Abstract
Melanoma is an aggressive cancer that is highly resistance to therapies once metastasized. We studied microRNA (miRNA) expression in clinical melanoma subtypes and evaluated different miRNA signatures in the background of gain of function somatic and inherited mutations associated with melanoma. Total RNA from 42 patient derived primary melanoma cell lines and three independent normal primary melanocyte cell cultures was evaluated by miRNA array. MiRNA expression was then analyzed comparing subtypes and additional clinicopathologic criteria including somatic mutations. The prevalence and association of an inherited variant in a miRNA binding site in the 3′UTR of the KRAS oncogene, referred to as the KRAS-variant, was also evaluated. We show that seven miRNAs, miR-142-3p, miR-486, miR-214, miR-218, miR-362, miR-650 and miR-31, were significantly correlated with acral as compared to non-acral melanomas (p < 0.04). In addition, we discovered that the KRAS-variant was enriched in non-acral melanoma (25%), and that miR-137 under expression was significantly associated with melanomas with the KRAS-variant. Our findings indicate that miRNAs are differentially expressed in melanoma subtypes and that their misregulation can be impacted by inherited gene variants, supporting the hypothesis that miRNA misregulation reflects biological differences in melanoma.
Key words: melanoma, microRNA profiling, biomarker, acral, KRAS-variant, SNP
Background
Melanoma is one of the most aggressive human cancers. It arises from uncontrolled melanocyte proliferation most commonly associated with exposure to UV radiation, which induces genetic damage resulting in tumor formation. In contrast to other skin cancers (i.e., squamous and basal cell cancers), melanomas may spread in an unpredictable manner to involve virtually any organ of the body. Additionally, melanoma exhibits considerable resistance to current therapies, particularly once it has metastasized. Identifying those at highest risk of developing melanoma, and accurate pathologic diagnosis of suspicious early lesions can be challenging, even to the most experienced pathologist, due to variability in cytomorphology and architecture and the similarity of some melanomas to benign nevomelanocytic lesions. There are four distinct histologic patterns in cutaneous melanoma: superficial spreading, nodular malignant, lentigo maligna and acral lentiginous malignant melanoma.1 The fundamental biological differences between the subtypes have not been well defined, and they are grouped together and treated the same, although it is likely that there are different mechanisms of oncogenesis for each.
Until recently, most identified tumor-suppressor genes and oncogenes were thought to exert their effects at the protein level. However, research into small, non-protein-coding RNA molecules, called microRNAs (miRNAs), has revealed potentially novel mechanisms for oncogenesis.2,3 MicroRNAs have been proven to play a key role in regulating gene expression by interacting with messenger RNA (mRNA), either by inhibiting mRNA translation or by causing mRNA degradation.4 Growing evidence is showing the capability of miRNA expression profiles to unequivocally distinguish between normal and neoplastic tissues, as well as different sub-types of the same cancer, leading to the identification of new diagnostic and/or prognostic molecular markers.5,6 In addition, miRNA disruptions through inherited variants in their binding sites or coding sequences are strong genetic markers of cancer risk.7–12 One of the first such variants, located in the 3′ untranslated region (3′UTR) of the KRAS oncogene and referred to as the KRAS-variant, has been associated with altered miRNA signatures in tumors7 as well as poor outcome and response to treatment in several cancers (Ratner et al. submitted),13,14 while a variant in the KIT 3′UTR is associated with acral melanoma.9 These findings suggest that miRNA 3′UTR-binding variants may be associated with melanoma and can also lead to miRNA expression alterations and altered biology in the tumors that they are associated with.
Our study sought to examine microRNA expression profiles of the different subtypes of melanomas to further elucidate characteristics of this cancer. Furthermore, we examined the association of miRNA signatures with acquired gene mutations, as well as inherited miRNA variants associated with cancer risk. Our findings suggest that miRNA expression patterns in melanoma can be used to help distinguish melanoma subtypes and can be influenced by the presence of inherited miRNA variants.
Results
MiRNA expression signatures in melanoma compared to normal melanocytes.
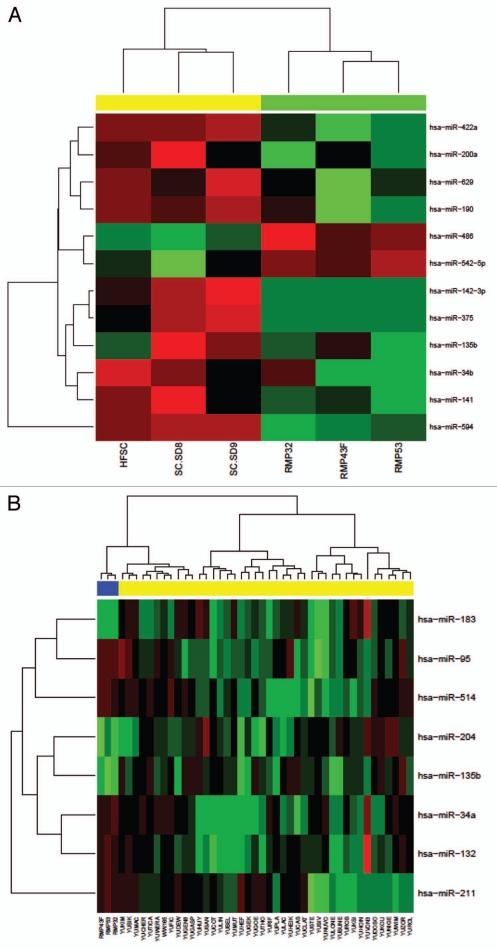
MiRNA expression profiles were compared between normal melanocytes derived from adult skins or newborn foreskins to determine variations within the control group. No statistically significant differences were found although a number of miRNAs did exhibit contrasting expression levels (Fig. 1A). The adult melanocytes displayed slightly more uniform expression between samples and were used as the control group for all subsequent analysis.
Figure 1.
Melanoma microRNA expression profiles. (A) Newborn foreskin melanocyte and normal adult melanocyte samples are arranged in columns on the horizontal axis with newborn melanocytes represented in three yellow columns on the left, and adult melanocytes in the three green columns on the right. (B) The top most differentially expressed miRNAs between normal melanocytes and melanoma cell lines. The miRNA profiles of 42 melanoma samples plus three control samples were assayed by Taqman miRNA expression array. Cell lines are arranged in columns on the horizontal axis with normal control melanocytes represented in blue.
We analyzed the expression of a panel of 384 miRNAs between 42 patient derived primary melanoma samples (including five acral melanomas) and three normal melanocyte control samples (details provided in Sup. Table 1) by microarray. We found eight miRNAs that were differentially expressed between the normal melanocytes and patient melanoma samples; miR-34a, miR-95, miR-132, miR-135b, miR-183, miR-204, miR-211 and miR-514 (Fig. 1B). None of the miRNAs found differentially expressed are clustered together and therefore not thought to be co-transcribed. After correction for multiple testing the differences in expression were not found to be significant (values provided in Table 1).
Table 1.
MicroRNAs exhibiting the greatest differential expression between normal and all melanomas
| Normal vs. melanoma samples | ||||
| miRNA ID | FC | t | p value | FDR |
| hsa-miR-135b | −6.76967 | −3.70559 | 0.000486 | 0.073173 |
| hsa-miR-204 | −6.5188 | −3.54629 | 0.0008 | 0.073173 |
| hsa-miR-95 | 4.531309 | 3.060823 | 0.003391 | 0.206851 |
| hsa-miR-514 | 7.058103 | 2.708594 | 0.008955 | 0.342949 |
| hsa-miR-211 | 8.82686 | 2.691491 | 0.00937 | 0.342949 |
| hsa-miR-132 | 6.011097 | 2.551931 | 0.013479 | 0.411118 |
| hsa-miR-183 | −4.18891 | −2.39334 | 0.020085 | 0.444838 |
Values determined after multiple test correction between normal (n = 3) and all melanoma samples (n = 42). FC, fold change; t, t-statistic; FDR, multiple test corrected p-value.
Melanoma subtypes.
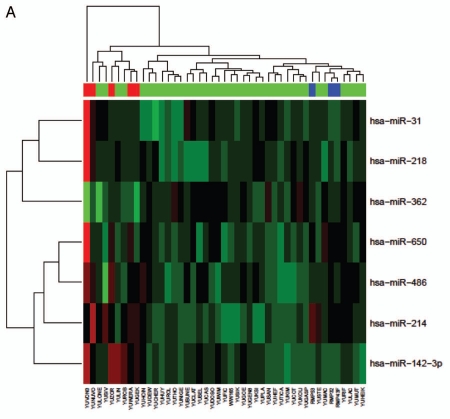
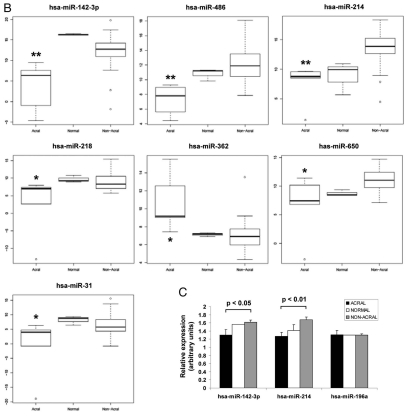
We next evaluated the differential expression of miRNAs in the melanoma subtypes, focusing on acral versus non-acral melanomas. All of our primary melanoma cell lines (acral and non-acral) were derived from patients segregated by race (Caucasian), without a family history of melanomas or significant UV-exposure. We found that the acral tumors exhibited a different miRNA signature compared to non-acral melanoma types (Fig. 2A and Table 2). Seven miRNAs had significantly different expression in acral tumors and were significant after multiple testing analyses, including; miR-142-3p, miR-486, miR-214, miR-218, miR-362, miR-650 and miR-31 (p < 0.04, Table 2). Again, none of these miRNAs are thought to be co-transcribed. All of the miRNAs were upregulated in acral versus non-acral melanomas with the exception of miR-362, which was downregulated in acral melanomas (Fig. 2B). The samples of normal melanocytes also clustered with each other for this subset of miRNAs. Secondary validation of miR-142-3p and miR-214 expression, along with miR-196 expression as a control was done by quantitative real-time PCR (qRT-PCR) for nine randomly selected normal melanocytes (n = 2), acral (n = 4) and non-acral (n = 6) melanoma samples normalized to RNU44 (as shown in Fig. 2C). When compared with the normal melanocytes, miR-214 expression was similarly upregulated as in the acral subtype. This is in contrast to all of the other miRNAs in this differentially expressed subset, where expression is generally more similar between the normal melanocytes and the non-acral samples.
Figure 2.
Acral melanomas exhibit significantly differential expression of seven miRNAs relative to other melanomas. (A) Clustered expression of miR-31, miR-218, miR-362, miR-650, miR-486, miR-214 and miR-142-3p in melanomas. Cell lines are arranged in columns on the horizontal axis with acral melanomas represented in red and normal melanocytes in blue. (B) Box-whisker plots of normalized miRNA expression in acral (n = 5), non-acral (n = 37) and normal melanocytes (n = 3). The vertical axis represents average comparative CT values, where higher values correspond to low relative expression. The top and bottom lines of the boxes represent the upper and lower quartile respectively, the center line represents the median, and the whiskers show the max/min range. Multiple testing corrected p-values for acral versus nonacral melanomas are shown, **p < 0.01, *p < 0.04. (C) Quantitative RT-PCR secondary validation of miRNA expression. The expression of miR-142-3p and miR-214, in addition to miR-196a as a control was validated in a random subset of normal (n = 2), acral (n = 4) and non-acral melanomas (n = 6). All expression values (arbitrary units) were standardised to RNU44 and the standard error is shown. The p-value between acral and non-acral melanomas was calculated by unpaired t-test.
Table 2.
MicroRNAs exhibiting the greatest differential expression between acral and non-acral melanomas
| Acral vs. non-acral melanoma samples | ||||
| miRNA ID | FC | t | p-value | FDR |
| hsa-miR-142-3p | 8.600406 | 4.741608 | 1.64E-05 | 0.002993 |
| hsa-miR-486 | 4.624591 | 4.497611 | 3.78E-05 | 0.003457 |
| hsa-miR-214 | 5.984218 | 4.286853 | 7.68E-05 | 0.004688 |
| hsa-miR-218 | 6.645029 | 3.917333 | 0.000258 | 0.011796 |
| hsa-miR-362 | −3.75799 | −3.74084 | 0.000452 | 0.016534 |
| hsa-miR-650 | 4.329809 | 3.604888 | 0.00069 | 0.021044 |
| hsa-miR-31 | 7.409108 | 3.399871 | 0.001287 | 0.033657 |
Values determined after multiple test correction between acral (n = 5) and non-acral melanoma (n = 37) samples. FC, fold change; t, t-statistic; FDR, multiple test corrected p-value.
MiRNA profiles and tumor somatic mutations in melanomas.
The miRNA expression profiles were also analyzed with respect to various known somatic mutations common in melanomas, such as BRAF and NRAS, and with survival outcomes post-tumor removal15 (as listed in Sup. Table 1). The acral melanomas were not included in this analysis because they express a unique miRNA signature and also rarely harbor BRAF or NRAS mutations.16 Of this subset, 13 samples harbored the common activating BRAF mutations at exon 15 (V600K and V600E), while eight samples harbored the NRAS Q61 mutations. We did not identify any significant differences in miRNA expression among the BRAF, NRAS or wild-type melanoma cell lines. Similarly, no significant differences in miRNA expression in relation to patient survival outcomes post-tumor removal could be found (data not shown).
The KRAS-variant, melanoma and miRNA profiles.
We next evaluated the association of the KRAS-variant with melanoma. Of the 32 non-acral melanoma samples genotyped for the KRAS-variant (as listed in Sup. Table 1), eight were positive for the KRAS-variant (25%). This prevalence is significantly higher than in any previously tested control populations,7,11,13 suggesting that the KRAS-variant may be associated with an increased risk of developing melanoma, as it does for developing non-small cell lung cancer,7 ovarian cancer11 and breast cancer.10 Interestingly, the KRAS-variant was more common in male (29%, n = 17) than in female (20%, n = 15) melanoma patients.
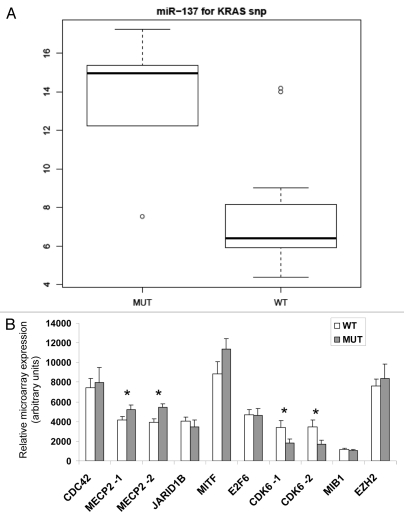
We next evaluated differences in miRNA expression between the KRAS-variant group (MUT) and the non-KRAS-variant group (WT) and identified significant downregulation of miR-137 in samples from melanoma patients with the KRAS-variant (p < 0.011 after multiple testing correction) (Fig. 3A). This miRNA was not differentially expressed between the different groups of melanomas or melanocytes in any of the previous analyses.
Figure 3.
MiR-137 differentially associates with the KRAS-variant in melanomas. (A) Box-whisker plot of miR-137 expression in melanoma samples with (MUT, n = 8) and without (WT, n = 29) the KRAS-variant, p < 0.011 after multiple testing correction. The vertical axis represents average comparative CT values, where higher values correspond to low relative expression. The top and bottom lines of the boxes represent the upper and lower quartile respectively, the center line represents the median, and the whiskers show the max/min range. (B) Microarray gene expression of miR-137 associated genes, *p < 0.05, -1 transcript variant 1, -2 transcript variant 2.
A number of genes regulate or are regulated by miR-137 in melanoma17,18 and in other cell types.19–24 We therefore evaluated the expression levels of seven targets of miR-137 (MITF, MIB1, CDK6, CDC42, JARID1B, EZH2 and E2F6) and one upstream regulator of miR-137 (MECP2) employing the NimbleGen whole-genome expression array25 in a subset of the WT and MUT samples (n = 19 and 5, respectively). Two transcript variants were spotted on the array for MECP2 (NM_004992 and NM_001110792) and CDK6 (NM_001259 and NM_001145306). Unpaired t-test analysis determined that there was differential expression for both transcripts of MECP2 and CDK6 between WT and MUT melanoma groups (Fig. 3B). MECP2 (Methyl CpG binding protein 2) the negative upstream regulator of miR-137,23 was significantly underexpressed in the WT group, where miR-137 is highly expressed. However, so far, neither MECP2 nor CDK6 and their association with miR-137 have been reported specifically in melanoma cells. One known target of miR-137 in melanomas is micropthalmia-associated transcription factor (MITF),17 a master regulator of melanocyte specific genes. Though there was a trend for lower MITF expression in our WT group where miR-137 expression was high, this was not statistically significant.
Discussion
In this study, we have identified a unique miRNA signature for acral melanomas, showing for the first time a miRNA signature that can potentially be used to differentiate between melanoma subtypes. In addition, we find enrichment of an inherited miRNA-binding site disrupting variant, the KRAS-variant, in non-acral melanomas, and a miRNA misexpression pattern that is specifically associated with the KRAS-variant. These findings support the hypothesis that miRNAs are important biomarkers of biological differences in melanoma subtypes and in tumor development in individual patients.
Acral lentiginous melanoma is the most common type of skin malignancy in darker-skinned individuals: Asians, Africans and North American Blacks. However, the lesions are found on palmar, plantar and subungual skin, where skin has no pigment. Few publications report differential expression of the seven miRNAs that we have identified in melanomas. This may largely be due to the absence or lack of segregation of acral melanomas, given its rare occurrences, within any profiled melanoma group, and may also be an indication of the unique expression patterns of these miRNAs to the acral subtype. Despite the differences in starting material, one group similarly observed an upregulation of miR-142-3p and a downregulation of miR-214 in the blood samples of melanoma patients.26 However, the differences we observed were more pronounced when we considered the acral subtype separately from the main melanoma group. In our studies the expression of miR-214 in the acral subtype closely resembled that of normal melanocytes, while miR-142-3p was highly upregulated in acral over both other types of melanomas and the normal groups. While an independent test set will be necessary to validate the potential of these seven miRNAs to act as an acral melanoma subtype classifier, given the unique miRNA expression pattern observed, it is reasonable to consider that the acral subtype likely represents a separate clinicopathological entity.
Primary cell lines derived from patient tumors are often used for miRNA profiling studies, and many of the miRNAs found to be discriminatory in melanomas match those derived directly from fresh tumor tissue indicating that many regulatory networks are conserved after culture.27 Our sample size of 42 independent patient derived primary cell lines was considerably larger than previously published studies on discerning miRNA signatures in melanoma against matched controls.27–30 There was overall good agreement between the expressions of our most discerning miRNAs with those reported to discriminate between melanocytes and melanomas,27,28 but the results did not reach statistical significance upon multiple testing in our study. This could be attributed to the differences in our sample size that alters our statistical power. Variations have also been observed between melanoma samples obtained from primary tumor derived cell lines versus metastasis.27,28 Nevertheless, the expression levels of miR-204, miR-132, miR-211 and miR-34a have generally been reported to be significantly downregulated in melanomas27,31,32 and are also downregulated in our melanoma samples. Similarly, miR-135b and miR-183 are upregulated in our melanomas and these miRNAs are also reportedly upregulated in primary melanoma cell lines compared with normal human melanocytes.28
In contrast to a previous report of miRNAs associated with common BRAF and NRAS mutations,27 we did not detect any influences of mutation status on miRNA expression in melanoma cell lines. However, we note that our cohort contained fewer BRAF mutations, 38% compared to >50% in previous studies,33,34 thus lowering our statistical power. Similarly, no miRNAs were significantly correlated with patient outcome post tumor removal. This may be due to the differences in tumor size and clinical profiles between our patient cohorts. Specifically in the analysis of post-recurrence survival, Segura et al. utilized a large cohort with a median time of 20 months and observed upregulation of miRNAs associated with the longer survival time.35 Hence, our inability to discern the same discriminating miRNAs may be due to our cohort's shorter median post-operative survival time of 13.5 months, and it is possible that a similar signature may arise after a longer follow-up time.
We evaluated for the first time the association of an inherited miRNA binding site variant, the KRAS-variant, in melanomas, as well as the association of the KRAS-variant with miRNA expression in melanomas. It is well documented that BRAF and NRAS mutations are commonly associated with melanomas, indicating that the RAS-RAF pathway plays an important role.33,36 Although coding mutations in KRAS are not commonly observed in melanomas, the implications of cancer risk from the presence of the germline KRAS-variant has been shown in other types of cancers with similar lack of associated KRAS mutations,10,11 suggesting that there may be important, as of yet unidentified, roles of KRAS overexpression in other tumor types. Our results show that 25% of the non-acral melanoma samples harbor the variant, suggesting that this inherited variant may be a risk factor for developing melanomas. However, larger studies will be needed to further evaluate the association of the KRAS-variant with melanoma risk. In addition, we show that one miRNA, miR-137, was significantly reduced in the presence of the KRAS-variant, and that a potential upstream regulator (MECP2) and a downstream target (CDK6) of miR-137 were also differentially expressed between the non-variant and KRAS-variant groups. While high expression of MECP2 in the KRAS-variant group corroborates its function as a negative regulator of miR-137,23 the unexpectedly low expression of CDK6 in this group implies that it may not be targeted by miR-137 in melanomas. The relationship of these genes with the KRAS-variant in melanoma warrants further investigation.
Our findings support the hypothesis that miRNA signatures can identify biological differences in melanoma, as well as differences in tumors that develop in association with diverse genetic backgrounds such as miRNA-disrupting variants. Based on these unique miRNA signatures, our findings may ultimately give insight into the fundamental biology of melanoma.
Methods
Samples.
Normal melanocytes from newborn and adult skins and primary melanoma cell cultures were provided by the Specimen Resource Core of the Yale SPORE in Skin Cancer. Normal human melanocytes were maintained in OptiMEM (Invitrogen, Carlsbad, CA) supplemented with penicillin-streptomycin (100 U/ml), 5% fetal calf serum FBS (all from GIBCO BRL), termed basal medium, that was further enriched with several ingredients required for optimal proliferation.25 The primary melanoma cells were grown from surgical specimens in basal medium. The specimens were collected with participants' informed consent according to Health Insurance Portability and Accountability Act (HIPAA) regulations with Human Investigative Committee protocol.
RNA extraction and RT-PCR.
Total RNA was isolated from cells (∼1 × 106/sample) using the mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's recommendations. Purity of the RNA was assessed using the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies). cDNA was synthesized from ∼800 ng of total RNA using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The conditions for running the samples were 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C.
microRNA profiling.
The expression of 384 mature miRNAs among 42 cutaneous melanoma samples, which included five acral, plus three normal adult (details provided in Sup. Table 1) and three newborn melanocytes, was analyzed using the Taqman Low-Density Array (TLDA) assay and the Applied Biosystems 7900HT Fast Real-Time PCR machine in accordance with manufacturer's instructions. Fold changes in miRNA expression between normal and malignant samples as well as between different malignant subtypes were determined by delta-delta CT values. Normalization was done to RNU44 and RNU48, two internal small nucleolar RNA used as controls.
Statistical analysis.
The SDS software associated with the Applied Biosystems 7900HT Fast Real-Time PCR System calculated relative levels of miRNA expression using the Comparative CT Method of data analysis. All sample CT values were normalized against the small nuclear RNA controls supplied with the TLDA assay. Relative intensities were scaled to have similar distributions across the entire series of samples to have the same median absolute deviation across samples. Differential expression was assessed using a moderated t-statistic; p values were adjusted for multiple testing based on all the miRNAs that were expressed in samples (excluding control and unexpressed miRNAs) according to the method of Benjamini and Hochberg37 to control the false discovery rate. Hierarchical clustering was performed with Pearson correlation and average linkage, based on miRNAs selected for differential expression between any of the groups of interest.
Acknowledgments
This work was supported by the Yale SPORE in Skin Cancer funded by the National Cancer Institute grant number 1 P50 CA121974 (R. Halaban, PI) and by a generous gift from Roslyn and Jerry Meyer. J.B.W. was supported by a K08 (CA124484) and an R01 (CA131301) from the NIH.
Supplementary Material
References
- 1.Liu V, Mihm MC. Pathology of malignant melanoma. Surg Clin North Am. 2003;83:31–60. doi: 10.1016/S0039-6109(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–298. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godshalk SE, Paranjape T, Nallur S, Speed W, Chan E, Molinaro AM, et al. A Variant in a MicroRNA complementary site in the 3′ UTR of the KIT oncogene increases risk of acral melanoma. Oncogene. 2010;30:1542–1550. doi: 10.1038/onc.2010.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paranjape T, Heneghan H, Lindner R, Keane FK, Hoffman A, Hollestelle A, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12:377–386. doi: 10.1016/S1470-2045(11)70044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelletier C, Weidhaas JB. MicroRNA binding site polymorphisms as biomarkers of cancer risk. Expert Rev Mol Diagn. 2010;10:817–829. doi: 10.1586/erm.10.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003–1007. doi: 10.1093/carcin/bgp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graziano F, Canestrari E, Loupakis F, Ruzzo A, Galluccio N, Santini D, et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacogenomics J. 2010;10:458–464. doi: 10.1038/tpj.2010.9. [DOI] [PubMed] [Google Scholar]
- 15.Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 2010;23:190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saldanha G, Potter L, Daforno P, Pringle JH. Cutaneous melanoma subtypes show different BRAF and NRAS mutation frequencies. Clin Cancer Res. 2006;12:4499–4505. doi: 10.1158/1078-0432.CCR-05-2447. [DOI] [PubMed] [Google Scholar]
- 17.Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 18.Haflidadottir BS, Bergsteinsdottir K, Praetorius C, Steingrimsson E. miR-148 regulates Mitf in melanoma cells. PLoS One. 2010;5:11574. doi: 10.1371/journal.pone.0011574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q, et al. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest, and inhibits invasion in colorectal cancer cells. Int J Cancer. 2010;128:1269–1279. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- 21.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarantino C, Paolella G, Cozzuto L, Minopoli G, Pastore L, Parisi S, et al. miRNA 34a, 100 and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 2010;24:3255–3263. doi: 10.1096/fj.09-152207. [DOI] [PubMed] [Google Scholar]
- 25.Halaban R, Krauthammer M, Pelizzola M, Cheng E, Kovacs D, Sznol M, et al. Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: Clinical implications. PLoS One. 2009;4:4563. doi: 10.1371/journal.pone.0004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leidinger P, Keller A, Borries A, Reichrath J, Rass K, Jager SU, et al. High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer. 2010;10:262. doi: 10.1186/1471-2407-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caramuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, et al. MicroRNA Expression Profiles Associated with Mutational Status and Survival in Malignant Melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 28.Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 29.Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, et al. Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS One. 2010;5:9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, et al. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 31.Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan D, Zhou X, Chen X, Hu DN, Dong XD, Wang J, et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci. 2009;50:1559–1565. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- 33.Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: A study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16:471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 34.Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- 35.Segura MF, Belitskaya-Levy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.