Abstract
This paper describes the detection of a cardiac biomarker, cardiac troponin I (cTnI), spiked into depleted human serum using cationic isotachophoresis (ITP) in a 3.9 cm long poly(methyl methacrylate) (PMMA) microfluidic channel. The microfluidic chip incorporates a 100x cross-sectional area reduction, including a 10x depth reduction and a 10x width reduction, to increase sensitivity during ITP. The cross-sectional area reductions in combination with ITP allowedvisualization of lower concentrations of fluorescently labeled cTnI. ITP was performed in both “peak mode” and “plateau mode” and the final concentrations obtained were linear with initial cTnI concentration. We were able to detect and quantify cTnI at initial concentrations as low as 46 ng mL−1 in the presence of human serum proteins and obtain cTnI concentrations factors as high as ~ 9000. In addition, preliminary ITP experiments including both labeled cTnI and labeled protein kinase A (PKA) phosphorylated cTnI were performed to visualize ITP migration of different phosphorylated forms ofcTnI. The different phosphorylated states of cTnI formed distinct ITP zones between the leading and terminating electrolytes. To our knowledge, this is the first attempt at using ITP in a cascade microchip to quantify cTnI in human serum and detect different phosphorylated forms.
Introduction
Cardiovascular disease is the leading cause of death in the world today claiming 17.1 million lives per year1 resulting in estimated direct and indirect costs of over $500 billion in the United States alone.2 It is projected that by 2030, the number of deaths that will occur due to cardiovascular diseases will increase to 23.6 million lives lost per year.3 In addition, myocardial infarction is misdiagnosed or unrecognized ~ 25% of the time4 resulting in myocardial infarction being the largest source of malpractice payments issued today.5
Currently, cardiac troponin I (cTnI) is the most sensitive and preferred biomarker in determining myocardial damage,6 in part, because cTnI is specific to only the heart muscle. Current assays measure cTnI concentration in the blood using immunoassay based principles.6–21 The i-STAT by Abbott Diagnostics is the preferred point-of-care technique in detecting cTnI levels in hospital facilities18,19,21 and requires 16 µL of blood at a concentration above ~ 0.02 ng mL−1 to accurately detect cTnI.22 Typically, cTnI concentrations above 0.08 ng mL−1 are considered elevated.22 In healthy individuals, cTnI levels are usually negligible and are at concentrations lower than the limits of detection of the diagnostic test.23 In addition, these assays only measure total cTnI concentration. However, total cTnI concentration may not be the best indicator of overall heart health and functionality. For example, there is growing evidence in the literature that up or down regulation of the phosphorylation state of the cTnI molecule is a better indicator of the overall health and functionality of the heart muscle.24–29 Recently, Lin and co-workers30 developed the first immunoassay to sequentially detect phosphorylated and unphosphorylated cTnI in real time, however, these results were only qualitative. Further, a recent study by Messer et al.31 showed a 6x higher phosphorylation state of cTnI in healthy heart samples compared to failing heart samples. More importantly, Solaro and van der Velden32 indicate that protein kinase A (PKA) phosphorylation at Ser23/Ser24 residues are perhaps the only phosphorylated sites relevant to control cardiac function. If this is the case, then only two different forms of the cTnI phosphorylated state would need to be monitored. However, further research needs to be performed to validate this.
cTnI concentrations can also be elevated in the blood of elderly individuals (> 65 yrs of age),33 patients with renal failure,34 and critically ill patients35 without having any actual heart related illness. Further, diagnosing myocardial infarction in intensive care unit patients is difficult because cTnI elevation may occur for alternative reasons.35 Therefore, a better indicator for overall heart health for these individuals would be an assay that measured both phosphorylated and unphosphorylated cTnI in the blood. Thus, there is a need for an assay that can differentiate and quantify phosphorylated and unphosphorylated cTnI in blood samples to determine whether elevated troponin levels are a result of cardiac events or such factors as age, sex, race, and medical history.
Current detection strategies for measuring phosphorylation levels in cTnI include non-equilibrium isoelectric focusing,36,37 mass spectrometry,38,39 and phosphate affinity SDS-PAGE;29,31 however these techniques are generally slow, cumbersome, have low throughput, and are not compatible with point-of-care instrumentation. Thus, there are currently no point-of-care instruments available that differentiate and quantify the phosphorylation states of cTnI in symptomatic patients.
The current limitation in detecting differences in phosphorylation states using point-of-care instrumentation is that cTnI is a low abundant protein in blood with concentrations in the sub ng mL−1 range. Thus, quantifying different phosphorylation states of an already low abundant protein becomes more challenging than quantifying the total concentration of all the states. As a result, lower limits of detection are necessary to not only quantify different phosphorylation states, but, also, to quantify cTnI in individuals that have concentrations below current limits of detection. For instance, an increase in analytical and diagnostic sensitivity should allow more accurate early diagnosis.40 This could provide tending physicians with baseline concentrations and, more importantly, the ratio of phosphorylated cTnI to unphosphorylated cTnI regardless of age, sex, race, and medical history etc., to determine during routine office visits whether or not myocardial damage has occurred. Monoclonal antibodies41 have been developed that recognize unphosphorylated cTnI and PKA phosphorylated cTnI and are also commercially available through Abcam and other vendors. A preconcentration technique coupled to an immunoassay with specific antibodies can be used in combination with preconcentration methods to lower detection limits, increase sensitivity, and quantify the phosphorylation states of cTnI.
This work describes the preconcentration of clinically relevant concentrations of cTnI (ng mL−1) spiked into depleted human serum using cationic isotachophoresis (ITP) in a cascade microchip to demonstrate the potential of this approach to preconcentrate cardiac biomarkers. The high abundant proteins, albumin and IgG, were removed from the human serum prior to experimentation using a commercially available depletion column.42 The resulting serum sample will be referred to as depleted human serum in the remainder of this paper. In addition, preliminary experiments using both cTnI and PKA phosphorylated cTnI were spiked into leading electrolyte at high initial concentrations to easily visualize the ITP of both labeled proteins. The two cTnI states formed distinct and separated ITP zones between the leading and terminating electrolyte. Work in progress includes coupling this preconcentration technique with an immunoassay to increase sensitivity, and capture and differentiate phosphorylation states of cTnI at different positions in the cascade microchip.
ITP is a nonlinear, electrophoresis technique that concentrates sample components into nearly pure zones between a leading electrolyte and a terminating electrolyte in order of their electrophoretic mobilities. The leading and terminating electrolyte are chosen so that their electrophoretic mobilities are faster and slower with respect to the sample components of interest, in this case cTnI. In addition, ITP has the power to concentrate sample components by several orders of magnitude43–45 and to be coupled to other analytical techniques such as capillary zone electrophoresis,44–46 isoelectric focusing,47 gel electrophoresis,48 mass spectrometry,49 or an on-chip sandwich immunoassay.50 This work uses ITP in combination with reductions in cross-sectional area across the axial length of a microchannel to increase concentration.
Dolnik and co-workers51 first demonstrated how low abundant analytes could be concentrated by ITP in microfluidic capillaries that included reductions in cross-sectional area. Building off of this work, our group has demonstrated the concentration of proteins in buffers by over four orders of magnitude using ITP in microchips.52,53 More recently, Bahga and co-workers have used ITP in a variable cross-section geometry to increase sensitivity, detect small molecules at concentrations as low as 100 nM, and to decrease assay time compared to uniform cross-section channels.54 This work uses microchips that have a 10x depth and a 10x width reduction across the axial length of the microchannel to concentrate and quantify cTnI spiked into depleted human serum.
Fluorescently tagged cTnI at different initial concentrations ranging from 46 to 2300 ng mL−1 were detected by UV induced fluorescence in a poly(methyl methacrylate) (PMMA) microchip. The microchip includes a 10x reduction in both the width and depth so that the cross-sectional area from the inlet to outlet is decreased by a factor of 100 (Fig. 1). As long as ITP is running in “peak mode,” which indicates a Gaussian-like peak shape, and will be the case for low abundant proteins, the concentration of sample will increase in direct proportion to the decrease in cross-sectional area52,53 as indicated by eqn (1),
| (1) |
where ci is the concentration of sample component,Mi is the total mass load of species i which is controlled using a tee channel, wi is the peak width of species i, and A is the cross-sectional area of the channel. Therefore, with a 100x reduction in cross-sectional area, we should expect a 100x increase in concentration according to eqn (1). In addition to the reductions in cross-sectional area, the proteins will also be concentrated by the ITP stacking effect which, as mentioned previously, can increase concentrations by several orders of magnitude. Using this technique, we are able to detect fluorescently labeled cTnI spiked into depleted human serum at concentrations as low as 46 ng mL−1.
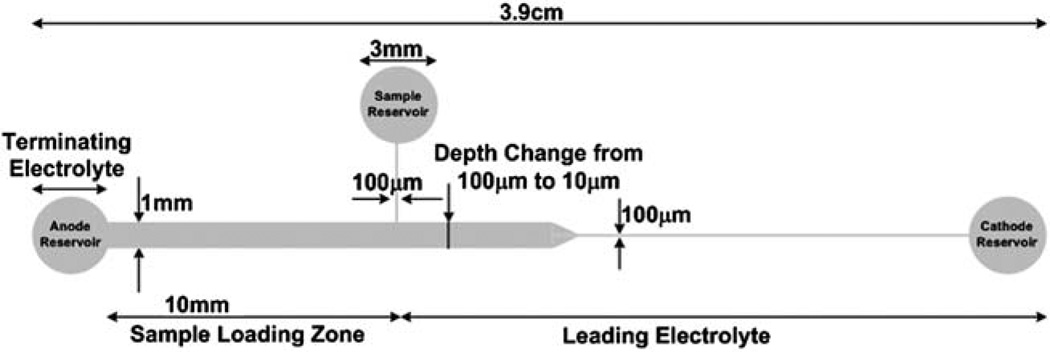
Fig. 1.
Schematic of PMMA microfluidic chip geometry that includes a 10x depth reduction occurring 1.3 cm from the anode reservoir and a 10x width reduction occurring 1.6 cm from the anode reservoir along the length of the microchannel. Both cross-sectional area changes occur over a distance of ~ 1 mm. The channel length is ~ 3.3 cm with reservoirs that are ~ 3 mm in diameter. The microchip includes a tee channel that is 100 µm wide and 100 µm deep between the sample reservoir and the anode reservoir in order to control the initial mass load of the cTnI spiked into depleted human serum. Sample is injected from the sample reservoir to the anode reservoir so that the sample only occupies the 10 mm long sample loading zone. Therefore, the sample loading volume is 1 µL (10 mm × 1 mm × 100 µm).
Experimental
Chemicals
All chemicals, unless otherwise stated, were purchased from Sigma-Aldrich (St. Louis, MO, USA). Full-Range Rainbow Molecular Weight Markers were purchased from GE Healthcare (Piscataway, NJ, USA). Pacific Blue™ C5-maleimide and Pacific Orange™ succinimidyl ester triethylammonium salt were purchased from Molecular Probes (Carlsbad, CA, USA). Gel-Code Blue Stain Reagent was purchased from Thermo Fisher Scientific (Rockford, IL, USA).
Microfluidic chip fabrication
The microfluidic chip fabrication has been reported in our previous work.52 Briefly, the microfluidic chips include a 100x reduction in cross-sectional area along the axial length of the channel. The reduction in cross-sectional area consists of a 10x depth reduction that occurs 1.3 cm from the anode reservoir and a 10x width reduction that occurs 1.6 cm from the anode reservoir as shown in Fig. 1. Each reduction occurs over a distance of ~ 1 mm. The microfluidic chip includes a cathode reservoir, an anode reservoir, and a sample reservoir. A tee channel is also included in the microfluidic chip between the sample reservoir and the anode reservoir to control the amount of sample, i.e., Mi, loaded into the channel.
Human cardiac troponin I preparation
Human cardiac troponin I (cTnI) was purified and labeled as previously described.53 The purified cTnI was labeled with Pacific Blue™ C-5 maleimide to the two intrinsic thiol groups at residues 81 and 98. The final protein concentration after labeling of cTnI, and the degree of labeling, were determined with a Beckman Coulter DU 730 UV/Vis spectrophotometer (Beckman Coulter, Inc., Brea, CA, USA) to be ~ 0.46 mg mL−1 and ~ 2, respectively. The cTnI isoelectric point was checked by isoelectric focusing PAGE (IEF-PAGE) and found to be ~ 10. The labeling did not significantly alter the isoelectric point of the labeled cTnI compared to the native cTnI (data not shown).
Protein cTnI was phosphorylated by the catalytic subunit protein kinase A (PKA), using a cardiac troponin C affinity column as previously described.55 The extent of phosphorylation was quantified by both mass spectral analysis and treatment of the sample with alkaline phosphatase, followed by determination of inorganic phosphate using the EnzChek Phosphate Assay kit (Molecular Probes).56 Phosphorylation of the two PKA sites in cTnI was > 95%. The phosphorylated cTnI was then labeled with Pacific Orange™ succinimidyl ester triethylammonium salt according to the manufacturer’s instructions. The final protein concentration after labeling of PKA phosphorylated cTnI, and the degree of labeling, were determined to be ~ 1.17 mg mL−1 and ~ 2.6, respectively.
Electrolyte solutions
The leading electrolyte (LE) solution was prepared by adjusting the pH of 40 mM potassium hydroxide, 6M urea, 0.1% Triton X-100, 2% PVP to pH 4.4 with 10% (v/v) acetic acid. The terminating electrolyte (TE) solution consisted of 20 mM acetic acid with 0.1% Triton X-100, 6 M urea, and 2% PVP. Urea and Triton X-100 were added to the electrolyte solutions to keep precipitates from forming during ITP. In addition, PVP was added to electrolyte solutions in order to suppress electroosmotic flow.57 All solutions were prepared using nanopure water from a Barnstead Thermolyne Nanopure Infinity UV/UF system (Dubuque, IA, USA) and degassed with a CPS-8B vacuum pump (US Vacuum Pumps LLC, Canton, TX, USA).
Serum pre-processing
Serum was processed to remove the high abundance proteins, albumin and IgG, using a ProteoPrep® Immunoaffinity Albumin and IgG Depletion Kit (Sigma, St. Louis, MO, USA) according to manufacturer’s instructions. Briefly, 50 µL of serum was mixed with 50 µL of the provided equilibration buffer and added to the immunoaffinity spin columns to incubate for 10 min at room temperature. The IgG and albumin interact with and bind to the packed column. The remaining serum components were spun down at 8000 × g for 60 s. using a MiniSpin Plus centrifuge (Eppendorf North America, Hauppauge, NY) to elute the other serum components while albumin and IgG remained bound to the immunoaffinity column. The eluted serum components were then reapplied to the immunoaffinity column, incubated for 10 min at room temperature and again spun down so that the serum was now twice depleted, as per manufacture’s protocol, of IgG and albumin. Next, 125 µL of equilibration buffer was added to the immunoaffinity column to wash any unbound protein and centrifuged again as previously described. The total volume of the depleted serum sample was ~ 225 µL. The twice depleted serum was brought up to a final volume of 500 µL with 40 mM potassium acetate, pH 4.5, 1% Triton X-100, and 6 M urea so that the total serum dilution after depletion was 10x. This solution was used as the sample solution for ITP. In addition, the depletion step did not remove physiological salts, but rather diluted them by a factor of 10x.
The bound albumin and IgG were then extracted from the column by centrifugation as previously described using 300 µL of the provided extraction buffer. The final volume of the extracted albumin and IgG was brought up to 500 µL with 40 mM potassium acetate, pH 4.5, 1% Triton X-100, and 6 M urea so that the albumin and IgG dilution after depletion from human serum was 10x. These samples were then analyzed by SDS-PAGE to examine how effective the immunoaffinity column was at removing albumin.
SDS-PAGE of depleted human serum samples
8% SDS-PAGE gels were hand-cast according to the Mini-PROTEAN® 3 Cell Instruction Manual58 into Invitrogen gel cassettes (Carlsbad, CA, USA). Human serum was initially diluted by a factor of 10 with 40 mM potassium acetate, pH 4.5, 1% Triton X-100, and 6 M urea. Each sample for gel analysis from above (human serum, depleted human serum, IgG/albumin extracted from human serum) was mixed 1 : 1 with sample buffer58 and 2 µL of each sample were injected into adjacent gel lanes (see Fig. 2). In addition, 10 µL of Full-Range Rainbow Molecular Weight Markers were injected into one lane. Prior to adding the samples to the gel, 1X electrode running buffer58 was added to an Invitogen gel box that held the gel in the vertical position. Electrophoresis was then performed at 200 V for 50 min. The gel was then washed 3x for 5 min each with nanopure water, stained with GelCode Blue Stain Reagent for 1 h, and destained overnight in nanopure water. In a separate experiment using a 12% SDS-PAGE gel, cTnI was spiked into the human serum prior to depletion to determine if cTnI was extracted by the depletion column. The cTnI band showed up in the depleted serum sample and not in the extracted IgG/Albumin sample (data not shown) indicating that cTnI was not lost during the human serum sample pre-processing depletion step.
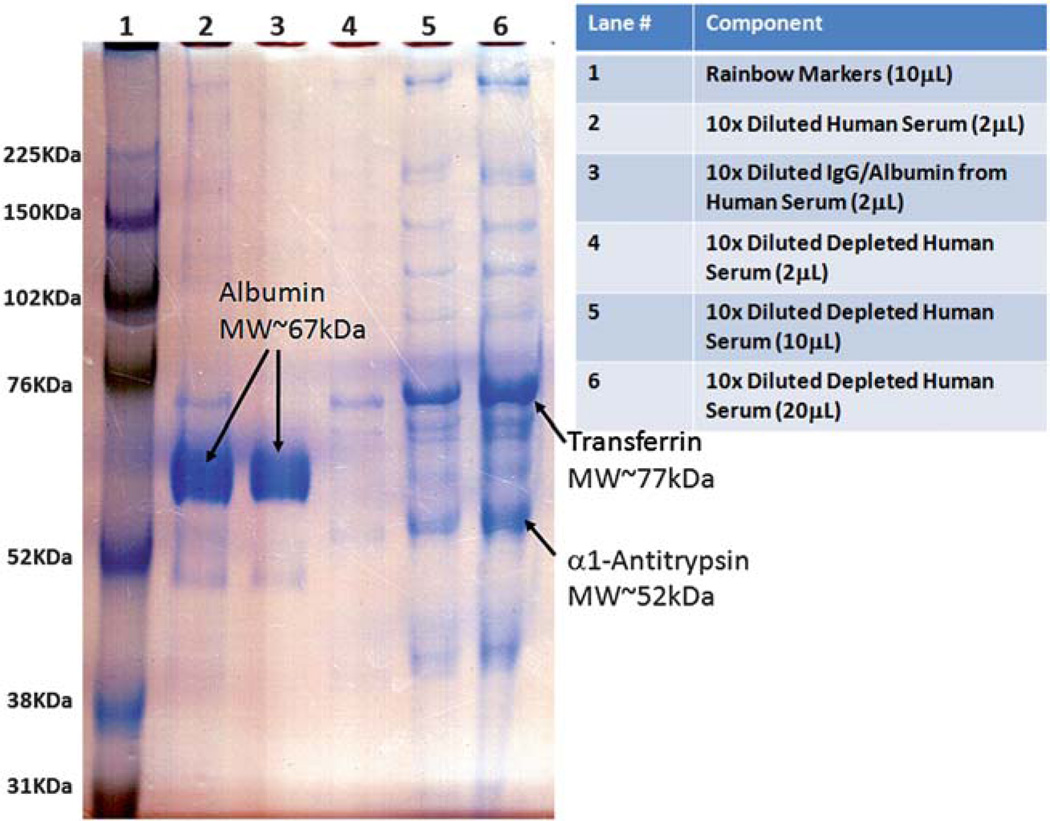
Fig. 2.
8% SDS-PAGE gel of serum components. Lane 1 shows the rainbow molecular weight markers. Lane 2 shows the original human serum sample. Lane 3 shows the albumin depleted sample from the original human serum sample extracted from the immunoaffinity column. Lanes 4–6 show increase loads of the depleted human serum sample confirming that most of the albumin from the original serum had been removed. The new major contaminants in the depleted serum are putatively transferrin (MW ~ 76 kDA) and α1-antitrypsin (MW ~ 52 kDa).
Loading the microchip
The sample solution consisted of labeled cTnI spiked into the depleted human serum sample at various concentrations ranging from 46 to 2300 ng mL−1. First, the microchip was filled from the cathode reservoir to the sample and anode reservoir with LE using a 3 mL disposable syringe. Next, the depleted serum containing cTnI was introduced into the sample reservoir. The sample was manually driven through the tee channel toward the anode reservoir using a 3 mL syringe. The hydrodynamic resistance between the sample reservoir and the cathode reservoir is 17 000 times higher than the hydrodynamic resistance between the sample reservoir and the anode reservoir as a result of the 100x difference in cross-sectional area and different lengths of the channel sections. Therefore, the authors do not expect a significant amount of sample to occupy more than the sample loading region of the microchip.
In addition, in the 1–2 min between loading the sample and applying the power to the chip, even small proteins in the sample will diffuse less than 50 µm. In either case, a higher mass load would result in higher final concentrations (see eqn (1)). Therefore, a conservative approach has been applied to the initial mass load calculations where any excess mass load as a result from the previously mentioned cases has been neglected. Furthermore, any LE solution previously in the region between the sample reservoir and the TE reservoir was flushed into the TE reservoir. At this point, only the sample solution occupied the region between the sample reservoir and the TE reservoir. Therefore, the total mass of cTnI loaded into the channel could be estimated from the concentration multiplied by the volume of the sample loading zone (Fig. 1). Next, the anode reservoir was washed several times with TE to remove sample solution and any remaining LE. Finally, the anode reservoir was filled with TE. At the end of each run, microchips were flushed twice with 1% Triton X-100 followed by nanopure water, blown out with house air, and refilled as previously described.
The loaded microchip was placed underneath the 5x objective lens of a Leica DM 2000 fluorescence microscope equipped with a DFC310 digital color camera (Leica Microsystems Inc., Bannockburn, IL, USA). The digital color camera was controlled with the provided Leica Application Suite (LAS) V3.6 software to collect images of the labeled cTnI as it migrated through the microfluidic chip by ITP. The labeled cTnI was excited with a Leica Microsystems EL 6000 light source using an A type filter cube. Platinum electrodes were then immersed in the anode reservoir and cathode reservoir.
The anode reservoir voltage was set to 300 V and the cathode reservoir was set to ground with an XHR 600-1 power supply (Xantrex technology Inc, Vancouver, Canada). Most of the proteins are positively charged, including cTnI, at the running pH and will migrate towards the cathode. Representative images of the protein migration though the microchannel were captured at different positions along the length of the channel and compiled using Windows Movie Maker 2.6. Images at the end of each experiment for each trial were also collected and imported into ImageJ (http://rsb.info.nih.gov/ij) software to produce electropherograms to be used for further analysis.
Results and discussion
Potassium ion was chosen as the leading ion because it has a higher effective electrophoretic mobility than all other positive ions including the salt ions in the serum sample, including sodium. Hydronium was chosen as the terminating ion because it migrates slower than the protein of interest, cTnI, due to its association with acetic acid. The sample was depleted human serum containing cTnI. In normal serum, albumin can make up as much as 70% of the total protein in human serum59 at an estimated concentration range between 34 and 54 mg mL−160 and was removed in order to significantly improve ITP results. When albumin was not removed from the serum, the cTnI band was very broad compared to when albumin was removed. The authors believe this could be due to 1) the albumin reaching concentrations above the solubility limit and precipitating out, or 2) the lack of separation power due to the short length of the microchannel. For instance, using information from Harrison and Ivory61 to calculate the band length of albumin in an ITP stack results in an albumin band length of ~ 3.4 cm (see Appendix 1). The total length of the separation channel described here is only 3.3 cm indicating a lack of separation power to fully resolve albumin from any of the other sample components. Therefore, the latter scenario seems more likely to explain the poor results obtained prior to albumin removal. As a result, albumin removal from the human serum was required to obtain satisfactory results. Therefore, future experiments may require an on-chip depletion step to fully automate the ITP protocol and to remove albumin from the sample prior to ITP in the microchip.
An 8% SDS-PAGE gel was prepared and used to analyze the depletion step. Fig. 2 shows the results obtained from the SDS-PAGE gel. The lane numbers and lane components are summarized in Fig. 2. It is observed that the depletion step was successful and that albumin was nearly completely removed from the human serum sample (see lanes 2–6). In addition, even at 10x mass load of the depleted serum sample (lane 6) compared to the original human serum sample (lane 1), only a trace amount of albumin could be visualized in the depleted serum sample (lane 6). From the depleted serum samples, we were also able to putatively identify the new major contaminants by comparing the molecular weight of the rainbow markers to other high abundant proteins in serum.62 These proteins are most likely transferrin (4.75 g/100 g of plasma proteins62) and α1-antitrypsin (2.95 g/100 g of plasma proteins62). The depleted human serum was spiked with different amounts of fluorescently labeled cTnI to perform ITP on the cascade microchip.
The total amount of depleted human serum sample spiked with fluorescently labeled cTnI was controlled by the addition of a tee channel between the sample reservoir and the anode reservoir. The initial mass load (Mi) of cTnI was calculated by multiplying the initial concentration (2300, 460, 115, and 46 ng mL−1) by the volume of the sample loading region (1 µL). Therefore, the total mass loads of cTnI ranged from 2.3 ng to 46 pg. These total mass loads will be used later in determining the final cTnI concentrations calculated from eqn (1) and will be discussed later. After, loading the microchip with sample and electrolyte solutions, the microchip was mounted on the x–y translation stage of the microscope platform and platinum electrodes were submerged in the anode and cathode reservoirs to produce the electric field required for analyte migration. The cathode was set to ground and the anode was set to 300 V. Each concentration was run in triplicate.
At high concentrations of cTnI (2300 and 460 ng mL−1) the protein began to collect and form a cohesive band just to the right of the anode reservoir. At a concentration of 115 ng mL−1, the cTnI band was not visible until after the tee channel where sufficient mass of cTnI had accumulated. At a concentration of 46 ng mL−1, the cTnI band was not visible until after the 1st reduction in cross-sectional area and was difficult to see until after the 2nd cross-sectional area reduction. Fig. 3 depicts the protein migration over time for the case of 460 ng mL−1 where the cTnI band could be easily visualized in all parts of the channel. In this example, the cTnI continued to accumulate mass through the sample loading zone and the fluorescence intensity increased as the protein migrated through the microchannel (Fig. 3(a) and Fig. 3(b)). At this point, the cTnI had now formed a tight cohesive band in the microchannel. The fluorescence intensity increased again as the cTnI passed the 10x depth reduction (Fig. 3(c)).
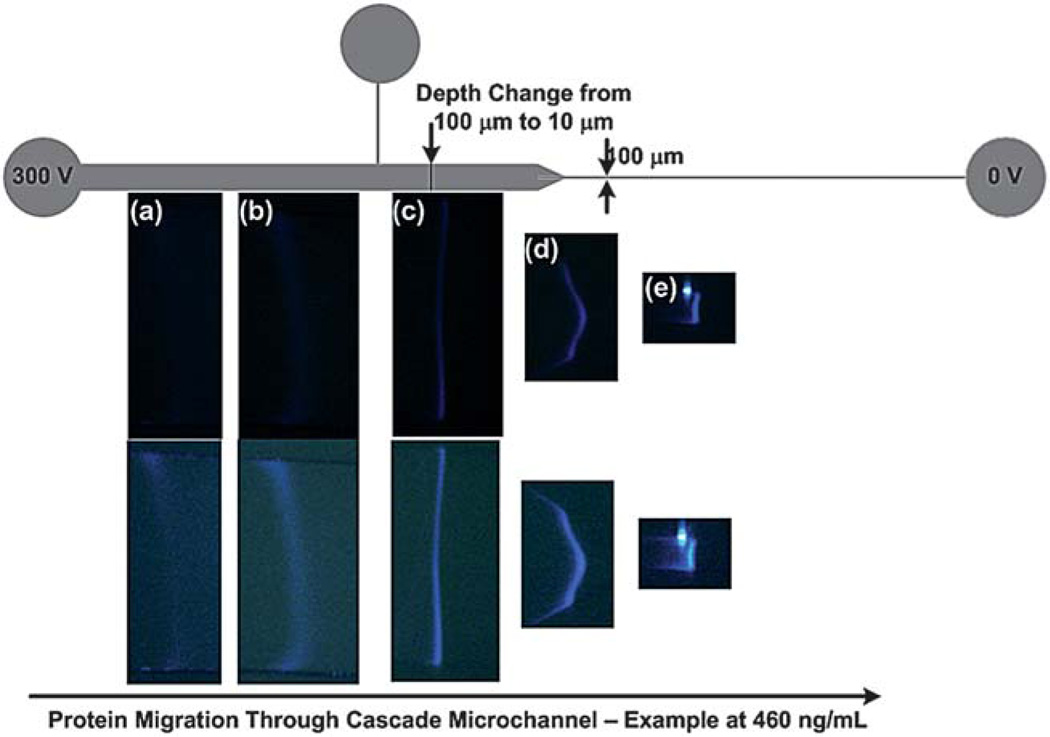
Fig. 3.
An example of protein migration of labeled cTnI spiked into depleted human serum at an initial concentration of 460 ng mL−1 through a cascade microchannel (a) cTnI begins to stack at the terminating electrolyte interface but is difficult to visualize. (b) After collecting sufficient mass the cTnI begins to become visible and continues to acquire additional mass until migrating past the tee channel. (c) The labeled cTnI has migrated past the 1st cross-sectional area reduction (10x depth change) and the signal intensity has increased to a point where the ITP band can be easily visualized. (d) The labeled cTnI begins to enter the 2nd cross-sectional area reduction (10x width change) where a slight distortion occurs; however the proteins become much brighter and easier to visualize as the protein begins to migrate into the final leg of the microfluidic channel. (e) The labeled cTnI has now migrated into the final leg of the microfluidic channel and concentrated into a relatively pure zone. The bottom row of figures are identical to figures in the middle row but have been enhanced in Adobe Photoshop using the brightness/contrast function to better visualize the bands.
Very little, if any, band distortion was observed through the depth reduction; however, as the protein began to migrate through the 10x width reduction, a slight distortion was observed (Fig. 3(d)). The authors believe that the distortion occurs in both cases, but is only visible at the width reduction because we can only see distortions that occur in the x-y plane and cannot see distortions that may occur in the z-direction. Distortion through cross-sectional area reductions are common;51,63 however these distortions are quickly eliminated by ITP’s self-sharpening effect as the band migrates into the last leg of the microchannel (Fig. 3 (e)). The cTnI band has clearly stacked into a relatively pure ITP zone. Each picture in the bottom row of Fig. 3 was modified using the brightness/contrast function in Adobe Photoshop to more easily visualize the cTnI band formation and progression through the cascade microchannel; the pictures in the middle row are untouched. Experiments could be performed in ~ 10 min. The captured images were compiled for each trial using Windows Movie Maker 2.6. An example video for each concentration can be viewed as a movie file found in the supplementary information.
Electropherograms for three trials at each concentration were obtained using ImageJ software. Each electropherogram was obtained from images of the cTnI located in the last leg of the microchannel where the fluorescence intensity was at a maximum. The resulting electropherograms yielded distance (mm) relative to the field of view of the camera versus average intensity over the entire width of the channel for the fluorescent protein, cTnI. The raw data was then transferred to Microsoft Excel and plotted for each case. Due to the dynamic nature of ITP and the capturing of images at different positions in the channel, the peak heights for each trial did not align. Therefore, the plots were translated along the x-axis (distance (mm) relative to the field of view of the camera) so that maximum peak heights occurred at the same x-position. The intensity plots were then averaged for each concentration and the average intensity as a function of position is shown in Fig. 4. The signal intensity was proportional to initial cTnI concentration.
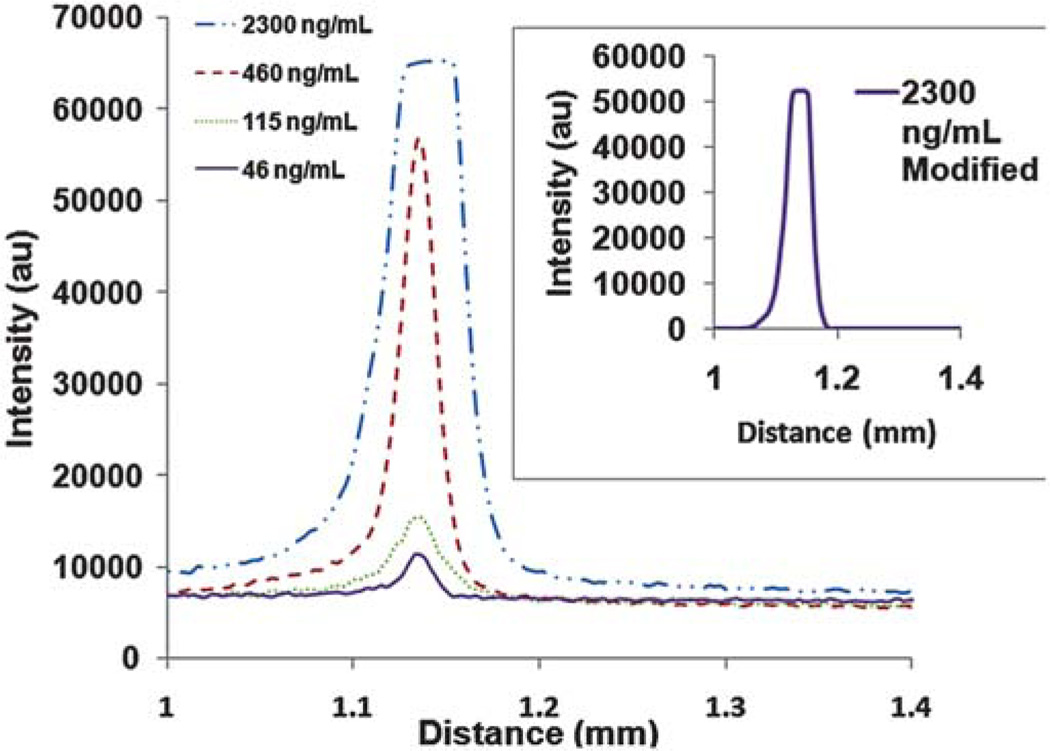
Fig. 4.
This is the average of the electropherograms extracted from ImageJ software for each of the four concentrations. The electropherograms were obtained by plotting distance (mm) relative to the field of view of the camera versus average intensity over the entire width of the channel and translating the peaks in the x-direction so that all peaks were aligned by peak height. The distance is based on the field of view of the camera and does not reflect the total migration distance through the microfluidic channel. The peaks at 460, 115, and 46 ng mL−1 are Gaussian-like indicating that ITP is being performed in “peak mode.” At 2300 ng mL−1, the intensity saturates the ImageJ software and yields a peak that looks “plateau-like.” However, the inset shows a modified average electropherogram for the case of 2300 ng mL−1 where the intensity was first decreased in Adobe Photoshop using the brightness function and then re-plotted below the software saturation threshold. The resulting peak demonstrates that the peak was “plateau shaped” and had reached its maximum allowable concentration, the plateau concentration (13.7 ± 0.24 mg mL−1).
The electropherograms for all three trials at each concentration were needed to perform a statistical analysis of band attributes such as peak width and to sequentially determine final protein concentration and concentration factors obtained by ITP in the cascade microchip. This was done by using moment analysis as previously described.53 Briefly, the nth moment for discrete position values using the trapezoidal rule64 is given by
| (2) |
where I(x) is the intensity signal, assumed to be proportional to the concentration,65 and x is the spatial position. The variance (σ2) and the resulting peak width (Wi) can then be derived using nth moments to the following equations
| (3) |
| (4) |
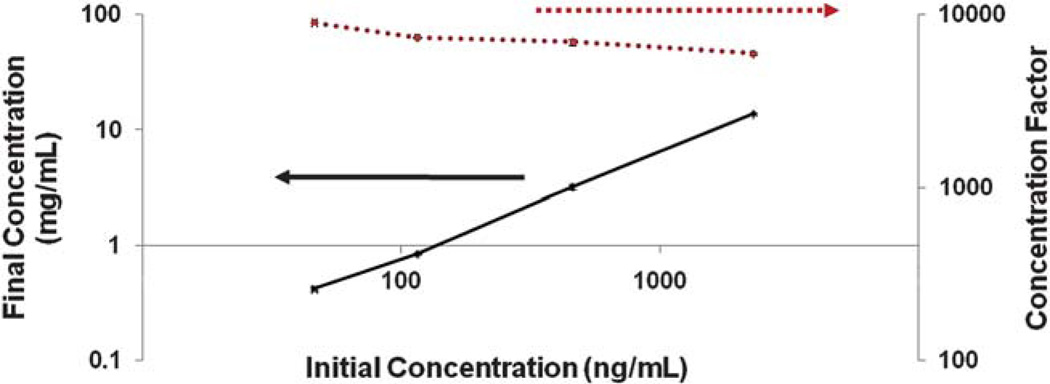
where σ is the standard deviation. The final concentrations for each trial are calculated from eqn (1) where Wi is the peak width of each protein calculated from eqn (4), A is the cross-sectional area of the last leg in the microfluidic chip (0.001 mm2), and Mi is the initial mass load of cTnI ranging from 2.3 ng to 46 pg. The final concentrations calculated from eqn (1) range from 13.7 ± 0.24 to 0.42 ± 0.02 mg mL−1 and show a linear relationship with initial concentration (Fig. 5). This is consistent with results obtained by Kaigala and co-workers65 who observed a linear response with initial analyte concentration in the nanomolar range, i.e., “peak mode” using ITP on fluorescently labeled dye molecules.
Fig. 5.
Final concentrations and concentration factors as a function of initial concentration. There is a linear relationship between final concentration and initial concentration; indicating that the ITP is running in “peak mode.” The concentration factor increased with decreasing initial concentration. Final concentrations ranged from 13.7 to 0.24 mg mL−1 and concentration factors ranged from ~ 6000 to 9000.
ITP in “peak mode” indicates a near-Gaussian peak shape which is consistent with most of the results obtained here and indicates that further preconcentration is possible. At the highest initial concentration (2300 ng mL−1), we were able to concentrate cTnI to its plateau concentration of 13.7 ± 0.24 mg mL−1 which is the maximum allowable concentration for this particular analyte. Although not apparent from the electropherogram due to intensity saturation of the ImageJ software, the resulting peak from an initial concentration of 2300 ng mL−1 was plateau shaped. This was only apparent after decreasing the signal intensity in Adobe Photoshop using the brightness function to values that did not saturate the ImageJ software. This is shown in the inset of Fig. 4 where the peaks are not Gaussian, but rather plateau shaped at this concentration. The plateau concentration is derived from the Kohlrausch regulating function,66 and converting to units of mg mL−1, is shown below as
| (5) |
where cLE and ccTnI are the concentrations of the leading electrolyte (40 mM) and the plateau concentration of cTnI (13.7 mg mL−1), μcTnI, μLE (76.2 × 10−9 m2/(V s)),67 and μC (−42.4 × 10−9 m2/(V s))67 are the electrophoretic mobilities of the cTnI, leading electrolyte, and counterion, respectively, zLE (+1) and zcTnI are the charge on the leading electrolyte and cTnI, respectively and MWcTnI is the molecular weight of cTnI (24.1kDa). The theoretical charge of cTnI was +29.4 in the leading electrolyte at pH 4.4 and was determined by inserting the amino acid sequence of cTnI68 into the Protein Calculator V3.3 (http://www.scripps.edu/~cdputnam/protcalc.html). After rearrangement of eqn (5), the electrophoretic mobility of labeled cTnI at our experimental conditions can be calculated. The electrophoretic mobility of labeled cTnI was ~ 15.6 × 10−9 m2/(V s)).
The concentration factors were determined by dividing the final concentration from the initial concentration and the results are shown in Fig. 5. At lower initial concentrations, a higher concentration factor was observed. At the low cTnI concentration we observe a concentration factor increase of ~ 9000. The average peak widths, final concentrations, and concentration factors for each initial concentration are shown in Table 1.
Table 1.
Summary of average experimental peak widths determined from moment analysis, final concentrations from eqn (1), and concentration factors for cTnI with standard deviations
| Initial concentration (ng mL−1) | Peak Width (µm) |
Final Concentration (mg mL−1) | Concentration Factor |
|---|---|---|---|
| 2300 | 167.93 ± 2.96 | 13.7 ± 0.24 | 5956 ± 104 |
| 460 | 143.92 ± 6.34 | 3.2 ± 0.14 | 6957 ± 303 |
| 115 | 135.67 ± 0.97 | 0.85 ± 0.01 | 7371 ± 53 |
| 46 | 108.78 ± 4.27 | 0.42 ± 0.02 | 8984 ± 16 |
This demonstrates the detection of cTnI in depleted human serum at clinically relevant concentrations without the use of antibodies and relying solely on ITP in a cascade microchip. The authors acknowledge that increased sensitivity is required to stand up to current point-of-care instruments for cTnI detection which have a detection limit of ~ 0.02 ng mL−1 and use antibody amplification. Antibody signal amplification can lower limits of detection by more than 1000 fold69 meaning that, by incorporating antibodies into our assay, we can reduce our limit of detection to below 0.05 ng mL−1 which puts us in a position to compete with current point-of-care instruments. Therefore, we are in the process of incorporating antibodies for immobilization of the cTnI molecule to significantly increase the sensitivity of our assay. In addition, sensitivity can be enhanced further using only ITP by using an infinite sample loading technique65 or stationary ITP with unlimited volume stacking,70 or by fabricating further cross-sectional area reductions in the microchannel, but these techniques were not analyzed here.
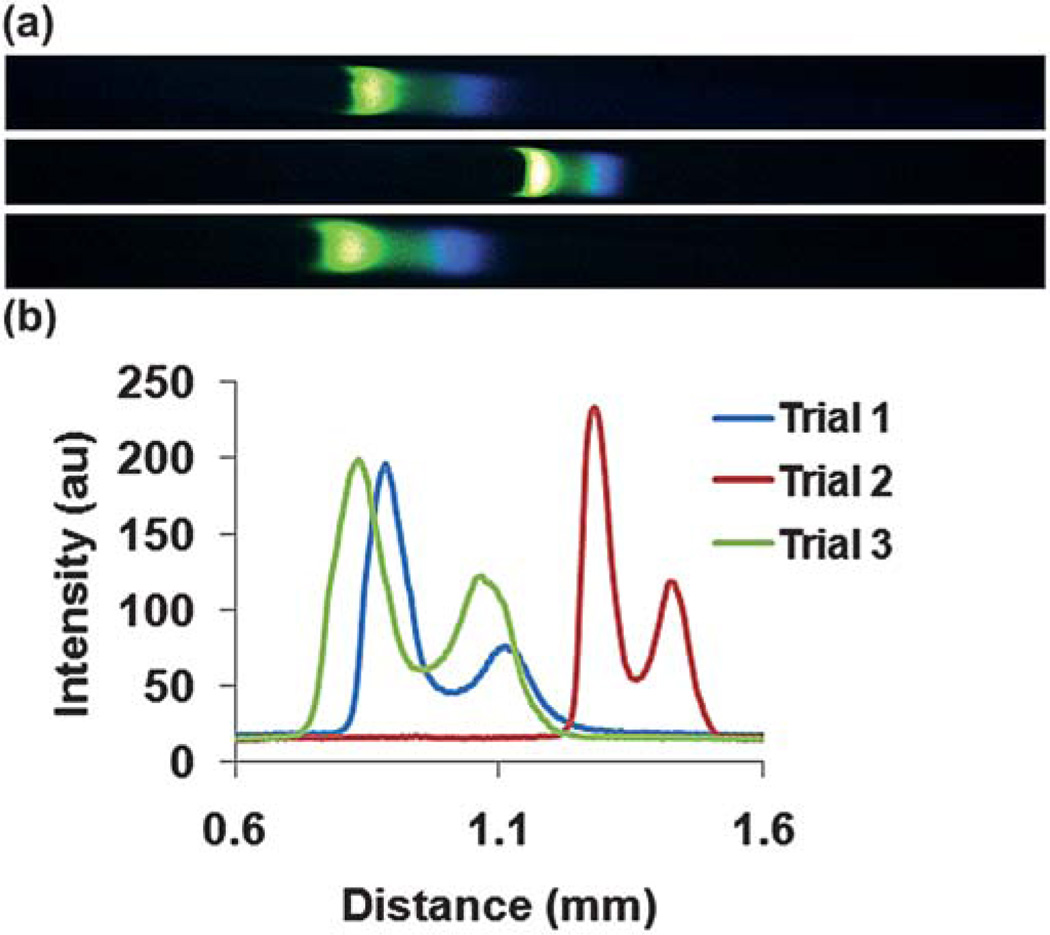
In a separate experiment, labeled PKA phosphorylated cTnI and labeled cTnI were both spiked into leading electrolyte at concentrations of 585 ng mL−1 and 920 ng mL−1, respectively. ITP was performed in the same manner on a similar, but different, microchip. All geometrical attributes of the microchip were identical except that the side channel for these experiments was 1 mm in width (Fig. 1). Experiments were performed in triplicate and representative images for each trial in the last section of the microchannel and corresponding electropherograms are shown in Fig. 6. The cTnI (blue) ran in front of the PKA phosphorylated cTnI (green) indicating that the electrophoretic mobility decreases with increasing phosphorylation and is consistent with results obtained by non-equilibrium isoelectric focusing gel electrophoresis.37 In addition, our preliminary results indicate the formation of two distinct ITP zones. An example video showing the migration of the two different phosphorylation states of cTnI in distinct zones can be viewed as a movie file found in the supplementary information.
Fig. 6.
(a) Representative images of ITP for each trial taken in the small-cross sectional area portion of the channel just before the cathode reservoir. Labeled cTnI (blue) migrates in front of the labeled PKA phosphorylated cTnI (green) indicating that the electrophoretic mobility of cTnI decreases with increasing phosphorylation. (b) The electropherograms for each trial represented in part (a) are shown.
The technique described here, ITP as a preconcentration platform, could be implemented as a 1st dimension to preconcentrate the different forms of cTnI and transfer them via ITP to a 2nd dimension that has the ability to capture, distinguish, and quantify different phosphorylation states that all current point-of-care assays lack. In addition, Kaigala and co-workers65 have recently demonstrated how ITP systems can be miniaturized to device designs as small as a cell phone for portable low-cost ITP instruments demonstrating that the technique described here is a promising approach for point-of-care applications.
The authors’ on-going research includes creating an on-chip ITP immunoassay that includes both capture antibodies and secondary detection antibodies to increase limits of detection by immobilizing the cTnI with the former and amplifying the signal with the latter. The capture antibodies will need to be specific to the phosphorylation state of cTnI allowing differentiation of phosphorylated and unphosphorylated cTnI. The detection antibodies, which only need to be cTnI specific, can then quantify the amount of each cTnI form present. We believe that by delivering ITP bands to an immunoassay we can lower the limit of detection, increase sensitivity, and differentiate and quantify different phosphorylation states of cTnI. Also, the authors are attempting to perform the removal of albumin on-chip so that this approach could potentially be used to concentrate and fractionate cTnI in clinical applications. This would be an invaluable tool for attending physicians to quickly monitor cTnI levels and correctly diagnose patients. Future experimentation will need to be carried out to test these hypotheses.
Conclusions
This report demonstrates the detection of 46 ng mL−1 of fluorescently labeled cTnI spiked into depleted human serum using cationic ITP in a PMMA microfluidic chip with two 10x reductions in cross-sectional area. In addition, concentration factors as high as ~9000 were obtained as a direct result of the ITP concentrating effect and a 100x reduction in cross-sectional area from inlet to outlet along the axial length of the microchannel.
This is the first attempt at using ITP to preconcentrate clinically relevant concentrations of a cardiac biomarker, in particular, cTnI spiked into depleted human serum, and determine sensitivity limits in a cascade microchip. Also, there is reason to believe that further reductions in cross-sectional area would improve the limit of detection. In addition, ITP could be performed on different phosphorylation states of cTnI, in this case the PKA phosphorylated cTnI and the unphosphorylated form of cTnI to produce distinct, concentrated zones. This experimental technique could be applied to a multidimensional diagnostic tool for the detection of cTnI and its phosphorylated variants in an immunoassay-based platform to improve limits of detection, increase sensitivity, and quantify different cTnI phosphorylation states. On-going research is being carried out to test these hypotheses. In addition, this technique could be applied to the detection of other relevant or low abundant biomarkers to quickly and accurately determine concentrations in complex matrices such as blood, saliva, urine, or cerebrospinal fluid.
Supplementary Material
Acknowledgements
The research was supported by Life Science Discovery Fund and the Washington State University National Institutes of Health Protein Biotechnology Training Program Award Number T32GM008336 from the NIGMS.
Appendix
-
ZLE = 1
Charge of Leading Electrolyte
-
ZAlbumin = 13
Charge of Albumin at pH 4.071
-
CLE =40mM
Concentration of Leading Electrolyte
-
Electrophoretic Mobility of Leading Electrolyte67
-
Electrophoretic Mobility of Counterion67
-
Electrophoretic Mobility of Albumin at pH 4.072
-
A = 10−9m2
Cross-Sectional Area of Small Portion of Microchannel
-
Concentration of Albumin after 10x Dilution in Leading Electrolyte
-
V = 1µL
Sample Loading Volume
-
Calculated Length of Albumin; Adapted from reference 61.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c1lc20469f
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
References
- 1.World Health Organization. Cardiovascular diseases. [Accessed 6/8/2010]; http://www.who.int/cardiovascular_diseases/en/
- 2.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, Ferguson TB, Ford E, Furie K, Gillespie C, Go A, Greenlund K, Haase N, Hailpern S, Ho M, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott MM, Meigs J, Mozaffarian D, Mussolino M, Nichol G, Roger VL, Rosamond W, Sacco R, Sorlie P, Stafford R, Thom T, Wasserthiel-Smoller S, Wong ND, Wylie-Rosett J, Stroke AHASC. Circulation. 2010;121:948–954. doi: 10.1161/CIRCULATIONAHA.109.192666. [DOI] [PubMed] [Google Scholar]
- 3.Wolrd Heart Federation. Cardiovascular disease: types and symptoms. [Accessed April 4, 2011]; http://www.world-heart-federation.org/fileadmin/user_upload/documents/Fact_sheets/2011/Cardiovascular%20disease_%20types%20and%20symptoms.pdf.
- 4.Sheifer SE, Manolio TA, Gersh BJ. Ann. Intern. Med. 2001;135:801–811. doi: 10.7326/0003-4819-135-9-200111060-00010. [DOI] [PubMed] [Google Scholar]
- 5.Storrow AB, Gibler WB. Ann. Emerg. Med. 2000;35:449–461. [PubMed] [Google Scholar]
- 6.Bock JL, Singer AJ, Thode HC. Am. J. Clin. Pathol. 2008;130:132–135. doi: 10.1309/NVXH8DL5HWFDNB74. [DOI] [PubMed] [Google Scholar]
- 7.Hamm CW, Goldmann BU, Heeschen C, Kreymann G, Berger J, Meinertz T. N. Engl. J. Med. 1997;337:1648–1653. doi: 10.1056/NEJM199712043372302. [DOI] [PubMed] [Google Scholar]
- 8.Di Serio F, Lovero R, Leone M, De Sario R, Ruggieri V, Varraso L, Pansini N. Clin. Chem. Lab. Med. 2006;44:768–773. doi: 10.1515/CCLM.2006.125. [DOI] [PubMed] [Google Scholar]
- 9.Jossi S, Gordon SL, Legge MA, Armstrong GP. Int. Med. J. 2006;36:325–327. doi: 10.1111/j.1445-5994.2006.01073.x. [DOI] [PubMed] [Google Scholar]
- 10.Lam Q, Black M, Youdell O, Spilsbury H, Schneider HG. Clin. Chem. 2006;52:298–300. doi: 10.1373/clinchem.2005.057216. [DOI] [PubMed] [Google Scholar]
- 11.Caulum MM, Murphy BM, Ramsay LM, Henry CS. Anal. Chem. 2007;79:5249–5256. doi: 10.1021/ac070452v. [DOI] [PubMed] [Google Scholar]
- 12.Cho I-H, Paek E-H, Kim Y-K, Kim J-H, Paek S-H. Anal. Chim. Acta. 2009;632:247–255. doi: 10.1016/j.aca.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Kiely J, Hawkins P, Wraith P, Luxton R. IET Sci., Meas. Technol. 2007;1:270–275. [Google Scholar]
- 14.Masson JF, Obando L, Beaudoin S, Booksh K. Talanta. 2004;62:865–870. doi: 10.1016/j.talanta.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Purvis D, Leonardova O, Farmakovsky D, Cherkasov V. Biosens. Bioelectron. 2003;18:1385–1390. doi: 10.1016/s0956-5663(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 16.Stringer RC, Hoehn D, Grant SA. IEEE Sens. J. 2008;8:295–300. [Google Scholar]
- 17.Dittmer WU, Evers TH, Hardeman WM, Huijnen W, Kamps R, de Kievit P, Neijzen JHM, Nieuwenhuis JH, Sijbers MJJ, Dekkers DWC, Hefti MH, Martens MFWC. Clin. Chim. Acta. 2010;411:868–873. doi: 10.1016/j.cca.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Apple FS, Ler R, Chung AY, Berger MJ, Murakami MM. Clin. Chem. 2006;52:322–325. doi: 10.1373/clinchem.2005.060293. [DOI] [PubMed] [Google Scholar]
- 19.Apple FS, Murakami MM, Christenson RH, Campbell JL, Miller CJ, Hock KG, Scott MG. Clin. Chim. Acta. 2004;345:123–127. doi: 10.1016/j.cccn.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 20.La’ulu SL, Roberts WL. Clin. Chim. Acta. 2010;411:1095–1101. doi: 10.1016/j.cca.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Singh J, Akbar MS, Adabag S. Clin. Chim. Acta. 2009;403:259–260. doi: 10.1016/j.cca.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Christenson RH, Azzazy HME. Clin. Biochem. 2009;42:150–157. doi: 10.1016/j.clinbiochem.2008.09.105. [DOI] [PubMed] [Google Scholar]
- 23.Babuin L, Jaffe AS. Can. Med. Assoc. J. 2005;173:1191–1202. doi: 10.1503/cmaj.050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messer AE, Jacques AM, Marston SB. J. Mol. Cell. Cardiol. 2007;42:247–259. doi: 10.1016/j.yjmcc.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 25.van der Velden J, de Jong JW, Owen VJ, Burton PBJ, Stienen GJM. Cardiovasc. Res. 2000;46:487–495. doi: 10.1016/s0008-6363(00)00050-x. [DOI] [PubMed] [Google Scholar]
- 26.Lamberts RR, Hamdani N, Soekhoe TW, Boontje NM, Zaremba R, Walker LA, de Tombe PP, van der Velden J, Stienen GJM. J. Physiol. 2007;582:695–709. doi: 10.1113/jphysiol.2007.134486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff MR, Buck SH, Stoker SW, Greaser ML, Mentzer RM. J. Clin. Invest. 1996;98:167–176. doi: 10.1172/JCI118762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zakhary DR, Moravec CS, Stewart RW, Bond M. Circulation. 1999;99:505–510. doi: 10.1161/01.cir.99.4.505. [DOI] [PubMed] [Google Scholar]
- 29.Gallon CE. J. Muscle Res. Cell Motil. 2008;29:169–172. doi: 10.1007/s10974-009-9170-4. [DOI] [PubMed] [Google Scholar]
- 30.Lin YX, Fu Q, Zhu J, Miller JM, Van Eyk JE. Clin. Chem. 2010;56:1307–1319. doi: 10.1373/clinchem.2009.135186. [DOI] [PubMed] [Google Scholar]
- 31.Messer AE, Gallon CE, McKenna WJ, Dos Remedios CG, Marston SB. Proteom. Clin. Appl. 2009;3:1371–1382. doi: 10.1002/prca.200900071. [DOI] [PubMed] [Google Scholar]
- 32.Solaro RJ, van der Velden J. J. Mol. Cell. Cardiol. 2010;48:810–816. doi: 10.1016/j.yjmcc.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myint PK, Al-Jawad M, Chacko SM, Chu GS, Vowler SL, May HM. Cardiology. 2008;110:62–67. doi: 10.1159/000109408. [DOI] [PubMed] [Google Scholar]
- 34.Apple FS, Murakami MM, Pearce LA, Herzog CA. Circulation. 2002;106:2941–2945. doi: 10.1161/01.cir.0000041254.30637.34. [DOI] [PubMed] [Google Scholar]
- 35.Lim W, Cook DJ, Griffth LE, Crowther MA, Devereaux PJ. Am. J. Crit. Care. 2006;15:280–288. [PubMed] [Google Scholar]
- 36.Scruggs SB, Walker LA, Lyu T, Geenen DL, Solaro RJ, Buttrick PM, Goldspink PH. J. Mol. Cell. Cardiol. 2006;40:465–473. doi: 10.1016/j.yjmcc.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi T, Yang XF, Walker LA, Van Breemen RB, Solaro RJ. J. Mol. Cell. Cardiol. 2005;38:213–218. doi: 10.1016/j.yjmcc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Bunk DM, Dalluge JJ, Welch MJ. Anal. Biochem. 2000;284:191–200. doi: 10.1006/abio.2000.4710. [DOI] [PubMed] [Google Scholar]
- 39.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Mol. Cell. Proteomics. 2008;7:1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Clerico A, Giannoni A. Clin. Sci. 2010;119:203–205. doi: 10.1042/CS20100234. [DOI] [PubMed] [Google Scholar]
- 41.Al-Hillawi E, Chilton D, Trayer IP, Cummins P. Eur. J. Biochem. 1998;256:535–540. doi: 10.1046/j.1432-1327.1998.2560535.x. [DOI] [PubMed] [Google Scholar]
- 42.Sigma-Aldrich Inc. ProteoPrep® Immunoaffinity Albumin and IgG Depletion Kit. [Accessed August 22, 2011]. http://www.sigmaaldrich.com/etc/medialib/docs/Sigma/Bulletin/protiabul.Par.0001.File.tmp/protiabul.pdf. [Google Scholar]
- 43.Gebauer P, Mala Z, Boček P. Electrophoresis. 2009;30:29–35. doi: 10.1002/elps.200800425. [DOI] [PubMed] [Google Scholar]
- 44.Jung B, Bharadwaj R, Santiago JG. Anal. Chem. 2006;78:2319–2327. doi: 10.1021/ac051659w. [DOI] [PubMed] [Google Scholar]
- 45.Wainright A, Williams SJ, Ciambrone G, Xue QF, Wei J, Harris D. J. Chromatogr., A. 2002;979:69–80. doi: 10.1016/s0021-9673(02)01246-3. [DOI] [PubMed] [Google Scholar]
- 46.Jung BG, Zhu YG, Santiago JG. Anal. Chem. 2007;79:345–349. doi: 10.1021/ac060949p. [DOI] [PubMed] [Google Scholar]
- 47.Mohan D, Lee CS. Electrophoresis. 2002;23:3160–3167. doi: 10.1002/1522-2683(200209)23:18<3160::AID-ELPS3160>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 48.Mohamadi MR, Kaji N, Tokeshi M, Baba Y. Anal. Chem. 2007;79:3667–3672. doi: 10.1021/ac0623890. [DOI] [PubMed] [Google Scholar]
- 49.Tomás R, Koval M, Foret F. J. Chromatogr., A. 2010;1217:4144–4149. doi: 10.1016/j.chroma.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 50.Kawabata T, Wada HG, Watanabe M, Satomura S. Electrophoresis. 2008;29:1399–1406. doi: 10.1002/elps.200700898. [DOI] [PubMed] [Google Scholar]
- 51.Dolnik V, Deml M, Boček P. J. Chromatogr., A. 1985;320:89–97. doi: 10.1016/s0021-9673(01)90483-2. [DOI] [PubMed] [Google Scholar]
- 52.Bottenus D, Jubery TZ, Dutta P, Ivory CF. Electrophoresis. 2011;32:550–562. doi: 10.1002/elps.201000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bottenus D, Jubery TZ, Ouyang YX, Dong WJ, Dutta P, Ivory CF. Lab Chip. 2011;11:890–898. doi: 10.1039/c0lc00490a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bahga SS, Kaigala GV, Bercovici M, Santiago JG. Electrophoresis. 2011;32:563–572. doi: 10.1002/elps.201000338. [DOI] [PubMed] [Google Scholar]
- 55.Finley N, Abbott MB, Abusamhadneh E, Gaponenko V, Dong WJ, Gasmi-Seabrook G, Howarth JW, Rance M, Solaro RJ, Cheung HC, Rosevear PR. FEBS Lett. 1999;453:107–112. doi: 10.1016/s0014-5793(99)00693-6. [DOI] [PubMed] [Google Scholar]
- 56.Dong WJ, Xing J, Chandra M, Solaro J, Cheung HC. Proteins: Struct., Funct., Genet. 2000;41:438–447. [PubMed] [Google Scholar]
- 57.Kaneta T, Ueda T, Hata K, Imasaka T. J. Chromatogr., A. 2006;1106:52–55. doi: 10.1016/j.chroma.2005.08.062. [DOI] [PubMed] [Google Scholar]
- 58.BioRad. Mini-PROTEAN® 3 Cell Instruction Manual. [Accessed March 1, 2011]. http://www.proteomicsnijmegen.nl/FTMS_pages/Documents/protean3.pdf. [Google Scholar]
- 59.Rengarajan K, deSmet MD, Wiggert BB. Biotechniques. 1996;20:30–32. doi: 10.2144/96201bm05. [DOI] [PubMed] [Google Scholar]
- 60.Nlm.nih.gov. Albumin - serum: MedlinePlus Medical Encyclopedia. [Accessed 08/17/2011]; http://www.nlm.nih.gov/medlineplus/ency/article/003480.htm.
- 61.Harrison SLM, Ivory CF. J. Sep. Sci. 2007;30:3255–3261. doi: 10.1002/jssc.200700243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diem K, Lentner C. Scientific Tables-Documenta Geigy. (7th Ed.) 1975:580–582. [Google Scholar]
- 63.Slais K. Electrophoresis. 1995;16:2060–2068. [PubMed] [Google Scholar]
- 64.Kreyszig E. Advanced engineering mathematics. Hoboken, NJ: John Wiley; 2006. [Google Scholar]
- 65.Kaigala GV, Bercovici M, Behnam M, Elliott D, Santiago JG, Backhouse CJ. Lab Chip. 2010;10:2242–2250. doi: 10.1039/c004120c. [DOI] [PubMed] [Google Scholar]
- 66.Everaerts FM, Mikkers FEP, Verheggen TPEM. Analytical isotachophoresis: proceedings of the 2nd International Symposium on Isotachophoresis, Eindhoven, September 9–11, 1980; New York: Elsevier Scientific Pub. Co.; distributors for the U.S. and Canada, Elsevier/North Holland, Amsterdam; 1981. [Google Scholar]
- 67.Boček P, Deml M, Gebauer P, Dolnik V. Analytical isotachophoresis, VCH, Weinheim, Federal Republic of Germany. New York, NY, USA: 1988. [Google Scholar]
- 68.Kaya Z, Goser S, Buss SJ, Leuschner F, Ottl R, Li J, Volkers M, Zittrich S, Pfitzer G, Rose NR, Katus HA. Circulation. 2008;118:2063–2072. doi: 10.1161/CIRCULATIONAHA.108.788711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wild D. The Immunoassay handbook. London: Nature Pub. Group; 2001. [Google Scholar]
- 70.Breadmore MC. Electrophoresis. 2008;29:1082–1091. doi: 10.1002/elps.200700728. [DOI] [PubMed] [Google Scholar]
- 71.Mosher RA, Saville DA, Thormann W. The dynamics of electrophoresis. New York: VCH, Weinheim; 1992. [Google Scholar]
- 72.Douglas NG, Humffray AA, Pratt HRC, Stevens GW. Chem. Eng. Sci. 1995;50:743–754. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.