SUMMARY
Several lines of evidence support genetic links between ovary size and division of labor in worker honey bees. However, it is largely unknown how ovaries influence behavior. To address this question, we first performed transcriptional profiling on worker ovaries from two genotypes that differ in social behavior and ovary size. Then, we contrasted the differentially expressed ovarian genes with six sets of available brain transcriptomes. Finally, we probed behavior-related candidate gene networks in wild-type ovaries of different sizes. We found differential expression in 2151 ovarian transcripts in these artificially selected honey bee strains, corresponding to approximately 20.3% of the predicted gene set of honey bees. Differences in gene expression overlapped significantly with changes in the brain transcriptomes. Differentially expressed genes were associated with neural signal transmission (tyramine receptor, TYR) and ecdysteroid signaling; two independently tested nuclear hormone receptors (HR46 and ftz-f1) were also significantly correlated with ovary size in wild-type bees. We suggest that the correspondence between ovary and brain transcriptomes identified here indicates systemic regulatory networks among hormones (juvenile hormone and ecdysteroids), pheromones (queen mandibular pheromone), reproductive organs and nervous tissues in worker honey bees. Furthermore, robust correlations between ovary size and neuraland endocrine response genes are consistent with the hypothesized roles of the ovaries in honey bee behavioral regulation.
KEY WORDS: honey bee, ovary size, division of labor, social behavior, artificial selection, ovarian transcriptome, ecdysteroid signaling
INTRODUCTION
Ovaries play important roles in behavioral regulation in animals as diverse as mammals (Kemnitz et al., 1989) and insects (Cupp and Collins, 1979; Hancock and Foster, 2000; Klowden, 1990). Recently, it was suggested that ovaries also affect complex behavioral patterns in functionally sterile honey bee (Apis mellifera) workers (Amdam et al., 2006; Wang et al., 2009; Wang et al., 2010). The honey bee is the most-studied social insect and is a model organism for understanding the regulation of behavior (Amdam et al., 2006; Evans and Wheeler, 1999; Giray and Robinson, 1996; Hunt et al., 1995; Lindauer, 1953; Pennisi, 2006). Worker honey bees progress through an age-associated behavioral sequence. They primarily conduct within-nest tasks during their first 2–3 weeks of adult life and then transition to performing foraging tasks outside the colony. The timing (age) of foraging onset varies among individuals. Foraging workers also show different behaviors; some bees prefer nectar collection and others prefer pollen collection (Page and Fondrk, 1995).
This behavioral plasticity is influenced by ovary size (Amdam et al., 2006; Wang et al., 2009; Wang et al., 2010). Workers that receive an implant of additional ovaries, i.e. surgery to enhance total ovary mass, initiate foraging earlier than sham controls (Wang et al., 2010). Bees with naturally large ovaries also transition from nest tasks to foraging at younger ages, and they collect a greater amount of pollen than bees with small ovaries. These associations are found within wild-type (unselected) European honey bees (Amdam et al., 2006), in crosses between unselected European and Africanized honey bees (Siegel, 2011), and between strains of two European honey bee genotypes that were bidirectionally bred for differential foraging behavior (Hunt et al., 2007; Page et al., 1995). These artificially selected genotypes are referred to as High vs Low pollen hoarding strains and differ in several traits: the High strains have larger ovaries, forage earlier in life and collect more pollen than the Low strain bees. Thus, ovary size and foraging behavior are heritable traits in honey bees (Amdam et al., 2009), and variation in both of these phenotypes maps to overlapping genomic regions [quantitative trait loci (QTL)] (Graham et al., 2011; Linksvayer et al., 2009b; Wang et al., 2009). Linkage mapping of these traits has been performed repeatedly and independently in crosses derived from different sources, i.e. from pollen-hoarding strain bees as well as Africanized and European wild-type bees (Graham et al., 2011; Linksvayer et al., 2009b; Wang et al., 2009).
The mapping studies have provided positional candidate genes for phenotypic variation in worker ovary size and behavior. Expression levels of some of these candidate genes have been examined in worker brains and fat bodies (abdominal adipose tissue). Fat body expression of hormone receptor-like in 46 (HR46) and phosphoinositide-dependent kinase-1 (PDK1) correlates with ovary size as well as worker division of labor (Wang et al., 2009). HR46 is a possible nuclear hormone receptor for ecdysteroid hormones and PDK1 is an element in the nutrient sensing target of rapamycin-, epidermal growth factor (Egf)and insulin-signaling pathways (Kamakura, 2011). However, although previous studies have documented phenotypic, functional genomic and genetic evidence for the links between honey bee division of labor and reproductive physiology (Amdam et al., 2006; Wang et al., 2009; Wang et al., 2010), less is known about how ovarian tissue affects worker behavior per se.
Here, we studied shared features of worker ovary and brain transcriptomes in order to begin to tease apart how ovaries may function in honey bee division of labor. First, we analyzed gene expression globally, as well as locally in co-expressed gene clusters between ovaries obtained from High and Low pollen-hoarding strain bees. This analysis was contrasted with six published brain transcriptional studies. Two of these studies described associations between gene expression and division of labor in the honey bee (Whitfield et al., 2003) and in paper wasps (Toth et al., 2010). Four others examined the pheromonal and endocrine effects on worker behaviors: the delay of foraging onset by queen mandibular pheromone (QMP) (Grozinger et al., 2003; Kocher et al., 2010); the acceleration of foraging onset by methoprene, a juvenile hormone (JH) mimic (Whitfield et al., 2006); and honey bee aggression by alarm pheromone (Alaux et al., 2009b). Our comparative analysis generated a list of candidate biological processes for shared behavioral and ovarian gene regulation in honey bees. Finally, as variation in ovary size is associated with worker behavioral differences (Amdam et al., 2006), we independently tested whether a subset of genes was also differentially expressed in wild-type worker ovaries of different sizes. Our results show that gene expression patterns in worker ovaries mirror brain transcriptional responses triggered by socio-environmental factors, and identify several physiological mechanisms that may facilitate the link between ovarian activity and division of labor in honey bee workers, including differences in metabolic pathways and endocrine physiology.
MATERIALS AND METHODS
Microarrays
Sample collection and RNA extraction
Honey bees, Apis mellifera L., from the High and Low strains were maintained at Arizona State University, Phoenix, AZ, USA, and at the University of California, Davis, CA, USA. Two High strain colonies and two Low strain colonies were used as donors for experimental workers for the microarray study. Ovaries were collected from newly emerged worker bees (<24 h old) and used for transcriptional profiling. Newly emerged workers have encountered few adult stimuli that could affect their ovary physiology, thus potential differences in the ovary between strains should be primarily due to genotype or ovariole number. Both ovaries from each worker were carefully dissected in sterile Ringers solution (on ice). For both strains, ovaries of 10 individual bees (five bees from each of the two colonies) were pooled to form a biological sample. Four biological samples were prepared from each genotype.
Ovaries were flash-frozen in liquid nitrogen (N2) and stored at –80°C. RNA was extracted using TRIzol Reagent (Invitrogen, cat. no. 15596-018, Carlsbad, CA, USA) (Kobayashi et al., 2006; Michaut et al., 2003). DNase (Qiagen, Valencia, CA, USA) treatment was used to remove residual genomic DNA. The integrity of extracted RNA was tested by capillary electrophoresis on an RNA6000 BioAnalyzer (Agilent Technologies, Palo Alto, CA, USA), and only high-quality samples were used for subsequent analyses.
Identification of significantly regulated transcripts
RNA (200 ng) was amplified with the MessageAmp II aRNA Amplification Kit (Ambion, Austin, TX, USA). Amplified RNA (2μg) was directly labeled with Cy3 or Cy5 dye using a Kreatech labeling kit (BioMicro, Salt Lake City, UT, USA). Labeled probe (120 pmol) from two samples was then hybridized to the whole-genome oligonucleotide arrays supplied by the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois, Urbana-Champaign. The High and Low ovaries were hybridized with dye-swaps (N=4 for each group). Arrays were scanned using the Axon Genepix 4000B scanner (Molecular Devices, Sunnyvale, CA, USA) and GENEPIX software (Agilent Technologies, Santa Clara, CA, USA), and raw data were imported into SAS (SAS Institute, Inc., Cary, NC, USA) for analysis.
Features with an intensity below 300 (the average background intensity on the arrays) were removed from the analysis. Genes with less than six observations (out of a possible 16) were also removed. Data were log-transformed and normalized using a mixed-model ANOVA (SAS) with the following model: y = dye array + block + dye×array + ε, where y is expression, ε is error, dye and block are fixed effects, and array and its interactions are random effects. Detection of significance for differential expression of residuals was performed using a mixed-model ANOVA with the model: y μ + genotype + spot + dye + array + ε, where y is the residual from the previous model, genotype and dye are fixed effects, and array is a random effect. P-values were corrected for multiple testing using a false discovery rate (FDR) adjustment (proc MULTTEST, SAS). We chose an FDR threshold of ≤0.05, suggesting that less than 5% of the significant transcripts should be false positives.
Functional analysis and mapping differentially expressed gene clusters
Gene ontology (GO) enrichment analysis was performed only on differentially expressed genes in either High and Low strain ovaries. Only transcripts (N=1534) identified as having orthologs in Drosophila were included in the analysis to exclude multiple transcripts from the same gene and transcripts for which the genomic location has not been assigned. Genes were included as being significantly differentially expressed if the FDR-corrected P-values were ≤0.05. Enrichment was determined using GO-getter (http://chem.colorado.edu/knightgroup/) (Argast et al., 2009).
The differentially expressed genes were then mapped onto honey bee chromosomes for detecting genomic clusters. We used transcripts (1794) that have been assigned for genomic locations and tested whether these genes were clustered more than expected and whether such clusters were located within the previously mapped QTL for foraging behavior (Hunt et al., 2007; Hunt and Page, 1995), sucrose sensitivity (Rueppell et al., 2006) and ovary size (Linksvayer et al., 2009b; Wang et al., 2009). Clustering was assessed across each chromosome by calculating a chi-square value for the observed number of significant genes within a sliding window of 15 loci and a step size of three loci, relative to the expected number of significant genes within that window, given the number of significant differentially expressed genes for each chromosome. We established conservative 0.05 chromosome-specific significance thresholds for the chi-square statistic using 10,000 random permutations of the data set with respect to significance of differential expression.
Contrasting transcriptomes from ovaries and brains
We contrasted our list of 2151 differentially expressed ovarian genes to four previously published brain transcriptomes from wild-type worker bees, which were exposed to different behavioral regulators (Grozinger et al., 2003; Kocher et al., 2010; Whitfield et al., 2006; Whitfield et al., 2003). We also contrasted our ovarian transcriptomes to brain transcriptomes from paper wasps, which were related to foraging behavior and reproduction (Toth et al., 2010). Finally, we used the results of a study that identified brain transcriptional changes associated with exposure to alarm pheromone as a negative control (Alaux et al., 2009b). As there is no evidence that alarm pheromone affects reproductive physiology in honey bee workers, we did not expect overlap between our results and those of Alaux et al. (Alaux et al., 2009b). Significantly overlapping genes, i.e. those that were differentially expressed in both the ovarian and brain transcriptomes, were identified using a right-tailed Fisher's exact test. Furthermore, GO analyses were performed on the overlapping genes within each pollen-hoarding strain, using GOToolBox (http://genome.crg.es/GOToolBox/) (Alaux et al., 2009a).
Verification of expression differences in candidate genes in High vs Low strain bees
Five genes [HR46, ftz transcription factor 1 (ftz-f1), tyramine receptor (TYR), major royal jelly protein 1 (MRJP1) and juvenile hormone-inducible protein 26 (JHi-26)] were chosen for verification of gene expression in samples that were used for the array. HR46 and ftz-f1 are nuclear receptors that may bind ecdysteroid hormones produced by the ovary of adult female insects, whereas TYR is a G protein-coupled receptor (GPCR) predicted to bind tyramine neurotransmitter. Both HR46 and TYR are also positional candidate genes in QTL regions that influence foraging behavior. MRJP1, a protein component in royal jelly, affects queen–worker differentiation (Kamakura, 2011) and potentially influences ovary physiology in workers. JHi-26 is a JH-inducible gene and can be triggered by methoprene, a JH analog. Ftz-f1, TYR, MRJP1 and JHi-26 were selected for profiling because they were differentially expressed between High and Low ovaries in the microarray assay (see Results). HR46 signals on the array were too low to pass the threshold for the analysis, but HR46 candidacy was inferred from prior testing of HR46 expression in ovaries from newly emerged bees of the High and Low strains [for which each biological sample consisted of 20 ovaries (pooled) from 10 individual bees from two colonies per strain, N=12, Student's t-test: t1,22=–2.4310, P=0.0237; supplementary material Fig. S1].
Characterizing candidate gene expression in large and small ovaries of wild-type bees
Both ovary size and behavioral phenotype affect ovarian physiology and gene expression. Using qRT-PCR, we tested whether relative quantification (RQ) of the candidate genes for ovary and behavioral regulation (HR46, ftz-f1, TYR, MRJP1 and JHi-26) were associated with ovary size in wild-type, newly emerged bees. For these bees, we used five wild colony sources for testing the association between candidate genes and ovary size. After ovary dissections, ovariole number was counted using a dissecting microscope (Leica, MZ 125, Buffalo Grove, IL, USA). In order to maximize the difference in gene expression between big and small ovaries, we used five and 12 as cut-off thresholds for the ovariole number. Ovaries from bees in which both ovaries had either many ovarioles (13±0.11, i.e. summing over the two ovaries) or few ovarioles (4±0.06 in total) were flash-frozen in liquid N2 and stored at –80°C. Biological samples were made by pooling 10 pairs of ovaries within the same size category, and these were assigned to either a large ovary (LAO) or a small ovary (SMO) group. We prepared 18 samples for each of the LAO and SMO groups.
Protocols for RNA extraction and DNase treatment were identical to those described above for the microarray study. cDNA was synthesized using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA, USA). Two-step qRT-PCR (real-time) was performed in triplicate using ABI Prism 7500 (Applied Biosystems), and the data were analyzed using the Delta-Delta CT method (Pfaffl, 2001) with actin (GenBank: XM_623378) as a reference gene. This gene is stably expressed across honey bee tissues, and is standardly used for gene expression studies in the bee (Lourenço et al., 2008). By monitoring negative control samples (without reverse transcriptase) and melting curves, we verified that the qRT-PCR assay was not confounded by DNA contamination or primer dimer (Vandesompele et al., 2002).
The primer sequences of the five target genes and actin were developed using Primer3 (http://frodo.wi.mit.edu/primer3/) (see supplementary material Table S1).
Expression data of candidate genes were log transformed to confer approximate normality (Wang et al., 2009). The resulting values conformed to assumptions of Student's t-test and ANOVA as assessed by normal probability plots of residuals, as well as by Bartlett's and Levene's tests for homogeneity of variance. Student's t-test was used to compare the candidate gene expressions in the ovary between High and Low strain bees. One-way ANOVA was used to compare the expression level of candidate genes between LAO and SMO groups. The log-transformed expression level of candidate genes was a dependent variable and ovary size (LAO or SMO) was an independent variable. Fisher's least significant difference (LSD) test was used for post hoc comparisons.
RESULTS
Comparison of ovarian transcriptomes between honey bee strains, and chromosome expression-mapping of genomic gene clusters
We compared ovarian transcript patterns from newly emerged High and Low pollen-hoarding strain bees using an established microarray approach (Grozinger et al., 2003; Whitfield et al., 2002). We found that 2151 transcripts, 20.3% of the 10,586 transcripts of the array, were differentially expressed between the High and Low strain sources at an FDR of 0.05 (supplementary material Table S2). Thus, the ovarian transcriptomes of the two pollen-hoarding strains were measurably different during the first hours of adult life.
GO analysis revealed the functional distribution pattern of differentially regulated genes in High and Low strain ovaries annotated against the Drosophila genome (supplementary material Fig. S2). A number of functions were ascribed, including the category ‘sensory perception’ with 10 olfactory receptors and two gustatory receptors expressed in ovaries. It is possible that the corresponding gene products incorporate into ovarian membranes and receive signals by binding molecules that circulate in the insects' hemolymph (blood). Such molecules can be internalized from the environment or secreted from tissues.
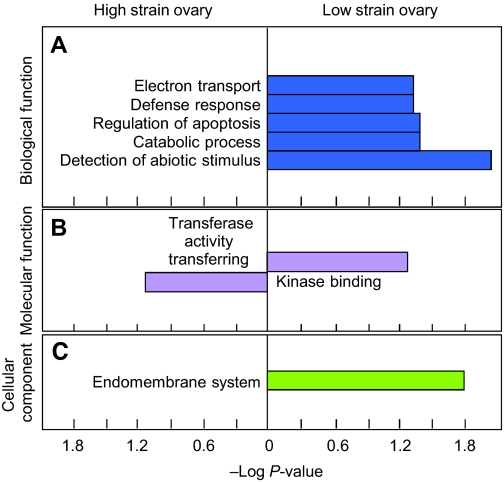
All differentially expressed genes were assigned to function groups within three functional domains of the GO analysis – ‘biological process’, ‘molecular function’ and ‘cellular component’ (Fig. 1). This comparison between genotypes demonstrates that ovarian gene expression can differ dramatically between young worker bees. Transcripts putatively involved in defense responses (immune response) (P=0.033), regulation of apoptosis (P=0.041), detection of abiotic stimulus (P=0.010) and electron transport (P=0.041) were significantly enriched in Low strain ovaries, while kinase binding (P=0.016) was significantly higher in High strain ovaries (Fig. 1, see supplementary material Table S3 for details on statistics). These results indicate that Low strain worker ovaries may be more sensitive to stimuli and show higher levels of apoptosis compared with High strain worker ovaries. This interpretation matches the finding that Low strain workers are more sensitive to ovarian inhibition and less likely to complete egg development than High strain workers (Amdam et al., 2006). Thus, ovarian gene expression differences detected early in life (Fig. 1) may have physiological implications for honey bee workers.
Fig. 1.
Comparison of gene ontology (GO) in the differentially regulated genes between High and Low strains of worker honey bees in terms of (A) biological process, (B) molecular function and (C) cellular component. The x-axis represents –Log P-value (Shamir et al., 2005) and P≤0.05 was used as a cut-off for the statistically enriched category. The corresponding GO term is given beside each horizontal bar. Detailed results for P-values ≤0.10 are given in supplementary material Table S3. Low strain ovaries are typically smaller than High strain ovaries and, interestingly, there was a significant overrepresentation of ‘regulation of apoptosis’ in the Low strain ovaries.
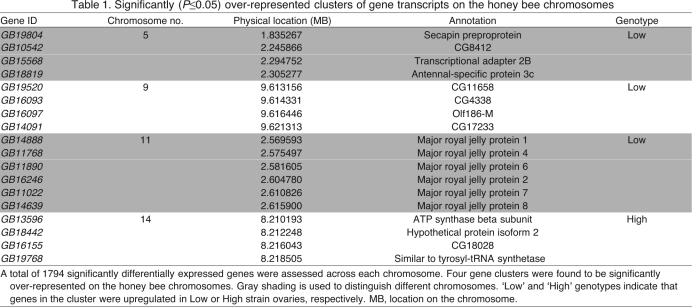
Using chromosome-wide thresholds of significance (P≤0.05), we found clusters of differentially expressed genes in four genomic regions, one on each of chromosomes 5, 9, 11 and 14 (Table 1). None of these clusters overlapped with previously identified QTL for ovary size or behavior. Interestingly, on chromosome 11, the cluster contained six transcripts (GB14888, GB11786, GB11890, GB16246, GB11022 and GB14639) of the yellow protein family, which putatively encode the major royal jelly proteins 1, 4, 6, 2, 7 and 8 (MRJP1, MRJP4, MRJP6, MRJP2, MRJP7 and MRJP8, respectively). MRJPs are abundant in royal jelly, a food secreted from hypopharyngeal (head) glands of workers. Yet, transgenic fruit flies with MRJP1 monomer expressed internally (in fat) show activation of Egf signaling and phenotypes (Kamakura, 2011), indicating internal roles of MRJP transcripts – perhaps in a variety of tissues including ovaries.
Table 1.
Significantly (P 0.05) over-represented clusters of gene transcripts on the honey bee chromosomes
Contrasting transcriptomes from ovaries and wild-type brains
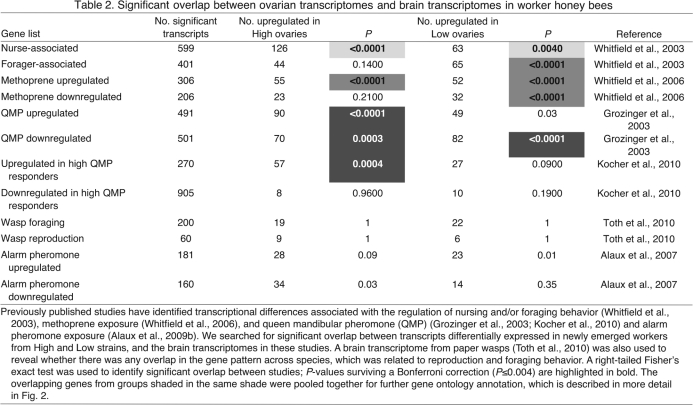
The genes that were differently expressed between ovaries from the High and Low strains were compared with previously published wild-type brain transcriptomes regulated by QMP, nurse/foraging behavior and treatment with methoprene (a JH mimic) (Grozinger et al., 2003; Kocher et al., 2010; Whitfield et al., 2006; Whitfield et al., 2003). We found significant overlap between ovarian and brain data (right-tailed Fisher's exact test; Table 2). P-values less than 0.001 were significant after Bonferroni correction for multiple testing (P<0.004).
Table 2.
Significant overlap between ovarian transcriptomes and brain transcriptomes in worker honey bees
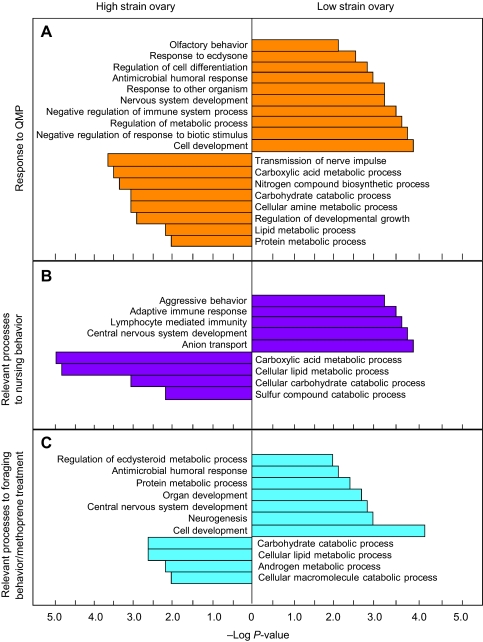
We selected genes for GO analysis that were differently regulated in ovaries of High or Low strain bees as well as regulated by QMP, nurse/foraging behavior or JH mimic in the brain of wild-type worker bees (Table 2, groups with light, medium or dark shading, respectively; Fig. 2). Similar differences in biological processes were found between High and Low strain ovaries in each of the three comparisons. The overlap between ovarian gene expression and brain transcriptomes was biased towards carbohydrate, lipid and amine metabolism in High strain bees, whereas the corresponding overlap for Low strain ovaries was neural network development and responses to different stimuli, including ecdysone hormone response (Fig. 2). Thus, we found that selection on pollen hoarding has changed ovarian expression of genes involved in metabolic control and signaling, sensory physiology and hormone responses, and the same genes respond in the brain during behavioral plasticity in wild-type bees.
Fig. 2.
Different patterns of GO biological process between High and Low strains of worker honey bees in the ovary–brain overlapping genes. (A) Response to queen mandibular pheromone (QMP), which inhibits the activation of the worker ovary and affects worker behavior (Grozinger et al., 2003; Kocher et al., 2010). (B) Relevant processes to nursing behavior, which is regulated by systemic regulators including QMP, juvenile hormone (JH), ecdysteroid and neural systems (Whitfield et al., 2003). (C) Relevant processes to foraging behavior/methoprene treatment, which accelerates the age at onset of foraging (Whitfield et al., 2006). P≤0.01 was used as a cut-off for the statistically enriched category. The GO term of each category is given by the corresponding bar. Overall, the overlap between brain transcriptomes and ovarian gene expression in the High strain bees was biased towards carbohydrate, lipid and amine metabolism, whereas the corresponding overlaps of Low strain ovaries were neural network development and responses to different stimuli, including the ecdysone hormone response.
There was no significant overlap between ovarian transcriptome in this study and the alarm-pheromone brain transcriptome selected as our negative control (P>0.004; Table 2). We also contrasted our ovarian gene list to the paper wasp brain transcriptomes related to foraging behavior and reproduction (Toth et al., 2010). We did not find significant overlap in gene expression pattern related to either foraging or reproduction (Table 2). These results are perhaps unsurprising because paper wasps (Polistes) and honey bees have fundamental difference in reproductive physiologies (Toth et al., 2010) that could differentially affect behavior. However, the common mechanisms between honey bees and wasps may be better revealed in the future by increasing the coverage of the wasp genome (Toth et al., 2010).
Verification of expression differences in candidate genes in High vs Low strain bees
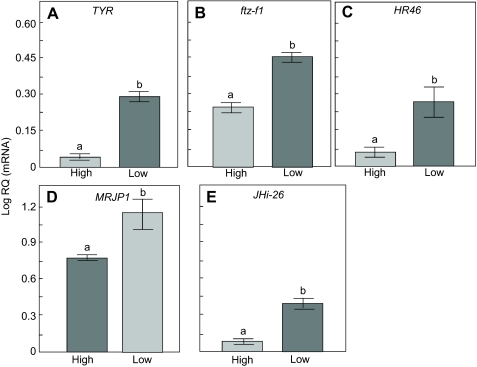
We selected five genes for expression verification that are involved in metabolic control and signaling, sensory physiology and hormone responses (i.e. HR46, Ftz-f1, JHi-26, MRJP1 and TYR; for a summary on function, see Materials and methods). The results of all five genes were consistent with the results from the array (Fig. 3) – all have significantly (P≤0.05) higher expression in Low strain ovaries than in High strain ovaries (one-tailed Student's t-test, TYR, P=0.039; ftz-f1, P=0.0458; HR46, P=0.0225; MRJP1, P=0.0004; JHi-26, P=0.0069).
Fig. 3.
Verification of expression differences in candidate genes in High vs Low strain worker honey bees using relative quantification (RQ). The same samples used in the microarray were used for gene expression verification. (A) TYR, a receptor of the neural transmitter tyramine; (B) ftz-f1, an ecdysteroid nuclear receptor; (C) HR46, an ecdysteroid nuclear receptor; (D) MRJP1, a protein component in royal jelly; (E) JHi-26, a JH-inducible gene that can be triggered by methoprene. Bars represent means ± s.e.m.; different letters (a or b) indicate significant differences (one-tailed Student's t-test, P≤0.05). All candidate genes showed a higher expression level in Low strain ovaries than in High strain ovaries, as was the case with the microarray.
Characterizing candidate gene expression in large and small ovaries of wild-type bees
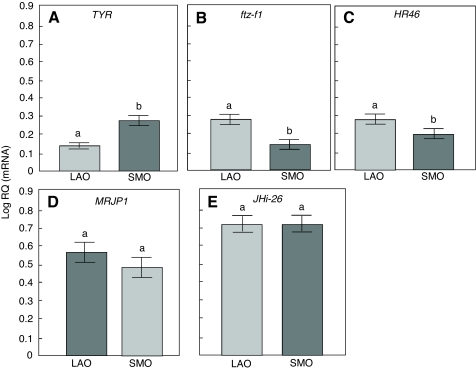
Gene expression was quantified in wild-type worker ovaries of different size. These ovary size groups had significant effects on overall gene expression (one-way ANOVA, F5,26=13.7682, P<0.0001, N=16; Fig. 4). TYR mRNA levels were significantly higher in the SMO group (Fisher's LSD, P=0.0016; Fig. 4A), whereas ftz-f1 and HR46 were more strongly expressed in the LAO group (Fisher's LSD, P=0.0102 and 0.0416, respectively; Fig. 4B,C). MRJP1 and JHi-26 expression, in contrast, did not vary by ovary size (Fisher's LSD, P=0.3844 and 0.9697, respectively; 3). These outcomes suggest that the ovarian expression levels of HR46, ftz-f1, MRJP1 and JHi-26 are best explained by selection (genotype) in this study, whereas the amount of TYR transcript can be robustly associated with ovary size in the wild-type as well as the pollen-hoarding strain bees.
Fig. 4.
Characterization of candidate gene expression in large and small ovaries of wild-type honey bees using relative quantification (RQ). Transcript abundances of four candidate genes were measured in large ovaries (LAO) and small ovaries (SMO) of wild-type bees. (A) TYR; (B) ftz-f1; (C) HR46; (D) MRJP1; (E) JHi-26. Bars represent means ± s.e.m.; different letters (a or b) indicate significant differences (post hoc Fisher's least significant difference, P≤0.05). Ftz-f1 and HR46 were more highly expressed in LAO than in SMO. TYR was more highly expressed in SMO than in LAO. There was no difference in MRJP1 and JHi-26 expression between LAO and SMO.
DISCUSSION
Here, we describe transcriptional differences between the ovaries of newly emerged bees from honey bee strains bidirectionally selected for behavior. A number of genes were associated with the biological process ‘sensory perception’ or the functional category ‘reproduction’ (GO analysis, supplementary material Fig. S2). Differentially expressed genes included ecdysone-inducible nuclear hormone receptors and olfactory and gustatory receptors. Taken together, these results suggest that the honey bee worker ovary is a dynamic organ (Wang et al., 2010), although it is often presumed to be inactive under normal social conditions (Foster and Ratnieks, 2001). Furthermore, the significant overlap observed between the present study and previously published brain transcriptomes suggests that both ovary and brain physiologies are affected by systemic factors such as JH, ecdysteroids and QMP, and further supports the hypothesis that each of these factors are key participants in regulatory networks for social behavior in this species.
Ecdysteroid hormones and worker ovarian physiology
Honey bee worker ovaries produce ecdysteroids during the first day of adult life (Amdam et al., 2010), and the ecdysone receptor gene (EcR) is expressed in worker and queen ovaries (Takeuchi et al., 2007). Ecdysteroids signal oogenesis in Tribolium, Drosophila and mosquitoes (Culex pipiens) (Bernardi et al., 2009; Khater et al., 1994; Xu et al., 2010), but oogenesis is uncommon in worker bees because yolk uptake and oocyte maturation is suppressed pheromonally when a queen is present in the colony (Atkins et al., 1975).
We found that three genes associated with the ecdysteroid cascade [HR46, ftz-f1 and ecdysone-induced protein 75 (E75)] were more highly expressed in Low pollen-hoarding strain ovaries than in the High strain. At adult emergence, Low strain bees also have the higher ecdysteroid titers, and the plausible source of this hormone is the ovary (Amdam et al., 2010). Yet, elevated ecdysteroid signaling and/or sensing in Low strain ovaries does not correlate with oogenesis (Amdam et al., 2006). Rather, ecdysteroids can induce oocyte apoptosis and follicle atresia (Paul et al., 2005; Soller et al., 1999; Uchida et al., 2004), and can be supported by increased apoptosis regulation in Low strain ovaries (Fig. 1A). This hypothesis is consistent with Low strain workers being more readily inhibited from activating their ovaries for egg laying (Amdam et al., 2006), perhaps because of heightened sensitivity to social inhibition and increased rates of follicle apoptosis (Hunt et al., 2007). Future studies may answer whether ecdysteroids regulate apoptosis in honey bee ovaries.
Ecdysteroid titers are very low in the hemolymph of adult bees, suggesting that ecdysteroids have lost their function in adult workers (Hartfelder et al., 2002). Recently, many studies have indicated that ecdysteroid cascades in the fat body and brain may be involved in social behavior (Velarde et al., 2009; Wang et al., 2009; Wang et al., 2010). In this study, we found a significant overlap of ecdysteroid-related processes between the Low ovarian and brain transcriptomes associated with QMP exposure (Fig. 2A) and foraging behavior (Fig. 2C). This implies that ecdysteroids may be involved in both brain and ovary physiologies related to QMP and foraging behavior. However, further investigation is needed to test the role of ecdysteroids in honey bee brain regarding social behavior.
QMP, JH-ecdysteroid hormone axes and the worker ovary
Our comparison between ovarian and brain transcriptomes relates biological processes of the ovary to brain mRNA expression profiles associated with exposure to QMP, methoprene and the nursing or foraging behavioral state. In every comparison, similar patterns of ovary–brain overlap were found (Fig. 2). Was this surprising? Worker behavior, including nursing and foraging, is modulated by QMP (Pankiw et al., 1998), JH (Huang et al., 1991) and ovarian physiology (Amdam et al., 2006; Nelson et al., 2007; Robinson, 1992). Therefore, the overlap we revealed (Fig. 2) may be a signature of (largely) systemically acting gene networks associated with the division of labor. Moreover, the clear separation between pollen-hoarding strains in their overlap of ovary and brain transcriptomes supports the hypothesis that the bidirectional selection on these strains acted on gene networks that connect behavior to physiology (Amdam et al., 2010; Page et al., 1998).
QMP emitted by honey bee queens inhibits ovary activation (Hoover et al., 2003), influences age-related division of labor (Pankiw et al., 1998) and affects brain development in worker bees (Morgan et al., 1998). The brain's transcriptional response to QMP is also negatively correlated with worker ovariole number (Kocher et al., 2010). This correlation points to unexplained connections between brain function and ovary size. We found that the functional category ‘olfactory behavior’ was over-represented in the overlap between Low strain ovaries and brains regulated by QMP (Fig. 2A). Moreover, we identified 10 olfactory receptor (OR) genes differentially expressed between High and Low strain ovaries. ORs belong to the GPCR gene family, which is also enriched in ovaries of Low strain genotype bees (P=0.0673; supplementary material Table S3). GPCRs comprise a large protein family of trans-membrane receptors that bind light-sensitive compounds, odors, pheromones, hormones and neurotransmitters. ORs are generally expressed in cell membranes of olfactory receptor neurons (antennae) (Getz and Akers, 1994; Murphy et al., 2003; Vosshall et al., 1999). However, additional functions of OR proteins have been suggested (Feingold et al., 1999; Itakura et al., 2006; Xu et al., 2000) because of their localization to different ectopic tissues (non-olfactory tissues), such as germ cells and testis tissue in mammals (Fukuda et al., 2004; Marchand, 2003; Spehr et al., 2003; Vanderhaeghen et al., 1997; Ziegler et al., 2002). Recent experiments also show that one OR in honey bee antennal neurons can respond to a component of QMP (Wanner et al., 2007). It is possible that connections between brain function and ovarian traits in worker bees could be caused by correlated expression of ORs. This hypothesis can be tested in future studies.
JH is a systemic hormone with effects on insect metabolic biology, reproductive physiology and adult behavior. JH and ecdysteroid titers are often negatively correlated in insects, with opposing regulation demonstrated in Drosophila melanogaster (reviewed by Riddiford, 1996), mosquitoes (Aedes atropalpus) L (Birnbaum et al., 1984) and honey bees (Rembold, 1987). Using brain transcriptome data from exposure to methoprene (a JH mimic), we found that Low strain ovaries bias functional processes towards ecdysteroid and neural regulation in contrast to the High strain bees (Fig. 2C). It has previously been demonstrated that worker sensitivity towards JH responses is conditional on their Low vs High genotypes (Amdam et al., 2007; Amdam et al., 2010), i.e. Low strain worker bees show reduced JH sensitivity, perhaps because of the genetic bias towards ecdysteroid signaling that is suggested by our results.
Genomic gene clusters
Our study presents a genomic mapping approach that can be used on large-scale transcript data. This method can enhance functional genomic studies by adding spatial information as to where significantly expressed genes are located in the genome. Using this method, we found four genomic clusters of differentially expressed genes, suggesting that, in these regions of the genome, nearby genes are consistently and differentially expressed between High and Low genotypes (Table 1, supplementary material Fig. S3). Similar gene clusters are often co-regulated (Cohen et al., 2000; Spieth et al., 1991; Takadera et al., 1996). According to our conservative chromosome-wide thresholds, previously mapped QTL for worker behavior and ovary size do not contain putatively co-regulated gene clusters, as would be expected if the QTL led to changes in cis regulation of groups of genes. More quantitative measures of differential gene expression, i.e. with RNA sequencing and subsequent genome mapping, may better reveal genomic patterns of gene expression and the degree to which gene clusters are co-regulated. Alternatively, the identified QTL may affect ovariole number during ovary development in late larval stages. Because we used newly emerged bees in this study, we might have missed the developmental window when the QTL affect the ovariole number.
One of the clusters we detected contained MRJP-encoding genes. A few MRJP transcripts are generally expressed in honey bee reproductive tissues, suggesting roles in sex-specific reproductive maturity (Drapeau et al., 2006). We found the gene cluster to be upregulated in Low strain ovaries. Ecdysteroid signaling, which presumably is also upregulated in Low strain ovaries, enhances the royal-jelly-producing (and strongly MRJP-expressing) hypopharyngeal glands of workers in addition to influencing ovarian physiology (Wegener et al., 2009). Cross-fostering studies indicate that low strain nurse workers provide different nutritional environments to developing larvae than high strain nurse workers (Linksvayer et al. 2009b, 2011), perhaps as a result of differential MRJP1 expression. The elevated expression of MRJP genes in Low strain ovaries might, in other words, be an ecdysteroid-driven response. An MRPJ1 monomer influences honey bee development when secreted in larval food (Kamakura, 2011). MRJP1 was not differentially expressed in wild-type ovaries of different sizes (Fig. 3D), suggesting that it is not involved in associations between ovary size and behavior in worker bees. Yet, candidate gene expression is a relative quantity, an amount corrected towards housekeeper genes that are equally present per unit total RNA. Between individuals, however, the summed total RNA of variably sized tissues can correlate with organ size, e.g. if the larger-sized organs have more cells. Large ovaries have more cells that make up the ovariole count. This consideration could be relevant for translated proteins such as MRJP1, as larger ovaries might release more product than small ovaries.
Genotype, ovary size and specific patterns of gene expression
Ovariole number plays a role in social behavior (Amdam et al., 2006; Wang et al., 2010) and ovarian physiology (Makert et al., 2006) in honey bees. However, High and Low pollen-hoarding genotypes can also be factors influencing the ovarian transcriptomes. Therefore, we tested the association between ovary size and the expression of five candidate genes in wild-type bees: HR46, ftz-f1, TYR, MRJP1 and JHi-26. For the two latter genes, no association was supported. For HR46 and ftz-f1, differences were significant but levels were opposite to those predicted by the average ovariole number of High and Low strain genotypes, suggesting that differential expression of these genes can be influenced by the genetic background. TYR, however, showed consistent relationships with ovary size between wild-type and the selected stocks.
It was suggested previously that selection for pollen-hoarding strains has influenced relationships between HR46, Ftz-f1 and ovary size in newly emerged bees (Wang et al., 2009). HR46 and ftz-f1 mRNA levels coincide with signaling by ecdysteroid hormones (Velarde et al., 2009), and ecdysteroid titers are shifted in late pupal stages and during early adult life between High and Low pollen-hoarding strain bees (Amdam et al., 2010). This shift may also change the induction of HR46 and ftz-f1 relative to ovary size when selected strains are compared with wild type. Similar phenomena are known from other systems (Toledo-Rodriguez et al., 2004).
TYR, which is activated by the neurotransmitter tyramine, was consistently negatively associated with ovariole number in the two selected strains and in wild-type bees. TYR is a positional candidate gene in the QTL Pln2 that is genetically linked to foraging behavior and ovary size (Hunt et al., 2007; Linksvayer et al., 2009b). Tyramine, furthermore, influences honey bee sucrose responsiveness, which diverges between pollen-hoarding strains, and predicts foraging behavioral traits in wild-type bees (Pankiw and Page, 2003; Scheiner et al., 2002). These connections might point to systemic patterns in TYR-dependent signaling. Yet, the modality of TYR expression differs between the brain and ovary of the pollen-hoarding strains (supplementary material Fig. S4), which suggests a more complex, tissue-specific function of TYR and tyramine that are supported by previous studies (e.g. Huang et al., 2004; Nykamp and Lange, 2000; Thompson et al., 2007). Taken together, our results support TYR as a candidate gene for associations between ovary size and behavior in worker bees.
Conclusions
Ovary size is affected by genetic and environmental factors and influences social behavior in honey bee workers. Social environmental signals (such as QMP) and physiological systems (such as the ecdysteroid and JH axes) regulate worker ovarian development and physiology. Yet, the worker ovary may also be an active endocrine signaling system and transcriptional feedback generator (Fig. 5). Therefore, our study supports the hypothesis that both ovaries and brains participate in the regulatory networks, which respond to the systemic stimuli such as hormones and pheromones, and modulate honey bee division of labor. These insights provide a platform for further functional studies that can determine how ovarian processes influence brain function and social behavior.
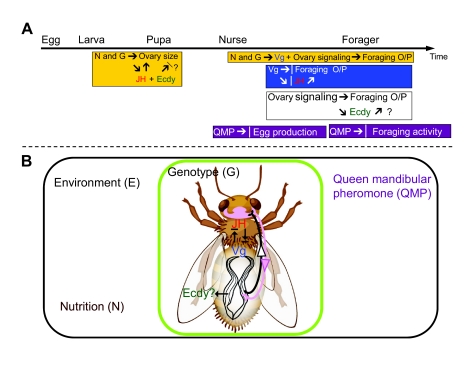
Fig. 5.
Model to explain roles of ovaries in a regulatory network of social behavior. Ovary size is determined by nutrition (N) status and colony genotype (G) at the last larval stage, which is affected by a systemic hormone, JH (Bloch et al., 2000; Schmidt-Capella and Hartfelder, 1998). Results from the present study and those from Wang et al. suggest that ecdysteroids (Ecdy) may be involved in apoptosis for shaping ovary size (Wang et al., 2009). Queen mandibular pheromone (QMP) inhibits egg production in nurses and slows workers to forage in their later lives. Nutrition and genotype also play important roles in regulating ovary signaling and vitellogenin (Vg) titers in adult workers, which affect age of foraging onset (O) and foraging preference (P). Vg regulates foraging O/P directly or through a feedback loop with JH (Amdam et al., 2004; Amdam and Omholt, 2003; Ihle et al., 2010). Worker ovaries can accelerate foraging O/P, in which ecdysteroids may function. There are also neural regulations between the ovary and brain, which may be involved in these behavioral networks.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Mutti and M. K. Fondrk for experimental assistance and we are also grateful to Z. Simões, C. S. Brent, A. Dolezal, K. Traynor and K. Dolezal for helpful comments on the manuscript.
LIST OF ABBREVIATIONS
- Egf
epidermal growth factor
- E75
ecdysone-induced protein
- 75
FDR false discovery rate
- ftz-f1
ftz transcription factor-1
- GO
gene ontology
- GPCR
G protein-coupled receptor
- HR46
hormone receptor-like in 46
- JH
juvenile hormone
- JHi-26
juvenile hormone-inducible protein 26
- LAO
large ovary
- LSD
least significant difference
- MRJP
major royal jelly protein
- MRJP1
major royal jelly protein 1
- OR
olfactory receptor
- PDK1
phosphoinositide-dependent kinase-1
- QMP
queen mandibular pheromone
- QTL
quantitative trait loci
- RQ
relative quantification
- SMO
small ovary
- TYR
tyramine receptor gene
FOOTNOTES
Supplementary material available online at http://jeb.biologists.org/cgi/content/full/215/1/124/DC1
FUNDING
This research was supported by the Research Council of Norway [180504, 185306] and The PEW Charitable Trust [to G.V.A.], the National Institute on Aging [NIA P01 AG22500] and the Wissenschaftskolleg zu Berlin [to G.V.A., T.A.L. and R.E.P.] and the National Science Foundation-CAREER grant [NSF 0746338 to C.M.G.]. Deposited in PMC for release after 12 months.
REFERENCES
- Alaux C., Robinson G. E. (2007). Alarm pheromone induces immediate-early gene expression and slow behavioral response in honey bees. J. Chem. Ecol. 33, 1346-1350 [DOI] [PubMed] [Google Scholar]
- Alaux C., Le Conte Y., Adams H. A., Rodriguez-Zas S., Grozinger C. M., Sinha S., Robinson G. E. (2009a). Regulation of brain gene expression in honey bees by brood pheromone. Genes Brain Behav. 8, 309-319 [DOI] [PubMed] [Google Scholar]
- Alaux C., Sinha S., Hasadsri L., Hunt G. J., Guzman-Novoa E., DeGrandi-Hoffman G., Uribe-Rubio J. L., Southey B. R., Rodriguez-Zas S., Robinson G. E. (2009b). Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl. Acad. Sci. USA 106, 15400-15405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Omholt S. W. (2003). The hive bee to forager transition in honeybee colonies: the double repressor hypothesis. J. Theor. Biol. 223, 451-464 [DOI] [PubMed] [Google Scholar]
- Amdam G. V., Norberg K., Fondrk M. K., Page R. E., Jr (2004). Reproductive ground plan may mediate colony-level selection effects on individual foraging behavior in honey bees. Proc. Natl Acad. Sci. USA 101, 11350-11355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Csondes A., Fondrk M. K., Page R. E., Jr (2006). Complex social behaviour derived from maternal reproductive traits. Nature 439, 76-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Nilsen K. A., Norberg K., Fondrk M. K., Hartfelder K. (2007). Variation in endocrine signaling underlies variation in social life history. Am. Nat. 170, 37-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V., Ihle K. E., Page R. E. (2009). Regulation of honey bee (Apis mellifera) life-histories by vitellogenin. In Hormones, Brain and Behavior, Vol. 4 (ed. Pfaff D., Arnold A., Etgen A., Fahrbach S., Rubin R.), pp. 1003-1025 San Diego, CA: Elsevier Academic Press; [Google Scholar]
- Amdam G. V., Page R. E., Jr, Fondrk M. K., Brent C. S. (2010). Hormone response to bidirectional selection on social behavior. Evol. Dev. 12, 428-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argast G. M., Croy C. H., Couts K. L., Zhang Z., Litman E., Chan D. C., Ahn N. G. (2009). Plexin B1 is repressed by oncogenic B-Raf signaling and functions as a tumor suppressor in melanoma cells. Oncogene 28, 2697-2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins E. L., Banker R., Butler C. G., Cale G. H., Cale G. H., Jr, Crane E., Dadant C. C. (1975). The Hive and the Honey Bee. Hamilton, IL: Datant & Sons; [Google Scholar]
- Bernardi F., Romani P., Tzertzinis G., Gargiulo G., Cavaliere V. (2009). EcR-B1 and Usp nuclear hormone receptors regulate expression of the VM32E eggshell gene during Drosophila oogenesis. Dev. Biol. 328, 541-551 [DOI] [PubMed] [Google Scholar]
- Birnbaum M. J., Kelly T. J., Woods C. W., Imberski R. B. (1984). Hormonal regulation of ovarian ecdysteroid production in the autogenous mosquito, Aedes atropalpus. Gen. Comp. Endocrinol. 56, 9-18 [DOI] [PubMed] [Google Scholar]
- Bloch G., Borst D. W., Huang Z., Robinson G. E., Cnaani J., Hefetz A. (2000). Juvenile hormone titers, juvenile hormone biosynthesis, ovarian development and social environment in Bombus terrestris. J. Insect Physiol. 46, 47-57 [DOI] [PubMed] [Google Scholar]
- Cohen B. A., Mitra R. D., Hughes J. D., Church G. M. (2000). A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 26, 183-186 [DOI] [PubMed] [Google Scholar]
- Cupp E. W., Collins R. C. (1979). The gonotrophic cycle in Simulium ochraceum. Am. J. Trop. Med. Hyg. 28, 422-426 [DOI] [PubMed] [Google Scholar]
- Drapeau M. D., Albert S., Kucharski R., Prusko C., Maleszka R. (2006). Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 16, 1385-1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. D., Wheeler D. E. (1999). Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc. Natl. Acad. Sci. USA 96, 5575-5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold E. A., Penny L. A., Nienhuis A. W., Forget B. G. (1999). An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics 61, 15-23 [DOI] [PubMed] [Google Scholar]
- Foster K. R., Ratnieks F. L. (2001). Convergent evolution of worker policing by egg eating in the honeybee and common wasp. Proc. R. Soc. Lond. B 268, 169-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N., Yomogida K., Okabe M., Touhara K. (2004). Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J. Cell Sci. 117, 5835-5845 [DOI] [PubMed] [Google Scholar]
- Getz W. M., Akers R. P. (1994). Honeybee olfactory sensilla behave as integrated processing units. Behav. Neural. Biol. 61, 191-195 [DOI] [PubMed] [Google Scholar]
- Giray T., Robinson G. E. (1996). Common endocrine and genetic mechanisms of behavioral development in male and worker honey bees and the evolution of division of labor. Proc. Natl. Acad. Sci. USA 93, 11718-11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A. M., Munday M. D., Kaftanoglu O., Page R. E., Jr, Amdam G. V., Rueppell O. (2011). Support for the reproductive ground plan hypothesis of social evolution and major QTL for ovary traits of Africanized worker honey bees (Apis mellifera L.). BMC Evol. Biol. 11, 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger C. M., Sharabash N. M., Whitfield C. W., Robinson G. E. (2003). Pheromone-mediated gene expression in the honey bee brain. Proc. Natl. Acad. Sci. USA 100 Suppl. 2, 14519-14525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. G., Foster W. A. (2000). Exogenous Juvenile hormone and methoprene, but not male accessory gland substances or ovariectomy, affect the blood/nectar choice of female Culex nigiripalpus mosquitoes. Med. Vet. Entomol. 14, 373-382 [DOI] [PubMed] [Google Scholar]
- Hartfelder K., Bitondi M. M. G., Santana W. C., Simões Z. L. P. (2002). Ecdysteroid titer and reproduction in queens and workers of the honey bee and of a stingless bee: loss of ecdysteroid function at increasing levels of sociality? Insect Biochem. Mol. Biol. 32, 211-216 [DOI] [PubMed] [Google Scholar]
- Hoover S. E. R., Keeling C. I., Winston M. L., Slessor K. N. (2003). The effect of queen pheromones on worker honey bee ovary development. Naturwissenschaften 90, 477-480 [DOI] [PubMed] [Google Scholar]
- Huang Z. Y., Robinson G. E., Yaoi S., Strambi C., Strambi A., Stay B. (1991). Hormonal regulation of behavioural development in the honey bee is based on changes in the rate of juvenile hormone biosynthesis. J. Insect Physiol. 37, 733-741 [Google Scholar]
- Huang Z. Y., Hanley A. V., Pett W. L., Langenberger M., Duan J. J. (2004). Field and semifield evaluation of impacts of transgenic canola pollen on survival and development of worker honey bees. J. Econ. Entomol. 97, 1517-1523 [DOI] [PubMed] [Google Scholar]
- Hunt G. J., Page R. E., Jr (1995). Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics 139, 1371-1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt G. J., Page R. E., Fondrk M. K., Dullum C. J. (1995). Major quantitative trait loci affecting honey-bee foraging behavior. Genetics 141, 1537-1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt G. J., Amdam G. V., Schlipalius D., Emore C., Sardesai N., Williams C. E., Rueppell O., Guzman-Novoa E., Arechavaleta-Velasco M., Chandra S., et al. (2007). Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften 94, 247-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle K. E., Page R. E., Frederick K., Fondrk M. K., Amdam G. V. (2010). Genotype effect on regulation of behaviour by vitellogenin supports reproductive origin of honeybee foraging bias. Anim. Behav. 79, 1001-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura S., Ohno K., Ueki T., Sato K., Kanayama N. (2006). Expression of Golf in the rat placenta: possible implication in olfactory receptor transduction. Placenta 27, 103-108 [DOI] [PubMed] [Google Scholar]
- Kamakura M. (2011). Royalactin induces queen differentiation in honeybees. Nature 21, 510-512 [DOI] [PubMed] [Google Scholar]
- Kemnitz J. W., Gibber J. R., Lindsay K. A., Eisele S. G. (1989). Effects of ovarian hormones on eating behaviors, body weight, and glucoregulation in rhesus monkeys. Horm. Behav. 23, 235-250 [DOI] [PubMed] [Google Scholar]
- Khater H. M., Gad A. M., Hassan A. N. (1994). Effect of juvenile hormone and ecdysone on initiating blood meal-dependent vitellogenic ovarian cycles in autogenous and anautogenous Culex pipiens (Diptera: Culicidae). J. Egypt. Public Health Assoc. 69, 213-225 [PubMed] [Google Scholar]
- Klowden M. J. (1990). The endogenous regulation of mosquito reproductive behavior. Experientia 46, 660-670 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Michaut L., Ino A., Honjo K., Nakajima T., Maruyama Y., Mochizuki H., Ando M., Ghangrekar I., Takahashi K., et al. (2006). Differential microarray analysis of Drosophila mushroom body transcripts using chemical ablation. Proc. Natl. Acad. Sci. USA 103, 14417-14422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher S. D., Ayroles J. F., Stone E. A., Grozinger C. M. (2010). Individual variation in pheromone response correlates with reproductive traits and brain gene expression in worker honey bees. PLoS ONE 5, e9116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindauer M. (1953). Division of labour in the honeybee colony. Bee World 34, 63-90 [Google Scholar]
- Linksvayer T. A., Fondrk M. K., Page R. E., Jr (2009a). Colony-level selection in honey bees produces coevolved socially interacting gene complexes. American Naturalist 173, 99-107 [DOI] [PubMed] [Google Scholar]
- Linksvayer T. A., Rueppell O., Siegel A., Kaftanoglu O., Page R. E., Jr, Amdam G. V. (2009b). The genetic basis of transgressive ovary size in honeybee workers. Genetics 183, 693-707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer T. A., Kaftanoglu O., Akyol E., Blatch S., Amdam G. V., Page R. E., Jr (2011). Larval and nurse worker control of developmental plasticity and the evolution of honey bee queen–worker dimorphism. J. Evol. Biol. 24, 1939-1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço A. P., Mackert A., Santos C., Simões Z. L. P. (2008). Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39, 372-385 [Google Scholar]
- Makert G. R., Paxton R. J., Hartfelder K. (2006). Ovariole number-a predictor for differential reproductive success among worker subfamilies in queenless honey bee (Apis mellifera L.) colonies. Behav. Ecol. Sociobiol. 60, 815-825 [Google Scholar]
- Marchand A. P. (2003). Chemistry. Diamondoid hydrocarbons-delving into nature’s bounty. Science 299, 52-53 [DOI] [PubMed] [Google Scholar]
- Michaut L., Flister S., Neeb M., White K. P., Certa U., Gehring W. J. (2003). Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc. Natl. Acad. Sci. USA 100, 4024-4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan S. M., Butz Huryn V. M., Downes S. R., Mercer A. R. (1998). The effects of queenlessness on the maturation of the honey bee olfactory system. Behav. Brain Res. 91, 115-126 [DOI] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277-284 [DOI] [PubMed] [Google Scholar]
- Nelson C. M., Ihle K., Amdam G. V., Fondrk M. K., Page R. E. (2007). The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 5, 673-677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykamp D. A., Lange A. B. (2000). Interaction between octopamine and proctolin on the oviducts of Locusta migratoria. J. Insect Physiol. 46, 809-816 [DOI] [PubMed] [Google Scholar]
- Page R. E., Fondrk M. K. (1995). The effects of colony-level selection on the social organization of honey bee (Apis mellifera L.) colonies: colony-level components of pollen hoarding. Behav. Ecol. Sociobiol. 36, 135-144 [Google Scholar]
- Page R. E., Robinson G. E., Fondrk M. K., Nasr M. E. (1995). Effects of worker genotypic diversity on honey bee colony development and behavior (Apis mellifera L.). Behav. Ecol. Sociobiol. 36, 387-396 [Google Scholar]
- Page R. E., Jr, Erber J., Fondrk M. K. (1998). The effect of genotype on response thresholds to sucrose and foraging behavior of honey bees (Apis mellifera L.). J. Comp. Physiol. A 182, 489-500 [DOI] [PubMed] [Google Scholar]
- Pankiw T., Page R. E., Jr. (2003). Effect of pheromones, hormones, and handling on sucrose response thresholds of honey bees (Apis mellifera L.). J. Comp. Physiol. A 189, 675-684 [DOI] [PubMed] [Google Scholar]
- Pankiw T., Huang Z.-Y., Winston M. L., Robinson G. E. (1998). Queen mandibular gland pheromone influences worker honey bee (Apis mellifera L.) foraging ontogeny and juvenile hormone titers. J. Insect Physiol. 44, 685-692 [DOI] [PubMed] [Google Scholar]
- Paul R. K., Takeuchi H., Matsuo Y., Kubo T. (2005). Gene expression of ecdysteroid-regulated gene E74 of the honeybee in ovary and brain. Insect Mol. Biol. 14, 9-15 [DOI] [PubMed] [Google Scholar]
- Pennisi E. (2006). Honey bee genome illuminates insect evolution and social behavior. Science 314, 578-579 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembold H. (1987). Caste specific modulation of juvenile hormone titers in Apis mellifera. Insect Biochem. 17, 1003-1006 [Google Scholar]
- Riddiford L. M. (1996). Molecular Aspects of Juvenile Hormone Action in Insect Metamorphosis. San Diego, CA: Academic Press; [Google Scholar]
- Robinson G. E. (1992). Regulation of division of labor in insect societies. Annu. Rev. Entomol. 37, 637-665 [DOI] [PubMed] [Google Scholar]
- Rueppell O., Chandra S. B., Pankiw T., Fondrk M. K., Beye M., Hunt G., Page R. E. (2006). The genetic architecture of sucrose responsiveness in the honeybee (Apis mellifera L.). Genetics 172, 243-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppell O., Metheny J. D., Linksvayer T., Fondrk M. K., Page R. E., Amdam G. V. (2011). Genetic architecture of ovary size and asymmetry in European honey bee workers. Heredity 106, 894-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner R., Pluckhahn S., Oney B., Blenau W., Erber J. (2002). Behavioural pharmacology of octopamine, tyramine and dopamine in honey bees. Behav. Brain Res. 136, 545-553 [DOI] [PubMed] [Google Scholar]
- Schmidt-Capella I. C., Hartfelder K. (1998). Juvenile hormone effect on DNA synthesis and apoptosis in caste-specific differentiation of the larval honey bee (Apis mellifera L.) ovary. J. Insect Physiol. 44, 385-391 [DOI] [PubMed] [Google Scholar]
- Shamir R., Maron-Katz A., Tanay A., Linhart C., Steinfeld I., Sharan R., Shiloh Y., Elkon R. (2005). EXPANDER – an integrative program suite for microarray data analysis. BMC Bioinformatics 6, 232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel A. (2011). Ovarian regulation of honey bee (Apis mellifera) foraging division of labor. PhD thesis, Arizona State University, Tempe, AZ, USA: [Google Scholar]
- Soller M., Bownes M., Kubli E. (1999). Control of oocyte maturation in sexually mature Drosophila females. Dev. Biol. 208, 337-351 [DOI] [PubMed] [Google Scholar]
- Spehr M., Gisselmann G., Poplawski A., Riffell J. A., Wetzel C. H., Zimmer R. K., Hatt H. (2003). Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299, 2054-2058 [DOI] [PubMed] [Google Scholar]
- Spieth J., Nettleton M., Zuckeraprison E., Lea K., Blumenthal T. (1991). Vitellogenin motifs conserved in nematodes and vertebrates. J. Mol. Evol. 32, 429-438 [DOI] [PubMed] [Google Scholar]
- Takadera K., Yamashita M., Hatakeyama M., Oishi K. (1996). Similarities in vitellin antigenicity and vitellogenin mRNA nucleotide sequence among sawflies (Hymenoptera: Symphyta: Tenthredinoidea). J. Insect Physiol. 42, 417-422 [Google Scholar]
- Takeuchi H., Paul R. K., Matsuzaka E., Kubo T. (2007). EcR-A expression in the brain and ovary of the honeybee (Apis mellifera L.). Zool. Sci 24, 596-603 [DOI] [PubMed] [Google Scholar]
- Thompson G. J., Yockey H., Lim J., Oldroyd B. P. (2007). Experimental manipulation of ovary activation and gene expression in honey bee (Apis mellifera) queens and workers: testing hypotheses of reproductive regulation. J. Exp. Zool. A Ecol. Genet. Physiol. 307, 600-610 [DOI] [PubMed] [Google Scholar]
- Toledo-Rodriguez M., Blumenfeld B., Wu C., Luo J., Attali B., Goodman P., Markram H. (2004). Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb. Cortex 14, 1310-1327 [DOI] [PubMed] [Google Scholar]
- Toth A. L., Varala K., Henshaw M. T., Rodriguez-Zas S. L., Hudson M. E., Robinson G. E. (2010). Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proc. R. Soc. Lond. B 277, 2139-2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K., Nishizuka M., Ohmori D., Ueno T., Eshita Y., Fukunaga A. (2004). Follicular epithelial cell apoptosis of atretic follicles within developing ovaries of the mosquito Culex pipiens pallens. J. Insect Physiol. 50, 903-912 [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen P., Schurmans S., Vassart G., Parmentier M. (1997). Specific repertoire of olfactory receptor genes in the male germ cells of several mammalian species. Genomics 39, 239-246 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Paepe A., Speleman F. (2002). Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 303, 95-98 [DOI] [PubMed] [Google Scholar]
- Velarde R. A., Robinson G. E., Fahrbach S. E. (2009). Coordinated responses to developmental hormones in the Kenyon cells of the adult worker honey bee brain (Apis mellifera L.). J. Insect Physiol. 55, 59-69 [DOI] [PubMed] [Google Scholar]
- Vosshall L. B., Amrein H., Morozov P. S., Rzhetsky A., Axel R. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725-736 [DOI] [PubMed] [Google Scholar]
- Wang Y., Amdam G. V., Rueppell O., Wallrichs M. A., Fondrk M. K., Kaftanoglu O., Page R. E., Jr (2009). PDK1 and HR46 gene homologs tie social behavior to ovary signals. PLoS ONE 4, e4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kaftanoglua O., Siegela A. J., Page R. E., Jr, Amdama G. V. (2010). Surgically increased ovarian mass in the honey bee confirms link between reproductive physiology and worker behavior. J. Insect Physiol. 56, 1216-1224 [DOI] [PubMed] [Google Scholar]
- Wanner K. W., Nichols A. S., Walden K. K., Brockmann A., Luetje C. W., Robertson H. M. (2007). A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc. Natl. Acad. Sci. USA 104, 14383-14388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener J., Huang Z. Y., Lorenz M. W., Bienefeld K. (2009). Regulation of hypopharyngeal gland activity and oogenesis in honey bee (Apis mellifera) workers. J. Insect Physiol. 55, 716-725 [DOI] [PubMed] [Google Scholar]
- Whitfield C. W., Band M. R., Bonaldo M. F., Kumar C. G., Liu L., Pardinas J. R., Robertson H. M., Soares M. B., Robinson G. E. (2002). Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 12, 555-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. W., Cziko A. M., Robinson G. E. (2003). Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296-299 [DOI] [PubMed] [Google Scholar]
- Whitfield C. W., Ben-Shahar Y., Brillet C., Leoncini I., Crauser D., Leconte Y., Rodriguez-Zas S., Robinson G. E. (2006). Genomic dissection of behavioral maturation in the honey bee. Proc. Natl. Acad. Sci. USA 103, 16068-16075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Tan A., Palli S. R. (2010). The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J. Insect Physiol. 56, 1471-1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. L., Stackhouse B. G., Florence K., Zhang W., Shanmugam N., Sesterhenn I. A., Zou Z., Srikantan V., Augustus M., Roschke V., et al. (2000). PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 60, 6568-6572 [PubMed] [Google Scholar]
- Ziegler A., Dohr G., Uchanska-Ziegler B. (2002). Possible roles for products of polymorphic MHC and linked olfactory receptor genes during selection processes in reproduction. Am. J. Reprod. Immunol. 48, 34-42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.