Abstract
Apoptosis has been widely accepted as the primary mechanism of drug-induced cell death. Recently, a second type of cell death pathway has been demonstrated: autophagy, also called programmed type II cell death. Autophagy is a highly regulated process, by which selected components of a cell are degraded. It primarily functions as a cell survival mechanism under stress. However, persistent stress can also promote extensive autophagy leading to cell death. Forkhead Box M1 (FoxM1), an oncogenic transcription factor that is abundantly expressed in a wide range of human cancers. Here, we evaluated the role of FoxM1 in sensitivity of human cancer cells to proteasome inhibitor-induced apoptosis and autophagy. We found that FoxM1 knockdown sensitized the human cancer cells to apoptotic cell death induced by proteasome inhibitors, such as, MG132, bortezomib and thiostrepton, while it did not affect the levels of autophagy following treatment with these drugs.
Key words: FoxM1, apoptosis, autophagy, cancer, proteasome inhibitor, caspase-3
Programmed cell death involves an intracellular mediated, highly regulated form of death of a cell. There are three kinds of programmed cell death. These include the apoptosis also known as the type I, autophagy or type II programmed cell death and necrosis or type III programmed cell death respectively. Apoptosis involves condensation and fragmentation of nucleus, cleavage of chromosomal DNA into oligonucleosomal fragments and packaging of the dead cell into apoptotic bodies without plasma membrane breakdown. Autophagy involves formation of a double membrane vesicle called an autophagosome. The autophagosome fuses with a lysosome forming an autolysosome, which release hydrolytic enzymes to degrade the cellular components.
Forkhead Box M1 (FoxM1) is a member of the Forkhead Box family of transcription factors. Its expression is limited to normal dividing cells and most solid tumors, while quiescent cells that exit the cell cycle show no detectable levels of FoxM1 expression.1 FoxM1 regulates expression of genes involved in DNA repair, mitosis and chromatin. Activity of FoxM1 is regulated by the Ras-mitogen-activated protein kinase (MAPK) pathway and CDK-dependent phosphorylation during the cell cycle. We have previously demonstrated that FoxM1 may be involved in a positive autoregulatory loop, where FoxM1 activates its own mRNA and protein expression.2 Additionally p53 negatively regulates the expression of FoxM1.3
The proteasome is a multiple-subunit protease complex that targets ubiquitintagged proteins for degradation in an ATP-dependent manner.4 The 20S catalytic proteasome subunit binds to 19S regulatory particles and facilitates the formation of 26S and 30S proteasome, which recognize and eliminate ubiquitinated proteins. The proteasome-mediated protein degradation is critical for regulation of a variety of cellular processes, including cell cycle, cell death, differentiation and immune response.5 Recent progress in the understanding of proteasome function has led to the development of proteasome inhibitors (PIs) as anticancer Bortezomib (Velcade) was the first PI approved for the treatment of human cancer (multiple myeloma) in 2003, with probable benefits against other types of cancer.6,7 It has been shown that bortezomib may synergize with other anticancer drugs.8–10 Following that, a number of PIs have been developed as anticancer agents.11
While impairment of proteasome activity leads to cell cycle arrest and apoptotic cell death, it also leads to activation of autophagy. Autophagy generally plays dual roles in cellular death or survival; one is to induce type II programmed cell death,12 different from apoptosis, while the other is to salvage cellular components to continue metabolism and to prevent the accumulation of damaged proteins and organelles during stress.12 It has been shown that nuclear, but not cytoplasmic p53 may stimulate autophagy by transactivation of pro-autophagic genes.13 It was demonstrated PIs such as MG132, bortezomib induce autophagy and inhibition of autophagy by autophagy inhibitor 3-MA partially inhibited or augmented apoptotic cell death in different cancer cell lines.13,14 These observations suggest that autophagic cell death may contribute in part to the PI-induced apoptosis and a crosstalk exists among the ubiquitin-proteasome system and the autophagylysosome system.12 Manipulation of autophagy may provide a useful way to prevent cancer development, limit tumor progression, and increase the efficacy of cancer treatments.15,16
In our previous studies, we also demonstrated that FoxM1 inhibitors thiazole antibiotics Siomycin A and thiostrepton induce apoptosis in human cancer cell, suppress FoxM1 expression and act as PIs.17,18 In addition, we have previously demonstrated that PIs such as MG115, MG132 and bortezomib inhibit FoxM1 transcriptional activity and FoxM1 expression.17 The oncogenic transcription factor forkhead box M1 (FoxM1) is upregulated in a wide range of different cancers, where as its expression is turned off in terminally differentiated cells. Recent studies have reported that aberrant expression of FOXM1 in a variety of human cancers is associated with their aggressive behavior.19,20 While targeting FoxM1 is a valid strategy for developing novel anticancer drugs, overexpression of FoxM1 exhibits resistance to anticancer therapy.20 It has been demonstrated that overexpression of FoxM1 specifically protects against apoptotic cell death (caspase-3 cleavage) induced by anticancer agents.20
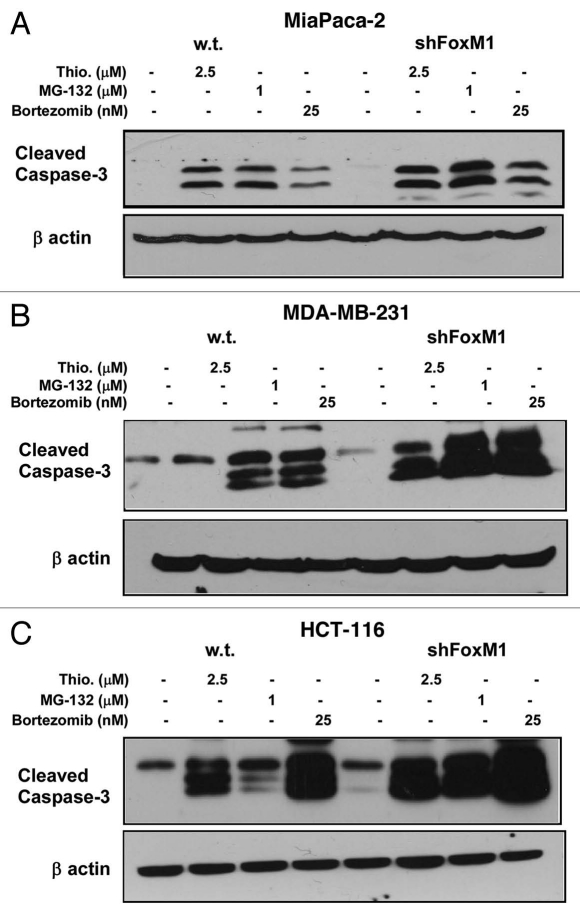
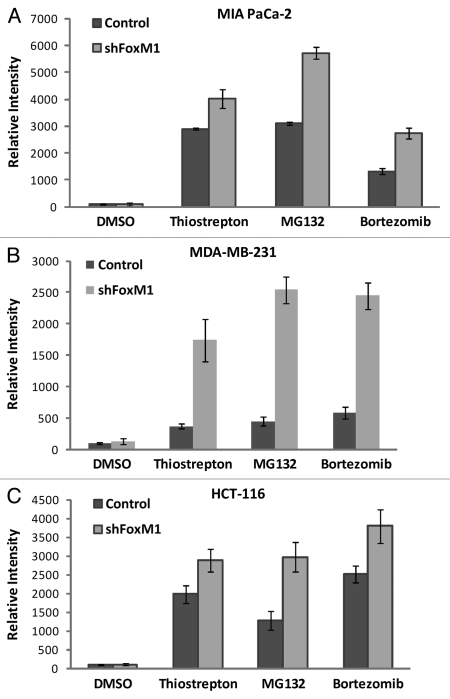
Since overexpression of FoxM1 protects cancer cells from apoptosis, we were interested to see the effect of knockdown FoxM1 expression in sensitivity to apoptosis and autophagy induced by PIs. In order to study the effect of FoxM1 expression on human cancer cells following treatment with PIs, we used human cancer control and FoxM1-knockdown cells and subjected those to treatment with two bona-fide PIs (bortezomib and MG132) and a novel PI described by our lab (thiostrepton) for 24 h following which cells were harvested for immunoblotting and flow cytometry. All three PIs induced stronger apoptosis (cleavage of caspase-3) in FoxM1-knockdown cells compared with control MIA PaCa-2 cells containing empty vector (Fig. 1A). To validate the phenomenon that knocking down of FoxM1 sensitized cancer cells to PI-induced apoptosis, we treated MDA-MB-231 control and FoxM1-knockdown human breast and HCT-116 wt and FoxM1-knockdown human colon cancer cells with PIs for 24 h and performed immunoblotting with extracts from these cells with antibody to cleaved caspase-3 (Fig. 1B and C). We found that knockdown of FoxM1 sensitized breast and colon cancer cells to PI-induced apoptosis. The induction of apoptosis following knockdown of FoxM1 in the cancer cells (Fig. 1A–C) was up to 2-fold that of control cells (Fig. 2A–C shown). These data suggest that suppressing FoxM1 in human cancer cells could make them more susceptible to PI-induced apoptosis.
Figure 1.
PIs sensitized the human cancer cells to apoptotic cell death following knockdown of FoxM1. MIA PaCa-2 vector control and short hairpin (sh) FoxM1 pancreatic human cancer cell lines were grown in DMEM medium (Invitrogen). MDA-MB-231 vector and shFoxM1 breast human cancer cell lines were grown in RPMI medium (Invitrogen). Stable cell lines were generated by transduction of control and FoxM1 shRNA lentiviral particles (Sigma) followed by selection with puromycin (Sigma). Actively dividing cells were seeded into a 100 mm plate at a density of 7.5 × 105 cells. Cells were treated with thiostrepton, MG132 and bortezomib for 24 h following which the cells were lysed. Cells were lysed in IP buffer (20 mM HEPES, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 100 mM NaF, 10 mM Na4P2O7, 1 mM sodium orthovanadate, 0.2 mM PMSF supplemented with protease inhibitor tablet Roche Applied Sciences) and the protein concentration was determined using the Bio-Rad protein assay reagent. Fifty micrograms of the cell lysates were separated by electrophoresis on SDS-polyacrylamide mini gel and transferred to PVDF membrane. Immunoblotting was performed with specific antibodies for cleaved caspase-3 (9664 cell signaling) and β-actin (A5441, Sigma). (A) Control and shFoxM1-k/d MIA PaCa-2 pancreatic cancer cells. (B) MDA-MB-231 breast cancer cells. (C) HCT-116 colon cancer cells were treated with thiostrepton, MG132 and bortezomib as shown for 24 h, total cell lysates were extracted and immunoblotted with antibodies for cleaved caspase-3. β-actin was used as loading control.
Figure 2.
FoxM1 knockdown cells undergo stronger apoptosis induced by PIs. Densitometry was performed on scanned immunoblot images using the ImageJ gel analysis tool. The gel analysis tool was used to obtain the absolute intensity (AI) for each experimental cleaved caspase-3 band for control and shFoxM1-k/d pancreatic (A) MIA Paca-2, breast (B) MDA-MB-231 and colon (C) HCT-116 cancer cells. Relative intensity (RI) for each experimental band was calculated by normalizing the experimental AI to the corresponding loading control β-actin AI. Columns, mean of three separate experiments; bars, SD.
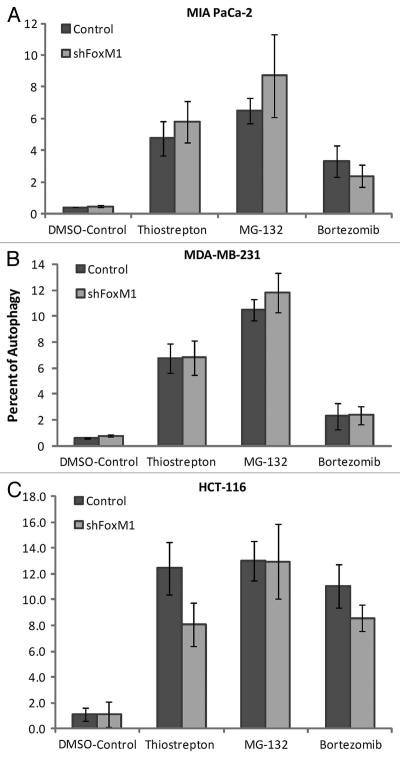
On subjection to stress such as nutrient deprivation, eukaryotic cells undergo a lysosome mediated degradation pathway for digesting their own contents by a process known as autophagy. Autophagy is typically low under basal conditions, but can be markedly upregulated by a number of physiological stimuli such as nutrient deprivation, hypoxia and pharmacological drug treatment, etc.12 Conversely, inhibition of autophagy can occur in certain kinds of cancers or following treatment with autophagy inhibitors.15,16 Proteasome inhibitors induce autophagy in multiple types of cancer.14 By using a green fluorescent probe used to detect autophagy that accumulates in spherical vacuoles and vesicles produced during autophagy (kit 51031-K200-Enzo Life Sciences), we compared the percent of control and FoxM1-knockdown cells undergoing autophagy following treatment with PIs. We found that between the control and FoxM1-knockdown human pancreatic MIA PaCa-2 cells, the percent of autophagy positive cells was similar following treatment with thiostrepton, MG132 and bortezomib (Fig. 3A). In addition to MIA PaCa-2 cells, MDA-MB-231 (Fig. 3B) and HCT-116 (Fig. 3C) control and FoxM1-knockdown cells exhibited similar percentage of autophagy positive cells following PI treatment. Serum starvation was used as positive control while co-treatment with 3-MA served as negative control in the flow cytometry.
Figure 3.
Knockdown of FoxM1 does not affect autophagy induced by PIs. Cells were stained using Enzo Cyto-ID (ENZ-51031-K200Enzo Life Sciences), according to the manufacturer's recommendations. Briefly 3–4 × 105 cells were plated in 60 mm culture dishes and allowed to grow overnight. Cells were treated with thiostrepton, MG132 and bortezomib. Following overnight incubation of drugs, cells were stained with Cyto-ID autophagy detection dye and submitted for flow cytometry analysis. Control and shFoxM1-k/d (A) MIA PaCa-2 pancreatic, (B) MDA-MB-231 breast and (C). HCT-116 colon cancer cells were treated with PIs as shown for 24 h stained with Cyto-ID autophagy detection green fluorescent dye and analyzed by using FL-1 channel of flow cytometer with excitation wavelength of 488 nM. Columns demonstrate percentage of cells undergoing autophagy. (Columns, mean of three separate experiments; bars, SD).
Overall, we demonstrated here that knockdown of FoxM1 in multiple human cancer cells sensitizes them to apoptosis, but not to autophagy induced by PIs. These data suggest that FoxM1 plays different roles in PI-induced apoptosis and autophagy. Further experiments are needed to explain the possible mechanisms behind these effects.
Acknowledgments
We would like to thank Marianna Halasi for the development of FoxM1-knockdown cancer cells.
Grant Support
NIH grants 1RO1CA1294414 and 1R21CA134615 (A.L. Gartel).
References
- 1.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 2011;10:396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halasi M, Gartel AL. A novel mode of FoxM1 regulation: positive auto-regulatory loop. Cell Cycle. 2009;8:1966–1967. doi: 10.4161/cc.8.12.8708. [DOI] [PubMed] [Google Scholar]
- 3.Pandit B, Halasi M, Gartel AL. p53 negatively regulates expression of FoxM1. Cell Cycle. 2009;8:3425–3427. doi: 10.4161/cc.8.20.9628. [DOI] [PubMed] [Google Scholar]
- 4.Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/S0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 5.Adams J. Proteasome a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 6.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) Cancer Invest. 2004;22:304–311. doi: 10.1081/CNV-120030218. [DOI] [PubMed] [Google Scholar]
- 7.Duensing S, Duensing A. Bortezomib: killing two birds with one stone in gastrointestinal stromal tumors. Oncotarget. 2010;1:6–8. doi: 10.18632/oncotarget.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandit B, Gartel AL. Thiazole antibiotic thiostrepton synergize with bortezomib to induce apoptosis in cancer cells. PLoS ONE. 2011;6:17110. doi: 10.1371/journal.pone.0017110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neznanov N, Komarov AP, Neznanova L, Stanhope-Baker P, Gudkov AV. Proteotoxic stress targeted therapy (PSTT): induction of protein misfolding enhances the antitumor effect of the proteasome inhibitor bortezomib. Oncotarget. 2011;2:209–221. doi: 10.18632/oncotarget.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fulda S. Novel insights into the synergistic interaction of Bortezomib and TRAIL: tBid provides the link. Oncotarget. 2011;2:418–421. doi: 10.18632/oncotarget.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazi A, Lawrence H, Guida WC, McLaughlin ML, Springett GM, Berndt N, et al. Discovery of a novel proteasome inhibitor selective for cancer cells over non-transformed cells. Cell Cycle. 2009;8:1940–1951. doi: 10.4161/cc.8.12.8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 13.Galluzzi L, Kepp O, Kroemer G. Tp53 and MTOR crosstalk to regulate cellular senescence. Aging. 2010;2:535–537. doi: 10.18632/aging.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge PF, Zhang JZ, Wang XF, Meng FK, Li WC, Luan YX, et al. Inhibition of autophagy induced by proteasome inhibition increases cell death in human SHG-44 glioma cells. Acta Pharmacol Sin. 2009;30:1046–1052. doi: 10.1038/aps.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janku F, McConkey DJ, Hong DS, Kurzroc R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 16.Puissant A, Robert G, Auberge P. Targeting autophagy to block hematopoietic malignancies. Cell Cycle. 2010;9:3470–3478. doi: 10.4161/cc.9.17.13048. [DOI] [PubMed] [Google Scholar]
- 17.Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS ONE. 2009;4:5592. doi: 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PLoS ONE. 2009;4:6593. doi: 10.1371/journal.pone.0006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. 2010;70:5054–5063. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]