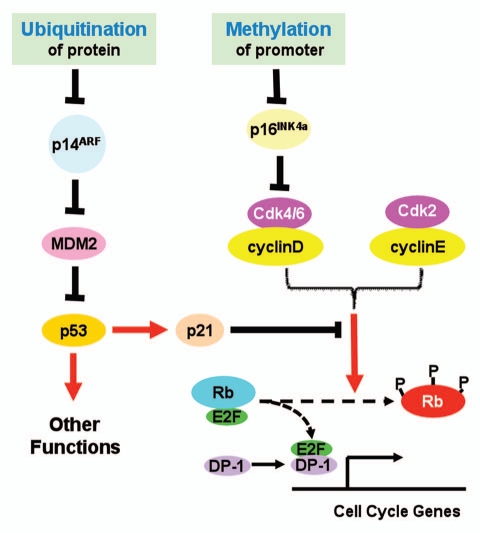
The INK4b/INK4a/ARF locus encodes three tumor suppressors: p15INK4b, p16INK4a and p14ARF.1 These three proteins play important roles in cell cycle control and tumor suppression. Both p15INK4b and p16INK4a serve as inhibitors of cyclinD-Cdk4/6 activity, and prevent the Rb family tumor suppressors from hyperphosphorylation, thus repress E2F-transactivated cell cycle genes. p14ARF antagonizes MDM-mediated ubiquitination and subsequent degradation of p53. As a transcription regulator, p53 transcriptionally promotes cell apoptosis and growth arrest, thus functions as a tumor suppressor. ARF also has been reported to suppress tumor growth by p53-independent pathway.2
Given the importance of the INK4b/INK4a/ARF locus, mechanisms underlying its regulation in normal cells, and more importantly, its inactivation in cancer cells have been intensively studied. One established mechanism that silences the whole locus involves CDC6, which may represent the coordinated control of DNA replication and transcriptional repression during cell division.3 Genetic alterations including deletion and missense mutations have been reported in a variety of tumors. Interestingly, INK4a and ARF, each has a unique promoter and exon 1, share the other two exons but using alternative reading frames. This genetic architecture increases the complexity of individualized regulation of expression. However, it has been known that some stimuli may specifically regulate either p16INK4a or p14ARF. Promoter-specific methylation has been reported to silence either INK4a or ARF.4
In a previous issue of Cell Cycle, Roberti et al. provided data to suggest another layer of regulation of INK4a/ARF locus in Burkitt's lymphoma cell lines (Fig. 1).5 They reported that in those cell lines the promoter of INK4a was heavily methylated but that of ARF was not. Accordingly, they found that the mRNA levels of INK4a were ubiquitously down-regulated whereas those of ARF, up-regulated. These up-regulated levels of ARF mRNA, however, apparently failed to result in elevated levels of p14ARF proteins. To explain this discrepancy, the authors explored the protein turnover in those cell lines. They were able to show that inhibition of proteasomal activity by incubating cells with MG132, a well known inhibitor of proteasomes, increased the protein levels of p14ARF. Furthermore, ubiquitinated forms of p14ARF were detected in protein samples from MG132-treated cells. Taken together, these data provided strong evidence to support that in Burkitt's lymphoma cell lines used in this study, INK4a was mainly repressed by promoter methylation, whereas p14ARF may be down-regulated by accelerated degradation by the ubiquitination-proteasome system.
Figure 1.
Proposed roles of protein ubiquitination and promoter methylation in control of INK4a/ARF expression. See text for detailed explanation.
As perhaps all other excellent studies, this interesting one also raises more questions than it has answered. Since p14ARF lacks lysyl residue, its ubiquitination has been reported to be mediated by the N-terminal α-amino group, instead of the more commonly reported ε-amino group of lysyl residues.6 For p14ARF is mainly localized in nucleolus and is stabilized by its interaction with NPM/B23,7 its degradation by the proteosomes is slow in most cell lines. It would be interesting to explore the molecular and biochemical mechanisms underlying this cell type-specific instability of p14ARF in those Burkitt's lymphoma cell lines. A particularly interesting question would be if this accelerated decay results from a mutation-driven p14ARF misfolding or disruption of its interaction with NPM/B23. It is also possible that a mutation of NPM/B23 may alter p14ARF function and subcellular localization. In addition, ubiquitination-independent degradation of regulatory proteins such as HIF-1α, p53 and p27 in tumor cells can be triggered by various chemotherapeutics or other stresses.8,9 While the ubiquitination of p14ARF was demonstrated, an interesting question would be whether such ubiquitination is a bona fide prerequisite for p14ARF degradation, or simply a consequence of accumulation of p14ARF when proteasomal activity was blocked. Future investigations stimulated by this report surely will significantly advance our understanding of the regulation of p14ARF and growth suppression.
In conclusion, these interesting new findings, together with published data from other researchers, depict an updated view of the regulation of tumor suppressive function of this locus. Both promoter methylation and accelerated ubiquitination may play roles in individualized control of INK4a and ARF expression, at least, in those Burkitt's lymphoma cell lines. The insight and perspectives brought by this new study may facilitate the identification of novel drug targets for the development of novel cancer therapy.
References
- 1.Dominguez-Brauer C, et al. Cell Cycle. 2010;9:86–89. doi: 10.4161/cc.9.1.10350. [DOI] [PubMed] [Google Scholar]
- 2.Kuo M-L, et al. Cancer Res. 2003;63:1046–1053. [PubMed] [Google Scholar]
- 3.Gonzalez S, et al. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M, et al. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- 5.Roberti A, et al. Cell Cycle. 2011;10:127–134. doi: 10.4161/cc.10.1.14446. [DOI] [PubMed] [Google Scholar]
- 6.Kuo M-L, et al. Genes Dev. 2004;18:1862–1874. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enomoto T, et al. J Biol Chem. 2006;281:18463–18472. doi: 10.1074/jbc.M602788200. [DOI] [PubMed] [Google Scholar]
- 8.Liang D, et al. Cell Cycle. 2006;5:2430–2435. doi: 10.4161/cc.5.21.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asher G, et al. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]