [Google Scholar]
- 6.Tang Y, Urs S, Liaw L. Hairy-related transcription factors inhibit Notch-induced smooth muscle alpha-actin expression by interfering with Notch intracellular domain/CBF-1 complex interaction with the CBF-1-binding site. Circ Res. 2008;102:661–668. doi: 10.1161/CIRCRESAHA.107.165134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakata Y, Xiang F, Chen Z, Kiriyama Y, Kamei CN, Simon DI, Chin MT. Transcription Factor CHF1/Hey2 Regulates Neointimal Formation In Vivo and Vascular Smooth Muscle Proliferation and Migration In Vitro. Arterioscler Thromb Vasc Biol. 2004;24:2069–2074. doi: 10.1161/01.ATV.0000143936.77094.a4. [DOI] [PubMed] [Google Scholar]
- 8.Shirvani SM, Mookanamparambil L, Ramoni MF, Chin MT. Transcription Factor CHF1/Hey2 Regulates the Global Transcriptional Response to Platelet-Derived Growth Factor in Vascular Smooth Muscle Cells. Physiol Genomics. 2007;30:61–68. doi: 10.1152/physiolgenomics.00277.2006. [DOI] [PubMed] [Google Scholar]
- 9.Sprague AH, Khalil RA. Inflammatory cytokines in vascular dysfunction and vascular disease. Biochemical pharmacology. 2009;78:539–552. doi: 10.1016/j.bcp.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardo A, Selman M, Kaminski N. Approaching the degradome in idiopathic pulmonary fibrosis. The international journal of biochemistry & cell biology. 2008;40:1141–1155. doi: 10.1016/j.biocel.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 12.Gill JH, Kirwan IG, Seargent JM, Martin SW, Tijani S, Anikin VA, Mearns AJ, Bibby MC, Anthoney A, Loadman PM. MMP-10 is overexpressed, proteolytically active, and a potential target for therapeutic intervention in human lung carcinomas. Neoplasia. 2004;6:777–785. doi: 10.1593/neo.04283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin MT, Maemura K, Fukumoto S, Jain MK, Layne MD, Watanabe M, Hsieh C-M, Lee M-E. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. Journal of Biological Chemistry. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Molecular & Cellular Biology. 2001;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: an overview. Molecular and cellular biochemistry. 2003;253:269–285. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Zhang Q, Zhang DE, Zhou C, Xing H, Tian Z, Rao Q, Wang M, Wang J. Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis, Leukemia : official journal of the Leukemia Society of America. Leukemia Research Fund, U.K. 2009;23:739–745. doi: 10.1038/leu.2008.350. [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa O, McFadden DG, Nakagawa M, Yanagisawa H, Hu TH, Srivastava D, Olson EN. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer A, Klattig J, Kneitz B, Diez H, Maier M, Holtmann B, Englert C, Gessler M. Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Mol Cell Biol. 2005;25:8960–8970. doi: 10.1128/MCB.25.20.8960-8970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang F, Sakata Y, Cui L, Youngblood JM, Nakagami H, Liao JK, Liao R, Chin MT. Transcription factor CHF1/Hey2 suppresses cardiac hypertrophy through an inhibitory interaction with GATA4. Am J Physiol Heart Circ Physiol. 2006;290:H1997–H2006. doi: 10.1152/ajpheart.01106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirvani S, Xiang F, Koibuchi N, Chin MT. CHF1/Hey2 Suppresses SM-MHC Promoter Activity Through an Interaction with GATA-6. Biochem Biophys Res Commun. 2006;339:151–156. doi: 10.1016/j.bbrc.2005.10.190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Increased MMP10 expression leads to increased MMP-10 enzymatic activity in CHF1/Hey2 knockout SMCs. (a) Western blot analysis of MMP-10 protein from mouse aortic SMCs. GAPDH was used as a loading control. (b) SMCs were lysed and total proteins were used for measuring MMP-10 activity. Fluorescence signal was recorded at Excitation/Emission=490nm/520nm. Experiments were done in triplicate. Results are shown as mean±SEM.*p<0.05.