Abstract
EAPII (also called TTRAP, TDP2), a protein identified a decade ago, has recently been shown to function as an oncogenic factor. This protein was also proven to be the first 5′-tyrosyl-DNA phosphodiesterase. EAPII has been demonstrated to have promiscuous protein associations, broad responsiveness to various extracellular signals and pleiotropic functions in the development of human diseases including cancer and neurodegenerative disease. Emerging data suggest that EAPII is a multi-functional protein: EAPII repairs enzyme (topoisomerase)-mediated DNA damage by removing phosphotyrosine from DNA adducts; EAPII is involved in multiple signal transduction pathways such as TNF-TNFR, TGF? and MAPK, and EAPII is responsive to immune defense, inflammatory response, virus infection and DNA toxins (chemo or radiation therapy). This review focuses on the current understanding of EAPII biology and its potential relations to many aspects of cancer development, including chromosome instability, tumorigenesis, tumor metastasis and chemoresistance, suggesting it as a potential target for intervention in cancer and other human diseases.
Key words: EAPII, transcription regulation, DNA repair, signal transduction, cancer development, chemoresistance
Introduction
EAPII (ETS1-Associated Protein II) was identified as a new protein interacting with ETS1, a transcription factor involved in tumorigenesis and metastasis,1–3 and this association modulates the transcriptional activity of ETS1.4 EAPII was also independently identified and designated as TTRAP (TRAF and TNF receptor-associated protein) through association with the cytoplasmic domain of CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs); this association inhibits NFκB activation.5 More recently, EAPII was recognized as the first enzyme that removes topoisomerase-mediated adducts at the 5′-phosphotyrosyl bond, called TDP2 (tyrosyl-DNA phosphodiesterase 2). TDP2 restores 5′-phosphate termini at double strand breaks, preparing them for ligation and overexpression of TDP2 in yeast caused resistance to camptothecin, a topoisomerase I (Top1) inhibitor.6 Emerging evidence delineates the physiological and pathological roles of EAPII in many biological processes including embryonic development, neuronal development, and cancer development, progression and chemoresistance. This review summarizes the current understanding of EAPII biology and its biological significance, including gene structure, protein interaction networks, biological functions and related human diseases mainly focusing on cancer. For clarity, we will hereafter use the name EAPII.
EAPII Genomic Organization and Protein Expression
The EAPII gene is conserved in chimpanzee, dog, cow, mouse, rat, chicken, zebrafish, mollusca and C. elegans. In humans, the EAPII cytogenetic band is located at 6p22.3–p22.1. Seven EAPII exons are spread on the reverse strand of 17 kb genomic DNA. A single 2.3 kb transcript of the EAPII gene is ubiquitously expressed in most tissues and cell lines examined, including normal human kidney, lung, liver, thymus, breast, ovary and prostate.4,5,7 EAPII mRNA encodes a 362-amino acid protein, migrating at an apparent molecular weight of 49 kD. The protein is detectable in most cancer cell lines examined. Interestingly, a variant form of EAPII protein migrating at 43 kD also appears in many cancer cell lines.8 Forced expression of exogenous EAPII also produces two bands migrating at 49 kD and 43 kD. Both forms can be specifically recognized by multiple EAPII antibodies and can be knocked down by a variety of short hairpin RNA (shRNA) targeting either the coding sequence or the untranslated region (UTR) of the EAPII transcript.8 It is clear that the smaller molecule (43 kD) is a short form of EAPII protein, which exists in mammalian cells. It will be interesting to determine how the short form is produced (cleavage or alternative splicing) and whether it functions differently.
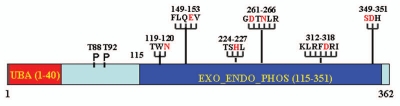
Structure of EAPII protein and potential structural-function correlation.
Protein sequence alignment of EAPII indicates that EAPII contains a conserved domain retaining all six motifs that are hallmarks of an endonuclease/exonuclease/phosphatase super-family (Exo_endo_phos, PF03372) (Fig. 1).9 This large family of proteins includes magnesium-dependent endonucleases and phosphatases involved in intracellular signaling. Within the Exo_endo_phos family, EAPII is most similar to human HAP1/REF-1/APE1, which functions as an endonuclease involved in recognizing and cleaving apurinic/apyrimidinic (AP) sites during DNA repair.10 HAP1/REF-1/APE1 is also involved in the modulation of transcriptional activation through regulating redox signaling.11,12 Although EAPII shares low overall primary sequence identity with the Exo_endo_phos family members (∼12%), the functionally and structurally critical residues are absolutely conserved: E152, G261, D262, N264, D350 and H351 are critical for enzymatic activity and Q151 and S349 are necessary for orienting and/or stabilizing catalytic residues.13–16 Based on these conserved motifs, EAPII was predicted to be a phosphodiesterase.9 Consistent with this prediction, EAPII was discovered as the first enzyme that is capable of removing topoisomerase II-mediated adducts at the 5′-phosphotyrosyl bond.17 In addition, EAPII protein contains an ubiquitin-associated protein-like domain (UBA-like domain) at the N-terminus (Fig. 1). The UBA-like domain is a sequence motif present in multiple enzyme classes of the ubiquitination pathway,18 and this domain provides a binding surface for EAPII to ubiquitin or its relatives, suggesting that EAPII may potentially be involved in the ubiquitin-proteasome pathway. Determination of whether these domains are essential or sufficient to the roles of EAPII is important for exploring the mechanisms underlying its functions.
Figure 1.
Schematic representation of EAPII protein structure. The UBA domain is indicated in red and the Exo_endo_phos domain in solid blue. Six exo_endo_phos motifs are indicated by the amino acid sequence and the starting and ending number and the functional critical residues are highlighted in red. The potential function of the motifs are: Asn hydrogen bonds of TWN to catalytic Asp of the GDXN motif; Glu of LQE coordinates Mg2+ or Mn2+; GDXN catalytic Asp and Asn hydrogen bonds to scissile phosphate in substrate; and the His of SDH paired with catalytic Asp forms a hydrogen bond to scissile phosphate. The phosphorylation sites (T88 and T92) are also indicated. All amino acid residues are indicated by a one-letter symbol.
Biochemical Functions
Signal transduction and the network of EAPII-associated proteins.
Originally, EAPII was identified as a protein that interacts with multiple components of signaling transduction pathways, including signaling adapters such as CD40, TNFR-75 and TRAFs5 and signaling effectors such as ETS1, ETS2, etc.4 So far, more than 20 proteins have been identified as being associated with EAPII (Table 1). Two major biological functions are implicated by the network of EAPII-associated proteins: transcriptional regulation and signal transduction.
Table 1.
EAPII-associated proteins and related biological functions
| Associated Protein | Interacting Regions in EAPII | Experiment Approaches | Related Functions | Ref. |
| ETS1 | 36–362 (N-terminal and central regions) | Yeast two-hybrid, IP, co-localization | Transcription modulator | 4 |
| ETS2 | ||||
| FLI1 | ||||
| TNFRSF8/CD30 | (230–245 aa) | Yeast two-hybrid | TNFα signaling | 5 |
| TNFRSF5/CD40 | ||||
| TNFRSF1B/TNF-R75 | ||||
| TRAF2 | ||||
| TRAF3 | ||||
| TRAF5 | ||||
| TRAF6 | ||||
| SMAD3 | Yeast two-hybrid, IP, co-localization | TGFβ signaling | 19 | |
| ACVR1B/ALK4 | ||||
| Hanta virus NP | 20–362 | Yeast two-hybrid | Virus response | 42 |
| SUMO1 | ||||
| UBE2I | ||||
| ΦC31 integrase | 60–180 | Yeast two-hybrid, IP | Virus response | 24 |
| HIV-1 integrase | N-terminus | Yeast two-hybrid, IP, co-localization | Virus response | 43 |
| DJ-1 | C-terminus | Yeast two-hybrid, IP, co-localization | Parkinson disease | 33 |
| PML | n/a | Yeast two-hybrid, co-localization | NBs function | 44 |
| DAXX | ||||
| Sp100 |
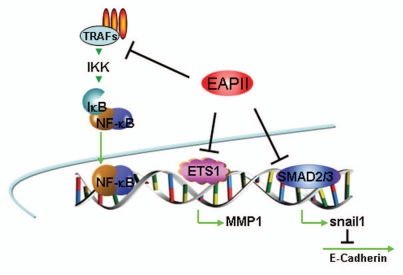
Transcriptional regulation. The first major group of EAPII partners are transcription factors including Ets family members [ETS1 (v-ets erythroblastosis virus E26 oncogene homolog 1), ETS2 (v-ets erythroblastosis virus E26 oncogene homolog 2), FLI1 (Friend leukemia virus integration 1)],4 and smad3.19 Ets family members control important biological processes, including cellular proliferation, differentiation, lymphocyte development, angiogenesis and tumor metastasis.1,20 Smad3 is a receptorregulated transcription factor that transduces extracellular signals activated by TGFβ, a multifunctional cytokine involved in the regulation of survival/or apoptosis, proliferation, differentiation and epithelial to mesenchymal transition (EMT) of epithelial cells.21–23 Through direct protein associations, it has been demonstrated that EAPII inhibits the transactivation by ETS1 4 and SMAD3 19 in reporter assays. EAPII and SMAD3 cooperation results in the de-repression of E-cadherin through inhibiting the expression of Snail1a in the embryonic development of zebrafish.19 EAPII itself is concluded to have no intrinsic transcriptional activity because a Gal4 DNA binding domain-fused EAPII (GAL4-DBD-EAPII) does not transactivate the Gal4 reporter (our unpublished observation). Therefore, although the molecular mechanism is still elusive, EAPII can either positively or negatively modulate the transcriptional activity of other transcription factors. In addition, EAPII may modulate the transcriptional activity of NFκB through regulating the upstream events of the signaling cascade, since no direct associations were identified. Similar repressive effects of EAPII on AP-1 transactivation were also demonstrated.4,5,24 Another possible mechanism of inhibition is through interfering with the transcriptional activity of ETS1, since ETS1 widely participates in combinatorial transcriptional controls through interaction with AP-1 or NFκB25–27 (Fig. 2).
Figure 2.
Model of EAPII -mediated transcription modulation: (1) EAPII represses NFκB activity through regulating upstream events of the TRAF-NFκB cascade; (2) EAPII represses transactivation by ETS1 on the MMP1 promoter; and (3) EAPII represses transactivation by Smad3 on Snail1, which is a transcriptional repressor of E-cadherin, subsequently leading to de-repression of E-cadherin transcription. Green lines and arrows denote transactivation and black lines indicate transcriptional repression.
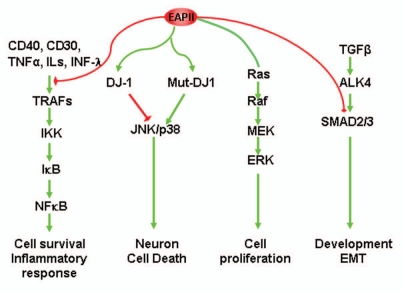
Signal transduction. A large cluster of receptor or adaptor proteins is associated with EAPII. These proteins include TGFβ receptor ACVR1B/ALK4 (activin A type 1B receptor), TNF superfamily receptor and adaptors: TNFRSF8/CD30 (tumor necrosis factor receptor superfamily, member 8), TNFRSF5/CD40 (CD40 molecule, TNF receptor superfamily member 5), TNFRSF1B/TNF-R75 (tumor necrosis factor receptor superfamily, member 1B), and TRAFs (TNF receptor-associated factors: TRAF2, TRAF3, TRAF5 and TRAF6).5 The region of the CD40 cytoplasmic domain required for EAPII association (230–245 aa) overlaps with the TRAFs recognition motif. CD40 ligand treatment significantly increases the EAPII-CD40 receptor association and overexpression of EAPII represses receptor-initiated NFκB transactivation but has no effect on that induced by p65 or IKKα, suggesting that EAPII acts on the upstream components of the signaling cascade.5 TRAFs participate in diverse signaling pathways, which involve cellular activation, differentiation, cell survival and immune responses,28,29 and TRAFs play critical roles in the cross-talk among these signaling pathways.30,31 EAPII is differentially associated with TRAFs, with greatest affinity for TRAF6,5 suggesting a complexity of signaling regulation by EAPII. We recently found that EAPII is involved in the modulation of MAPK-ERK signaling in lung cancer cells. EAPII overexpression significantly activates Raf-1 and ERK1/2 but not the JNK and p38 pathways, subsequently regulating downstream targets including MYC and cyclin D1.8 These observations indicate that EAPII contributes to lung cancer development through deregulation of EGFR signaling, which has been implicated in the development and progression of NSCLC.32 The phosphorylation of EAPII at threonine 88 or/and 92 (T88 or T92) by TGFβ receptor (ALK4) is essential for the function of EAPII, as demonstrated by the inability of EAPIIT88A/T92A to rescue the defects caused by EAPII knockout during zebrafish gastrulation.19 This finding suggests that EAPII is likely one of the direct downstream targets of TGFβ receptor (ALK4) in zebrafish. In neuroblastoma cells, interestingly, EAPII represses JNK activity in the presence of wild-type DJ-1. However, mutant DJ-1, which strongly binds to EAPII, induces cell death via the JNK and p38 MAPK pathways,33 suggesting that EAPII can act as a switch to turn on/off the specific signaling. It is no doubt that these protein associations provide the foundation for the EAPII-related signaling network (Fig. 3).
Figure 3.
Model of EAPII-mediated signal transduction: (1) EAPII negatively modulates TNFα signaling; (2) EAPII facilitates or represses the JNK-mediated apoptosis pathway, depending on the genotype of DJ-1 protein; (3) EAPII activates MAPK-ERK signaling; and (4) EAPII negatively modulates Nodal signaling through Smad3 association.
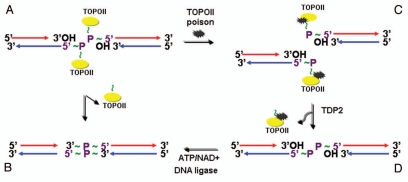
Tyrosyl-DNA phosphodiesterase (TDP) activity and DNA repair.
Using an elegant experimental design, Ledesma et al. discovered that EAPII possesses enzymatic activity that removes topoisomerase protein from 5′-tyrosyl-DNA adducts in mammalian cells.17 Topoisomerases are nuclear enzymes needed for virtually every process that requires movement of DNA within the nucleus or the opening of the double helix.34 The enzyme creates breaks in DNA and thereby allows the DNA strands to unknot and untangle. In order to carry out its critical physiological functions, topoisomerase generates transient topoisomerase-DNA cleavage complexes, so-called cleavable complexes, in DNA. Oxidation, ionizing radiation or chemotherapeutic agents can stabilize the complex and prevent the enzyme from resealing the DNA break it creates, resulting in enzyme-mediated DNA damage.35 Tyrosyl-DNA phosphodiesterase 1 (TDP1) was identified to specifically remove the phosphodiester bond from topoisomerase I-3′ tyrosyl-DNA while EAPII (also called TDP2) removes the phosphodiester bond from topoisomerase II-5′ tyrosyl-DNA and generates free 5′-phosphate ready for ligation, providing an error-free DNA repair (Fig. 4). The structure of EAPII shares no common functional domain with TDP1, which is a member of phospholipase D superfamily.36 The Exo_endo_phos domain of EAPII suggests it has nuclease activity (removes the phosphodiester bond between nucleotides). Indeed, cfEAPII, an EAPII homolog from C. farreri, was recently identified and was shown to have prominent endonuclease activity by cleaving the genomic DNA from C. farreri, but not from E. coli or Bacillus subtilis.37 Repairing protein-DNA complexes can also be achieved via nonspecific nucleolytic pathways which include the double-strand break repair (DSBR) pathway mediated by Ku70/Ku80 and/or Rad1/Rad1038 and the single-strand break repair (SSBR) pathway mediated by the XRCC-1 complex.39 In addition, Rad32 (Mre11) nuclease or Ctp1 (CtIP) can remove covalently bound topoisomerase I and II from DNA, and this topoisomerase removal contributes significantly to resistance against topoisomerase-trapping drugs in yeast.40 The removal of protein-DNA adducts is essential for maintaining cell survival, and it has been shown that EAPII provides the major 5′-TDP activity, at least, in chicken DT40 cells.41 However, genetic depletion of EAPII did not change the proliferation rate of these cells.41 By contrast, EAPII knockdown in human lung cancer cells resulted in apoptotic cell death.8 Therefore, how physiologically prominent EAPII-mediated-DNA repair activity is in cellular homeostasis in mammalian cells and how this activity modulates cell behavior during cancer development are critical questions for the study of EAPII. In addition, EAPII was shown to associate with exogenous pathogens that attack human DNA, including hantavirus (HTNV) nucleocapsid proteins (NPs),42 ΦC31 integrases24 and HIV-1 integrases.43 EAPII association with ΦC31 integrase inhibits the efficiency of phage C31 integrase-mediated site-specific recombination, but the association with lentivirus HIV-1 integrase results in an enhanced viral integration, suggesting that EAPII is indeed involved in the processes of these pathogenic attacks. The potential models could be the following: (1) the TDP activity of EAPII helps to resolve the topological catenanes produced by the integrase during the recombination; (2) EAPII associates with PML, Daxx, Sp100 and SUMO1, which are colocalized in the promyelocytic leukemia protein nuclear bodies (PML-NBs).44 PML-NBs are matrix-associated domains that have been hypothesized to control key pathways for growth suppression, apoptosis and host anti-viral defense through regulating posttranslational modifications of partner proteins like sumoylation, ubiquitination, phosphorylation or acetylation.45–50 The role of EAPII in the pathogenesis of these infections is warranted for further study.
Figure 4.
Model of error-free DNA repair function of EAPII: Transit DNA break (cleavable complex) is re-ligated and DNA topological entanglement is resolved (A→B); Stabilizing the transit cleavable complex by TopoII poison results in enzyme-mediated double strand breaks (A→C); TDP2 removes TopoII from protein-DNA adducts and leaves a free phosphate at 5′ of DNA, which is ready for ligation (C→D) and double strand DNA break is repaired (D→B). The solid yellow ovals represent topoisomerase II , and “∼” indicates ester bond. Phosphodiester bond links nucleotide of DNA.
Biological Significance of EAPII and Relevance for Human Diseases
Embryogenesis.
EAPII expression was found in multiple human fetal organs including the lung, brain and thymus.4 In mice, EAPII expression can be identified as early as E7 of embryonic development, and the level decreases along the course of fetal development.5 In situ hybridization of EAPII showed that EAPII mRNA appears ubiquitously at E12.5. This widespread expression is reduced at E15.5, but the signals from kidney, small intestine, testis, liver and lung remain strong; in particular, the highest expression levels are in the thymus and discrete brain regions,5 suggesting an important role of EAPII in embryonic development. In support of these observations, mouse blastocysts with homozygous deficiency of EAPII (EAPII-/-) can be identified, but EAPII-/- embryos die before 7.5 d post coitum (dpc) (our unpublished observation), demonstrating that EAPII is a critical gene, at least, for mouse embryogenesis. Consistent with this observation, it was also demonstrated that in zebrafish development, EAPII modulates Nodal signaling, which is critical for mesoderm formation, positioning and left-right asymmetric development of the heart and viscera.51 EAPII controls gastrulation movements through regulating expression of Snail1a and E-cadherin,19 both of which are epithelial-mesenchymal transition (EMT) markers involved in both developmental and cancer-related EMT.52 DNA repair activity of EAPII might be important for the maintenance of cellular homeostasis of embryos, and the involvement of EAPII in the regulation of cell migration makes this gene more critical for embryonic development.
Cancer.
Modulation of apoptotic induction. Evasion of apoptosis is one of the hallmarks of cancer.53 EAPII may protect cells from apoptosis or promote apoptosis in a cell-specific manner, and controversial observations have been obtained from different models. EAPII protects neuroblastoma cells from apoptosis induced by MG132, a proteasome inhibitor.33 EAPII knockdown results in apoptosis in H460, H522 and H1975 lung cancer cells.8 However, EAPII plays a pro-apoptotic role in some other cell lines. EAPII promotes apoptosis of HL-60 cells induced by hydroquinone, a cytotoxic agent.54 The mutant of DJ-1, an oncogene in cooperation with Ras,55 promotes apoptosis through JNK and p38 MAPK pathways in an EAPII-dependent manner.33 Furthermore, EAPII is upregulated in FOXO3a-induced apoptosis, in which FOXO3a turns TNF receptor signaling to a pro-apoptotic JNK-dependent pathway.56 The inconsistency of findings regarding the apoptosis regulation of EAPII may result from the complexity of the EAPII interacting-protein network and the EAPII-associated signaling pathways such as TNF and TGFβ, which involve either pro- and/or anti-apoptosis events,57–59 depending on the integrity of the cascade of specific pathways and the status of the protein(s) in the signaling cascade.60–62
Cancer development and progression. Although the involvement of EAPII in apoptosis was observed in cultured cells, its biological significance in cancer development remained unknown until recently. Immunohistochemical (IHC) studies showed that EAPII protein is significantly elevated in most non-small cell lung cancer (NSCLC) patients (90%) and that the expression level and localization of EAPII change along the course of lung cancer development: while the nuclear staining of EAPII can be seen in highly proliferative tissue, the majority of lung carcinomas showed cytoplasmic or cytoplasmic and nuclear staining.8 This finding suggests that EAPII may contribute to both early and advanced stages of lung cancer development. Indeed, an oncogenic role of EAPII expression was observed in a xenograft model of NSCLC in mice.8 This evidence supports our hypothesis that EAPII is an oncogenic factor for lung cancer development. EAPII expression is increased in Barrett's esophagus, a premalignant condition whereby the normal stratified squamous esophageal epithelium undergoes a transdifferentiation program resulting in a simple columnar epithelium reminiscent of the small intestine, and in esophageal adenocarcinoma compared with normal esophagus.63,64 EAPII is upregulated in cervical cancer and head and neck cancer patients with human papillomavirus (HPV) infection, which is a well-known pathogenic factor for these cancers.65 This evidence supports the hypothesis that EAPII contributes to the pathogenesis of cancer. On the other hand, it is noteworthy that EAPII reduction was observed in several types of cancer or precancerous conditions: primary lymphoma cells, primary cutaneous anaplastic large cell lymphoma and primary lymphoma cells of classical Hodgkin lymphoma showed a significant reduction of EAPII expression compared with that in non-neoplastic T- and NK-lymphocytes.66 EAPII is downregulated in precancerous adenoma, from which colorectal cancers are believed to predominantly arise, compared with normal mucosa from the same patients.67–69 Comparison of blood-derived gene-expression profiling indicated that the level of the EAPII transcript was lower in patients with xeroderma pigmentosum (XP)-like syndrome, in which multiple spinocellular carcinomas appear, than in healthy control individuals.70 It is known that loss of TDP1, which repairs Top1-mediated DNA damages, may contribute to genomic instability in cancer cells because of the transcription or replication-blocking cleavable complex formation.71,72 Therefore, it is plausible that DNA repair deficiency due to loss of EAPII disrupts DNA integrity and stability, subsequently resulting in susceptibility of cells to cancer. Altered EAPII expression was also observed in the late stage of cancers: remarkable decreases of EAPII expression were found in a mouse invasive colon cancer cell line (vs. non-invasive cell line) and in human metastatic prostate tumor (vs. the primary prostate tumor),73,74 consistent with our previous observations that expression of EAPII in prostate cancer cells inhibits cell migration.4 Significantly, EAPII may play sophisticated roles in a variety of cancers or cancer developmental stages although the gene arraybased data need to be experimentally validated in the future.
Chemoresistance. Topoisomerase II poisons are widely used in oncology,75 and tumor cells may be resistant to these DNA damages due to increased DNA repair.76–78 Emerging evidence supports that EAPII might be a significant factor involved in chemoresistance. Genetic deletion of EAPII in chicken DT40 cells results in specific and severe sensitivity to Top2 poison etoposide (VP-16), but not to Top1 poison camptothecin (CPT), while EAPII suppresses the sensitivity of yeast cells to CPT.6,41 In lung cancer A549 cells, which have an elevated EAPII expression, EAPII knockdown results in hypersensitivity to VP-16,6,41 implying that elevated EAPII levels in cancer cells render them chemoresistant, although a systematic related clinical study has not yet been conducted. Therefore, determining whether altered expression of EAPII could have an impact on the outcome of chemo- or radiation-therapy will have significant implications for cancer therapy.
Neuronal development (reading disability, RD) and Parkinson disease.
High EAPII expression in the developing mouse brain and fetal human brain indicates the functional role of EAPII in neuronal development. Reading disability (RD) or developmental dyslexia (DD) is a condition in which a sufferer displays difficulty reading resulting primarily from neurological factors.79 It was hypothesized that impaired neuronal migration is a cellular neurobiological antecedent to RD.80 Genetic linkage studies have identified Chromosome 6p23–21.3, where EAPII is located, as one of the potential loci related to RD.7,81–87 Within this locus, a 77-kilobase region spans the EAPII gene, portions of KIAA0319 and the upstream region of THEM2.88–91 Current results have converged on KIAA0319 as one of the likeliest RD susceptibility genes at this locus.92,93 Although a significant association of a haplotype spanning the EAPII gene with reading ability was observed,91 data are currently inconclusive as to whether EAPII could be another RD susceptibility gene. However, EAPII has shown to be related to the pathogenesis of Parkinson disease. EAPII associates with DJ-1, mutations in which have been demonstrated to cause autosomal recessive Parkinson disease.94 The mutant DJ-1 forms aggresome structures in the cytoplasm through recruiting EAPII, facilitating apoptosis of neurons.33 Since many neurodegenerative diseases are characterized by the accumulation of misfolded proteins that adversely affect neuronal connectivity and plasticity and trigger cell death signaling pathways,95–100 the appearance of EAPII in the aggresome suggests that EAPII may contribute to Parkinson disease through a similar mechanism.
Regulation of EAPII
EAPII expression.
Understanding the regulation of EAPII expression will reveal its biological significance and define how EAPII coordinates its diverse functions in physiological or pathological conditions. Although such studies are currently limited, array data from public databases allowed us to sketch the processes controlling EAPII regulation. In Table 2, we summarize the available data, albeit the data need to be further validated. It seems that EAPII regulation is controlled by many aspects: (1) transcriptional modulators such as Myb, HOXA9, DEAF1, MDM2 and histone deacetylases (HDAC8), which may directly or indirectly control the transcription of EAPII gene; (2) inflammatory or immune cytokines including IL-19, IL-24, IL-4, GM-CSF, TREM-1 and TRAIL. All of these factors increase EAPII expression, suggesting that EAPII is not only a component of the TNF-TNFR signaling cascade, as evidenced by EAPIIprotein interacting networks, but also an important responsive gene for immune/inflammatory stimuli. Interestingly, significant downregulation of EAPII expression by bacterial lipopolysaccharide (LPS) was found in mammalian (mouse) cells101–103 and invertebrate (mollusc) cells.37 LPS is one of the most important inflammatory mediators, which activates a potential immune response through Toll-like receptor 4 (TLR4)-TRAF6-p38/JNK/NFκB pathway.104 Significantly, the inhibition of EAPII expression is important for the activation of key inflammatory pathways such as NFκB. Therefore, a possible feedback loop mediated by EAPII could be a key switch that coordinates immune defense and inflammatory responses. It is noteworthy that EAPII expression can be downregulated by PCDH24 and SERPINA1. The former is related to cell contact inhibition,105 and the latter is a serine protease inhibitor which represses protease enzymes such as elastase, plasmin, thrombin and plasminogen activator.106 The substrates of these enzymes are part of the extracellular matrix, implying another potential link of the tumor microenvironment to the function of EAPII.
Table 2.
Regulation of EAPII expression
| Modulator | Function | EAPII changes | System/Models | References |
| knockdown HDAC8 (Histone deacetylases) | Transcription Modulator | ↓ | prostate cancer cells | 107 |
| FOXO3a | Transcription Modulator | ↑ | Apoptosis | 56 |
| HOXA9 knockdown | Transcription Modulator | ↓ | human AML cells | 108 |
| Myb | Transcription Modulator | ↑ | human monocytes | 109, 110 |
| Knockout DEAF1 | Transcription Modulator | ↑ | knockout mice | 111 |
| Knockdown MDM2 | Transcription Modulator | ↑ | cancer cell lines: A549, TOV-21G | 112 |
| Chronic antigen stimulation | ↑ | Regulatory T cell | 113 | |
| TRAIL | TNF-TNFR signaling | ↑ | T42 breast cancer cells | |
| IL-19 | Inflammatory response | 114 | ||
| IL-24 | ↑ | |||
| TRE M-1 activation | inflammatory responses | ↑ | 115 | |
| IL-4, GM-CSF | T cell differentiation/inflammatory response | ↑ | Monocyte to DC differentiation | 116 |
| LPS | Inflammatory mediator | ↓ | ATF3-/- mouse wild-type mouse | 101–103 |
| Insulin | Glucose metabolism | ↓ | 117 | |
| Knockout SIRT6 | glucose metabolism/histone deacetylase | ↓ | liver of SIR T6 knockout mice | 118 |
| Protocadherin LKC (PCDH24) | Contact inhibition | ↓ | Colon cancer cells | Na, 105 |
| α1antitrypsin (SERPINA1) | Serine protease inhibitor | ↓ | Transgenic mice | 119 |
| Knockdown UPF1 | eukaryotic surveillance | ↑ | 120 | |
| IFNγ | eukaryotic surveillance | ↑, NB-localized | 44 |
Data presented in this table are derived from NextBio (www.nextbio.com), which is based on multiple sources including NCBI GEO and EBI ArrayExpress. Original articles are reviewed and cited in the references if the array data have been published.
Nucleus-cytoplasm translocation.
In agreement with the function of EAPII in signaling transduction, the cytoplasmic translocation of EAPII is observed under certain circumstances. Overexpressed EAPII was found in the nucleus,4 but endogenous EAPII is expressed in both the nucleus and the cytoplasm: the elevated nuclear staining of EAPII occurs in the highly proliferative bronchial epithelium, and the cytoplasmic staining of EAPII can be observed in many NSCLC tissues.8 Therefore, it is possible that an elevated nuclear expression of EAPII favors enhanced proliferative activity in both normal bronchial epithelium and tumor cells, and the cytoplasmic EAPII may be more closely related to its oncogenic role, suggesting that EAPII plays different roles in the cytoplasm and nucleus. Upon proteasome impairment, EAPII relocalizes to the cytoplasm of neuroblastoma cells and forms aggresome-like structures.33 Furthermore, accumulation of EGFP-EAPII can be found at sites of laser-induced DNA damage,6 and occasionally, EAPII also appears in PML-NBs.44 These observations indicate that the compartmentalization of EAPII protein may determine its specific targets and unique function, and the regulation of nucleus-cytoplasm translocation is critical for the modulation of EAPII function. In many cases, EAPII interacts with both receptor and endeffector, but it is unknown whether EAPII alone or together with its partner(s) shuttles around in the cytosol or between the cytoplasm and nucleus. Further study of how EAPII localization is regulated and how its localization is correlated with specific functions of EAPII would be helpful in understanding its pleiotropic role. In addition, the interactions of EAPII with UBE2I (ubiquitin-conjugating enzyme E2I) and SUMO1 (SMT3 suppressor of mif two 3 homolog 1) 42 suggested that EAPII is potentially SUMO-modified although there is as yet no experimental evidence for this.
Conclusions and Further Directions
Evidence strongly supports that EAPII is a multi-functional protein that plays important roles in many aspects of cell biology, including topological entanglement of DNA catenanes, cellular responsiveness to microenvironments, cell survival and cell migration. These functions are essential for either embryonic or neuronal development or involve many aspects of cancer development, including chromosome instability, tumorigenesis, tumor metastasis and chemoresistance (Fig. 5). Particularly, EAPIImediated signals, including immune defense, inflammatory response and cellular stress such as hypoxia and DNA toxicity, are key players in the orchestration of the tumor microenvironment. The involvement of EAPII in multiple steps of cancer development and chemoresistance implicates it as a potential target for cancer intervention. In future studies, determination of EAPII-mediated signaling pathway(s) and its mechanism is critical for understanding the role of EAPII in cancer development. Other important open questions in the study of EAPII include: (1) Does EAPII have other types of substrate besides DNA or protein-DNA in mammalian cells, and is this related to EAPII's function in signal transduction? The phosphoester bond can be found in nucleic acid (DNA and RNA), protein-DNA, phosphoprotein, protein phosphoglycosylation, phospholipid and second message cAMP and cGMP. (2) How are the signal transduction, transcription and DNA repair aspects of EAPII functions related? Theoretically, the 5′TDP activity of EAPII may generally enhance transcriptional activity by facilitating the opening of the DNA template at the transcription bubble. However, EAPII appears to have specific transcriptional repression activity depending on its interacting partners. (3) Does EAPII play differential functions in embryonic development and cancer progression? How is EAPII reactivated during tumorigenesis and cancer progression? (4) How does the promiscuous protein association of EAPII transduce signals and then change the fate of cancer cells and how is EAPII activity regulated by the tumor microenvironment?
Figure 5.
EAPII-related functions.
Acknowledgments
We thank Dr. Anthea Hammond for assistance in editing and Dr. Arun Seth for critical reading of the manuscript. This work was supported in part by National Institutes of Health grants K22CA109577 (R.L.), Kennedy award from the Winship Cancer Institute (R.L.), DRP award of NCI P50 CA128613 (R.L.) and a start-up fund from the Department of Hematology and Medical Oncology, Emory University (R.L.).
References
- 1.Hahne JC, Okuducu AF, Sahin A, Fafeur V, Kiriakidis S, Wernert N. The transcription factor ETS-1: its role in tumour development and strategies for its inhibition. Mini Rev Med Chem. 2008;8:1095–1105. doi: 10.2174/138955708785909934. [DOI] [PubMed] [Google Scholar]
- 2.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Pei H, Li C, Adereth Y, Hsu T, Watson DK, Li R. Caspase-1 is a direct target gene of ETS1 and plays a role in ETS1-induced apoptosis. Cancer Res. 2005;65:7205–7213. doi: 10.1158/0008-5472.CAN-04-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK, Li R. EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene. 2003;22:2699–2709. doi: 10.1038/sj.onc.1206374. [DOI] [PubMed] [Google Scholar]
- 5.Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JG, Dewulf N, et al. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappaB activation. J Biol Chem. 2000;275:18586–18593. doi: 10.1074/jbc.M000531200. [DOI] [PubMed] [Google Scholar]
- 6.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 7.Londin ER, Meng H, Gruen JR. A transcription map of the 6p22.3 reading disability locus identifying candidate genes. BMC Genomics. 2003;4:25. doi: 10.1186/1471-2164-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, Fan S, Owonikoko TK, Khuri FR, Sun SY, Li R. Oncogenic role of EAPII in lung cancer development and its activation of the MAPK-ERK pathway. Oncogene. 2011;30:3802–3812. doi: 10.1038/onc.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodrigues-Lima F, Josephs M, Katan M, Cassinat B. Sequence analysis identifies TTRAP, a protein that associates with CD40 and TNF receptor-associated factors, as a member of a superfamily of divalent cation-dependent phosphodiesterases. Biochem Biophys Res Commun. 2001;285:1274–1279. doi: 10.1006/bbrc.2001.5328. [DOI] [PubMed] [Google Scholar]
- 10.Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsujishita Y, Guo S, Stolz LE, York JD, Hurley JH. Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate-5-phosphatase. Cell. 2001;105:379–389. doi: 10.1016/S0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- 14.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 15.Whisstock JC, Romero S, Gurung R, Nandurkar H, Ooms LM, Bottomley SP, et al. The inositol polyphosphate-5-phosphatases and the apurinic/apyrimidinic base excision repair endonucleases share a common mechanism for catalysis. J Biol Chem. 2000;275:37055–37061. doi: 10.1074/jbc.M006244200. [DOI] [PubMed] [Google Scholar]
- 16.Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 17.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 18.Grabbe C, Dikic I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem Rev. 2009;109:1481–1494. doi: 10.1021/cr800413p. [DOI] [PubMed] [Google Scholar]
- 19.Esguerra CV, Nelles L, Vermeire L, Ibrahimi A, Crawford AD, Derua R, et al. Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development. 2007;134:4381–4393. doi: 10.1242/dev.000026. [DOI] [PubMed] [Google Scholar]
- 20.Turner DP, Findlay VJ, Moussa O, Watson DK. Defining ETS transcription regulatory networks and their contribution to breast cancer progression. J Cell Biochem. 2007;102:549–559. doi: 10.1002/jcb.21494. [DOI] [PubMed] [Google Scholar]
- 21.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGFbeta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 22.Kaminska B, Wesolowska A, Danilkiewicz M. TGFbeta signalling and its role in tumour pathogenesis. Acta Biochim Pol. 2005;52:329–337. [PubMed] [Google Scholar]
- 23.Xu J, Lamouille S, Derynck R. TGFbeta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang BY, Xu GL, Zhou CH, Tian L, Xue JL, Chen JZ, et al. PhiC31 integrase interacts with TTRAP and inhibits NFkappaB activation. Mol Biol Rep. 2010;37:2809–2816. doi: 10.1007/s11033-009-9829-3. [DOI] [PubMed] [Google Scholar]
- 25.Li R, Pei H, Watson DK. Regulation of Ets function by protein-protein interactions. Oncogene. 2000;19:6514–6523. doi: 10.1038/sj.onc.1204035. [DOI] [PubMed] [Google Scholar]
- 26.Babayeva ND, Wilder PJ, Shiina M, Mino K, Desler M, Ogata K, et al. Structural basis of Ets1 cooperative binding to palindromic sequences on stromelysin-1 promoter DNA. Cell Cycle. 2010;9:3054–3062. doi: 10.4161/cc.9.14.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R, Pei H, Watson DK, Papas TS. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000;19:745–753. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Zhang P, Liu Y, Cheng G. TRAF-mediated regulation of immune and inflammatory responses. Sci China Life Sci. 2010;53:159–168. doi: 10.1007/s11427-010-0050-3. [DOI] [PubMed] [Google Scholar]
- 29.Lee NK, Lee SY. Modulation of life and death by the tumor necrosis factor receptor-associated factors (TRAFs) J Biochem Mol Biol. 2002;35:61–66. doi: 10.5483/BMBRep.2002.35.1.061. [DOI] [PubMed] [Google Scholar]
- 30.Bradley JR, Pober JS. Tumor necrosis factor receptorassociated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 31.Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Stiegler AL, Boggon TJ, Kobayashi S, Halmos B. EGFR-mutated lung cancer: a paradigm of molecular oncology. Oncotarget. 2010;1:497–514. doi: 10.18632/oncotarget.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zucchelli S, Vilotti S, Calligaris R, Lavina ZS, Biagioli M, Foti R, et al. Aggresome-forming TTRAP mediates pro-apoptotic properties of Parkinson's disease-associated DJ-1 missense mutations. Cell Death Differ. 2009;16:428–438. doi: 10.1038/cdd.2008.169. [DOI] [PubMed] [Google Scholar]
- 34.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci USA. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Qiu L, Wang L, Huang M, Zhang H, Song LA. TRAF and TNF receptor-associated protein (TTRAP) in mollusk with endonuclease activity. Dev Comp Immunol. 2011;35:827–834. doi: 10.1016/j.dci.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Sabourin M, Nitiss JL, Nitiss KC, Tatebayashi K, Ikeda H, Osheroff N. Yeast recombination pathways triggered by topoisomerase II-mediated DNA breaks. Nucleic Acids Res. 2003;31:4373–4384. doi: 10.1093/nar/gkg497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connelly JC, Leach DR. Repair of DNA covalently linked to protein. Mol Cell. 2004;13:307–316. doi: 10.1016/S1097-2765(04)00056-5. [DOI] [PubMed] [Google Scholar]
- 40.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Z, Cortes-Ledesma F, ElKhamisy SF, Caldecott KW. TDP2/TTRAP is the major 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J Biol Chem. 2011;286:403–409. doi: 10.1074/jbc.M110.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee BH, Yoshimatsu K, Maeda A, Ochiai K, Morimatsu M, Araki K, et al. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 2003;98:83–91. doi: 10.1016/j.virusres.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Zhang JQ, Wang JJ, Li WJ, Huang L, Tian L, Xue JL, et al. Cellular protein TTRAP interacts with HIV-1 integrase to facilitate viral integration. Biochem Biophys Res Commun. 2009;387:256–260. doi: 10.1016/j.bbrc.2009.06.153. [DOI] [PubMed] [Google Scholar]
- 44.Xu GL, Pan YK, Wang BY, Huang L, Tian L, Xue JL, et al. TTRAP is a novel PML nuclear bodies-associated protein. Biochem Biophys Res Commun. 2008;375:395–398. doi: 10.1016/j.bbrc.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 45.Van Damme E, Laukens K, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci. 2010;6:51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saffert RT, Kalejta RF. Promyelocytic leukemia-nuclear body proteins: herpesvirus enemies, accomplices or both? Future Virol. 2008;3:265–277. doi: 10.2217/17460794.3.3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salomoni P. Stemming out of a new PML era? Cell Death Differ. 2009;16:1083–1092. doi: 10.1038/cdd.2009.63. [DOI] [PubMed] [Google Scholar]
- 49.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Regad T, Saib A, Lallemand-Breitenbach V, Pandolfi PP, de The H, Chelbi-Alix MK. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 2001;20:3495–3505. doi: 10.1093/emboj/20.13.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:3459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Micalizzi DS, Farabaugh SM, Ford HL. Epithelialmesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 54.Zhang ZB, Hu J, Bi YY, Zhao ZW, Tao N, Yan H, et al. Effect of TTRAP expression on apoptosis induced by hydroquinone in HL-60 cells in vitro. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2007;25:654–656. [PubMed] [Google Scholar]
- 55.Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, Iguchi-Ariga SM, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 56.Lee HY, Youn SW, Kim JY, Park KW, Hwang CI, Park WY, et al. FOXO3a turns the tumor necrosis factor receptor signaling towards apoptosis through reciprocal regulation of c-Jun N-terminal kinase and NFkappaB. Arterioscler Thromb Vasc Biol. 2008;28:112–120. doi: 10.1161/ATVBAHA.107.153304. [DOI] [PubMed] [Google Scholar]
- 57.Bachman KE, Park BH. Duel nature of TGFbeta signaling: tumor suppressor vs. tumor promoter. Curr Opin Oncol. 2005;17:49–54. doi: 10.1097/01.cco.0000143682.45316.ae. [DOI] [PubMed] [Google Scholar]
- 58.Tian M, Schiemann WP. The TGFbeta paradox in human cancer: an update. Future Oncol. 2009;5:259–271. doi: 10.2217/14796694.5.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes? Acta Pharmacol Sin. 2008;29:1275–1288. doi: 10.1111/j.1745-7254.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Donnell MA, Ting AT. Chronicles of a death foretold: dual sequential cell death checkpoints in TNF signaling. Cell Cycle. 2010;9:1065–1071. doi: 10.4161/cc.9.6.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirley S, Micheau O. The heme oxygenase-1 and c-FLIP in acute myeloid leukemias: two non-redundant but mutually exclusive cellular safeguards protecting cells against TNF-induced cell death? Oncotarget. 2010;1:317–319. doi: 10.18632/oncotarget.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rushworth SA, Zaitseva L, Langa S, Bowles KM, MacEwan DJ. FLIP regulation of HO-1 and TNF signalling in human acute myeloid leukemia provides a unique secondary anti-apoptotic mechanism. Oncotarget. 2010;1:359–366. doi: 10.18632/oncotarget.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stairs DB, Nakagawa H, Klein-Szanto A, Mitchell SD, Silberg DG, Tobias JW, et al. Cdx1 and c-Myc foster the initiation of transdifferentiation of the normal esophageal squamous epithelium toward Barrett's esophagus. PLoS ONE. 2008;3:3534. doi: 10.1371/journal.pone.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao Y, Triadafilopoulos G, Sahbaie P, Young HS, Omary MB, Lowe AW. Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology. 2006;131:925–933. doi: 10.1053/j.gastro.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eckerle S, Brune V, Doring C, Tiacci E, Bohle V, Sundstrom C, et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23:2129–2138. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]
- 67.Gyorffy B, Molnar B, Lage H, Szallasi Z, Eklund AC. Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS ONE. 2009;4:5645. doi: 10.1371/journal.pone.0005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galamb O, Spisak S, Sipos F, Toth K, Solymosi N, Wichmann B, et al. Reversal of gene expression changes in the colorectal normal-adenoma pathway by NS398 selective COX2 inhibitor. Br J Cancer. 2010;102:765–773. doi: 10.1038/sj.bjc.6605515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 70.Vahteristo P, Kokko A, Saksela O, Aittomaki K, Aaltonen LA. Blood-derived gene-expression profiling in unravelling susceptibility to recessive disease. J Med Genet. 2007;44:718–720. doi: 10.1136/jmg.2007.051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tuduri S, Crabbe L, Tourriere H, Coquelle A, Pasero P. Does interference between replication and transcription contribute to genomic instability in cancer cells? Cell Cycle. 2010;9:1886–1892. doi: 10.4161/cc.9.10.11539. [DOI] [PubMed] [Google Scholar]
- 72.Sordet O, Nakamura AJ, Redon CE, Pommier Y. DNA double-strand breaks and ATM activation by transcription-blocking DNA lesions. Cell Cycle. 2010;9:274–278. doi: 10.4161/cc.9.2.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosell R, Lord RV, Taron M, Reguart N. DNA repair and cisplatin resistance in non-small-cell lung cancer. Lung Cancer. 2002;38:217–227. doi: 10.1016/S0169-5002(02)00224-6. [DOI] [PubMed] [Google Scholar]
- 77.Rünger TM, Emmert S, Schadendorf D, Diem C, Epe B, Hellfritsch D. Alterations of DNA repair in melanoma cell lines resistant to cisplatin, fotemustine or etoposide. J Invest Dermatol. 2000;114:34–39. doi: 10.1046/j.1523-747.2000.00844.x. [DOI] [PubMed] [Google Scholar]
- 78.Hawtin RE, Stockett DE, Wong OK, Lundin C, Helleday T, Fox JA. Homologous recombination repair is essential for repair of vosaroxin-induced DNA double-strand breaks. Oncotarget. 2010;1:606–619. doi: 10.18632/oncotarget.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCandliss BD, Noble KG. The development of reading impairment: a cognitive neuroscience model. Ment Retard Dev Disabil Res Rev. 2003;9:196–204. doi: 10.1002/mrdd.10080. [DOI] [PubMed] [Google Scholar]
- 80.Gabel LA, Gibson CJ, Gruen JR, LoTurco JJ. Progress towards a cellular neurobiology of reading disability. Neurobiol Dis. 2010;38:173–180. doi: 10.1016/j.nbd.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaplan DE, Gayan J, Ahn J, Won TW, Pauls D, Olson RK, et al. Evidence for linkage and association with reading disability on 6p21.3–22. Am J Hum Genet. 2002;70:1287–1298. doi: 10.1086/340449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gayán J, Olson RK. Reading disability: evidence for a genetic etiology. Eur Child Adolesc Psychiatry. 1999;8:52–55. doi: 10.1007/PL00010695. [DOI] [PubMed] [Google Scholar]
- 83.Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability on chromosome 6. Science. 1994;266:276–279. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- 84.Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, et al. Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet. 1997;60:27–39. [PMC free article] [PubMed] [Google Scholar]
- 85.Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, et al. A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet. 1999;64:146–156. doi: 10.1086/302190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gayán J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, et al. Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet. 1999;64:157–164. doi: 10.1086/302191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grigorenko EL, Wood FB, Meyer MS, Pauls DL. Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. Am J Hum Genet. 2000;66:715–723. doi: 10.1086/302755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, et al. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet. 2004;75:1046–1058. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, et al. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet. 2004;115:128–138. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- 90.Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–591. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luciano M, Lind PA, Duffy DL, Castles A, Wright MJ, Montgomery GW, et al. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry. 2007;62:811–817. doi: 10.1016/j.biopsych.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 92.Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 93.Harold D, Paracchini S, Scerri T, Dennis M, Cope N, Hill G, et al. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol Psychiatry. 2006;11:1085–1091. doi: 10.1038/sj.mp.4001904. [DOI] [PubMed] [Google Scholar]
- 94.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 95.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 96.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 97.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 98.Nakamura T, Lipton SA. Cell death: protein misfolding and neurodegenerative diseases. Apoptosis. 2009;14:455–468. doi: 10.1007/s10495-008-0301-y. [DOI] [PubMed] [Google Scholar]
- 99.Gregersen N, Bolund L, Bross P. Protein misfolding, aggregation and degradation in disease. Mol Biotechnol. 2005;31:141–150. doi: 10.1385/MB:31:2:141. [DOI] [PubMed] [Google Scholar]
- 100.Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 102.Ramsey SA, Klemm SL, Zak DE, Kennedy KA, Thorsson V, Li B, et al. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLOS Comput Biol. 2008;4:1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carlson BA, Yoo MH, Sano Y, Sengupta A, Kim JY, Irons R, et al. Selenoproteins regulate macrophage invasiveness and extracellular matrix-related gene expression. BMC Immunol. 2009;10:57. doi: 10.1186/1471-2172-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 105.Ose R, Yanagawa T, Ikeda S, Ohara O, Koga H. PCDH24-induced contact inhibition involves downregulation of beta-catenin signaling. Mol Oncol. 2009;3:54–66. doi: 10.1016/j.molonc.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ekeowa UI, Marciniak SJ, Lomas DA. alpha(1)-antitrypsin deficiency and inflammation. Expert Rev Clin Immunol. 2011;7:243–252. doi: 10.1586/eci.10.95. [DOI] [PubMed] [Google Scholar]
- 107.Welsbie DS, Xu J, Chen Y, Borsu L, Scher HI, Rosen N, et al. Histone deacetylases are required for androgen receptor function in hormone-sensitive and castrate-resistant prostate cancer. Cancer Res. 2009;69:958–966. doi: 10.1158/0008-5472.CAN-08-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu F, Lei W, O'Rourke JP, Ness SA. Oncogenic mutations cause dramatic, qualitative changes in the transcriptional activity of c-Myb. Oncogene. 2006;25:795–805. doi: 10.1038/sj.onc.1209105. [DOI] [PubMed] [Google Scholar]
- 110.Lei W, Rushton JJ, Davis LM, Liu F, Ness SA. Positive and negative determinants of target gene specificity in myb transcription factors. J Biol Chem. 2004;279:29519–29527. doi: 10.1074/jbc.M403133200. [DOI] [PubMed] [Google Scholar]
- 111.Yip L, Su L, Sheng D, Chang P, Atkinson M, Czesak M, et al. Deaf1 isoforms control the expression of genes encoding peripheral tissue antigens in the pancreatic lymph nodes during type 1 diabetes. Nat Immunol. 2009;10:1026–1033. doi: 10.1038/ni.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chau BN, Diaz RL, Saunders MA, Cheng C, Chang AN, Warrener P, et al. Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Res. 2009;69:1368–1374. doi: 10.1158/0008-5472.CAN-08-2742. [DOI] [PubMed] [Google Scholar]
- 113.Hansen W, Westendorf AM, Reinwald S, Bruder D, Deppenmeier S, Groebe L, et al. Chronic antigen stimulation in vivo induces a distinct population of antigen-specific Foxp3 CD25 regulatory T cells. J Immunol. 2007;179:8059–8068. doi: 10.4049/jimmunol.179.12.8059. [DOI] [PubMed] [Google Scholar]
- 114.Sa SM, Valdez PA, Wu J, Jung K, Zhong F, Hall L, et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- 115.Dower K, Ellis DK, Saraf K, Jelinsky SA, Lin LL. Innate immune responses to TREM-1 activation: overlap, divergence and positive and negative cross-talk with bacterial lipopolysaccharide. J Immunol. 2008;180:3520–3534. doi: 10.4049/jimmunol.180.5.3520. [DOI] [PubMed] [Google Scholar]
- 116.Szatmari I, Torocsik D, Agostini M, Nagy T, Gurnell M, Barta E, et al. PPARgamma regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood. 2007;110:3271–3280. doi: 10.1182/blood-2007-06-096222. [DOI] [PubMed] [Google Scholar]
- 117.Rome S, Clement K, Rabasa-Lhoret R, Loizon E, Poitou C, Barsh GS, et al. Microarray profiling of human skeletal muscle reveals that insulin regulates approximately 800 genes during a hyperin-sulinemic clamp. J Biol Chem. 2003;278:18063–18068. doi: 10.1074/jbc.M300293200. [DOI] [PubMed] [Google Scholar]
- 118.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NFkappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hidvegi T, Mirnics K, Hale P, Ewing M, Beckett C, Perlmutter DH. Regulator of G Signaling 16 is a marker for the distinct endoplasmic reticulum stress state associated with aggregated mutant alpha1-antitrypsin Z in the classical form of alpha1-antitrypsin deficiency. J Biol Chem. 2007;282:27769–27780. doi: 10.1074/jbc.M704330200. [DOI] [PubMed] [Google Scholar]
- 120.Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res. 2007;35:4542–4551. doi: 10.1093/nar/gkm461. [DOI] [PMC free article] [PubMed] [Google Scholar]