Abstract
Metabolic diseases are characterized by the failure of regulatory genes or enzymes to effectively orchestrate specific pathways involved in the control of many biological processes. In addition to the classical regulators of metabolic homeostasis, recent discoveries have shown the remarkable role of small non-coding RNAs (microRNAs) in the post-transcriptional regulation of a number of genes, and their involvement in many pathological states, such as diabetes, atherosclerosis and cancer. Of note is microRNA-33 (miR-33), an intronic microRNA (miRNA) located within the sterol regulatory element-binding protein (SREBP) genes, one of the master regulators of cholesterol and fatty acid metabolism. We have recently shown that miR-33 regulates cholesterol efflux and high-density lipoprotein (HDL) formation, as well as fatty acid oxidation and insulin signaling. These results describe a model in which miR-33 works in concert with its host genes to ensure that the cell's metabolic state is balanced, thus highlighting the clinical potential of miRNAs as novel therapeutic targets for treating cardiometabolic diseases.
Key words: microRNA, miR-33, lipid metabolism, cholesterol, HDL, ABCA1
Regulation of Cholesterol Metabolism
Cellular dysregulation of lipid homeostasis is a primary perturbation associated with the development of many diseases such as atherosclerosis and type II diabetes.1 Cholesterol, a main constitutive of cell membranes and the precursor of steroids and bile acids, is synthesized endogenously from acetyl-CoA in a highly regulated enzymatic pathway2 or obtained from the circulation via apolipoprotein B-containing lipoproteins, such as the lowdensity lipoprotein (LDL).3 The intracellular levels of cholesterol are highly regulated by a series of feedback mechanisms mediated by the ER-bound sterol regulatory element-binding proteins (SREBPs) 4–6 and the liver X receptors (LXRs).7 Vertebrates have two Srebp genes. Srebp-2 preferentially controls the synthesis and uptake of cholesterol through the regulation of both the LDL-receptor (LDLR) and 3-hydroxy-3-methylglutaryl coenzyme-A reductase (HMGCR; the rate-limiting enzyme in the cholesterol biosynthesis), whereas Srebp-1 regulates genes involved in the synthesis of fatty acids and, more recently, cell cycle regulation.4–6,8 Therefore, an increase in SREBP activity leads to cholesterol and fatty acid accumulation and the downregulation of their own processing pathway.4–6 The other major transcription factors that contribute to the regulation of lipid metabolism are the oxysterol-activated nuclear receptors, namely LXRs. When intracellular cholesterol levels are high, LXRs activate the expression of genes involved in cholesterol efflux including the ATP-binding cassette (ABC) transporters, ABCA1 and ABCG.7 ABCA1 primarily functions in macrophages and hepatocytes, where it promotes cholesterol efflux, and in the liver, as an essential player in the production of HDL.9,10 Cholesterol efflux to HDL and its associated apolipoprotein, Apo-A1, is considered a critical step in the initiation of reverse cholesterol transport (RCT), the process that delivers cholesterol excess to the liver for its excretion into bile.9,10 Because HDL levels correlate inversely with atherosclerosis susceptibility, there is an increasing interest in studying the regulation, mechanism of action, and suitability of ABCA1 as a target to increase HDL levels for the treatment and prevention of atherosclerosis.11
microRNAs
In addition to the classical transcriptional regulators, SREBPs and LXRs, miRNAs have been shown to regulate the expression of key genes involved in lipid homeostasis. microRNAs are small (∼23 nucleotides), non-coding, single-stranded RNAs that control the expression of protein coding genes by primarily acting as sequence-specific inhibitors of mRNA (mRNA).12–14 miRNAs are transcribed in the nucleus by RNA polymerase II into primary transcripts (pri-miRNAs) that are then processed sequentially in the nucleus and cytoplasm by a complex of RNase III-endonucleases, Drosha and Dicer.13,14 Specifically, Drosha processes the pri-miRNA transcript to a 70–100 nt stem-loop precursor (pre-miRNA), which is then delivered to the cytoplasm by Exportin 5, where it is subsequently cleaved by Dicer to produce a miRNA duplex.13,14 The resulting duplex is then incorporated into the RNA-induced silencing complex (RISC) in association with an Ago family member. One of the strands (the passenger strand) is degraded, while the other strand (the mature miRNA) remains associated with the Ago protein and binds to partially complementary sites in mRNAS.13,14 In animals, miRNAs control gene expression by binding to the 3′-untranslated region (3′UTR) of their targets, causing mRNA destabilization or protein translation inhibition.14
miRNAs have emerged as key regulators of almost all cellular processes including cell differentiation and cell proliferation.15,16 Specifically, many micro-RNAs have been shown to play a role in the post-transcriptional regulation of lipid metabolism, including miR-122, miR-370, miR-378/378*, miR-335, miR-125a-5p and miR-33.17–20 Among these, miR-122 was the most widely studied miRNA and the first described for its role in regulating total serum cholesterol and hepatic metabolism.17,21 More recently, several groups have described the role of the miR-33 family as a key regulator of lipid homeostasis.22–26
miR-33 and Cholesterol Homeostasis
miR-33 is expressed in a variety of cell types and tissues and consists of two intronic microRNAs, miR-33a and miR-33b, which are encoded within the introns of the Srebp-2 and Srebp-1 genes, respectively.22,23 While miR-33a and miR-33b differ in only 2 nucleotides in the mature form and have the same targets, they differ in their pattern of evolutionary conservation. miR-33a is encoded within intron 16 of the human Srebp-2 gene and is conserved in many animal species. However, the conservation of miR-33b, which is found within intron 17 of the human Srebp-1 gene, is lost. The Srebp-1 genes of larger mammals contain miR-33b, but in the Srebp-1 genes of rodents and chickens, miR-33b is absent.23
Metabolic stimuli that activate Srebp genes expression also regulate miR-33a and -b expression, indicating that miR-33 is co-expressed with its host genes. For example, miR-33a and its host gene, Srebp-2, are upregulated in response to cellular cholesterol depletion and downregulated in response to cholesterol loading.22 In this condition, miR-33 represses genes involved in cholesterol trafficking and efflux, such as ABCA1, which has three highly conserved binding sites for miR-33 in its 3′UTR.22–24 Consistent with this finding, cultured mammalian cells that overexpress miR-33 show a decrease in cholesterol efflux to Apo.22–24 More interestingly, the opposite occurs when cells are transfected with “anti-miRs” or “antago-miRs,” RNA molecules that specifically reduce endogenous levels of miR-33. Antagonism of miR-33 in vitro and in vivo significantly increases ABCA1 expression, promotes cholesterol efflux to Apo-A1 and increases HDL plasma levels in mice.22–27 Furthermore, genetic deletion of miR-33 in mice causes an increase in hepatic ABCA1 expression and a 25% increase in circulating levels of HDL.25
miR-33 Regulates Fatty Acid Oxidation and Insulin Signaling
The intronic location of miR-33a is highly conserved in many species, including Drosophila melanogaster. Interestingly, these fruit flies do not synthesize sterols or express ABCA1; instead, their Srebp gene regulates fatty acid production, similar to the Srebp-1 gene of humans. This observation suggests additional functions for miR-33a and -b and led us to identify new targets involved in fatty acid metabolism, including carnitine palmitoyltransferase 1A (CPT1A), carnitine O-octanoyltransferase (CROT), and hydroxyacyl-CoA dehydrogenase/3-ketoacy-CoA thiolase/enoyl-CoA hydratase (trifunctional protein) and β subunit (HADHB).28 Consistent with bioinformatic analyses, we and Gerin et al. showed that miR-33 decreases the expression of CPT1a, CROT and HADHB at the mRNA and protein level.26,28 Furthermore, overexpression of miR-33a and -b reduces fatty acid oxidation and leads to the accumulation of triglycerides in human hepatic cells and in the fat body of miR-33 transgenic flies.28
Our previous work also revealed an interesting role for miR-33a and -b in glucose metabolism, as miR-33a and -b overexpression reduces IRS2 levels and inhibits the activation of downstream messenger cascades, including the PI3/AKT pathway.28 Consistent with these findings, miR-33b overexpression reduces insulin-induced 2-deoyxglucose (2-DOG) uptake in hepatic cells, suggesting that miR-33 plays a key role in regulating insulin signaling.28
Pivotal Role of miR-33 in Metabolism
In addition to modulating fatty acid and insulin signaling, miR-33 also regulates the expression of sirtuin-6 (SIRT6).28 Sirtuins are the mammalian homologs of the yeast histone deacetylase, Sir2, and have emerged as broad regulators of many important processes, including cell fate determination, organ metabolism and function, age-related diseases and tumorigenesis.29–34 Given their important regulatory function, many are highly controlled at the transcriptional and post-transcriptional level. For example, SIRT1 has recently been shown to be regulated by microRNAs.35,36 Additionally, our work now shows that another member of the sirtuin family, SIRT6 (which plays a significant role in regulating glucose homeostasis), is also subject to posttranscriptional regulation by these noncoding RNAs.37,38 Liver-specific deletion of SIRT6 in mice causes profound alterations in gene expression, leading to increased glycolysis, triglyceride synthesis and reduced β-oxidation,37 which correlates with our in vitro results in human hepatic cell lines transfected with miR-33.28
Another interesting finding is the inhibitory effect of miR-33 on the AMP-activated protein kinase (AMPK).28 This cellular energy sensor contributes to the regulation of the liver circadian clock,39 which coordinates hepatic lipid metabolism at the transcriptional and post-transcriptional level.40 In the liver, activation of AMPK promotes fatty acid β-oxidation, while inhibiting cholesterol and triglyceride synthesis. Taken together, miR-33a and -b appear to be fundamental modulators of lipid metabolism.
Future Directions: Implications in Metabolic Disease
Our data suggests a model in which miR-33a and -b work in concert with their host genes to ensure that the cell's metabolic state is balanced. During conditions in which SREBP-2 and -1 are activated, miR-33a and -b act in concert with their host genes to boost intracellular cholesterol levels by targeting ABCA1, reduce insulin signaling by targeting IRS2, and increase fatty acid levels by targeting a variety of fatty acid oxidation enzymes. Given that the abnormal regulation of these pathways leads to diseases such as atherosclerosis, metabolic syndrome and non-alcoholic fatty liver disease (NAFLD), miR-33 represents an ideal target for future therapies. Although much remains to be learned concerning the role of miRNAs in regulating lipid homeostasis and insulin signaling, these results highlight the potential of miRNA manipulations in the treatment of diseases.
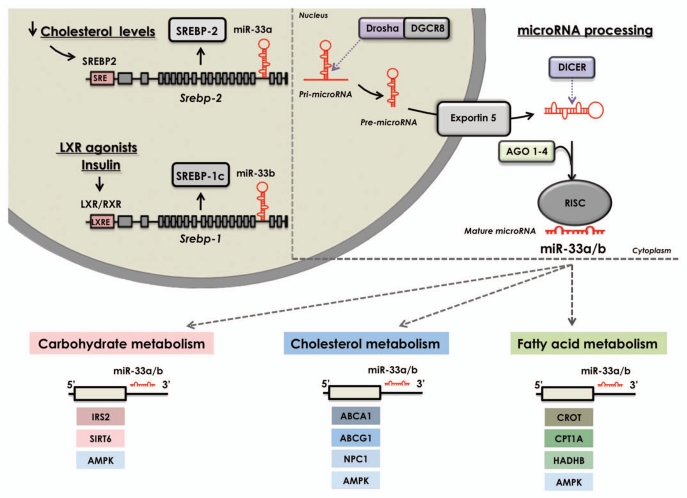
Figure 1.
miRNA-33: A key regulator of metabolic pathways. Transcriptional activation of Srebp-2 or Srebp-1 by low sterol levels or LXR ligands/insulin, respectively, induces the co-transcription of miR-33a and miR-33b from their host genes. These miRNAs simultaneously inhibit the expression of several targets involved in cholesterol transport, fatty acid oxidation and glucose metabolism by binding to complementary regions in their 3′UTRs. Upper right part: General view of microRNA biogenesis. miRNAs are transcribed in the nucleus as long primary transcripts (pri-miRNAs) and processed by Drosha to produce stem-loop-structured precursors (pre-miRNA), which are then exported to the cytoplasm via Exportin 5. Once in the cytoplasm, they are further processed by Dicer to generate a 22-bp miRNA duplex. One strand of the duplex is incorporated in the RNA-induced silencing complex (RISC complex) containing Ago proteins. These mature miRNAs bind to partially complementary sites in the 3′UTR of target genes to promote translational repression or mRNA degradation.
Acknowledgments
The Fernández-Hernando Lab is supported by grants from the American Heart Association (SDG-0835585D), and the National Institutes of Health (R01HL107953 and R01HL106063).
Abbreviations
- miRNA
microRNA
- SREBPs
sterol response element-binding protiens
- LXR
liver X receptor
- ABCA1
ATP-binding cassette transporter A1
- ABCG1
ATP-binding cassette transporter G1
- HDL
high-density lipoprotein
- LDL
low-density lipoprotein
- 3′UTR
3′ untranslated region
- CPT1a
carnitine palmitoyltransferase 1A
- CROT
carnitine O-octanoyltransferase
- HADHB
hydroxyacyl-CoA dehydrogenase/3-ketoacy-CoA thiolase/enoyl-CoA hydratase (trifunctional protein) and beta subunit
- HMGCR
3-hydroxy-3-methylglutaryl coenzyme-A reductase
- FASN
fatty acid synthase
- RCT
reverse cholesterol transport
References
- 1.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 2.Grundy SM. Absorption and metabolism of dietary cholesterol. Annu Rev Nutr. 1983;3:71–96. doi: 10.1146/annurev.nu.03.070183.000443. [DOI] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. Receptor-mediated control of cholesterol metabolism. Science. 1976;191:150–154. doi: 10.1126/science.174194. [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 5.Horton JD. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem Soc Trans. 2002;30:1091–1095. doi: 10.1042/BST0301091. [DOI] [PubMed] [Google Scholar]
- 6.Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 7.Peet DJ, Janowski BA, Mangelsdorf DJ. The LXRs: a new class of oxysterol receptors. Curr Opin Genet Dev. 1998;8:571–575. doi: 10.1016/S0959-437X(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 8.Eberhard Y, Gronda M, Hurren R, Datti A, MacLean N, Ketela T, et al. Inhibition of SREBP1 sensitizes cells to death ligands. Oncotarget. 2011;2:186–196. doi: 10.18632/oncotarget.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tall AR. Cholesterol efflux pathways and other potential mechanisms involved in the athero-protective effect of high density lipoproteins. J Intern Med. 2008;263:256–273. doi: 10.1111/j.1365-2796.2007.01898.x. [DOI] [PubMed] [Google Scholar]
- 10.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. HDL, ABC transporters and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Linsel-Nitschke P, Tall AR. HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat Rev Drug Discov. 2005;4:193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress and timing. Cell. 2003;113:673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrera-Merchan A, Cerrato C, Luengo G, Dominguez O, Piris MA, Serrano M, et al. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle. 2010;9:3277–3285. doi: 10.4161/cc.9.16.12598. [DOI] [PubMed] [Google Scholar]
- 16.Dang CV. MYC, microRNAs and glutamine addiction in cancers. Cell Cycle. 2009;8:3243–3245. doi: 10.4161/cc.8.20.9522. [DOI] [PubMed] [Google Scholar]
- 17.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299:198–206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 20.Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, Ishii K, et al. The upregulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun. 2009;385:492–496. doi: 10.1016/j.bbrc.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 21.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to upregulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci USA. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daválos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 30.Haigis MC, Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 31.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 32.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Kemper JK. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging. 2010;2:527–534. doi: 10.18632/aging.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging. 2010;2:415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. 2010;285:36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gachon F, Bonnefont X. Circadian clock-coordinated hepatic lipid metabolism: only transcriptional regulation? Aging. 2010;2:101–106. doi: 10.18632/aging.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]