Abstract
The evolution of colorectal cancer suggests the involvement of many genes. We performed insertional mutagenesis with the Sleeping Beauty (SB) transposon system in mice carrying germline or somatic Apc mutation. Analysis of common insertion sites (CISs) isolated from 446 tumors revealed many hundreds of candidate cancer drivers. Comparison to human datasets suggested that 234 CIS genes are also deregulated in human colorectal cancers. 183 CIS genes are candidate Wnt targets, and 20 are shown to be novel modifiers of canonical Wnt signaling. We also identified gene mutations associated with a subset of tumors containing an expanded number of Paneth cells, a hallmark of deregulated Wnt signaling, and genes associated with more severe dysplasia included members of the FGF signaling cascade. Some 70 genes showed pairwise co-occurrence clustering into 38 sub-networks that may regulate tumor development.
It is estimated that on average 7-15 somatic mutations are required for a normal human cell to undergo malignant transformation1-3. Colorectal cancer progression appears to have an extended clinical course, with mutation of genes including APC, K-RAS, TP53 and SMAD4, being frequent events, whilst genomic analysis has implicated many more genes in the pathogenesis of this disease4-6. The diversity of the mutagenic processes that create tumors in humans results in many different types of lesions in the cancer genome such that discriminating cancer driver genes from passengers is often not possible using genome analysis alone7. Recently the Sleeping Beauty (SB) DNA transposon system has been used to identify genes involved in the development of intestinal tumorigenesis8. Insertional mutagenesis provides the advantage of using engineered elements to initiate mutagenesis largely through the loss or gain of gene function, while tagging potential cancer genes9. In the intestine, loss of APC function is widely believed to be an initiating event in the formation of colorectal cancer, with 80% of sporadic tumors having an APC mutation, while germline APC mutations are associated with a predisposition to familial polyposis 4,6,10. APC normally functions to sequester and target β-catenin for degradation through phosphorylation by GSK-3β. In the absence of APC, β-catenin can freely translocate to the nucleus where it signals an intracellular cascade impacting upon the transcription of hundreds of genes6. Here, in mice with somatic or germline mutation of Apc, we performed large-scale insertional mutagenesis using the Sleeping Beauty transposon system to create a comprehensive catalog of new candidate drivers of intestinal tumorigenesis 8,9,11-17. Several hundred candidate driver genes are implicated, allowing novel genetic interactions between genes and conserved pathways to be explored.
RESULTS
Generation of Mice and Histological Analysis
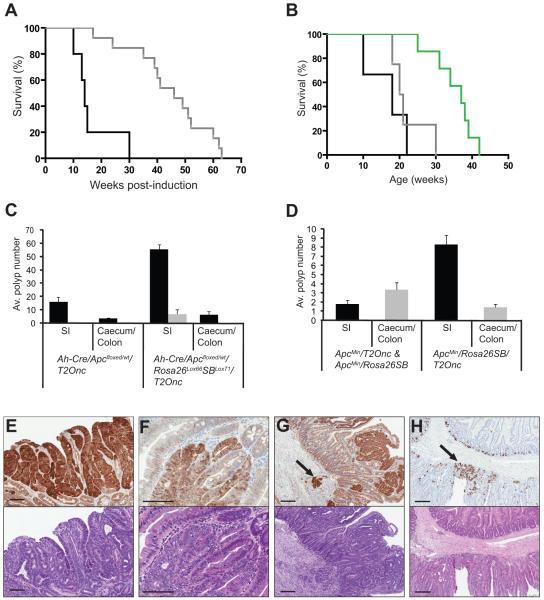
A cohort of 67 transgenic animals (AhCre;Apcfloxed/wt;Rosa26Lox66SBLox71;T2Onc) underwent concomitant somatic heterozygous loss of Apc and activation of SB transposase following Cre induction. Thirty-seven control mice were Apc deficient but unable to mobilize the transposons (AhCre;Apcfloxed/wt;T2Onc or AhCre;Apcfloxed/wt;Rosa26Lox66SBLox71) (Supplementary Fig. 1-2). All Apcfloxed/wt mice eventually developed intestinal tumors, but those with mobilized transposons showed a significantly earlier onset of disease compared to controls (Fig. 1). Tumors mostly occurred in the small intestine, with the occasional lesion found in the caecum and colon (Fig. 1c). In ApcMin mice the experimental group [ApcMin;RosaSBase;T2Onc (10 mice)] developed disease approximately 30 days earlier than control ApcMin littermates, and had a higher proportion of colonic lesions (Fig. 1b,d).
Figure 1. SB transposon mobilization increases morbidity and tumor burden of Apc-deficient mice.
(a) Kaplan-Meier survival curve comparing Apcfloxed experimental (Ah-Cre/Apcfloxed/wt/Rosa26Lox66SBLox71/T2Onc) (black line) and control mice (Ah-Cre/Apcfloxed/wt/T2Onc and Ah-Cre/Apcfloxed/wt/Rosa26Lox66SBLox71) (grey line) (log-rank P<0.001). (b) Kaplan-Meier survival curve comparing ApcMin experimental (ApcMin/RosaSBase/T2Onc) (black line), ApcMin control mice (ApcMin/RosaSBase and ApcMin/T2Onc) (grey line) and RosaSBase/T2Onc control mice (green line) (log-rank P<0.0057). (c) Average polyp size and number in Apcfloxed experimental mice (n=3), and matched littermate Apcfloxed control mice (n=3) at 18 weeks post-induction. All polyps visible under the dissecting microscope at 6x magnification were counted and grouped into those that were non-excisable (black bars) (<3mm) and those that could be excised for further analysis (>3mm) (grey bars). Average polyp number for small intestine and caecum/colon is shown. (d) Average number of excisable polyps (>3mm) from small intestine, caecum and colon of ApcMin experimental mice (n=10) and ApcMin control mice (n=7) at 10-22 and 17-30 weeks of age respectively. β-catenin immunohistochemistry (upper panel of each) and hematoxylin and eosin staining (lower panel of each) of representative tissue from Apcfloxed mice showing (e) a tubulovillous adenoma at 18 weeks post-induction, (f) an adenoma at 28 weeks post-induction, (g) an adenoma at 17 weeks post-induction with the arrow indicating the region of adenocarcinoma in the submucosa. (h) Lysozyme staining of representative tissue from an Apcfloxed/wt mouse at 22 weeks post-induction, with the arrow indicating Paneth cell differentiation. All scale bars = 100μm.
A total of 446 tumors (of which 367 underwent histopathological review) were excised for analysis of transposon insertion sites from the experimental groups; 391 and 55 from the somatic Apcfloxed and germline ApcMin models, respectively. In addition, 106 (46 somatic, 60 germline) tumors from animals without mobilized transposon were taken for histopathological review. Tumors were larger and more numerous in Apcfloxed animals with mobilized transposons (Fig. 1c). The majority of tumors were oligocryptal and tubulovillous adenomas (Supplementary Table 1). However, a proportion of tumors from the somatic model with mobilized transposons showed moderate or severe dysplasia (n=40, 23 mice), while 10 showed evidence of invasion (9 mice) (Fig. 1e-g). Both somatic and germline models produced microsatellite stable tumors (Supplementary Fig. 3).
Identification of Sleeping Beauty Common Insertion Sites (CISs)
In total, 146,249 non-redundant transposon insertion sites (an average of 328 per tumor) were identified after 454 sequencing and mapping to the mouse genome. We employed two statistical frameworks to determine common insertion sites (CISs): Monte Carlo (MC) simulations, and a Gaussian kernel convolution (GKC) algorithm that was optimized to take into account the TA bias of the Sleeping Beauty transposon8,18,19. To maximize statistical power, we pooled insertions from both the Apcfloxed and ApcMin models. The concordance between the two statistical methods was 60%, based on overlapping CIS regions; Monte Carlo analysis identified 749 CISs, while GKC analysis with 30 Kb and 120 Kb kernel sizes identified 919 and 641 CISs, respectively, equating to 997 unique CIS regions (867 genes) (Supplementary Tables 2a-d). The results of the GKC analysis were used to define a discovery set for downstream analyses and refinement by comparison to other datasets. Importantly the T2Onc transposon can generate gain-of-function mutations by integrating in or near genes and driving over-expression from the MSCV promoter in the transposon, and also loss-of-function mutations by integrating in genes and hijacking transcription by use of splice acceptors and polyA signals in the transposon. Insertions may also generate neomorphic alleles, or potentially disrupt regulatory elements around cancer gene loci. To identify CISs that may be the result of preferential insertion hotspots, we mobilized transposons as described above and collected a total of 48 tissue samples for insertion site analysis 2 weeks and 10 months after mobilization. Sequencing of the insertions from these tissues yielded 17,358 unique insertions and 35 CISs using GKC analysis. Only 1.8% of CISs called from the tumors overlapped with these control CISs (Supplementary Table 2e). Analysis of CISs associated with known cancer genes allowed us to establish a set of rules that were used to assign insertions to genes. These rules are outlined in the supplementary methods (Supplementary Fig. 4-9, Supplementary Tables 3-4).
Tumor Etiology
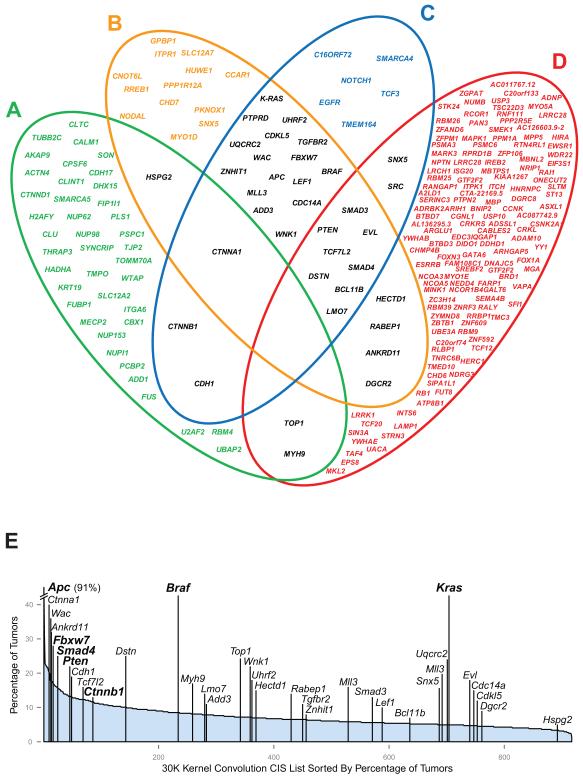
The implication that there is positive selection for hundreds of candidate cancer genes following insertional mutagenesis initially appeared surprising. Consideration of the tumor etiology is important in understanding this observation. First, we determined that multiple tumors obtained from individual mice were biologically independent because their insertions were no more likely to be in the same genomic locations as tumors from different mice (Supplementary Fig. 10). Therefore, separate lesions taken from the same mouse intestine evolve independently. Second, consideration of Apc (mutated in 91.3% of tumours) demonstrates not only that mutation of the second allele is a near obligate event in this model, but also that it is recurrently mutated (65% of tumors had 3 or more insertions). This must arise from selection for serial truncating mutations within Apc during tumor development, as has been shown for the attenuated form of FAP – the ‘three hit’ hypothesis20. This observation also suggests tumor oligoclonality, supported by the number of non-redundant insertions per tumor being 10-fold greater than the number of transposons available for mobilization (328 versus 30). A corollary is that transposons are inefficiently mobilized following Cre induction. This was confirmed by measuring the copy number of transposons retained at the donor locus: around 40% of the original 30 copies of T2Onc are retained (n=24, mean number of unmobilized transposons = 11.19 copies, SD = 2.611) (Supplementary Methods). Since 50% survival is reached at around 16 weeks post-induction (Fig 1a), the rate of insertional mutagenesis is less than 1 event/cell/week (the presence of contaminating stromal cells in which the transposase is not activated may mean that this rate is an underestimate). Thus, transposon mobilization is inefficient and ongoing throughout tumorigenesis, with the extensive accumulated mutational burden arising from selective forces. This leads to the development of oligoclonal tumours, and explains in part the number of insertions we identify. As a consequence many mutated genes make a relatively small contribution to tumor growth.
Overall, the average number of insertions per tumor that contributed to a CIS was 1.14, demonstrating that some tumors contained multiple insertions contributing to a CIS (Supplementary Tables 5a-d). For example, 5.4% of tumors had 2 or more insertions in Ctnnd1, 4.7% of tumors had 2 or more insertions in Kcnq1, and 4.0% of tumors had 2 or more insertions in Lass6. This may be due to the presence of multiple independent tumor sub-clones within the tumor bulk, suggesting that the mutation of these genes may represent a bottleneck in tumor evolution whereby individual cells within the tumor acquire different mutations in order to progress. This observation is also consistent with these genes functioning as tumor suppressors, requiring biallelic mutations before they become operative in cancer development21. Given the above, we cannot distinguish between these possibilities.
Other frequently mutated CIS genes previously implicated in intestinal cancer include Pten, Smad4 and Bmpr1a, which were mutated in a significant number of tumors (13.2%, 15.7% and 7.6% respectively (P<1.51 × 10−5)) demonstrating that transposons mutate the same genes that are disrupted in the human disease. In 48 tumors with single insertions in Pten, Smad4 and Bmpr1a we sequenced the coding regions of these genes and found no further spontaneous mutations, indicating that the insertions in these genes may be acting in a haploinsufficient manner (data not shown).
Transposons Drive Intestinal Tumorigenesis via Conserved Pathways
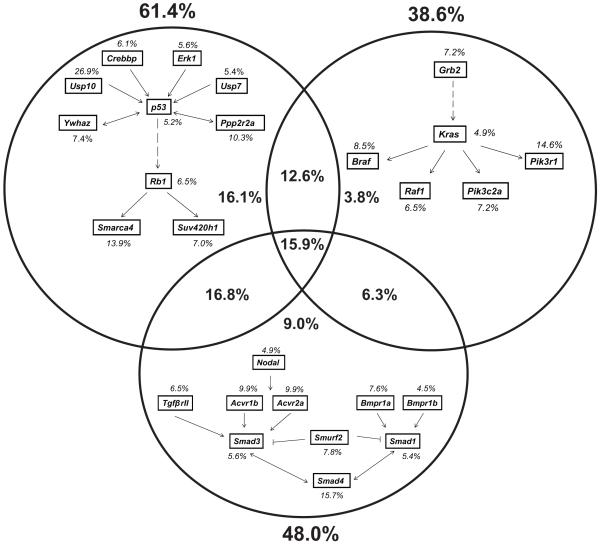
To evaluate the types of genes involved, and to facilitate the filtering of the discovery set of CIS genes, we used Ingenuity Pathway Analysis (IPA) (Ingenuity Systems Inc.). We determined that the CISs in this screen were significantly enriched in cancer-associated signaling pathways, accounting for 113 CIS genes. For example genes in the PI3K/Akt, ERK/MAPK, Wnt and integrin signaling pathways were mutated with P-values of 2.69 × 10−8, 2.76 × 10−8, 2.13 × 10−7 and 1.39 × 10−7, respectively (Supplementary Table 6). Loss of function mutations, characterized by intragenic transposon insertions in both orientations, appeared to predominate. Insertions near K-Ras, mutated in around 40% of sporadic human colorectal carcinomas, were present in only 5% of tumors22,23. In addition, mutations in p53 and Smad4 were found at a frequency of only 5.2% and 15.7%, respectively. To assess the ability of our insertional mutagenesis screen to exploit the K-Ras, p53 and TGFβ signaling pathways, we examined insertions in CISs associated with genes immediately up or downstream of these key cancer genes in each signaling cascade, and determined that they are frequently mutated (all with P ≤2.23 × 10−5) (Fig. 2). In total, 38.6% of tumors carried mutations in genes in the K-Ras pathway, 48.0% in the TGFβ pathway, and 61.4% in the p53 pathway. Just under half of all tumors carried mutations in CISs associated with genes in more than one of these pathways, with 15.9% carrying mutations in all three (Fig. 2). Consideration of tumor pathology with the number or type of pathways mutated showed no correlation with more dysplastic disease (data not shown). However, a proportion of CIS genes were over-represented in tumors carrying insertions in these pathways; around 40, 43 and 36 CIS genes for the K-Ras, p53 and TGFβ signaling pathways, respectively (Supplementary Tables 7-9). Therefore, transposon insertions drive tumorigenesis using the same conserved pathways as are operative in human cancers, and these are associated with additional gene insertions.
Figure 2. Comparison of Sleeping Beauty insertions in K-Ras, p53 and TGFβ signaling pathways.
CIS genes immediately upstream or downstream of K-Ras, p53 or Smad4 were identified and analyzed for the occurrence of transposon insertions. Percentages of tumors with insertions in genes are indicated in italics. Percentages in bold show the overall proportion of tumors with insertions in each pathway.
Cross-Species Comparison
Human cancer genomes acquire mutations that not only drive tumorigenesis, but that also reflect the signatures of the mutagenic processes that have driven their evolution7. Using cross-species oncogenomics, we compared the CISs identified in this study to human colorectal cancer datasets to implicate up to 234 CIS genes in the human disease (Fig. 3a-d, Supplementary Table 10-14). Genes overlapping in two or more of the datasets (34 genes) were distributed throughout the CIS list and include important known CRC genes such as Fbxw7, K-Ras and Ctnndb1 (Fig. 3e, Supplementary Tables 15-16). In addition to comparisons with human tumor studies, we also identified 55/77 of the CIS genes identified in a recent SB transposon screen in the intestine, but by using a significantly larger number of tumors (446 versus 135) and deeper sequencing, we extend this CIS list by 12-fold (Supplementary Table 17)8. The 55 CISs identified by Starr et al.8, were distributed throughout our entire CIS list (Supplementary Table 18), with 7 identified in the top 20 genes of the 30 Kb and 120 Kb GKC analyses (Supplemental Figure 11, Supplementary Table 19). Interestingly, an additional 10 proteins encoded by CIS genes overlap with over-expressed proteins and transcripts identified in a screen of adenomas from ApcMin mice, including Cul1 and Pdlim124,25. Furthermore, recent studies using intercrosses with ApcMin/+ mice have shown that Pparg, Pten, Rb1, Mapk1, Stat3, Ephb4, Myb, Cd44 and Smad3 co-operate with Apc loss in intestinal tumorigenesis, all of which were detected in our analysis26-34. Finally, comparison of our data with a CGH dataset from spontaneously evolving tumors from ApcMin mice showed a significant overlap of CIS genes with regions rearranged in these tumors (P = 6.75 × 10−7) (Supplementary Methods)35, suggesting that transposons identify the same genes that are disrupted in spontaneous cancers. Importantly, 224 genes were predicted to be disrupted in each of these spontaneous mouse tumors, a number 3-fold higher than the average number of CISs per tumor (76).
Figure 3. Cross-species comparison of CISs with cancer datasets.
(a) 47 CIS orthologs of gene products found to be up-regulated or down-regulated in a proteomic comparison of nuclear matrix fractions from human adenomas and adenocarcinomas (Supplementary Table 14) 59. (b) 43 CIS orthologs of genes identified through exon re-sequencing of human colorectal tumors, 13 of which were highlighted as potential drivers by the authors (P= 0.003) (Supplementary Table 13) 47. (c) 236 CIS orthologs of genes reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) as harboring mutations in human sporadic colorectal cancers (36 orthologues in colorectal cancer) (Supplementary Table 12) 60. (d) 143 CIS orthologs of genes found in deleted or amplified regions in 123 human sporadic colorectal cancers through CGH analysis (q<25%) (P = 0.025) (Supplementary Tables 10 & 11) 44. Genes in black indicate CIS orthologs that overlap in two or more datasets. (e) Distribution of CIS gene orthologs from the 30 Kb Gaussian Kernel Convolution analysis. Genes are shown in rank order according to the percentage of tumours containing a given CIS (and then by height of the CIS peak). Some genes such as Mll3 contain two CIS regions and are therefore shown twice. Genes in bold indicate well-known CRC genes.
Association of CIS with the Wnt Pathway
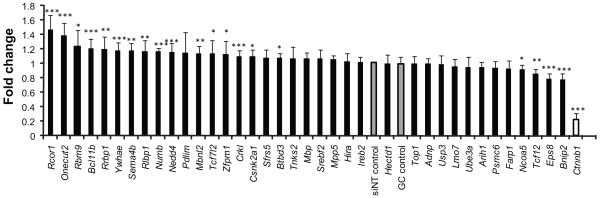
The observation that there was enrichment for subsets of CIS genes in tumors with mutations affecting the p53, K-Ras and Wnt pathways suggested that one explanation for so many potential cancer drivers is their modulation of known cancer pathways. To test this functionally, we explored one such pathway, namely the Wnt signaling pathway. The presence of TCF/β-catenin consensus binding sites in promoter regions of 183 CIS genes suggested that they are potential Wnt targets (P = 1.1×10−64) (TRANSFAC 2009.3, BIOBASE GmBH)36. Of these, 37 genes showed significant copy number changes in a CGH analysis of human colorectal cancers (Fig. 3; Supplementary Table 20), and were assessed to determine if they were potential modulators of Wnt signaling. The CellSensor Wnt reporter system in SW480 colorectal carcinoma cells was used in conjunction with siRNA knockdown, and revealed 16 genes as being negative regulators of Wnt signaling, and 4 genes as positive regulators (Fig. 4). To control for off-target effects, 9 randomly selected siRNAs were tested and were found to have no effect on Wnt reporter activity (Supplemental Fig. 12). Overall this represents a significant enrichment for genes that regulate canonical Wnt signaling in our screen (P<0.005, Fisher’s exact test) (Supplementary Methods). The E3 ubiquitin-protein ligase NEDD4, known to polyubiquinate the tumor suppressor PTEN, appears to negatively regulate Wnt signaling, suggesting a potential link between two distinct tumor suppressor mechanisms involving APC and PTEN. Rcor1, Onecut2 and Zfpm1 are transcription factors, knockdown of which caused a robust increase in Wnt signaling, thus confirming a regulatory role for these genes in the Wnt pathway.
Figure 4. Identification of novel Wnt targets with tumourigenic potential.
Potential regulators of Wnt signaling were identified by analysis with TRANSFAC 2009.3 (BIOBASE GmbH). The results were then cross-referenced with CIS corresponding to orthologs of genes identified from the CGH analysis of human sporadic colorectal cancers44 to generate a final set of CIS genes for further study. SW480 cells containing a stably integrated β-lactamase reporter gene under the control of the LEF/TCF binding consensus sequence were transiently transfected with siRNA (final concentration 20nM) and processed after 72 h. FRET read-out was measured using the manufacturer’s standard protocol. The fold change represented in the graph is calculated with respect to GC control. Error bars: s.d, n=4. Data were pooled from three independent experiments to determine statistical significance by Student’s t-test (***P<0.001, **P<0.01, *P<0.05).
A subset of human colorectal cancers containing an increased proportion of Paneth cells has been described previously 37. In the present study a significant number of tumors (83/367) contained an enlarged proportion of these cells (Fig. 1h). Mouse models in which germline Apc mutations are engineered to retain a single domain of β-catenin binding/degradation and thereby retain some residual functionality from the truncated allele (1322T mutation) have also been shown to carry tumors with a higher proportion of Paneth cells and a more dysplastic phenotype 38. Thirty-six CISs were identified as being over-represented in this group when compared to non-Paneth cell tumors (Table 1). Of these, eight have previously been identified as Wnt targets36. Interestingly Eps8, which we show to be a positive regulator of the Wnt pathway (Fig. 4), is also a CIS gene enriched in the Paneth cell-containing tumors. Three of the 36 genes (Usp10, Eps8 and Cask) have been shown to be upregulated in intestines showing an increase in Paneth cell number due to deletion of Klf939. These results further suggest an important role in the fine-tuning of Wnt signaling throughout tumor development, with one outcome being altered tumor pathology.
Table 1.
Genes over-represented in tumors containing differentiated Paneth cells.
| Gene ID | Total insertions |
Insertions in Paneth tumors |
Sample specific P-value |
|---|---|---|---|
| Usp10 | 13 | 8 | 0.001 |
| Tnpo3 | 25 | 12 | 0.002 |
| Slmap | 17 | 9 | 0.003 |
| Pak2 | 21 | 10 | 0.004 |
| Ppm1b | 38 | 15 | 0.005 |
| Mbnl1 | 26 | 11 | 0.008 |
| Ncoa4 | 26 | 11 | 0.008 |
| Ppp1r2 | 48 | 17 | 0.009 |
| Tax1bp1 | 30 | 12 | 0.010 |
| CIS7:143946337_30k | 14 | 7 | 0.012 |
| Bspry | 18 | 8 | 0.016 |
| Frs2 | 32 | 12 | 0.017 |
| Zfx | 12 | 6 | 0.019 |
| Sfrs7 | 26 | 10 | 0.023 |
| Spin1 | 19 | 8 | 0.023 |
| Ppp1r12a | 93 | 27 | 0.024 |
| 9130404D08Rik|Gatad2a | 41 | 14 | 0.024 |
| Fubp3 | 16 | 7 | 0.027 |
| Eps8 | 38 | 13 | 0.029 |
| Fat1 | 38 | 13 | 0.029 |
| Bclaf1|Mtap7 | 46 | 15 | 0.031 |
| Styk1 | 27 | 10 | 0.031 |
| Cask | 20 | 8 | 0.032 |
| Rps27 | 20 | 8 | 0.032 |
| Ash1l | 39 | 13 | 0.036 |
| CISX:39487624_30k | 24 | 9 | 0.036 |
| Nipbl | 47 | 15 | 0.037 |
| Mlxip | 28 | 10 | 0.039 |
| CIS9:21215498_30k | 21 | 8 | 0.043 |
| H2afy | 21 | 8 | 0.043 |
| Ocln | 21 | 8 | 0.043 |
| Nacc1 | 25 | 9 | 0.047 |
| Tomm70a | 25 | 9 | 0.047 |
| Akap9 | 29 | 10 | 0.049 |
| CIS11:50153019_30k | 29 | 10 | 0.049 |
| Nfib | 61 | 18 | 0.050 |
CIS that are not located within ± 150K base pairs of a gene are labeled identified as ‘CIS’ followed by the chromosome, the peak location of the Gaussian kernel and the kernel size.
We also compared tumors histopathologically classified as presenting with either mild or moderate atypia (n=332) against those with moderate dysplasia and severe dysplasia (n=35); 23 genes were over-represented in the tumors with a more severe phenotype suggesting that these genes contribute to disease progression (Table 2). Importantly, genes involved in FGF signaling (Frs2 and Grb2) feature; it has been proposed that co-activation of this pathway with Wnt signaling results in a more malignant phenotype40. From this list Uhrf2 has been associated with an increased risk of colon cancer41, while a gene fusion of Tmprss2 (Tmprss2-Erg1) has been linked with a poor prognosis in prostate cancer42. Slk is a serine-threonine protein kinase, which, when complexed with Asap1, promotes CRC metastasis43. Therefore, these over-represented genes may provide new insights into CRC progression.
Table 2.
Genes over-represented in tumors with severe pathology.
| Gene ID | Total insertions |
Insertions in severe tumors1 |
Sample specific P-value |
|---|---|---|---|
| Uhrf2 | 22 | 14 | 0.002 |
| Kdelc2 | 64 | 49 | 0.004 |
| Fgfr2|Oat | 12 | 7 | 0.007 |
| Frs2 | 28 | 20 | 0.010 |
| Msl2 | 23 | 16 | 0.011 |
| Tmed10 | 18 | 12 | 0.012 |
| Fnbp1l | 40 | 30 | 0.012 |
| Tpm1 | 24 | 17 | 0.014 |
| Atp1a1 | 37 | 28 | 0.020 |
| 6430598A04Rik | 20 | 14 | 0.020 |
| Chn2 | 15 | 10 | 0.021 |
| Grb2 | 26 | 19 | 0.022 |
| Agpat3 | 38 | 29 | 0.023 |
| Tmprss2 | 38 | 29 | 0.023 |
| Chd6 | 39 | 30 | 0.027 |
| C77080 | 11 | 7 | 0.028 |
| Cnot2 | 53 | 42 | 0.033 |
| Brwd1 | 28 | 21 | 0.033 |
| Rtn4rl1 | 28 | 21 | 0.033 |
| Tnrc6c | 23 | 17 | 0.038 |
| Bspry | 18 | 13 | 0.045 |
| CIS12:88752362_30k | 30 | 23 | 0.046 |
| Slk | 30 | 23 | 0.046 |
Tumors showing severe atypia, moderate dysplasia or severe dysplasia
Co-occurrence of Genes
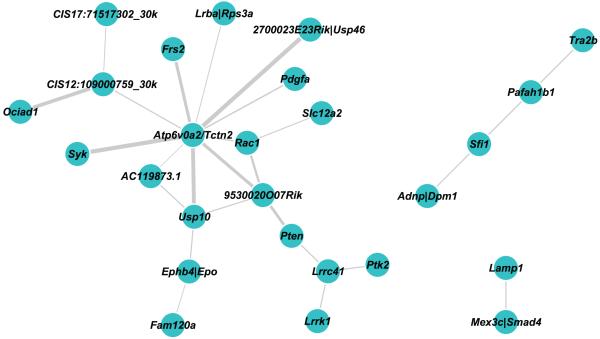
Co-occurring CISs (co-CISs), where two CISs were co-mutated at a higher frequency than expected by chance, were identified. As expected the majority of co-CISs we identified were with Apc, since mutation of the wild type allele of Apc is a near obligate event in adenomagenesis in our model. We focused on other co-CIS identifying 70 CIS genes involved in statistically significant pairwise co-occurrences constituting 38 networks, the largest containing 28 genes (Supplementary Table 21, Fig. 5). Several of the interactions identified were with CISs associated with known tumor suppressor genes, such as Lrrc41 with Pten, and Lamp1 with Smad4 (or Mex3c) (Fig. 4). Analysis of human CGH data provided independent support for two of the co-CISs: those between Mex3c/Smad4 and Lamp1 and between Sfi1 and Adnp (Supplementary Table 20)44. At the centre of the largest network we found the CIS Atp6v0a2/Tctn2; Tctn2 has previously been shown to be a gene mutated in both mouse and human colorectal cancers35. Intriguingly, these networks include genes that co-occur with well-established cancer drivers such as Pten, Smad4 and Rac1.
Figure 5. Co-occurring CISs can be grouped into interacting networks.
CISs generated from the CIMPLR 30 Kb and 120 Kb analyses were merged and pairwise comparisons between each CIS were performed. Contingency tables were constructed for each comparison and a Fisher-Exact test was performed. To account for multiple testing a q value was generated for each test and adjusted P values of P<0.05 were reported. This generated 70 co-occurring relationships in 38 networks, three of which are shown. CIS are represented by the nearest associated gene and the thickness of each interconnecting line between two genes represents the level of significance of the co-occurrence.
DISCUSSION
We have performed an extensive forward genetic screen in the intestinal tract using mouse models of somatic and germline neoplasia. These studies reveal hundreds of genes in addition to 38 genetic networks between these genes that are candidate drivers of carcinogenesis. The value of this discovery set will ultimately be established by the overlaying of high-throughput mutation data from human colon cancers. However, it is worth noting that our screen identified novel genes recently found to be mutated in human colorectal cancers including, MLL3 and CDK844,45. In line with previous commentaries that implicate selection pressures and not elevated mutation rates in driving tumorigenesis, we confirm that the Sleeping Beauty transposon is inefficiently mobilized, generating a very low rate of cellular mutagenesis46. The high number of accumulated mutations observed is thus a consequence of selection acting on increasingly diverse populations to create highly oligoclonal tumours.
Using cross-species oncogenomics, we can independently implicate around a third of our CIS genes in colorectal neoplasia from human studies. Functionally, we identify 20 novel regulators of Wnt signaling that implicates these genes in colorectal tumorigenesis through this pathway. Collectively, our analysis provides a rich catalogue of candidate genes that represent potential diagnostic, prognostic and therapeutic targets, and defines the breadth of genes that may contribute to cancer of the intestine. Our screen is in agreement with a previous exome re-sequencing study of human colorectal cancers, which predicted many hundreds of functional mutations in human cancer47.
The number of genes identified in our screen suggests that many more genes than previously thought can contribute to colorectal cancer development if appropriately altered. Others have suggested that around 10 genes in any one cancer may drive cancer development, with each cancer carrying common driver mutations and also rarer mutations that may also contribute to cancer growth1-3,7. Determining which genes are drivers is complicated by a lack of knowledge of the functional significance of the many hundreds of infrequent somatic alterations that are acquired during cancer development, and because at present no study has systematically analyzed the landscape of somatic mutations in a human cancer type in a large number of samples at base-pair resolution. In this screen, we use a transposon that must integrate proximally to a gene to function, and in this way loci-containing candidate cancer genes were identified when they were mutated at a frequency higher than expected by chance.
While genes that are infrequently mutated in human cancer and in our screen are less likely to be major players in tumorigenesis, population theory suggests that any genetic alteration that causes a fractional increase in the ability of a cell to outgrow other cells may contribute to tumorigenesis48,49. The classical model for this is yeast, in which as many as 6% of mutations that alter the growth kinetics of a population of cells can result in the outgrowth of a more vigorous population50. Likewise, in human studies there is evidence to suggest that mutations that fractionally increase the fitness of a cell within a population can predispose that cell to evolve into a cancer51. In this way, many of the genes that we have discovered in this screen may contribute independently or combinatorially to tumor development. This interpretation of our study is supported by several lines of evidence. First, we have shown that the majority of cancers in this screen carry mutations in Apc, and furthermore, that 20 genes can modify canonical Wnt signaling, even in tumor cell lines in which Apc is already disrupted. There is a long standing observation that the second somatic hit identified as affecting APC in sporadic and familial colorectal cancers is influenced by the nature of the first mutation, such that at least one β-catenin binding domain is retained in one allele. The interpretation is that the complete loss of Apc function is not tolerated by the normal epithelium (the ‘just-right’ phenomenon). Hence, the initial transformation event does not give ‘maximal’ dysregulation of the pathway 52. Loss of function mutations in SFRPs (Wnt antagonists) in human colorectal cancers have been found to increase Wnt signaling over that resulting from the initiating APC mutation 53,54. Moreover, one of the key effects of both Kras and Braf mutations is to synergize with Apc mutations by enhancing Wnt signaling 55. In short, it is well established that the dysregulation of Wnt signaling is not a single event, but is subject to modulation throughout cancer development. Fine-tuning of Wnt signaling is one likely substrate for achieving fractional increases in fitness.
Second, many of the genes discovered in this screen are associated with known colorectal cancer pathways, such as the Wnt, PI3K and Ras cascades. Genes associated with these pathways are significantly enriched for mutations over the level that would be expected by chance. This suggests that many of the genes recurrently mutated by transposons are functioning through these pathways to optimize tumor cell growth. Finally, many CIS genes can be clustered into other novel pathways using pathway analysis tools, suggesting in addition to the main oncogenic routes other pathways are also operative in bowel cancer, and that if appropriately altered can contribute to tumorigenesis. Follow-up functional analysis will help us gain a better understanding of how the CIS genes we have discovered contribute to CRC.
The co-occurring networks of mutated genes discovered from this screen suggest that the mutation of many genes is important in colorectal cancer development. The presence of co-mutated genes in a cancer does not however necessarily mean that mutations of these genes function in a cell autonomous manner to promote cancer growth. Each of the tumors we analyzed is composed of multiple tumor cell clones, and this heterogeneity may favor cellular interactions that result in some of the co-occurrences we observe.
In conclusion, we have demonstrated that insertional mutagenesis in the mouse, performed on a hitherto unprecedented scale, reveals hundreds of candidate cancer driver genes that promote colorectal cancer development.
METHODS
Generation of mice
All experiments were performed on a C57BL/6J background. ApcMin mice: ApcMin mice have been described previously56. Apcfloxed/wt mice: To model sporadic cancer formation we used Ah-Cre/Apcfloxed/wt mice in which the first initiating event (loss of exon 14 of Apc and a concomitant frameshift mutation in exon 15 is induced somatically following administration of β-naphthoflavone (bNF)17. In Ah-Cre mice the CYP1A1 promoter drives expression of Cre throughout the intestinal tract57. T2Onc mice: We used the mouse line LC76 with a transposon donor locus located on chromosome 19. RosaSBase mice: These mice have been described previously12. Conditional Sleeping Beauty transposase mice (Rosa26Lox66SBLox71): The conditional Sleeping Beauty transposase allele was generated by cloning the HSB5 variant of the Sleeping Beauty transposase in the antisense orientation between Lox66 and Lox71 sites. This cassette was targeted to the Rosa locus in E14Tg2a ES cells, transmitted onto a C57BL/6J background and then backcrossed to C57BL/6J. Expression of Cre in these mice catalyzes recombination between the Lox66 and Lox71 sites inverting the transposase cDNA so it can be expressed from the CAGGS promoter. The CAGGS promoter was used in the transposase allele because it drives reproducible expression in a broad range of cells and tissues. Ah-Cre(hom)/T2Onc(het) mice were maintained by crosses to Ah-cre(hom). Rosa26Lox66SBLox71/Apcfloxed/floxed mice were maintained as obligate homozygotes for both alleles. Experimental and control cohorts were obtained in a single generation by crossing Ah-Cre(hom)/T2Onc(het) mice to Rosa26Lox66SBLox71/Apcflox which gave obligate inheritance of Ah-Cre, Rosa26Lox66SBLox71, Apcfloxed alleles and segregation of the T2Onc allele.
For the germline model ApcMin mice were intercrossed with Rosa26SBase mice, which express the SB transposase ubiquitously, and offspring from this cross were in turn mated to T2Onc mice to obtain experimental and control cohorts.
Tumor Watch Analysis
Animals aged between 6-11 weeks received intraperitoneal injections of 80mg/kg β-naphthoflavone daily for 5 days. Experimental mice were monitored on ‘tumor watch’ and sacrificed when moribund. Normal and tumor tissue was collected from each mouse and samples were divided into two parts; one section was snap frozen in liquid nitrogen for splinkerette PCR, while the other section was stored in 10% NBF, transferred to 70% EtOH and processed for histopathological analysis. All animal procedures were approved by the UK Home Office.
Splinkerette PCR
Genomic DNA was extracted from snap frozen tumor samples using Gentra Puregene kits (Qiagen). Splinkerette PCRs were performed as previously described19.
Generation and Pre-processing of 454 Reads
Pools of transposon-genome junction PCRs from the 446 tumor samples were sequenced on the “454 FLX” platform generating a total of 1,763,374 DNA sequence reads. The raw reads were processed removing SB transposon sequence, DNA barcode tags, and sequence beyond the restriction enzyme site. Processed sequences shorter than 20 nucleotides were discarded from the subsequent genome mapping process since they have an increased tendency to map to multiple genomic locations. Following this filtering process 1,316,365 clipped sequences remained for alignment to the mouse genome.
Mapping Clipped Sequences to the Mouse Genome and Identifying Insertion Sites
The set of clipped sequences were mapped to the mouse genome (NCBI Build 37 assembly) using the ssaha2 alignment algorithm (version 2.5)58. Of the 1,316,365 clipped sequences aligned using ssaha, 898,082 unique alignments were reported (50.93% of the original reads). All aligned sequences were checked to confirm that the two nucleotides adjacent to the start of each alignment were TA. Aligned sequences from the same tumor, that were generated using the same restriction enzyme and that mapped to the same genomic location, were merged together to report a single insertion site. Chromosome 1 contains the donor site of the T2Onc transposon and insertion site reads mapping to this chromosome were removed to negate CIS calls resulting from “local hopping”. Further, insertions were filtered from the loci of the genes En2 (chr5:28492236-28498706) and Foxf2 (chr13:31717648-31723271) as elements from these loci are found in the T2Onc transposon. These filtering steps resulted in 146,249 insertion sites which were used for CIS analysis (Supplementary Materials Table S2a).
Identification of Common Insertion Sites
Common insertions sites (CISs) were identified using two complementary approaches:
(1) Gaussian Kernel Convolution: CIMPLR
The Common Insertion Mapping Platform (CIMPLR) is a Gaussian kernel convolution (GKC) method developed in a previous study to identify CISs in retroviral insertional mutagenesis screens18. An enhanced version of the method was developed for SB screens which considers the local density of TA sites within the genome. Kernel widths of 30 Kb and 120 Kb nucleotides were selected for CIS detection.
For each CIS, the genomic location of its kernel peak was used as the reference point to assign gene annotations. A CIS peak was associated with a gene if either it lay within the coding region of a gene or was within +/− 150 Kb nucleotides of its nearest gene. If the distance to the nearest gene was greater than 150 Kb nucleotides, no gene name was assigned. Using 30 Kb and 120 Kb kernels 919 and 641 CISs were reported, respectively (Supplementary Materials Tables S2a,b).
To summarise the CIS predictions from the two independent kernel widths, a method of combining predictions across the kernels was devised, such that where CISs from the two kernels overlapped, those CISs with the smallest “footprints” on the genome were reported. Using this procedure, a total of 997 cross-scale CISs were predicted (Supplementary Materials Table S2c). Depending on the size and location of CISs, genes may be associated with multiple CISs. Consequently the 997 cross-scales CISs are associated with 867 unique genes.
(2) Monte Carlo Simulations
In a previous study Starr et al. developed a Monte Carlo (MC) simulation method to identify CISs in a colorectal cancer screen8. This method determines the local density of insertions in a given window that exceed the expectation of observing this density by chance. The 146,429 insertions were analysed using a Perl script that implemented the Starr et al. MC method (personal communication: Kevin A.T. Silverstein, Univ of Minnesota). Five independent simulations were performed of 100 iterations each. The range of insertions to be detected in a CIS varied from 5 to 20, and the size of windows examined in the simulations ranged from 10 Kb to 150 Kb, in increments of 1 Kb.
Using a similar approach for gene annotation as in the CIMPLR method, described above, CISs were annotated according to the nearest mouse gene. A total of 749 CISs were reported by the MC simulation method.
Discovery Set vs Genome-wide adjusted CISs
The analysis described in our paper used a discovery set of CIS calls that were compared to other datasets to help filter passengers from drivers and to identify other collections of genes of biological interest. This approach (described above) identified statistically significant CISs on a chromosome-by-chromosome basis, and was motivated by the observation that there were significant differences in the number of insertions, and hence CISs, per chromosome. In Supplementary Table 2c we also present a set of CISs calculated using genome-wide GKC with more stringent genome-wide cut-offs.
Co-occurrence
Pairwise comparisons were made between the 997 combined CISs from the GKC analyses, recording counts for all 446 tumor samples for the following cases: shared insertions between the two CISs, insertions unique to each CIS and samples that had no insertions in either CIS (Fig. 5). Where tumor samples had multiple insertions within a CIS, a single occurrence was recorded. The counts were converted into contingency tables and Fisher Exact Tests were performed. To account for multiple testing on the 496,506 comparisons, a q value was generated for each test. CIS pairwise comparisons with adjusted p-values of P < 0.05 were used to construct the resultant co-occurrence networks.
Software
Cytoscape software (version 2.7.0, www.cytoscape.org) was used to create the CIS interaction network.
Cell Culture
CellSensor LEF/TCF-bla SW480 cell line (Invitrogen - further details included in Supplementary Methods) was cultured in RPMI medium 1640 with GlutaMAX (Invitrogen) supplemented with dialyzed FBS (Invitrogen), non-essential amino acids (Invitrogen), sodium pyruvate (Invitrogen), blasticidin (Invitrogen) and penicillin/streptomycin (Invitrogen) in a 37 °C, 5% CO2 incubator.
CellSensor Experiments
CellSensor LEF/TCF-bla SW480 cells (Invitrogen) were seeded at 2.5 × 105 cells in 96-well plates and transfected with 240nm siRNA (Dharmacon) or medium GC RNAi oligo negative control duplex (GC control) (Invitrogen) or AllStars Neg. Control siRNA (siNT control) (Qiagen) for a final concentration of 20 nm, using Lipofectamine RNAimax (Invitrogen). After 48 h cells were treated with Liveblazer FRET B/G loading kit (Invitrogen), incubated for 2 h at RT and read on a spectrophotometer (Tecan) at an excitation wavelength of 409 nm and emission wavelengths of 460 nm and 530 nm. Emission ratios were calculated from these values. Experiments were carried out in triplicate on three separate occasions and data were statistically analysed using a Student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
H.N.M, A.G.R, N.A.W, M.E., R.K, M.J.A, L.vdW, D.J.W and D.J.A are supported by Cancer Research-UK. D.J.A is also supported by the Wellcome Trust and L.v.d.W by the Kay Kendall Leukemia Foundation. A.G.U, L.W, J.t.H and J.d.R are supported by the NWO Genomics program and the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NWO). J.d.R. was also supported by the BioRange program of the Netherlands Bioinformatics Centre (NBIC), which is supported via a BSIK grant through the Netherlands Genomics Initiative (NGI). A.U. is also supported by the Cancer Genomics Centre through the Netherlands Genomics Initiative (NGI). The authors would like to thank the Cambridge Research Institute core service Biological Resources and Histopathology and the Sanger Institute sequencing services for their vital contributions to this study. We are grateful to V. Theodorou for critical reading of the manuscript.
Footnotes
COMPETING INTEREST STATEMENT
The authors declare they have no competing interests.
REFERENCES
- 1.Miller DG. On the nature of susceptibility to cancer. The presidential address. Cancer. 1980;46:1307–18. doi: 10.1002/1097-0142(19800915)46:6<1307::aid-cncr2820460602>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Beerenwinkel N, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schinzel AC, Hahn WC. Oncogenic transformation and experimental models of human cancer. Front Biosci. 2008;13:71, 84. doi: 10.2741/2661. [DOI] [PubMed] [Google Scholar]
- 4.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 6.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–38. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 7.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–50. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–6. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 10.Luchtenborg M, et al. APC mutations in sporadic colorectal carcinomas from The Netherlands Cohort Study. Carcinogenesis. 2004;25:1219–26. doi: 10.1093/carcin/bgh117. [DOI] [PubMed] [Google Scholar]
- 11.Collier LS, Largaespada DA. Hopping around the tumor genome: transposons for cancer gene discovery. Cancer Res. 2005;65:9607–10. doi: 10.1158/0008-5472.CAN-05-3085. [DOI] [PubMed] [Google Scholar]
- 12.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–6. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 13.Collier LS, et al. Whole-body sleeping beauty mutagenesis can cause penetrant leukemia/lymphoma and rare high-grade glioma without associated embryonic lethality. Cancer Res. 2009;69:8429–37. doi: 10.1158/0008-5472.CAN-09-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–6. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–74. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 17.Shibata H, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–3. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 18.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uren AG, et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc. 2009;4:789–98. doi: 10.1038/nprot.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segditsas S, et al. APC and the three-hit hypothesis. Oncogene. 2009;28:146–55. doi: 10.1038/onc.2008.361. [DOI] [PubMed] [Google Scholar]
- 21.Uren AG, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–41. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos JL, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–7. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 23.Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature. 1987;327:298–303. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 24.Hung KE, et al. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prev Res (Phila Pa) 2009;2:224–33. doi: 10.1158/1940-6207.CAPR-08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paoni NF, Feldman MW, Gutierrez LS, Ploplis VA, Castellino FJ. Transcriptional profiling of the transition from normal intestinal epithelia to adenomas and carcinomas in the APCMin/+ mouse. Physiol Genomics. 2003;15:228–35. doi: 10.1152/physiolgenomics.00078.2003. [DOI] [PubMed] [Google Scholar]
- 26.McAlpine CA, Barak Y, Matise I, Cormier RT. Intestinal-specific PPARgamma deficiency enhances tumorigenesis in ApcMin/+ mice. Int J Cancer. 2006;119:2339–46. doi: 10.1002/ijc.22115. [DOI] [PubMed] [Google Scholar]
- 27.Shao J, Washington MK, Saxena R, Sheng H. Heterozygous disruption of the PTEN promotes intestinal neoplasia in APCmin/+ mouse: roles of osteopontin. Carcinogenesis. 2007;28:2476–83. doi: 10.1093/carcin/bgm186. [DOI] [PubMed] [Google Scholar]
- 28.Kucherlapati MH, et al. Loss of Rb1 in the gastrointestinal tract of Apc1638N mice promotes tumors of the cecum and proximal colon. Proc Natl Acad Sci U S A. 2008;105:15493–8. doi: 10.1073/pnas.0802933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, et al. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat Med. 16:665–70. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musteanu M, et al. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology. 138:1003–11. e1–5. doi: 10.1053/j.gastro.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 31.Dopeso H, et al. The receptor tyrosine kinase EPHB4 has tumor suppressor activities in intestinal tumorigenesis. Cancer Res. 2009;69:7430–8. doi: 10.1158/0008-5472.CAN-09-0706. [DOI] [PubMed] [Google Scholar]
- 32.Ciznadija D, et al. Intestinal adenoma formation and MYC activation are regulated by cooperation between MYB and Wnt signaling. Cell Death Differ. 2009;16:1530–8. doi: 10.1038/cdd.2009.94. [DOI] [PubMed] [Google Scholar]
- 33.Zeilstra J, et al. Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 2008;68:3655–61. doi: 10.1158/0008-5472.CAN-07-2940. [DOI] [PubMed] [Google Scholar]
- 34.Sodir NM, et al. Smad3 deficiency promotes tumorigenesis in the distal colon of ApcMin/+ mice. Cancer Res. 2006;66:8430–8. doi: 10.1158/0008-5472.CAN-06-1437. [DOI] [PubMed] [Google Scholar]
- 35.Alberici P, et al. Aneuploidy arises at early stages of Apc-driven intestinal tumorigenesis and pinpoints conserved chromosomal loci of allelic imbalance between mouse and human. Am J Pathol. 2007;170:377–87. doi: 10.2353/ajpath.2007.060853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knuppel R, Dietze P, Lehnberg W, Frech K, Wingender E. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J Comput Biol. 1994;1:191–8. doi: 10.1089/cmb.1994.1.191. [DOI] [PubMed] [Google Scholar]
- 37.Joo M, Shahsafaei A, Odze RD. Paneth cell differentiation in colonic epithelial neoplasms: evidence for the role of the Apc/beta-catenin/Tcf pathway. Hum Pathol. 2009;40:872–80. doi: 10.1016/j.humpath.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Pollard P, et al. The Apc 1322T mouse develops severe polyposis associated with submaximal nuclear beta-catenin expression. Gastroenterology. 2009;136:2204–2213. e1–13. doi: 10.1053/j.gastro.2009.02.058. [DOI] [PubMed] [Google Scholar]
- 39.Simmen FA, et al. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1757–69. doi: 10.1152/ajpgi.00013.2007. [DOI] [PubMed] [Google Scholar]
- 40.Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5:1059–64. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 41.Poynter JN, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–32. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 42.Mosquera JM, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–11. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller T, et al. ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene. 29:2393–403. doi: 10.1038/onc.2010.6. [DOI] [PubMed] [Google Scholar]
- 44.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–51. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashktorab H, et al. Distinct genetic alterations in colorectal cancer. PLoS One. 5:e8879. doi: 10.1371/journal.pone.0008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomlinson I, Bodmer W. Selection, the mutation rate and cancer: ensuring that the tail does not wag the dog. Nat Med. 1999;5:11–2. doi: 10.1038/4687. [DOI] [PubMed] [Google Scholar]
- 47.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 48.Campbell PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–6. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciccarelli FD. The (r)evolution of cancer genetics. BMC Biol. 8:74. doi: 10.1186/1741-7007-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joseph SB, Hall DW. Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics. 2004;168:1817–25. doi: 10.1534/genetics.104.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein AM, Brash DE, Jones PH, Simons BD. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc Natl Acad Sci U S A. 107:270–5. doi: 10.1073/pnas.0909738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 54.Caldwell GM, et al. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–8. doi: 10.1158/0008-5472.can-03-1346. [DOI] [PubMed] [Google Scholar]
- 55.Janssen KP, et al. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology. 2006;131:1096–109. doi: 10.1053/j.gastro.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 56.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–70. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 57.Ireland H, et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–46. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 58.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–9. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albrethsen J, et al. Subnuclear proteomics in colorectal cancer: identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol Cell Proteomics. 9:988–1005. doi: 10.1074/mcp.M900546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10, Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.