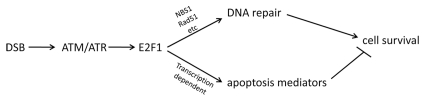
Over the past decades it has become increasingly clear that the DNA damage response (DDR) plays a crucial role in the prevention of cancer. Our DNA is extremely vulnerable to genotoxic stress. Genotoxic insults commonly arise from within as well as from without the organism. From within, genotoxic stress can be imposed by toxic byproducts of cellular metabolism, such as reactive oxygen species. From without, genotoxic stress can be imposed by numerous chemicals as well as by irradiation, such as exposure to ionizing radiation in medical practice. The estimated number of DNA lesions is as high as 105 per cell per day.1 To ensure genomic stability, it is critical that an appropriate DNA damage response (DDR) is mounted. The DDR is a complex signaling network and involves detection of the lesion, induction of transient cell cycle arrest to allow time for and activation of repair and the execution of a cell fate decision.2 Different cell fates include resumption of cell cycle progression, induction of irreversible cell cycle arrest (cellular senescence) or induction of cell death (apoptosis). Numerous factors can modulate cell fate decision, including the efficiency of DNA repair and the degree and nature of persistent lesions.
DNA double-strand breaks (DSBs) are genotoxic lesions that can be induced by exposure to ionizing radiation. Activation of the threonine kinase ataxia-telangiectasia mutated (ATM) comprises a first step in the DDR to DSBs. ATM can directly phosphorylate and stabilize the tumor suppressor p53 and indirectly regulate p53 phosphorylation by activating the cell cycle regulator CHEK2.3 Components of the DDR signaling pathway are mutated in several cancers, suggesting that the DDR must be overcome during the process of tumorigenesis. Moreover, previous work has demonstrated that germline mutations in genes encoding components of the DDR are at the basis of several human hereditary cancer-prone syndromes, including mutations in ATM [which are at the basis of ataxia telangiectasia (AT)]4 and mutations in TP53, which are at the basis of Li-Fraumeni syndrome (LFS)5. In the population at large, analysis of genetic variation in the DDR pathway in relation to cancer susceptibility has been a topic of much interest. In case of sporadic cancers, several common polymorphisms in components of the DDR have been associated with enhanced cancer susceptibility. However, cancer susceptibility will be influenced by the combined and often subtle effects of individual genetic variations in the DDR pathway, as well as in interacting pathways. Moreover, epigenetic variations and post-translational modifications will also affect the activity of the DDR pathway. In the April 1 2011 issue of Cell Cycle, Kabacik and colleagues propose to focus on an intermediate phenotype instead of genetic variations in the DDR pathway for individual cancer risk assessment. Kabacik et al. argue that, since most functions of the DDR pathway are attributable to its role as transcriptional activator in response to DNA damage, assessment of ionizing radiation (IR)-induced changes in the transcription of key p53 target genes could provide a simple readout of DDR pathway activity.
The study by Kabacik et al. convincingly shows, using mouse strains with different copy numbers of the DDR pathway components Atm, Trp53 (p53) or Check2 that the IR-induced changes in the transcription of the p53 target genes p21, Puma and Sens2 are strongly dependent on the DDR pathway component copy number. As expected, for the mouse strains with different copy numbers of the Trp53 gene, cancer incidence is also strongly dependent on the p53 copy number. Across the mouse strains with different copy numbers of the DDR pathway components, the most robust associations between gene copy number and IR-induced changes in transcription were observed for the p53 target gene Puma (p53 upregulated modulator of apoptosis), which plays a key role in execution of the cell fate apoptosis. Interestingly, the mean IR induced upregulation of Puma in fresh blood from wild-type mice was comparable to the mean IR induced upregulation of PUMA in fresh blood from healthy humans. Moreover, similarly to the DDR component copy number dependent increase in IR-induced Puma expression that was observed in mice, a linear increase in IR-induced expression of PUMA was observed across mitogen-stimulated T lymphocyte cultures from a human AT case, a group of AT heterozygotes and LFS heterozygotes and a group of healthy donors. Most importantly, in a limited sample of healthy donors, a range of IR induced PUMA upregulation was observed, suggesting that considerable variation of IR-induced upregulation of PUMA is present among healthy humans. Future studies, including large prospective studies on cancer incidence in the population at large should be directed at unraveling the potential functional significance of the observed variation in IR-induced PUMA upregulation to assess the value of the minimally invasive assay for individual assessment of the ATM/CHEK2/p53 pathway activity proposed by Kabacik for individualized prediction of human cancer susceptibility.
References
- 1.Hoeijmakers JH. N Eng J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 2.Zhou BBS, et al. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 3.Kastan MB, et al. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 4.Rotman G, et al. Hum Mol Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- 5.Hwang SJ, et al. Am J Hum Genet. 2003;72:975–983. doi: 10.1086/374567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabacik S, et al. Cell Cycle. 2011;10:1152–1161. doi: 10.4161/cc.10.7.15231. [DOI] [PMC free article] [PubMed] [Google Scholar]