Abstract
Background
Iron-sulfur [Fe-S] clusters are prosthetic groups required to sustain fundamental life processes including electron transfer, metabolic reactions, sensing, signaling, gene regulation and stabilization of protein structures. In plants, the biogenesis of Fe-S protein is compartmentalized and adapted to specific needs of the cell. Many environmental factors affect plant development and limit productivity and geographical distribution. The impact of these limiting factors is particularly relevant for major crops, such as soybean, which has worldwide economic importance.
Results
Here we analyze the transcriptional profile of the soybean cysteine desulfurases NFS1, NFS2 and ISD11 genes, involved in the biogenesis of [Fe-S] clusters, by quantitative RT-PCR. NFS1, ISD11 and NFS2 encoding two mitochondrial and one plastid located proteins, respectively, are duplicated and showed distinct transcript levels considering tissue and stress response. NFS1 and ISD11 are highly expressed in roots, whereas NFS2 showed no differential expression in tissues. Cold-treated plants showed a decrease in NFS2 and ISD11 transcript levels in roots, and an increased expression of NFS1 and ISD11 genes in leaves. Plants treated with salicylic acid exhibited increased NFS1 transcript levels in roots but lower levels in leaves. In silico analysis of promoter regions indicated the presence of different cis-elements in cysteine desulfurase genes, in good agreement with differential expression of each locus. Our data also showed that increasing of transcript levels of mitochondrial genes, NFS1/ISD11, are associated with higher activities of aldehyde oxidase and xanthine dehydrogenase, two cytosolic Fe-S proteins.
Conclusions
Our results suggest a relationship between gene expression pattern, biochemical effects, and transcription factor binding sites in promoter regions of cysteine desulfurase genes. Moreover, data show proportionality between NFS1 and ISD11 genes expression.
Background
[Fe-S] clusters may be the most ancient and versatile inorganic cofactors in biological systems. They can be found in all living organisms, participating in electron transfer, catalysis and regulatory processes. Besides, [Fe-S] clusters are involved in sensing environmental stimuli and regulation of protein expression [1-3]. In plants, the biogenesis of Fe-S proteins is compartmentalized and mostly adapted to the requirements of the green tissue, which carries out both photosynthesis and respiration, processes that require significant amounts of Fe-S proteins. Mitochondria and plastid have their own machineries for [Fe-S] cluster assembly, which differ in biochemical and genetic properties. Among the Fe-S proteins known in plant mitochondria are complexes I, II and III of the respiratory chain and aconitase of the citric acid cycle, and in plastids are cytochrome b6f complex, photosystem I and ferredoxin-thioredoxin reductase [4-7].
Three different systems for [Fe-S] clusters biosynthesis have been identified in bacteria, all of them share cysteine desulfurases and [Fe-S] cluster scaffold proteins. Those systems are referred to as NIF (nitrogen fixation system), ISC (iron-sulfur cluster assembly system) and SUF (sulfur mobilization system) [8-10]. There are several mitochondrial proteins homologous to the bacterial ISC system, including a group I NifS-like proteins, supporting the evolutionary relationship between α Proteobacteria and mitochondria [5]. In yeast, it has been shown that mitochondria are the primary site of [Fe-S] cluster formation; however, these organelles not only produce their own Fe-S proteins, but are also required for the maturation of cytosolic Fe-S proteins [11]. In the chloroplast, five different [Fe-S] cluster types are found in various proteins, and this organelle possesses its own machinery for [Fe-S] biosynthesis which is most similar to those found in cyanobacteria containing the SUF system and the cysteine desulfurase which is similar to the bacterial SufS, a group II NifS-like protein [12,13].
Cysteine desulfurase is a pyridoxal 5'-phosphate (PLP)-dependent enzyme that catalyzes the conversion of L-cysteine to L-alanine and sulfane sulfur. This occurs through the formation of a protein-bound cysteine persulfide intermediate on a conserved cysteine residue [14,15]. Considering that sulfide and free iron are toxic to the cell, intracellular concentrations are thought to be extremely low. Besides being involved in sulfur mobilization, cysteine desulfurase is proposed to be involved in cellular iron homeostasis [16-18]. ISD11 is an essential mitochondria matrix protein, a component of the ISC-assembly machinery, and is conserved in eukaryotes, but not found in prokaryotes. This protein forms a stable complex with NFS1, increases NFS1 activity, and is essential for the enzymatic activity of several Fe-S proteins [19,20]. Sulfur-containing defense compounds (SDCs) are involved in stress response and their synthesis involve several genes for sulfur assimilation [21].
It is hypothesized that soybean (Glycine max) has gone through at least two polyploidy and diploidization events, being considered a paleopolyploid [22], still presenting many gene duplications [23]. Various stresses can adversely affect plant growth and crop production, such as low temperature which modifies membrane lipid composition, thus affecting mitochondria respiratory function [24] and presumably photosynthesis. Expression of various plant genes is regulated by abiotic environmental stresses such as cold. Many cis-acting elements involved in stress response and stress-inducible genes contain cis-acting elements in their promoter regions have been described.
Here, we identified the soybean cysteine desulfurase genes by sequence comparison. Furthermore, we investigate the responsiveness of these genes under biotic and abiotic stresses, as well as transcript distribution in different tissues. Association between the high transcript level of mitochondrial genes, NFS1 and ISD11, and an increased expression of two cytosolic Fe-S proteins is showing here. Our data also demonstrate the relationship between the presence of specific cis-elements and regulation of transcript levels under various conditions.
Results
Sequence analysis
Comparative protein analyses showed that there are four cysteine desulfurase genes in G. max, corresponding to loci Glyma01g40510, Glyma09g02450, Glyma11g04800 and Glyma15g13350. These proteins can be classified into two groups: the first group is composed of IscS-like proteins, mitochondrial cysteine desulfurases, which are encoded by the genes located on chromosome 01 and 11 (NFS1_Chr01 and NFS1_Chr11); the second group encompasses SufS-like proteins, plastid cysteine desulfurases, which are encoded by genes located on chromosome 9 and chromosome 15 (NFS2_Chr09 and NFS2_Chr15). Soybean NFS1 genes share 94% nucleic acid similarity and 98% protein identity, while NFS2 genes share 96% nucleic acid similarity and 97% protein identity. When compared to Arabidopsis thaliana sequences, NFS1 proteins have 76% protein identity, whereas NFS2 have 77%. Pfam analysis demonstrated that all genes encode for an aminotransferase class-V motif and alignment analysis showed the location of a cysteine in the active site and a histidine and alanine in the cofactor binding site (Additional files 1 and 2). To find ISD11 genes, we used sequences from Saccharomyces cerevisiae and A. thaliana as queries against the Glyma1 genome. We found two loci that encode ISD11 orthologs, Glyma08g26490 and Glyma18g49970, showing 87% protein identity (Additional file 3). Soybean genes that encode NFS1, NFS2 and ISD11 appear at least twice on different chromosomes due to duplication events [25].
Phylogenetic analysis
Comparative amino acid analysis of IscS-like and SufS-like proteins of different plants and bacterial species showed that conserved regions varied from 54 to 98% and from 37 to 97% identity, respectively. A phylogenetic analysis of a wide range of organisms has shown that cysteine desulfurases form three independent clusters (Figure 1). One clade was composed by all sequences from the ISC system, and divided into monocots, dicots and bacteria, forming three subclades. The second cluster contained bacterial and algae sequences of cysteine desulfurases from the NIF system. The third clade was composed of proteins from the SUF system, and subdivided into three clades, showing the same branching as the ISC system. Some bacterial and all mitochondria located proteins clustered together; some bacterial, cyanobacterial and all plastid located proteins were also found in one cluster. This is in agreement with the endosymbiotic theory, which establishes a relationship between the endosymbiotic host and the bacterial ancestors [5].
Figure 1.
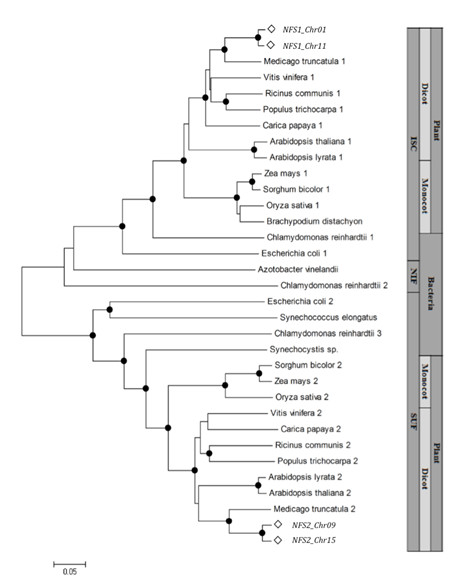
Phylogenetic analysis of cysteine desulfurase proteins. It is indicated to which [Fe-S] cluster biosynthesis systems (ISC, NIF and SUF) cysteine desulfurase belongs, and if this is a bacterial or plant (monocot or dicot) sequence. Black dots indicate bootstrap value higher than 80%.
Transcript analysis of cysteine desulfurases and ISD11 genes in soybean
Considering that Fe-S proteins are involved in environmental or cellular sensing [5], quantitative RT-PCR was performed in order to investigate transcript levels of cysteine desulfurases and ISD11 in soybean. We designed gene-specific primers for NFS1_Chr01, NFS1-_Chr11, NFS2_Chr09 and NFS2_Chr15 and analyzed the expression pattern of leaves and roots from non-treated plants and plants treated with salicylic acid (SA) and cold incubation. Further, to study whether ISD11 transcript levels are co-regulated with the NFS1 expression pattern, we performed a quantitative RT-PCR with non-treated and cold-treated plants. For all studied genes the transcript levels were normalized to the transcript levels of F-BOX and Metalloprotease [26].
In order to determine whether duplicated genes have differential expression profiles, we analyzed mRNA accumulation in control plants. These analyses showed that each cysteine desulfurase gene is individually expressed indicating a differential response to environmental stimuli. As the duplicated genes share a high degree amino acid identity, we summed expression levels from both copies to compare total NFS1 and NFS2 mRNA accumulation. While NFS1_Chr01 is predominantly expressed in roots, we found higher NFS1_Chr11 transcript levels in leaves. In sum, a higher level is found in roots (Figure 2a). NFS2_Chr15 shows a higher expression in both organs as compared to NFS2_Chr09. NFS2_Chr09 transcripts mostly accumulate in roots and those of NFS2_Chr15 in leaves (Figure 2b). In both organs, ISD11_Chr18 shows a higher expression level than ISD11_Chr08. Taken together, a higher level was found in roots (Figure 2c). As shown in Genevestigator database, NFS1 is highly expressed in roots than in leaves in Arabidopsis thaliana, while NFS2 is predominantly expressed in leaves [27].
Figure 2.
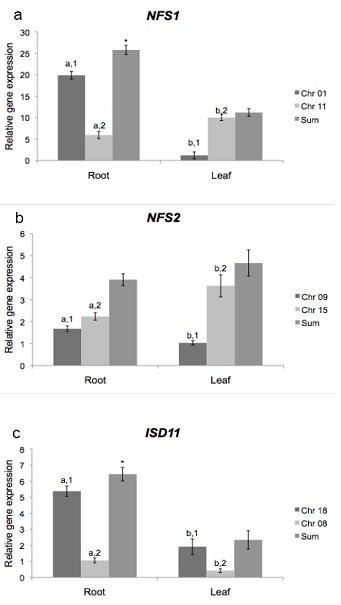
NFS1, NFS2 and ISD11 gene expression in root and leaf. Quantitative RT-PCR analysis of (a) NFS1, (b) NFS2 and (c) ISD11 gene expression in soybean tissues from total root and leaf RNA. Relative expression level was measured by performing PCR in four biological replicates and four technical replicates for each biological replicate per tissue with SE shown. Values were normalized against F-BOX and MET. a and b indicate difference between tissues for each gene. 1 and 2 indicate difference between genes in each tissue. * indicates difference in sum.
It appeared that cold-treated plants exhibited a differential response depending on the gene and tissue. In roots, NFS1_Chr01 showed a higher expression than NFS1_Chr11 during the whole treatment. While NFS1_Chr01 transcript level decreased upon cold treatment, those of NFS1_Chr11 increased (Figure 3a). Sum analysis showed that total NFS1 mRNA in roots did not respond to cold treatment (Figure 3). In leaves, NFS1_Chr11 was higher expressed than NFS1_Chr01 during the whole treatment. NFS1_Chr01 transcript level oscillated, and NFS1_Chr11 increased its expression during cold treatment (Figure 3b). Total NFS1 mRNA increased in leaves after cold incubation (Figure 3). These results corroborate with A. thaliana database, where leaves improve expression during cold treatment, while roots do not change [27]. Cold-treatment induced a decrease in both NFS2_Chr09 and NFS2_Chr15 transcript levels in roots reaching a comparable expression level at 5, 10 and 24 h (Figure 3c) (Figure 3). In leaves, NFS2_Chr15 showed a higher expression level than NFS2_Chr09 during the whole treatment. While NFS2_Chr09 transcript levels decreased, NFS2_Chr15 increased at 24 h (Figure 3d). Sum analysis showed that cold-treatment induced a decrease in NFS2 genes transcript levels (Figure 3). In roots, ISD11_Chr18 was higher expressed than ISD11_Chr08 during the whole treatment. While ISD11_Chr18 transcript levels increased were cold induced, ISD11_Chr08 did not show changes in expression (Figure 3e). Sum analysis revealed that total ISD11 mRNA initially decreased and then reached the former level in roots upon cold treatment (Figure 3). In leaves, ISD11_Chr18 showed a higher expression than ISD11_Chr08 during whole treatment, but both genes were upregulated upon cold treatment (Figure 3f).
Figure 3.
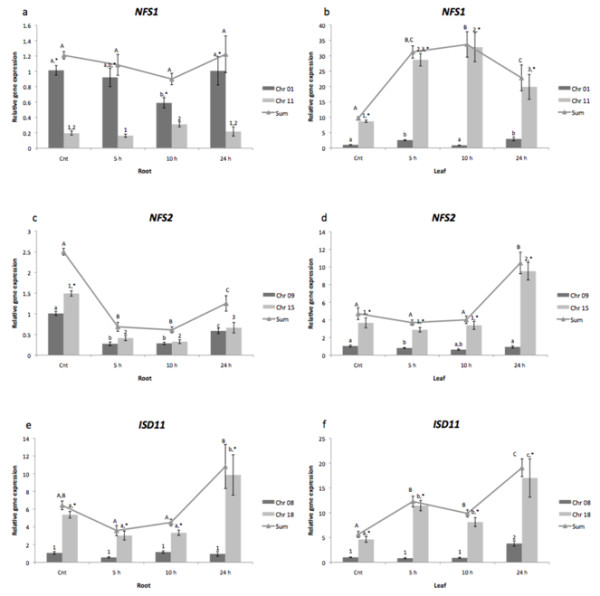
NFS1, NFS2 and ISD11 gene expression in cold-treated plants. Quantitative RT-PCR analysis of NFS1 gene expression in (a) root and (b) leaf, NFS2 gene expression in (c) root and (d) leaf and ISD11 gene expression in (e) root and (f) leaf from cold-treated plants. Relative expression level was measured by performing qPCR in four biological replicates and four technical replicates for each biological replicate per tissue with SE shown. Values were normalized against F-BOX and MET. Letters or numbers indicate difference in transcription level among time-points analyzed. * indicates difference in transcription level between duplicated genes at one point.
When treated with 2 mM SA, the response of cysteine desulfurase expression varies depending on tissue and gene. In roots, both NFS1 genes were upregulated upon SA incubation. Thus, sum analysis showed that NFS1 mRNA level after SA treatment was significantly higher than before (Figure 4a). In leaves, NFS1_Chr01 decreased expression, while NFS1_Chr11 did not show any changes. Total NFS1 mRNA level decreased due to SA treatment (Figure 4b). When A. thaliana were treated with 2 mM SA during 24 hours, NFS1 expression was induced [27]. NFS2 transcript levels did not change in roots (Figure 4c). In leaves, NFS2_Chr15 showed a significant decrease in expression, while NFS2_Chr09 did not change. Sum analysis, for leaf, did not show any changes in expression (Figure 4d).
Figure 4.
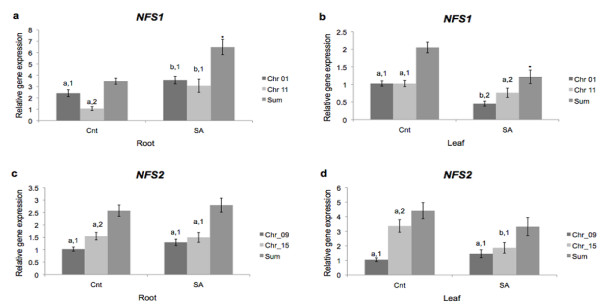
NFS1 and NFS2 gene expression in SA-treated plants. Quantitative RT-PCR analysis of NFS1 gene expression in (a) root and (b) leaf, and NFS2 gene expression in (c) root and (d) leaf from SA-treated plants. Relative expression level was measured by performing qPCR in four biological replicates and four technical replicates for each biological replicate per tissue with SE shown. Values were normalized against F-BOX and MET. a and b indicate difference between tissues for each gene. 1 and 2 indicate difference between genes in each tissue. * indicates difference in sum.
Cis-elements search in promoter regions
To identify putative cis-elements present in the NFS1 and NFS2 promoters, we inspected the sequences 1,500 bp upstream of the transcriptional start site of all genes using the Plant Cis-Acting Regulatory Elements (PlantCARE) database [28]. The analysis identified a total of 178, 168, 163 and 151 hits for potential cis-elements putative transcription factor binding sites in NFS1_Chr01, NFS1_Chr11, NFS2_Chr09 and NFS2_Chr15, respectively. While some of the predicted cis-elements were present multiple times in the promoters, others occurred only once. All putative transcription factor binding sites with known function are shown in Table 1. Comparative analysis among cysteine desulfurase promoter regions showed sequence similarity between 8 and 66%, and that the amount of shared cis-elements varies from 38.2 to 76.9% (Figure 5). Comparing duplicated genes, they have a high promoter region similarity, and NFS1 and ISD11 promoters diverged less than those of NFS2 genes (Figure 5). The relationship between some motifs and our quantitative RT-PCR results are shown in Table 2.
Table 1.
Transcription factor binding sites and motifs.
| Motifs |
NFS1 Chr01 |
NFS1 Chr11 |
NFS2 Chr09 |
NFS2 Chr15 |
Function |
|---|---|---|---|---|---|
| 3-AF1 binding site | x | light responsive element | |||
| 5UTR Py-rich stretch | x (1) | x (1) | x (2) | cis-acting element conferring high transcription level | |
| ABRE | x | cis-acting element involved in the abscisic acid responsiveness | |||
| ACE | x | x | cis-acting element involved in light responsiveness | ||
| AE-box | x | part of a module for light response | |||
| ARE | x (1) | x (1) | x (1) | cis-acting regulatory element essential for anaerobic induction | |
| as-2-box | x (1) | x (1) | involved in shoot-specific expression and light responsiveness | ||
| AT- rich element | x | binding site of AT-rich DNA binding protein (ATBP-1) | |||
| AT1-motif | x | part of a light responsive module | |||
| ATCT-motif | x | part of a conserved DNA module involved in light responsiveness | |||
| Box 4 | x | x | x | x | part of a conserved DNA module involved in light responsiveness |
| Box I | x | x | x | x | light responsive element |
| Box II | x | part of a light responsive element | |||
| Box III | x | protein binding site | |||
| Box W1 | x | x | fungal elicitor responsive element | ||
| CAAT-box | x | x | x | x | common cis-element in promoter and enhancer regions |
| CAT-box | x | x | cis-acting regulatory element related to meristem expression | ||
| CATT-motif | x | x | part of a light responsive element | ||
| CCAAT-box | x | x | MYBHV1 binding site | ||
| CGTCA-motif | x | x | cis-acting regulatory element involved in MeJA-responsiveness | ||
| chs-CMA2a | x | part of a light responsive element | |||
| circadian | x | x | x | cis-acting regulatory element involved in circadian control | |
| ERE | x | ethylene-responsive element | |||
| GAG-motif | x | x | part of a light responsive element | ||
| GA-motif | x | x | x | part of a light responsive element | |
| GARE-motif | x | x | gibberillin-responsive element | ||
| G-Box | x | x | cis-acting regulatory element involved in light responsiveness | ||
| G-box | x | x | cis-acting regulatory element involved in light responsiveness | ||
| GT1-motif | x | light responsive element | |||
| HSE | x | cis-element involved in heat stress responsiveness | |||
| LAMP-element | x | part of a light responsive element | |||
| LS7 | x | part of a light responsive element | |||
| MBS | x (2) | x (2) | x (1) | x (1) | MYB binding site involved in drought-inducibility |
| MBSI | x | MYB binding site involved in flavonoid biosynthetic genes regulation | |||
| MBSII | x | MYB binding site involved in flavonoid biosynthetic genes regulation | |||
| motif 1 | x | cis-acting regulatory element root specific | |||
| MRE | x | x | MYB binding site involved in light responsiveness | ||
| P-box | x | gibberillin-responsive element | |||
| sdOCT | x | cis-acting regulatory element related to meristem specific activation | |||
| Skn-1 motif | x | x | x | x | cis-acting regulatory element required for endosperm expression |
| Sp1 | x | x | x | light responsive element | |
| TATA-box | x | x | x | x | core promoter element around -30 of transcription start |
| TCA- element | x (1) | x (2) | x (2) | cis-acting element involved in salicylic acid responsiveness | |
| TC-rich repeats | x (2) | x (3) | x (2) | x (2) | cis-acting responsive element involved in defense and stress responsiveness |
| TCT-motif | x | x | x | part of a light responsive element | |
| TGACG-motif | x | x | cis-acting regulatory element involved in MeJA-responsiveness | ||
| TGA-element | x | x | auxin-responsive element | ||
| Total | 23 | 27 | 21 | 25 |
Transcription factor binding sites and number of motifs in each 1.500 bp upstream regions from transcription start site of soybean genes, according to PlantCARE database in default parameters.
* The motifs cited in Table 2 are marked in bold and the number inside parenthesis represent the time it appeared.
Figure 5.
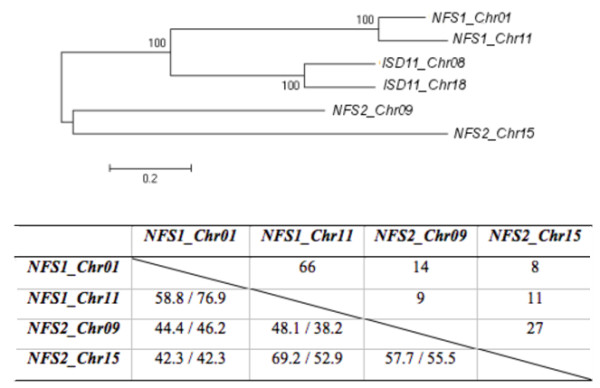
Phylogenetic analysis of promoter regions. Phylogenetic analysis of promoter regions of soybean NFS1, NFS2 and ISD11 genes. Numbers at branches indicate percentage of bootstrap values from 1,000 trials. As shown in the table below, sequence identity (%) between 1,500 bp upstream regions from transcriptional start site of soybean genes (top triangle) and percentage of common motifs between genes (bottom triangle).
Table 2.
Relationship between motifs and qPCR.
| Cis-element and organisma | Function | Gene | Correlation |
|---|---|---|---|
| 5UTR Py-rich stretch Lycopersicon esculentum | cis-acting element conferring high transcription levels | NFS1_Chr11, NFS2_Chr15 | NFS2_Chr15 is highly expressed in leaves and roots. NFS1_Chr11 is highly expressed in leaves. |
| ARE Zea mays | cis-acting regulatory element essential for the anaerobic induction | NFS1_Chr01 | It is highly expressed in roots, where the O2 availability is low. |
| MBS Arabidopsis thaliana | MYB binding site involved in drought-inducibility | All | Drought stress effects are related to cold stress effects. All genes respond to cold. |
| TC-rich repeats Nicotiana tabacum | cis-acting element involved in defense and stress responsiveness | All | All genes respond to cold. |
| TCA-element Brassica oleracea |
cis-acting element involved in salicylic acid responsiveness | NFS1 genes | Sum analysis showed that transcript level of NFS1 genes vary in SA treatment. |
| as-2-box Nicotiana tabacum | involved in shoot-specific expression and light responsiveness | NFS1_Chr11, NFS2_Chr15 | Both genes are highly expressed in leaves. |
Putative transcription factor binding sites within the NFS1 and NFS2 promoters that showed correlation to our qPCR data.
a Organism where the cis-element was described, according to PlantCARE.
Coincidence of increased NFS1/ISD11 transcript levels and activities of cytosolic Fe-S enzymes
Aldehyde oxidase (AO) catalyzes the conversion of an aldehyde to an acid and hydrogen peroxide in the presence of oxygen and water and Xanthine dehydrogenase (XDH) catalyzes the hydrogenation of xanthine to urate. Both enzymes require FAD, molybdenum and two [2Fe-2S] clusters as cofactors. Therefore, AO and XDH activities are directly dependent on the mitochondrial [Fe-S] cluster assembly machinery. Hence, we analyzed the activity of AO and XDH using an in-gel activity assay [29]. In comparison to unstressed leaves, XDH activities were clearly enhanced upon cold treatment while AO activities increased only moderately under these conditions. Crude extract were obtained from three independent treatment (Figure 6), indicating that NFS1/ISD11 are required for [Fe-S] cluster assembly on both proteins tested.
Figure 6.
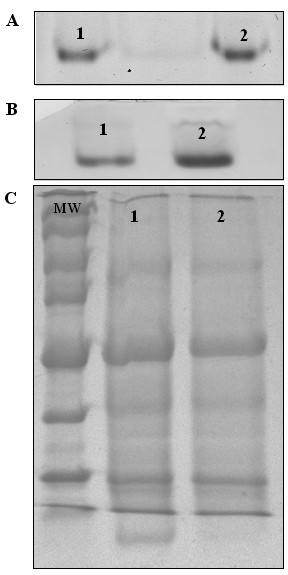
Cold stress effects on AO and XDH activity. (a) AO activity visualized by in situ staining after exposition of plants to cold stress for 18 h. Wells were loaded with 100 μg of protein of soybean wild type crude extracts of leaves from either untreated (1) or cold-stressed plants (2). Indole-3-carboxaldehyde plus 1-naphthaldehyde were used as substrate. (b) XDH activity visualized by in situ staining after exposition of plants to cold stress for 18 h. Wells were loaded with 100 μg of protein of soybean wild type crude extracts of leaves from either untreated (1) or cold-stressed plants (2). Hypoxanthine was used as substrate. (c) SDS PAGE gel 12% staining with Comassie blue. MW; molecular weight (Broad Range Protein Molecular Marked from Promega); wells were loaded with 100 μg of protein of soybean wild type crude extracts of leaves from either untreated (1) or cold-stressed plants (2).
Discussion
Soybean is a paleopolypoid plant, whose polyploidisation may have occurred in the common ancestor of the soybean and Medicago truncatula. In addition, it was suggested that a relatively recent polyploidy event occurred in the soybean lineage [22,30]. All analyzed genes are present in duplicate on different chromosomes showing a high degree of conservation and share important characteristics [25]. Due to the polyploidy events, mutations and gene rearrangements occurred, resulting in diversification of gene expression [31]. Here, we present the characterization of the promoters of soybean NFS1and NFS2 genes, and identified tissue- and stress-specific response in expression of cysteine desulfurase and ISD11 genes both involved in [Fe-S] cluster biosynthesis.
Three different systems responsible for [Fe-S] cluster biosynthesis have been described [3], and involved genes appear to be conserved in bacteria, fungi, animals and plants [6,32]. In our phylogenetic analysis (Figure 1), it was possible to identify three distinct groups, composed of proteins from ISC, NIF and SUF systems. G. max protein sequences were located in plant clades near to M. truncatula. Comparing our phylogenetic approach and the described polyploidy events [30], it was possible to hypothesize that analyzed cysteine desulfurase genes were duplicated after the divergence of soybean and M. truncatula. Thus, soybean has two copies of each cysteine desulfurase gene, while M. truncatula has only one copy (Figure 1). Both species contain duplicated ISD11 genes (data not shown). Therefore, this polyploidy event may have occurred prior to divergence of both lines.
The present results suggest that NFS1 and NFS2 soybean genes, which encode proteins involved in sulfur assimilation and [Fe-S] cluster biosynthesis [16], are involved in response to cold stress and SA. Sulfur is an essential macronutrient which is assimilated to cysteine [33,34], which will take part in the assembly of SDCs. When exposed to biotic and/or abiotic stress, synthesis of SDCs is induced via different signals, demonstrating their potential involvement in stress defense. There is an increased demand for cysteine as a precursor due to SDCs synthesis; therefore, the expression of genes for sulfur assimilation is induced [21]. Analyzing cold-treated plants, it is possible to observe that, in leaves, NFS1 and NFS2 genes increased transcript levels (Figure 3), perhaps due to SDCs stress response or due to its possible role in SDCs synthesis. When treated with SA, a simulator of biotic stress, NFS1 genes changed their expression pattern (Figure 4). In both experiments we observed a particular expression pattern, i.e. organs with primary contact to the stressor showed an increase in cysteine desulfurase transcript levels, while those less exposed showed a lower expression (Figures 3 and 4). This opposite profile may be due to a compensatory mechanism present in early stress response, and it may change if plants are exposed to longer stress periods.
When the plant cell are exposed to biotic or abiotic stress factors, modifications of the lipid composition of its membranes occur [35]. Soybean mitochondria show modifications in lipid content in response to low temperature [36], and this can alter respiratory properties and gene expression [24,37]. As many proteins involved in respiration, such as complexes I, II and III, are Fe-S proteins [4], a modification in the respiratory profile may change the requirement for proteins of the [Fe-S] cluster biosynthesis pathway, i.e. altering the expression of cysteine desulfurase genes. Stress dependent changes in gene expression occur in the cytoplasm as well as in chloroplasts. Whereas mitochondria developed an export system for [Fe-S] clusters that is essential for maturation of many nuclear and cytosolic proteins, [Fe-S] cluster biosynthesis in mitochondria has a direct impact on protein activity, such as for aldehyde oxidase and xanthine dehydrogenase [16,38,39] as shown in Figure 6. The chloroplast is extremely sensitive to abiotic stress factors, such as elevated temperature and light, both increasing reactive oxygen species. Glutathione is involved in protection against oxidative damage triggered by biotic and abiotic stress in the cytosol and other cellular compartments. Synthesis of this peptide depends on sulfur assimilation and cysteine synthesis [21,33,34], as this amino acid is the substrate of cysteine desulfurase [14,40], a change in cysteine content may lead to a modification in its catalytic properties.
SA and its methylated form are involved in development, and are also fundamental for hypersensitive response and for systemic acquired resistance under biotic stress [41,42]. SA can induce the formation of reactive oxygen species, and these can react with various molecules in the cell, including lipids. As the organelle is often exposed to strong oxidative stress, some antioxidant enzymes should be simultaneously upregulated. An alternative oxidase has been proposed to represent a functional marker for mitochondrial dysfunction during biotic stress, and its content is increased in SA-treated soybean [35,43]. The treatment with SA causes mitochondrial dysfunction via oxidative stress causing changes in the cysteine desulfurase expression. This enzyme transfers electrons from reduced ubiquinone to molecular oxygen, bypassing complexes III and IV [24], and complex III contains [Fe-S] cluster [4]. In addition, SA-treated soybean altered the fatty acid composition of its mitochondria. As these organelles modified their membranes upon SA treatment, and cellular respiration involves Fe-S proteins, the expression of cysteine desulfurase may be altered under biotic stress.
The present quantitative RT-PCR results revealed a relationship between NFS1 and ISD11 transcript contents in roots and leaves as both genes showed a similar expression pattern (Figure 2). Moreover, to analyze whether an increase in NFS1 expression triggers an increase in ISD11 transcript levels, we studied ISD11 expression levels under cold stress. In roots, total ISD11 mRNA decreased during the treatment and recovered to the initial level after 24 h, while NFS1 transcript levels did not change. In leaves, both genes were upregulated (Figure 3). The similarity in expression pattern between these genes may be explained by their function. NFS1 is a cysteine desulfurase involved in [Fe-S] biosynthesis in mitochondria [6], whereas Isd11 was recently identified in yeast as a protein responsible for forming a stable complex with Nfs1 [19,20]. Besides interacting with the cysteine desulfurase, ISD11 showed in humans an important role in mitochondrial and cytosolic iron homeostasis [44] mediated by NFS1 [18]. Here, we demonstrated that interaction between mitochondrial genes NFS1/ISD11 increased expression and maturation of cytosolic enzymes XDH and AO. These results corroborate with data described for yeast, which associates the mitochondrial machinery for [Fe-S] cluster biosynthesis as being responsible for maturation of cytosolic Fe-S proteins. Also, results in Figure 6, show, for the first time, a direct relation among increase expression of both cytosolic Fe-S protein (XDH and AO) and mitochondrial cysteine desulfurase as a result of cold stress conditions. Moreover, our results are in agreement with the experiments involved co-expression of NFS1 and ISD11 of A. thaliana, which show a higher stability of NFS1 when co-expressed with ISD11 may suggesting that the interaction of NFS1/ISD11 promote the correct conformational structure of NFS1 (de Oliveira, LA and Frazzon, APG personal communication).
The soybean genome contains highly similar genes integrated in wider regulatory networks involved in differential regulation, including the presence of cis-acting regulatory elements in promoter regions [31]. Therefore, we analyzed DNA sequences to predict putative transcription factor binding sites located in the -1500 bp promoter regions. Duplicated genes have highly homologous promoter regions (Figure 5). When cis-elements were compared, all genes share high degree of common binding sites (Figure 5), suggesting that cysteine desulfurase genes share regulatory networks. In spite of this similarity, it has been shown that different environmental factors may trigger gene expression (Figures 2, 3 and 4). Since a complex molecular network is involved in regulation of gene expression and transcription factors are important components that lead to activation or repression of transcription [45], the differences observed may be due to the requirements of the corresponding factors in a particular tissue or organelle. The analysis of transcription factor binding sites provided an insight into transcript level data. Cis-elements related to quantitative RT-PCR experiments are shown in Table 2. A Py-rich element was found in NFS1_Chr11 and NFS2_Chr15 genes that showed high transcription levels in leaves and in both leaves and roots, respectively. An ARE element was found in NFS1_Chr01, which displayed higher expression in roots than in leaves, whereas promoters with an as-2-box element showed higher expression in leaves. The TCA-element, related to SA response, was found in NFS1 genes that changed transcription pattern under this stress. Besides, all genes had cis-elements related to defense and stress (TC-rich) and to drought response (MBS), and several genes are induced by both drought and cold stress, indicating a crosstalk between signaling pathways [46].
Conclusions
In this study, we carried out an analysis of cysteine desulfurase genes from soybean, which are involved in [Fe-S] cluster biosynthesis. This study suggests that NFS1 and NFS2 genes are involved in stress response, and that their differential expression may be due to the presence of different cis-elements (Figure 7). Furthermore, ISD11 displayed an expression pattern similar to NFS1 genes, supporting a positive correlation in their activity. Our results provide the first insight into differential expression of duplicated genes involved in [Fe-S] cluster pathway, but further research is needed to determine whether other genes involved in [Fe-S] cluster biogenesis follow this pattern.
Figure 7.
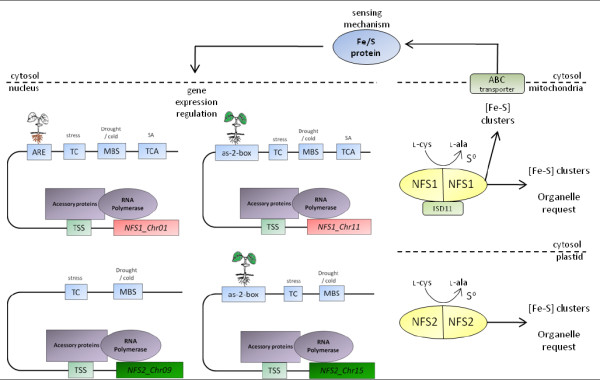
Overview of expression control of NFS1 and NFS2 genes. This model shows some cis-elements found within the promoter regions of cysteine desulfurases genes (green and red boxes). According to our qPCR data and in silico analysis those genes have particular expression triggers, indicated above the cis-elements (blue boxes). NFS1_Chr01 is modulated by stresses, such as cold and salicylic acid (SA), and induced in roots. NFS1_Chr11 is modulated by the same stimuli but induced in leaves. NFS2_Chr09 is modulated by cold, whereas NFS2_Chr15 is modulated by the same stress and induced in leaves. In plastid (bottom), cysteine desulfurase catalyses the release of sulfur from cysteine to [Fe-S] cluster biogenesis in order to organelle request. In mitochondria (top), the same process occurs to provide clusters to the organelle and cytosol/nucleus, being stimulated by a small protein called ISD11 (light green box). An export system, including an ABC transporter, is involved in maturation of Fe-S proteins outside mitochondria. Some Fe-S proteins in cytosol and nucleus are involved in cellular sensing, controlling gene expression.
Methods
Identification of [Fe-S] cluster genes in soybean
To identify cysteine desulfurase-encoding genes from the annotations of Glyma1 in the soybean genome, a similarity search method was performed. We used a protein sequence data set of known cysteine desulfurases from A. thaliana, Synechocystis sp. and Escherichia coli, and the modeled proteome data of annotated genes downloaded from Phytozome [47]. To confirm the protein identity, sequences were subjected to a profile search using Pfam [48]. Besides, the search results for each cysteine desulfurase were then applied to retrieve discovered regions as conserved active sites and cofactor binding amino acids. Other genes that encode proteins involved in [Fe-S] cluster biosynthesis, such as ISD11, were found using the strategy described above.
Phylogenetic analysis
Sequence alignments of cysteine desulfurase proteins were performed using ClustalX2 [49] with default parameters and then visualized with GeneDoc Program [50]. Phylogenetic analysis was made with the Molecular Evolutionary Genetic Analysis (MEGA) Package Version 4.0 [51] with Neighbor-Joining method. Pairwise deletion was used to analyze gap and missing data in the alignments. The bootstrap test was performed with 1000 replications, and only the bootstraps values higher than 80% are displayed as black dots on the nodes. The species and their respective access numbers in Phytozome or NCBI database are as follows: G. max (Glyma01g40510/Glyma11g04800/Glyma09g02450/Glyma15g13350), Ricinus communis (XP_002531989/XP_002523229), A. thaliana (At5g65720/At1g08490), Zea mays (ACF83040/NP_001130656), Oryza sativa (NP_001062914/EEC69110), Chlamydomonas reinhardtii (XP_001695008/XP_001701051/Au9.Cre12.g525650), Brachypodium distachyon (Bradi4g28300), Arabidopsis lyrata (XP_002864976/XP_002892460), Sorghum bicolor (Sb02g022360/Sb05g001270), Carica papaya (supercontig_129.49/supercontig_34.204), Vitis vinifera (XP_002274517/XP_002267920), M. truncatula (Medtr5g014960/Medtr3g105250), Populus trichocarpa (XP_002310363/XP_002314018), E. coli (YP_001458463/NP_289087), Azotobacter vinelandii (YP_002797401), Synechococcus elongates (NP_665776) and Synechocystis sp. (NP_442475).
DNA sequence analysis
To predict the transcription factor binding sites located in the -1500 bp promoter regions of each soybean analyzed gene, we retrieved the -1500 bp upstream sequence from the putative transcription start site for each gene from the Glyma1 annotation. After that, the analysis of each promoter region was performed using the Plant Cis-Acting Regulatory Elements (PlantCARE) database [28]. Phylogenetic analysis of promoter regions was performed as described above, and percentage of bootstrap values are shown at branches.
Plant material and treatments
To study the responsiveness of cysteine desulfurase genes under stress, two different experiments were performed. For cold and SA treatments, we used the soybean (G. max [L.] Merr.) cv. IAS-5 and cv. Conquista seeds, respectively. Plants were grown in plastic pots (3 seeds per pot), filled with vermiculite and solution of half-strength MS medium [52], in a controlled environment growth chamber at 16/8 h of day/night (22.5 μmol m-2 s-1) photoperiod at 28 ± 1°C. For cold treatment, after 11 days, seedlings were transferred to a plastic pot, under the same conditions, containing one seedling. At the age of 27 days, plants were submitted to cold stress at 4°C for 0, 5, 10 and 24 hours. For SA treatment, plants were kept for 14 days in vermiculite, and then transferred to a hydroponic solution (half-strength MS medium) with additional 2 mM SA for 48 h. Control plants were submitted to the same conditions without SA addition. After both treatments, root and leaf tissues were collected, frozen in liquid nitrogen and stored at -80°C till further use. All the analyses were performed in biological quadruplicates.
RNA isolation and quantitative RT-PCR
To study the expression of cysteine desulfurase genes, samples from control and treated plants were collected (n = 4 per each group). Total RNA was isolated from frozen tissues by extraction with Trizol (Invitrogen) according to the manufacturer's instructions and quality was evaluated by electrophoresis on a 1% agarose gel. Prior to quantitative RT-PCR, RNA samples were treated with DNase I (Promega) at 37°C for 30 min. Reverse transcription reactions were performed using the M-MLV reverse transcriptase (Invitrogen) following manufacturer's instructions.
Quantitative RT-PCR was conducted in an ABI 7500 Real-Time PCR System (Applied Biosystem) using SYBR Green I (Invitrogen) to detect double-strand cDNA synthesis. Soybean F-BOX (F-Box protein family) and MET (insulin-degrading enzyme, metalloprotease) genes were used as reference genes for data normalization and to calculate the relative mRNA levels. Reactions were done in a volume of 20 μL containing 10 μL of cDNA, 0.1 × SYBR Green I (Invitrogen), 0.025 mM dNTP, 1 × PCR Buffer, 3 mM MgCl2, 0.25 U Platinum Taq DNA Polimerase (Invitrogen), and 200 nM of each reverse and forward primers. A negative control without cDNA template was included for each primer combination. Primer sequences for quantitative RT-PCR were as follows: for NFS1_Chr01, NFS1_01R 5'-CCTCCCAATTCTCTCCATCGGT-3', and for NFS1_Chr11, NFS1_11R 5'-CCTCCCAATTTCCTCCATGGGC-3' with the same forward primer NFS1F 5'-CGGAGCACAAGTGCGTCC-3'; for NFS2_Chr09, NFS2_09R 5'-CCCGTGCACTTGAGCTGACA-3', and for NFS2_Chr15, NFS2_15R 5'-CACGTGCACTTGAGCTGACG-3' with the same forward primer NFS2F 5'-GTCGAACGAGCTGCCCTTTG-3'; for ISD11_Chr08, ISD11_08R 5'- CGCTGCGGAGCGGAGAAT-3', and for ISD11_Chr18, ISD11_18R 5'- TTGCGGAGCGGAGGGG -3' with the same forward primer ISD11F 5'-TCCACCGCCTTCGCCC-3'. These primers set generated amplicon sizes of 200, 157 and 91 and 89 bp for NFS1, NFS2 and ISD11, respectively (Additional file 4). The conditions were set as follows: an initial step for polymerase activation for 5 min at 94°C, 40 cycles of 15 s at 94°C for denaturation, 10 s at 60°C for annealing, and 15 s at 72°C for elongation, followed by a melting-curve analysis and fluorescence measured from 60 to 95°C. Four biological replicates and four technical replicates for each biological replicate were used.
Activity assays
Total proteins extract from untreated and cold stress treatment tissue were obtained from plants grown in the same conditions as described for quantitative RT-PCR. Protein quantification was performed by Bradford assay (BioRad) and equal amount (100 μg) of protein was applied in a 7.5% native PAGE gel, AO and XDH were detected by activity staining previously described [29,53]
Data analysis
Threshold and baselines were manually determined using the ABI 7500 Real-Time PCR SDS Software v2.0. To analyze the relative cysteine desulfurases mRNA expression relative to the constitutive genes, we used the 2-ΔΔCt method [54]. Student's t test was performed to compare pairwise differences in gene expression, following the two samples assuming unequal variances and two-tailed distribution parameters. For time-course treatment, 1-way ANOVA and Duncan post hoc analysis were performed using SPSS17. The means were considered significantly different when P < 0.05.
Authors' contributions
MDH carried out the quantitative RT-PCR analysis, sequence alignment, phylogenetic studies, cis-elements search, and performed the statistical analysis and drafted the manuscript. EMD performed the AO and XDS assays. LAO grow the plants and performed the RNA extraction and preparation of cDNA. APGF participated in the design of the study and coordination. RM performed the statistical analysis, participated in the design of the study and coordination. JF participated in the design and coordination of the study and gave the final approved. All authors read and approved the final manuscript.
Supplementary Material
Alignment of IscS-like. Alignment of soybean cysteine desulfurase homologue to IscS from Escherichia coli. * indicates residues from active and from cofactor binding sites. # indicates amino acids residues that differ between soybean duplicated genes.
Alignment of SufS-like. Alignment of soybean cysteine desulfurase homologue to SufS from Escherichia coli. * indicates residues from active and from cofactor binding sites. # indicates amino acids residues that differ between soybean duplicated genes.
Alignment of ISD11 proteins. Alignment of soybean, Arabidopsis thaliana and Saccharomyces cerevisiae ISD11 proteins.
Primer sequences. Primer sequences and amplicon characteristics for each gene.
Contributor Information
Marta D Heis, Email: martaheis@gmail.com.
Elisabeth M Ditmer, Email: elleditmer@gmail.com.
Luisa A de Oliveira, Email: lu_abruzzi@yahoo.com.br.
Ana Paula G Frazzon, Email: ana.frazzon@ufrgs.br.
Rogério Margis, Email: rogerio.margis@ufrgs.br.
Jeverson Frazzon, Email: jeverson.frazzon@ufrgs.br.
Acknowledgements
We are very grateful to Prof. Giancarlo Pasquali and Prof. Márcia P. Margis from UFRGS for laboratorial and technical support. We also thank Prof. Jörg Meurer (Ludwig-Maximilians-University, Munich, Germany) for his editing of the manuscript. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq- #302471/2009-0, #470388-2009-9 and #473769/2007-7) of Brazil, and the Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (PDEE/CAPES) of Brazil.
References
- Ayala-Castro C, Saini A, Outten FW. Fe-s cluster assembly pathways in bacteria. Microbiology and Molecular Biology Reviews. 2008;72(1):110. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Lill R. Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochimica Et Biophysica Acta-Bioenergetics. 2000;1459(2-3):370–382. doi: 10.1016/S0005-2728(00)00174-2. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annual Review of Biochemistry. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annual Review of Biochemistry. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460(7257):831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Balk J, Lobreaux S. Biogenesis of iron-sulfur proteins in plants. Trends in Plant Science. 2005;10(7):324–331. doi: 10.1016/j.tplants.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Schwenkert S, Netz DJA, Frazzon J, Pierik AJ, Bill E, Gross J, Lill R, Meurer J. Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochemical Journal. 2010;425:207–214. doi: 10.1042/BJ20091290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. Journal of Biological Chemistry. 2002;277(32):28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Biochemical and Genetic-Analysis of the Nifusvwzm Cluster from Azotobacter-Vinelandii. Molecular & General Genetics. 1989;219(1-2):49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- Zheng LM, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters - Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. Journal of Biological Chemistry. 1998;273(21):13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- Kushnir S, Babiychuk E, Storozhenko S, Davey MW, Papenbrock J, De Rycke R, Engler G, Stephan UW, Lange H, Kispal G. et al. A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell. 2001;13(1):89–100. doi: 10.1105/tpc.13.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Abdel-Ghany SE, Cohu CM, Herbert SK, Kugrens P, Pilon M, Pilon-Smits EAH. Chloroplast iron-sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(13):5686–5691. doi: 10.1073/pnas.0700774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Pilon M, Pilon-Smits EAH. CpNifS-dependent iron-sulfur cluster biogenesis in chloroplasts. New Phytologist. 2006;171(2):285–292. doi: 10.1111/j.1469-8137.2006.01751.x. [DOI] [PubMed] [Google Scholar]
- Zheng LM, White RH, Cash VL, Dean DR. Mechanism for the Desulfurization of L-Cysteine Catalyzed by the Nifs Gene-Product. Biochemistry. 1994;33(15):4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- Zheng LM, White RH, Cash VL, Jack RF, Dean DR. Cysteine Desulfurase Activity Indicates a Role for Nifs in Metallocluster Biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(7):2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazzon APG, Ramirez MV, Warek U, Balk J, Frazzon J, Dean DR, Winkel BSJ. Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Molecular Biology. 2007;64(3):225–240. doi: 10.1007/s11103-007-9147-x. [DOI] [PubMed] [Google Scholar]
- Godman J, Balk J. Genome analysis of Chlamydomonas reinhardtii reveals the existence of multiple, compartmentalized iron-sulfur protein assembly machineries of different evolutionary origins. Genetics. 2008;179(1):59–68. doi: 10.1534/genetics.107.086033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Tong WH. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nature Reviews Molecular Cell Biology. 2005;6(4):345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Urzica E, Guiard B, Muller H, Lohaus C, Meyer HE, Ryan MT, Meisinger C, Muhlenhoff U, Lill R. et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. Embo Journal. 2006;25(1):184–195. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam AC, Bornhovd C, Prokisch H, Neupert W, Hell K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. Embo Journal. 2006;25(1):174–183. doi: 10.1038/sj.emboj.7600905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Wachter A. Sulfur metabolism: a versatile platform for launching defence operations. Trends in Plant Science. 2005;10(10):503–509. doi: 10.1016/j.tplants.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Kim KD, Shin JH, Van K, Kim DH, Lee SH. Dynamic Rearrangements Determine Genome Organization and Useful Traits in Soybean. Plant Physiology. 2009;151(3):1066–1076. doi: 10.1104/pp.109.141739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter J, Doyle JJ. Paleopolyploidy and gene duplication in soybean and other legumes. Current Opinion in Plant Biology. 2006;9(2):104–109. doi: 10.1016/j.pbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Matos AR, Hourton-Cabassa C, Cicek D, Reze N, Arrabaca JD, Zachowski A, Moreau F. Alternative oxidase involvement in cold stress response of Arabidopsis thaliana fad2 and FAD3+ cell suspensions altered in membrane lipid composition. Plant and Cell Physiology. 2007;48(6):856–865. doi: 10.1093/pcp/pcm061. [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. In silico Analysis of Transcription Factor Repertoire and Prediction of Stress Responsive Transcription Factors in Soybean. DNA Research. 2009;16(6):353–369. doi: 10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Thibivilliers S, Bilgin D, Radwan O, Benitez M, Clough S, Stacey G. Identification of four soybean reference genes for gene expression normalization. Plant Genome. 2008;1:44–54. doi: 10.3835/plantgenome2008.02.0091. [DOI] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner F, Oreb M, Mendel RR. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. Journal of Biological Chemistry. 2001;276(44):40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ. Placing paleopolyploidy in relation to taxon divergence: A phylogenetic analysis in legumes using 39 gene families. Systematic Biology. 2005;54(3):441–454. doi: 10.1080/10635150590945359. [DOI] [PubMed] [Google Scholar]
- Yi JX, Derynck MR, Chen L, Dhaubhadel S. Differential expression of CHS7 and CHS8 genes in soybean. Planta. 2010;231(3):741–753. doi: 10.1007/s00425-009-1079-z. [DOI] [PubMed] [Google Scholar]
- Frazzon J, Dean DR. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Current Opinion in Chemical Biology. 2003;7(2):166–173. doi: 10.1016/S1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
- Saito K. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiology. 2004;136(1):2443–2450. doi: 10.1104/pp.104.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Raval M, Biswal U, Joshi P. Sulfur Assimilation and Abiotic Stress in Plants. Berlin/Heidelberg: Springer; 2008. Response of Photosynthetic Organelles to Abiotic Stress: Modulation by Sulfur Metabolism; pp. 167–191. [Google Scholar]
- Matos AR, Mendes AT, Scotti-Campos P, Arrabaca JD. Study of the effects of salicylic acid on soybean mitochondrial lipids and respiratory properties using the alternative oxidase as a stress-reporter protein. Physiologia Plantarum. 2009;137(4):485–497. doi: 10.1111/j.1399-3054.2009.01250.x. [DOI] [PubMed] [Google Scholar]
- de Virville JD, Cantrel C, Bousquet AL, Hoffelt M, Tenreiro AM, Pinto VV, Arrabaca JD, Caiveau O, Moreau F, Zachowski A. Homeoviscous and functional adaptations of mitochondrial membranes to growth temperature in soybean seedlings. Plant Cell and Environment. 2002;25(10):1289–1297. doi: 10.1046/j.1365-3040.2002.00901.x. [DOI] [Google Scholar]
- Caiveau O, Fortune D, Cantrel C, Zachowski A, Moreau F. Consequences of omega-6-oleate desaturase deficiency on lipid dynamics and functional properties of mitochondrial membranes of Arabidopsis thaliana. Journal of Biological Chemistry. 2001;276(8):5788–5794. doi: 10.1074/jbc.M006231200. [DOI] [PubMed] [Google Scholar]
- Mendel RR, Bittner F. Cell biology of molybdenum. Biochimica et Biophysica Acta. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Teschner J, Lachmann N, Schulze J, Geisler M, Selbach K, Santamaria-Araujo J, Balk J, Mendel RR, Bittner F. A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. The Plant Cell. 2010;22:468–480. doi: 10.1105/tpc.109.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Applied Microbiology and Biotechnology. 2002;60(1-2):12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annual Review of Phytopathology. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Dong XN. NPR1, all things considered. Current Opinion in Plant Biology. 2004;7(5):547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Amirsadeghi S, Robson CA, Vanlerberghe GC. The role of the mitochondrion in plant responses to biotic stress. Physiologia Plantarum. 2007;129(1):253–266. doi: 10.1111/j.1399-3054.2006.00775.x. [DOI] [Google Scholar]
- Shi Y, Ghosh MC, Tong WH, Rouault TA. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Human Molecular Genetics. 2009;18:3014–3025. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P. Recent advances in computational promoter analysis in understanding the transcriptional regulatory network. Biochemical and Biophysical Research Communications. 2003;309(3):495–501. doi: 10.1016/j.bbrc.2003.08.052. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6(5):410–417. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- Phytozome. http://www.phytozome.net
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K. et al. The Pfam protein families database. Nucleic Acids Research. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- The GeneDoc Program. http://www.nrbsc.org/gfx/genedoc/
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiologia Plantarum. 1962;15(3):473–&. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Koshiba T, Saito E, Ono N, Yamamoto N, Sato M. Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from coleoptiles of maize. Plant Physiology. 1996;110(3):781–789. doi: 10.1104/pp.110.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of IscS-like. Alignment of soybean cysteine desulfurase homologue to IscS from Escherichia coli. * indicates residues from active and from cofactor binding sites. # indicates amino acids residues that differ between soybean duplicated genes.
Alignment of SufS-like. Alignment of soybean cysteine desulfurase homologue to SufS from Escherichia coli. * indicates residues from active and from cofactor binding sites. # indicates amino acids residues that differ between soybean duplicated genes.
Alignment of ISD11 proteins. Alignment of soybean, Arabidopsis thaliana and Saccharomyces cerevisiae ISD11 proteins.
Primer sequences. Primer sequences and amplicon characteristics for each gene.


