The centromere is a unique chromosomal locus that ensures the accurate segregation of chromosomes during mitosis in each cell cycle by directing the assembly of a multiprotein kinetochore complex. The identity of the centromere is marked by a conserved conventional histone H3 variant termed CenH3 (Cse4 in budding yeast and CENP-A in human). One of the important questions in current centromere biology is how CenH3s are loaded to the centromere. Several studies have indicated that histone chaperones Scm3 (suppressor of chromsome missegregation) in yeast1-3 and HJURP (Holliday junction recognition protein) in humans,4,5 specifically recognize CenH3s and recruit them to the centromere. In addition, it is shown that the L1 loop and the α2 helix in the histone fold domain of CenH3, termed the CENP-A centromere targeting domain (CATD), are important for Scm3/HJURP recognition.4,6
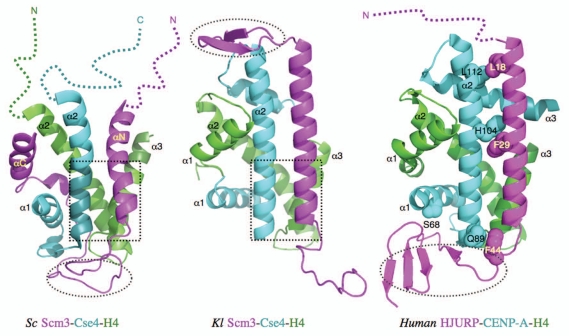
The structures of CenH3-binding domains (CBD) of budding yeast Scm3 (Saccharomyces cerevisiae and Kluyveromyces lactis) and HJURP in complex with their corresponding CenH3/H4 histones have now been determined by the Bai,7 Xu8 and Harrison9 groups, respectively (Fig. 1). All three chaperones directly block the sites of histones that would bind to DNA in the nucleosome structure. They also induce conformational changes in histones, including the formation of additional helical structures at the C-terminal region of histone H4 in all three structures, the bending of the α2 helix of CenH3Sc and the extension of the α3 helix in CENP-A. The direct blocking and induced conformational changes make histones in the complex unsuitable for DNA binding, providing the structural basis for the function of the histone chaperones.
Figure 1.
Comparison of the structures of the Kl and Sc Scm3-Cse4-H4 and human HJURP-CENP-A-H4 complexes. The dashed open ovals indicate the regions that are occupied by DNA in the nucleosome structures. The dashed open squares indicate the regions that have similar structures. In the case of the Sc complex, this region is necessary and sufficient for specific recognition between Scm3 and Cse4/H4. The colored dashed lines represent the disordered regions.
The specific recognition of CenH3Sc by the Scm3Sc chaperone is determined by the N-terminal region of the α2 helix, a subregion of the CATD, including four Cse4-specific residues (M181, M184, A189, S190) that are necessary and sufficient for Scm3Sc recognition in vitro.7 They interact with hydrophobic residues W107, I111, Y114 and I117 at the C-terminal region of the αN helix of Scm3Sc. These residues are essential for cell growth. The structure of this region is similar to the corresponding region in the Scm3Kl-CenH3Kl-H4Kl complex.8
In contrast, CENP-A-specific residues (Q89, H104, L112) in the α2 helix interact with the residues distributed throughout the helix of HJURP.9 In addition, there are interactions between the N-terminal region of the HJURP helix and the extended region of the α3 helix of CENP-A. The roles of these interactions in determining binding affinity have not been examined in vitro. However, it was shown that mutation of residue S68 in the α1 helix of CENP-A to the corresponding residue Q in H3h abolishes the binding between CENP-A/H4h and HJURP in a GST pull-down experiment, suggesting that the α1 helix, which is outside of the CATD in CENP-A, is also important for HJURP recognition.
Most surprisingly, the overall structures of the three complexes are strikingly different despite the homology they share.10 For example, although all chaperones form a long helix in the three complexes as predicted on the basis of their amino acid sequences, the helix in Scm3Sc (αN) is shorter than those in Scm3Kl and HJURP (Fig. 1). Scm3Sc, however, has an additional helix at the C-terminal region (αC) of the CBD, whose corresponding region is absent in the construct of Scm3Kl used for structure determination. The region N-terminal to the long helix in Scm3Kl forms a β-hairpin, whereas the corresponding region is disordered (unpublished results) and subsequently deleted in the construct of Scm3Sc used for the structure determination. Both the αC helix and β-hairpin regions are not conserved in HJURP. The regions C-terminal to the long helices of Scm3Sc and HJURP form an irregular structure and β-sheet, respectively, and interact with both CenH3 and H4, whereas the corresponding region in Scm3Kl forms an irregular structure that has no interactions with histones. The CenH3/H4 histones in complex with Scm3Kl and HJURP have the typical histone fold as in the corresponding (CenH3/H4)2 tetramers. In contrast, the regions corresponding to the α1 helix of H4Sc and the helices of α3 and the C-terminal region of α2 in CenH3Sc are disordered. Importantly, deletion of these disordered regions does not prevent the histones from binding to Scm3Sc in a pull-down experiment.7
The three structures will provide the basis for future investigations on the interactions that determine the specific recognition between CenH3s and their chaperones, and on their conserved/non-conserved features. Before we can have a full understanding of the interactions, however, two issues regarding the structures of the three complexes need to be solved. (1) The Scm3Sc-CenH3Sc-H4Sc structure is determined with solution NMR method at 35°C while the structures of the other two complexes are determined with X-ray crystallographic method at lower temperature. For the Scm3Kl-CenH3Kl-H4Kl complex, the conformation of the C-terminal region of Scm3Kl in the crystal structure is likely incorrect due to alternative crystal packing. Thus, the two crystal structures need to be examined with solution NMR or mutation studies to verify them. (2) The αC helix of Scm3Sc occupies the position of the α1 helix of H4 in the histone fold, preventing it from folding. The corresponding region is absent in the Scm3Kl construct. Therefore, the structure of a Scm3Kl construct that extends the one used by the Harrison group to include the corresponding region of the αC helix of Scm3Sc in complex with CenH3Kl/H4Kl needs to be determined.
Comment on: Zhou, et al. Nature. 2011;47:234–237. Hu, et al. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. Cho, et al. Proc Natl Acad Sci USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108.
References
- 1.Mizuguchi, et al. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 2.Camahort, et al. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Stoler, et al. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foltz, et al. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunleavy, et al. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 6.Shuaib, et al. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou, et al. Nature. 2011;47:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, et al. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, et al. Proc Natl Acad Sci USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Pulido, et al. Cell. 2009;137:173–174. doi: 10.1016/j.cell.2009.06.010. [DOI] [Google Scholar]
- 11.Sekulic, et al. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]