Abstract
Allatotropin is an insect neuropeptide with pleiotropic actions on a variety of different tissues. In the present work we describe the identification, cloning and functional and molecular characterization of an Aedes aegypti allatotropin receptor (AeATr) and provide a detailed quantitative study of the expression of the AeATr gene in the adult mosquito. Analysis of the tissue distribution of AeATr mRNA in adult female revealed high transcript levels in the nervous system (brain, abdominal, thoracic and ventral ganglia), corpora allata-corpora cardiaca complex and ovary. The receptor is also expressed in heart, hindgut and male testis and accessory glands. Separation of the corpora allata (CA) and corpora cardiaca followed by analysis of gene expression in the isolated glands revealed expression of the AeATr primarily in the CA. In the female CA, the AeATr mRNA levels were low in the early pupae, started increasing 6 hours before adult eclosion and reached a maximum 24 hours after female emergence. Blood feeding resulted in a decrease in transcript levels. The pattern of changes of AeATr mRNA resembles the changes in JH biosynthesis. Fluorometric Imaging Plate Reader recordings of calcium transients in HEK293 cells expressing the AeATr showed a selective response to A. aegypti allatotropin stimulation in the low nanomolar concentration range. Our studies suggest that the AeATr play a role in the regulation of JH synthesis in mosquitoes.
Keywords: Mosquito, allatrotopin, receptor, GPCR, juvenile hormone, Aedes
1. INTRODUCTION
Allatotropin (AT) was first identified as a 13-residue amidated peptide that stimulated in vitro JH synthesis in the corpora allata (CA) of the adult moth Manduca sexta [21]. In addition to stimulating JH biosynthesis, AT displays multifunctional roles in different insect species; including inhibition of ion transport in the midgut [25], stimulation of foregut contractions [9, 10] and acceleration of heart rate [22, 46, 54]. AT also plays a role in circuits relaying photic information from circadian photoreceptors to the central pacemaker in the cockroach Leucophea maderae [41]. Aedes aegypti allatotropin, first isolated and characterized by Veenstra and Costes [53], was shown later to stimulate JH synthesis on the mosquito CA [29].
The first insect AT receptor was described for the silkworm moth Bombyx mori (BmATr) [58]. The BmAT receptor is a member of the family of G-Protein-Coupled Receptors (GPCRs) and an orthologue to the vertebrate orexin/hypocretin receptors. BmATr is expressed in the corpora cardiaca (CC), not in the CA, and it was suggested that AT stimulates JH synthesis by turning off a short neuropeptide F-mediated (sNPF) inhibition of the CA [58]. Recently, AT receptors were also described for M. sexta [20] and Tribolium castaneum [56], but neither their expression in the CC-CA nor their role on JH synthesis were analyzed in detail.
In the present work we describe the identification and functional and molecular characterization of an A. aegypti AT receptor (AeATr) that is not expressed in the CC, but on the contrary it is preferentially expressed in the CA of the female A. aegypti mosquito. The receptor showed a selective response to A. aegypti allatotropin stimulation in the nanomolar concentration range. The pattern of changes of AeATr mRNA in the CA resembled the changes in JH biosynthesis. Our studies suggest that the AeATr might play a role in the regulation of JH synthesis in mosquitoes.
2. MATERIAL & METHODS
2.1. Insects
Aedes aegypti of the Rockefeller strain were reared at 28 °C and 80% relative humidity under a photoperiod of 16 h light: 8 h dark. Mated adults were offered a cotton pad soaked in 3% sucrose solution. The cotton pad sucrose-fed adults are referred to as sugar fed. Four-day-old female mosquitoes were fed porcine blood equilibrated to 37 °C. Adenosine triphosphate was added to the blood meal to a final concentration of 1 mM immediately before use [38].
2.2. Identification of the AT receptor
The A. aegypti allatotropin receptor (AeATr) was identified using a bioinformatic approach based on the expression of the A. aegypti GPCR orphan receptors in the CA of adult female mosquito. The outcomes of these studies are described in detail in the results section.
2.3. RNA extraction and molecular cloning
Mosquito tissues were dissected in a drop of sterile DNA-RNAse free phosphate buffered saline (PBS). Total RNA was isolated using RNA-binding glass powder as previously described [36]. Contaminating genomic DNA was removed using the DNA-free™ kit (Ambion, Austin, TX). First strand cDNA synthesis was carried out using SuperScript® III first strand synthesis system for reverse transcription-PCR (Invitrogen, Carlsbad, CA). Full 5′ and 3′ ends of the mRNA were obtained by RACE using the GeneRacer kit (Invitrogen). PCR was performed in a Mastercycler gradient (Eppendorf, Westerbury, NY) using Taq DNA polymerase (Promega, Madison, WI). PCR products were cloned in pCR®2.1-TOPO and sequenced by the DNA Core Sequence Facility at Florida International University (Miami, FL).
The amino acid sequence of the A. aegypti allatotropin receptor presented three putative translation start codons named AeATrM1, AeATrM2 and AeATrM3 (Fig. 1). Based on these sequences, primers were designed to amplify the full lengths of the three putative start codons. For functional experiments, the AeATrM1, AeATrM2 and AeATrM3 were cloned into the plasmid pcDNA5/FRT (Invitrogen). For immunocytochemical localization experiments we fused our target receptors with the Yellow Fluorescent Protein (YFP) using the vector pEYFP-N1 (BD Biosciences Clontech, San Jose, CA).
Fig. 1. AeATr amino acid sequence.
Deduced amino acid sequence of the AeATr. The seven transmembrane domains (TMs) are indicated by boxes. The three putative starting methionines (M) (ATtrM3, ATrM2 and ATrM1) are in bold and underlined. The orexin receptor signature (YANSCANPI/VLY) located in the 7TM domain is shown as italic and bold. Putative N-linked putative glycosylation sites relevant for ATrM1 functionality are shown in bold and indicated by a g.
2.4. Real Time-PCR
RNA samples were treated with rDNAseI using DNA-free™ kit (Ambion, Austin, TX) according to manufacturer’s recommendations. Reverse transcription was carried out using the SuperScript® III first strand synthesis kit. Real-time PCR was performed in a 7300 Real Time PCR System (Applied Biosystems, Foster City, CA) using TaqMan® Gene Expression Assays together with TaqMan® Universal PCR Master Mix (Applied Biosystems). The primers and probes for the housekeeping gene 60S ribosomal protein rpL32, the AeATr, adipokinetic hormone (AKH), crustacean cardioacceleratory peptide (CCAP), juvenile hormone acid methyltransferase (JHAMT) and epoxidase (EPOX) genes are included in the Supplementary table 1. Primer/probes were synthesized by Applied Biosystems and reactions were carried out in 20 μl volume according to the manufacturer’s recommendations for Custom TaqMan® gene expression assays. Reactions were run in triplicate using 1–4 μl of cDNA per reaction. Standard curves to quantify relative gene copy number were made from ten fold serial dilutions of plasmids containing rpL32 or the gene of interest (from 300,000 to 30 copies of a plasmid per reaction). Real-time data were collected by 7300 System SDS Software and analyzed in Microsoft Excel. Transcript levels were normalized with rpL32 mRNA levels in the same sample. Relative transcript levels are expressed as a number of copies of transcript per 10,000 copies of rpL32.
2.5. Functional expression of the AT receptors
For this purpose we used a HEK293 cell line stably expressing Gα16gust44. Gα16gust44 is a chimeric G protein that couples various Gαi-type G protein-preferring GPCRs to phospholipase C activity, phosphatidyl inositol breakdown, and elevation of cytosolic calcium [4, 6, 33, 40, 47]. Cells were seeded into 96-well black-wall, clear bottom microtiter plates (Greiner Bio One, Frickenhausen, Germany) and grown for 24 h to about 80% confluence. Then the cells were transiently transfected with the three AeATr encoding plasmids (M1, M2 and M3) using Lipofectamine 2000 (Invitrogen). After an additional 24 h, the cells were loaded with the calcium-sensitive dye Fluo4-AM (Invitrogen) and incubated for 1 h at 37 °C. Cells were then washed three times with buffer (130 mM NaCl, 5mM KCl, 10 mM HEPES, 2 mM CaCl2, 10 mM glucose, pH 7.4) and stimulated with the appropriate ligands in a Fluorometric Imaging Plate Reader (FLIPR) (Molecular Devices, Sunnyvale, CA). Calcium-dependent increase of Fluo4-fluorescence was recorded at 1 Hz and 510 nm simultaneously from each well after excitation at 488 nm [5]. Responses of three wells containing cells expressing the same receptor and receiving the same stimulus were averaged. For dose-response curve calculation, the changes in fluorescence after the ligand was added were corrected for fluorescence changes in mock-transfected cells (transfected with a plasmid without receptor DNA) and normalized to background fluorescence (ΔF/F = (F−F0)/F0). Our calculations were based on at least three independent transfection experiments
2.6. Test solutions
Custom made peptides were provided by Alpha Diagnostics International (San Antonio, Texas), purified by reverse phase liquid chromatography and assessed to be ≥ 99% pure by analytical reversed phase liquid chromatography, Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) and amino acid analysis. Stock solutions of A. aegypti allatotropin (APFRNSEMMTARGF) [29] and A. aegypti allatostatin-C (AS-C) were prepared at a concentration of 1 mM and stored in aliquots at −80 °C. For each assay, a new aliquot was used. Stock solutions of linear synthetic AS-C were oxidized overnight shaking at 4 °C [23]. Assay concentrations were ranging from 3 μM to 30 pM.
2.7. Immunocytochemistry
HEK293/Gα16gust44 cells were seeded on coverslips coated with poly-D-lysine and, after 24 h, transfected with constructs for AeATrM1-pEYFP-N1, AeATrM2-pEYFP-N1 or AeATrM3-pEYFP-N1 using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were washed with PBS, cooled on ice and incubated for 1 h on ice with 5 μg/mL biotin-labeled concanavalin A (Sigma). After washing with PBS five times, cells were fixed for 2 min with methanol:acetone (1:1). To reduce non-specific binding, the coverslips were incubated in 3% goat serum. We added streptavidin conjugated with Alexa Fluor 633 (1:1000) and incubated in darkness for 1 h at RT. After washing 3 times with PBS and once with double distilled water, cells were mounted using Fluorescent Mounting Medium (DakoCytomation, CA). Cells were analyzed using a Leica TCS SP2 Laser Scan Inverted microscope.
2.8. Phylogenetic analysis
Allatotropin receptor sequences were obtained from databases and used for the alignments and phylogenetic analysis. We aligned the allatotropin receptor sequences using ClustalW [17]. A neighbor-joining tree was built using MEGA software version 5 [50] with a bootstrapping of 1000. Analyses were performed using the full amino acid sequence. Pairwise deletion method was selected for the gap/missing data.
2.9. Statistical analysis
Results are expressed as the means ± S.E. Student’s T-test was performed using GraphPad Prism (GraphPad Software, San Diego, CA) and considered significantly different at p ≤ 0.05. Regression curves and EC50 values were obtained by fitting the data using non linear regression analysis using the GraphPad Prism software.
3. RESULTS
3.1. Identification of a putative A. aegypti allatotropin receptor
To identify the putative AT receptors we used a bioinformatics approach based on the expression of the A. aegypti GPCR orphan receptors in the CA of adult female mosquito. We first screened the A. aegypti genome to identify GCPRs that could be involved in regulation of JH synthesis. Candidate GCPRs genes were identified by mining literature that describes GPCRs in A. aegypti [35], Anopheles gambiae [18], Drosophila melanogaster [13] and Apis mellifera [14]; as well as our own searches of the A. aegypti genome data displayed in VectorBase [24]. We first selected putative relevant genes based on the following 2 criteria: 1) Predicted genes in VectorBase that contained transmembrane domains and homology to one of the four classes of GPCRs (A-rhodopsin, B-secretin, C-metabotropic glutamate, D-atypical 7TM proteins). 2) Predicted genes classified as orphan GPCRs in A. aegypti [35] or with an identified putative ligand that was not unmistakably experimentally validated in mosquitoes or other insect species.
Fifty genes fulfilled the first 2 criteria (Table 1). Primers were designed for most of the identified genes (47 GPCRs were tested and only three paralogue genes were omitted). Expression in corpora allata-corpora cardiaca complex (CA-CC) in A. aegypti was tested using RT-PCR. Thirty-five genes were expressed in the CA-CC. Among them, we selected putative allatotropin receptor genes based on the following 2 additional criteria: 1) Since JH synthesis increases during the first 24 h after eclosion and AT is believed to play a role in this process, we selected 11 GPCRs genes that are expressed in CA-CC and showed increased expression during that time. 2) As a gene encoding allatotropin has not been found in the D. melanogaster genome (or any other Drosophila species), it was reasonable to assume that neither an orthologue for its receptor will be present in the fruit fly. Therefore only those GPCRs without an orthologue in Drosophila were further considered as candidates to be the AeATr. That left us with only 4 candidate GPCRs. Only 2 of them belonged to the A-rhodopsin class, a widespread protein family that includes hormone, neurotransmitter and photoreceptors. One of them, AAEL007644, has low similarity to a serotonin receptor and the other, AAEL011680 is a member of the orexin family of GPCRs and the orthologue of an ATr described from B. mori (BmATr) [58] (Supplementary table 2), so we selected this A. aegypti GPCR for further functional analysis, although the possibility cannot be eliminated that an additional GPCR, perhaps expressed in a different tissue, might also respond to AT.
Table 1.
Screening for the identification of a putative Aedes aegypti allatotropin receptor
| Receptor Class | Identified Genes | Tested Genes | Expressed in CA | Up regulated 0–24h | No orthologue in Drosophila |
|---|---|---|---|---|---|
| A | 37 | 34 | 23 | 7 | 2 |
| B | 7 | 7 | 7 | 2 | 1 |
| C | 3 | 3 | 2 | 1 | 0 |
| D | 3 | 3 | 3 | 1 | 1 |
Receptor classes: A: rhodopsin, B: secretin, C: metabotropic glutamate, D: Atypical 7TM proteins
3.2. A. aegypti allatotropin receptor structural and phylogenetic analysis
After aligning the full length cDNA sequence of AAEL011680 with sequences from the A. aegypti genome in VectorBase, we found that the gene is located in the supercontig 1.150, and it has a paralogue gene (AAEL005310) located in the supercontig 1.602. In both locations only small parts of the genes were originally predicted, probably due to the presence of large introns in both genes. The AAEL011680 gene in A. aegypti is composed of 10 exons and spans across an impressive stretch of 352,118 bp in the supercontig 1.150 (Supplementary Table 3). The coding sequence is 1899 bp long and it translates into a 632 amino acid protein (accession number: JN030894). The predicted amino acid sequences of the AeATr receptor exhibit the characteristic seven transmembrane domains (TM1-TM7) of GPCRs (Fig. 1).
The sequence of the AeATr paralogue (AAEL005310) spans across 360,462 bp in the supercontig 1.602. The AAEL005310 sequence is not complete; exon 3 and part of exon 8 are currently missing due to incomplete database information (Supplementary Table 3). Our full length cDNA sequence of ATr most likely is the transcript from the gene located on supercontig 1.150, because it has a better sequence similarity to that gene. A BLASTn alignment of the entire genomic regions which contain the two ATr paralogues (Supplementary Fig. 1) suggests that these two genes are the result of a gene duplication event which occurred after A. aegypti separated from other mosquito species, since the A. gambiae and Culex pipiens genomes contain only one copy of the ATr gene.
Large introns exist in both AeATr paralogues that contain highly conserved as well as highly diverse regions. The locations of the introns in the two paralogues genes are identical, but they vary largely in size. On the other hand sequences of exons of both paralogues are almost identical. Taken together this comparative analysis suggests that while introns accumulated mutations, the exons stayed well conserved and it is beyond the scope of this analysis to distinguish if one or both paralogues are being expressed, since the diversity between the two paralogues sequences within one genome is probably similar to the diversity caused by polymorphisms among different A. aegypti strains.
Allatotropin receptor orthologues were searched in other species of insects. An alignment of the AeATr with related insect receptors is shown in the Supplementary Figure 2. A cladogram of the phylogenetic relationship of these sequences of the AT receptor family was generated (Supplementary Fig. 3).
3.3. Functional expression and characterization of the AeATr
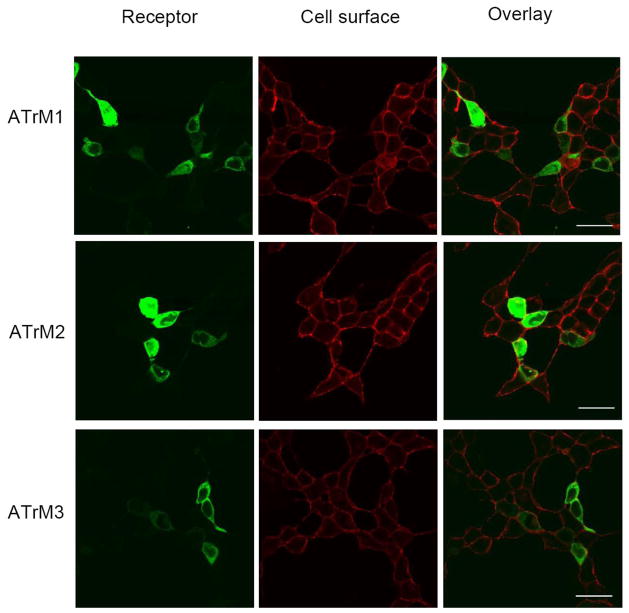
A mammalian cell line transiently expressing the putative AeATr constructs was used to functionally characterize the three putative receptor constructs (AeATrM1, AeATrM2 and AeATrM3) (Fig. 1). The localization of the three receptors at the cell surface in the transfected cells was assessed by expressing the AT receptor-YFP (Yellow Fluorescent Protein) fusion proteins and simultaneously marking the plasma membrane glycoproteins with biotin-conjugated concanavalin A and streptavidin-conjugated Alexa Fluor 633. Yellow color in the overlay indicates AT receptor expression at the cell membrane. All three fusion proteins were expressed in the cell membrane (Fig. 2).
Fig. 2. Membrane localization of AeATr in HEK293/Gα16gust44 cells.
Fluorescence images of HEK293/Gα16gust44 cells transfected with the three different AeATr-pEYFP-N1 DNAs. ATrM1 (top), ATrM2 (middle) and ATrM3 (bottom). The cell surface location of the AT receptor in transfected cells was recognized by expressing them fused to YFP (Yellow Fluorescent Protein) (first row) and marking the plasma membrane glycoproteins with biotin-conjugated concanavalin A and streptavidin-conjugated Alexa Fluor 633 (second row). Yellow color in the overlay indicates AT receptor expression at the cell membrane (third row). Scale bars, 10 μm. Similar results were obtained in two independent transfection experiments.
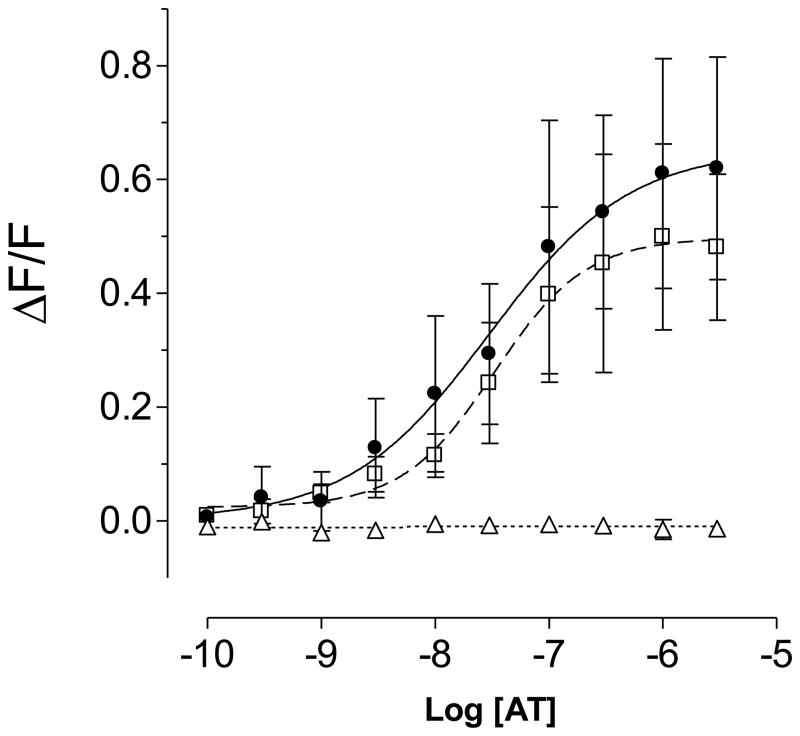
Activation of numerous GPCRs is coupled to the release of Ca2+ from intracellular stores, which can be measured using calcium-sensitive fluorescent dyes [5, 7, 51]. We measured the responses of the mosquito allatotropin receptors to two different A. aegypti peptides, AT and allatostatin-C (AS-C) in transient transfection assays. HEK293/Gα16gust44 cells expressing ATrM2 or ATrM3 receptors were robustly activated by AT in a dose-responsive manner (Fig. 3); but no signal was detected when AS-C (1 μM) or a buffer control with no peptides were used (results not shown). The minimum concentrations of Aedes allatotropin able to activate the receptors were in the low nanomolar range. EC50 values were 29.7 ± 6.2 nM for ATrM2 and 33.1 ± 4.4 nM for ATrM3. Cells expressing ATrM1 did not show any response when stimulated with AT.
Fig. 3. Functional characterization of the AT receptors by FLIPR experiments.
Dose–response curves of the effects of AT on the intracellular calcium concentration in cells expressing ATrM2 (filled circles), ATrM3 (open squares) and ATrM1 (open triangles). Vertical scale: ΔF/F ratio; horizontal scale: logarithmic concentration of AT. Data represent the means ± SEM of three independent experiments.
3.4. Tissue distribution and developmental expression of AT receptor mRNA
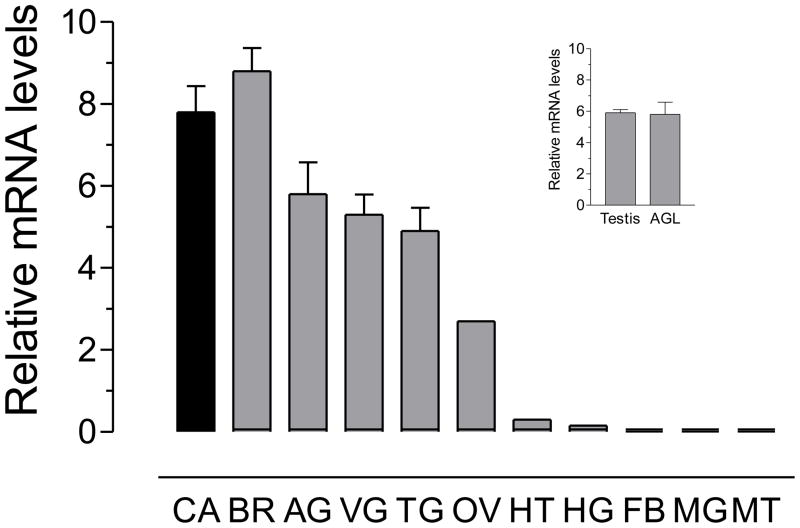
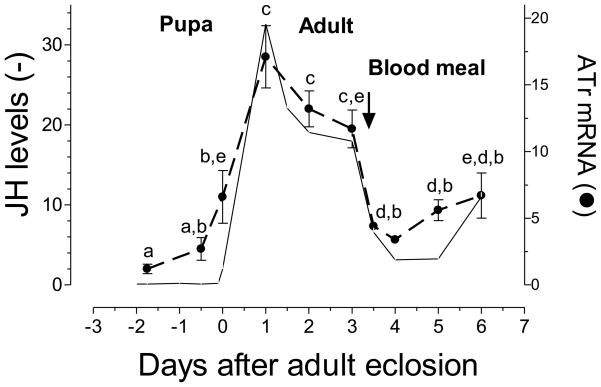
Real time PCR was used to analyze the transcript tissue specificity of the AeATr receptor in different female tissues and testis and accessory glands of male mosquitoes. The highest expression of AeATr mRNA was detected in the female nervous system (brain, abdominal ganglia, thoracic ganglia and ventral ganglia) and CA-CC complex, as well as in the female and male reproductive system (ovary, testis and accessory glands). Lower levels of mRNA transcripts were detected in female heart and hindgut. No transcripts were detected in the female midgut, fat body and Malphigian tubules (Fig. 4). In addition, the levels of the AT receptor transcripts were studied in the CA-CC during female pupal and adult development. In the female CA-CC, the AeATr mRNA levels were low in the early pupae, started increasing 6 hours before adult eclosion and reached a maximum 24 hours after female emergence. Blood feeding resulted in a decrease in transcript levels. The pattern of changes of AeATr mRNA resembled the changes in JH biosynthesis (Fig. 5).
Fig. 4. Tissue specific expression of AT receptor mRNA.
All tissues were dissected from three-day old sugar-fed females, except for testis (TE) and accessory glands (AGL) dissected from three-day old sugar-fed males. CA: corpora allata-corpora cardiaca; BR: brain; AG: abdominal ganglia; VG: ventral ganglia; TG: thoracic ganglia; OV: ovaries; HT: heart; HG: hindgut; FB: fat body; MG: midgut and MT: Malpighian tubules. The insert shows the males tissues analyzed. Receptor mRNA is expressed as copy number of receptor mRNA/10,000 copies of rpL32 mRNA. Each RT-PCR data point is mean ± SD of two independent biological replicates of 10–20 tissue samples.
Fig. 5. Developmental expression of AT receptors mRNA.
Expression of AT receptors mRNA in corpora allata-corpora cardiaca of pupa, sugar-fed and blood-fed adult female. AT mRNA is expressed as copy number of AaATr mRNA/10,000 copies of rpL32 mRNA. Each RT-PCR data point is mean ± SD of two independent biological replicates of 20 CA-CC complexes. Values labeled with the different letters denote significant differences (unpaired t-test P > 0.05). JH biosynthesis values are based on Li et al. (24), and are expressed as fmol/CA/h.
3.5. Analysis of expression of the AT receptor mRNA in the isolated CA and CC
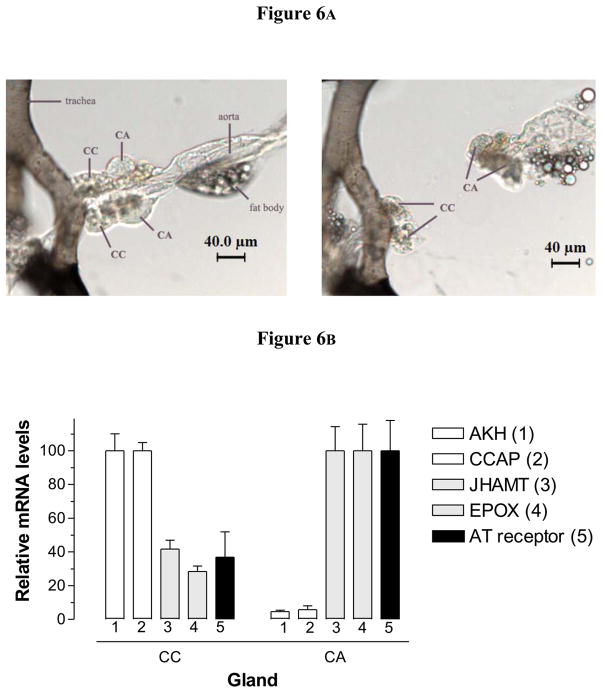
The very low level of expression of ATr mRNA in mosquito tissues made it difficult to use in situ hybridization as the method of choice to investigate the expression of the receptor in the CA and the CC. Alternatively, we decided to perform a surgical separation of the CA and CC, followed by analysis of expression of specific gene markers for CA and CC in the isolated glands. Images of a typical separation of the CA and CC are provided (Fig. 6A). The minute size of the CC made it difficult to obtain glands that were not contaminated with tissue from the larger CA gland; on the other hand our CA samples had only minor contaminations with CC tissue. As a result, we expected that genes normally expressed in the CC were going to be highly enriched in the CC samples, with a very low relative expression in the CA samples. On the other hand, transcripts expressed in the CA were going to be enriched in this gland, but also present in significant amounts in the CC sample. This is exactly what it was observed (Fig. 6B). CC markers such as adipokinetic hormone (AKH) [12] and crustacean cardioacceleratory peptide (CCAP) [12] had very low expression in the CA (about 5–6%) when compared with transcript levels in the CC. Juvenile hormone biosynthetic enzymes, such as juvenile hormone acid methyltransferase (JHAMT) and methyl farneosate epoxidase (EPOX), that are known to be expressed in the CA [56], were enriched in the CA, but also present as contaminants in the CC samples (about 35% of the CA levels). The expression of the AeATr was clearly distinct from the CC-specific genes (AKH and CCAP) and resembled that of the CA-specific genes (JHAMT and EPOX); although we could not reject the possibility that the AeATr is also expressed in the CC, our results strongly suggest that the ATr is expressed primarily in the CA.
Fig. 6. Analysis of expression of the AT receptor mRNA in the isolated CA and CC.
Corpora allata and CC were individually dissected from three-day old sugar-fed females. A) Images of a separation of the CA and CC. The left panel shows the glands after separation. B) Expression of the AT receptor and four additional marker genes in CC and CA. AKH (1): adipokinetic hormone; CCAP (2): crustacean cardioacceleratory peptide; JHAMT (3): juvenile hormone acid methyltransferase; EPOX (4): epoxidase; AT receptor (5): allatotropin receptor. Transcript levels are expressed as percentage of the maximum value found in the two glands. Each RT-PCR data point is mean ± SD of two independent biological replicates of 20 tissue samples.
4. DISCUSSION
4.1. Identification and characterization of the A. aegypti allatotropin receptor
We successfully used a combination of bioinformatics research and analysis of tissue and temporal gene expression to identify a putative A. aegypti AT receptor. The receptor is a GPCR that shares a high degree of sequence similarity with the AT receptors from other insects. A highly conserved sequence in the TM7 (YANSCANPI/VLY), which is considered a mammalian orexin receptor signature [55], is still recognisable in the AeATr receptors (Fig. 1). Although allatotropin and orexin are not structurally related, it is interesting that both peptides have been implicated with the regulation of food intake, starvation and energy metabolism [27, 31, 48, 49]. Our FLIPR results indicated that the AT receptor responded in the low nanomolar concentration range comparable to the values already reported for M. sexta [20] and T. castaneum [56], but lower than those published for B. mori [58]. Although, tests with cloned receptors in heterologous cell lines might not fully reflect the actual situation in the insect, the values match the biological concentration of AT measured in the head of mosquitoes [16], as well as the levels that activated the CA in vitro [29]. The ATrM1 clone was not functional probably because of missing 4 Asn-linked putative glycosylation sites. It has been shown that glycosylation is critical for GPCR function [8, 34, 44, 45]. In marked contrast, HEK293/Gα16gust44 cells expressing ATrM2 or ATrM3 receptors were both robustly activated by AT in a dose-responsive manner. The functional experiments did not help to distinguish the starting methionine of the receptor; although analysis of the sequence homology with the A. gambiae ATr suggests that it could be ATrM2 (Supplementary Fig. 2).
In A. aegypti, besides stimulating JH biosynthesis [29], addition of AT resulted in increases of motility of the hindgut, ovarioles, ovarian muscular sheets and heart (unpublished observations). Therefore those tissues were expected to express the AT receptor. In agreement with this hypothesis, we detected high expression in the CA, the nervous system and the reproductive organs; in addition, although at low levels, the receptor mRNA was also present in heart and hindgut.
4.2. Understanding the basis for allatotropin pleiotropic effects in insects
Allatotropin plays multiple neural, endocrine and myoactive roles, some of which are species specific. It has been proposed that during insect evolution AT functioned originally as myo- and/or neuroregulators and later was co-opted as a modulator of hormone synthesis [11]. There is a possibility that at least in some insect species these multiple functions are mediated by the presence of multiple peptides and/or multiple receptors. On the other hand, a single peptide could interact with a single receptor that signals to different transduction pathways; there is also an additional scenario where activation of the same receptor and the same transduction pathway might generate distinct biological responses in different cell types.
The idea that the multiple functions of AT are mediated by multiple AT receptors has not been experimentally supported. Searches in the genomes of several insects have found typically only one copy of the ATr. We found two paralogue genes in the A. aegypti genome; but the two genes encode very similar transcripts that would generate almost identical proteins; therefore it is improbable that these two receptor proteins could mediate different actions. Two AT receptors are indeed present in B. mori (BNGR-A16 and BNGR-A5). BNGR-A16 is the originally described functional AT receptor [58]; it exhibits 73% sequence identity with BNGR-A5 in the region between TM1 and TM7. The ligand binding specificity of BNGR-A5 has not yet been determined, so further analysis is necessary to determine if each of the 2 receptors are responsible for some of the different pleiotropic effects.
Genomes of most insect species encode a single AT peptide [11, 57], so the possibility that AT multiple functions are mediated by the presence of multiple peptides is also questionable. The exceptions again are some species of Lepidoptera, where alternative splicing of a single AT mRNAs yields a diverse set of AT-like peptides (ATL) that can vary in a dynamic manner during insect development [1, 19, 26, 47, 59]. In M. sexta it was recently shown that these different ATL peptides present overlapping biological activities and act with different potencies through a common receptor [20].
The structure of most AT and ATL is quite well conserved across insect, especially in the C-terminal portion; consequently antibodies against several AT peptides have been shown to cross react in heterologous species, or synthetic peptides to have biological effects in different species [11]. These results predicted that the AT receptor from one species could be activated by structurally related peptides from different species, and it has now been experimentally proved; the T. castaneum ATr was activated by T. castaneum ATL, Locusta migratoria AT, as well as by M. sexta AT [56].
Comprehensive tissue expression studies of receptors should also help to elucidate the different physiological roles of AT in their diverse target tissues. For the ATr these types of analysis have been limited to the larvae of B. mori [58] and M. sexta [20] and some tissues of adult T. castaneum [56]. The expression profile of the two B. mori receptors differs considerably [58]. In larvae, BNGR-A5 is expressed at the highest levels in the testis, while BNGR-A16 is expressed at the highest levels in several tissues including the brain, CC-CA, epidermis and testis. On the contrary, in M. sexta feeding 5th instar larvae, ATr mRNA is present at highest levels in the Malpighian tubules, followed by the midgut, hindgut and testes. In T. castaneum ATr mRNA levels were detected in the head, gut, fat body and reproductive system of sexually mature beetles.
Expression showed sexual dimorphism; with the highest transcript levels found in the male reproductive system followed by the head and the fat body. As stated before, our data showed that in adult females, AeATr mRNA expression was higher in the nervous system, CA and reproductive tissues. It was not possible to detect transcripts in the midgut, fat body and Malphigian tubules.
Transcripts levels are very low and might change during development or with physiological conditions, so tissues that did not express the ATr might do so in more detailed future studies. The actions of AT have been mostly determined using in vitro bioassays whereas virtually nothing is known of their roles in the intact insect [3]. Functional analysis in multiple cell lines suggest that the M. sexta AT receptor is coupled through elevated levels of Ca2+ and cAMP, showing that an activated receptor could signal to different transduction pathways [20]. AT effects are often described as fast and reversible [11] and have been often connected with mobilization of intracellular Ca2+; this could explain the multiple myotropic effects observed in experiments in vitro.
4.3. Role of AT on JH synthesis in mosquitoes
Here we report for the first time a detailed quantitative study of the expression of the AT receptor transcripts in the CA of an insect and their relationship with JH synthesis. The changes in receptor transcripts are strikingly in agreement with the changes in JH synthesis in the CA as previously described [28]. Prior research has shown that rates of JH biosynthesis closely reflect the hemolymph levels of JH in mosquitoes implying that there is a dynamic modulation of synthesis in the CA [28]. AeAT stimulates JH synthesis in the mosquito CA in vitro [29]. We reported AT immunoreactivity in cells in the brain [16] and ELISA studies revealed a 5-fold increase AT peptide levels in heads on the fourth day of adult life when compared with levels at adult eclosion [16].
The A. aegypti AT receptor was highly expressed in the CA-CC complex; but when we isolated both glands, it was clear that the receptor was expressed preferentially in the CA and not in the CC. These results suggested that the model described for B. mori [58], where AT might act through inhibition of release of the inhibitory sNPF, is not valid for mosquitoes. On the other hand, it has been previously described that inhibition of JH synthesis in mosquitoes is mediated by allatostatin-C [30].
At least two different scenarios could explain the similar patterns observed between JH levels and AeATr transcript levels: 1) AeATr is regulated by JH, and 2) AeATr regulates JH synthesis. Examples of genes matching those scenarios has been previously described; early trypsin is a mosquito midgut gene that is regulated by JH [37], Juvenile hormone methyl transferase and additional JH biosynthetic enzymes regulate JH synthesis [32; 39]. Further experiments are needed to clarify the relevance of the differential pattern of expression of the AT receptor in the CA as well as the various functions in the other tissues.
If AT is indeed regulating JH synthesis in mosquitoes a possible mechanism might be a regulation of CA intracellular Ca2+ levels. In insects, the rate of JH biosynthesis is often correlated with an increase in the intracellular Ca2+ concentration of the CA cells [2; 42; 43]; a Ca2+ chelator antagonized the stimulatory effects of M. sexta AT, suggesting that the peptide affects JH biosynthesis by increasing intracellular Ca2+ concentrations [43]. Additional experiments are needed to validate AT actions in vivo and elucidate its mechanism of action.
Final conclusion
In the present work we describe the functional and molecular characterization of the Aedes aegypti allatotropin receptor, and for the first time we provide a detailed quantitative study of the expression of AT receptor transcripts in the CA of an insect. The presence of an AT receptor in the CA of mosquitoes responding to AT stimulation in the nanomolar concentration range and the remarkable similarity between the pattern of changes of AeATr mRNA and the changes in JH biosynthesis suggest that the AeATr could play a role on JH synthesis modulation.
Supplementary Material
Highlights.
We characterized an allatotropin receptor expressed in the mosquito Aedes aegypti.
The receptor is stimulated by mosquito allatotropin in the low nanomolar concentration range.
The receptor is expressed in the corpora allata (CA), not in the corpora cardiaca.
The pattern of changes of receptor mRNA in the CA resembles the changes in JH biosynthesis.
Acknowledgments
The authors thank Mario Perez, Crisalejandra Rivera-Perez and Mark Clifton for critical reading of the manuscript. This work was supported by NIH grant AI 45545 to FGN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdel-latief M, Meyering-Vos M, Hoffmann KH. Molecular characterization of cDNAs from the fall armyworm Spodoptera frugiperda encoding Manduca sexta allatotropin and allatostatin preprohormone peptides. Insect Biochem Molec Biol. 2003;33:467–6. doi: 10.1016/s0965-1748(03)00005-5. [DOI] [PubMed] [Google Scholar]
- 2.Allen CU, Herman B, Granger NA. Fura-2 measurement of cytosolic free Ca2+ concentration in corpus allatum cells of larval Manduca sexta. J Exp Biol. 1992;166:253–66. doi: 10.1242/jeb.166.1.253. [DOI] [PubMed] [Google Scholar]
- 3.Audsley N, Matthews HJ, Price NR, Weaver RJ. Allatoregulatory peptides in Lepidoptera, structures, distribution and functions. J Insect Physiol. 2008;54:969–80. doi: 10.1016/j.jinsphys.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Behrens M, Brockhoff A, Batram C, Kuhn C, Appendino G, Meyerhof W. The Human Bitter Taste Receptor hTAS2R50 Is Activated by the Two Natural Bitter Terpenoids Andrographolide and Amarogentin. J Agric Food Chem. 2009;57:9860–6. doi: 10.1021/jf9014334. [DOI] [PubMed] [Google Scholar]
- 5.Bufe B, Hofmann T, Kraustwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 6.Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–27. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zucker CS, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–11. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 8.Davis D, Liu X, Segaloff DL. Identification of the sites of N-linked glycosylation on the follicle-stimulating hormone (FSH) receptor and assessment of their role in FSH receptor function. Mol Endocrinol. 1995;9:159–170. doi: 10.1210/mend.9.2.7776966. [DOI] [PubMed] [Google Scholar]
- 9.Duve H, East PD, Thorpe A. Regulation of lepidopteran foregut movement by allatostatins and allatotropin from the frontal ganglion. J Comp Neurol. 1999;413:405–16. [PubMed] [Google Scholar]
- 10.Duve H, Audsley N, Weaver RJ, Thorpe A. Triple co-localization of two types of allatostatin and an allatotropin in the frontal ganglion of the lepidopteran Lacanobia oleracea (Noctuidae): innervation and action on the foregut. Cell Tiss Res. 2000;300:153–63. doi: 10.1007/s004410000192. [DOI] [PubMed] [Google Scholar]
- 11.Elekonich MM, Horodyski FM. Insect allatotropins belong to a family of structurally-related myoactive peptides present in several invertebrate phyla. Peptides. 2003;24:1623–32. doi: 10.1016/j.peptides.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Gäde G, Goldsworthy GJ. Insect peptide hormones: a selective review of their physiology and potential application for pest control. Pest Manag Sci. 2003;59:1063–75. doi: 10.1002/ps.755. [DOI] [PubMed] [Google Scholar]
- 13.Hauser F, Williamson M, Cazzamali G, Grimmelikhuijzen CJP. Neuropeptides and neuropeptide receptors in the Drososphila melanogaster genome. Genome Res. 2006;11:1126–42. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJP. A review of neurohormone GPCR present in the fruitfly Drososphila melanogaster and the honey bee Apis mellifera. Progress Neurobiol. 2006;80:1–19. doi: 10.1016/j.pneurobio.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Hernández-Martínez S, Li Y, Rodriguez MH, Lanz-Mendoza H, Noriega FG. Allatotropin and PISCF- and YXFGL-amide-allatostatins distribution in Aedes aegypti and Anopheles albimanus mosquitoes. Cell Tissue Res. 2005;321:105–113. doi: 10.1007/s00441-005-1133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez-Martinez S, Mayoral JG, Li Y, Noriega FG. Role of juvenile hormone and allatotropin on nutrient allocation, ovarian development and survivorship in mosquitoes. J Insect Physiol. 2007;53:230–4. doi: 10.1016/j.jinsphys.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill CA, Fox AN, Pitts JR, Kent LB, Tan PL, Chrystak MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. G Protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–8. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 19.Horodyski FM, Bhatt SR, Lee K-Y. Alternative splicing of transcripts expressed by the Manduca sexta allatotropin (Mas-AT) gene is regulated in a tissue-specific manner. Peptides. 2001;22:263–9. doi: 10.1016/s0196-9781(00)00378-8. [DOI] [PubMed] [Google Scholar]
- 20.Horodyski FM, Verlinder H, Filkin N, Vandermissen HP, Fleury C, Reynolds SE, Vanden Broeck J. Isolation and functional characterization of an allatotropin receptor from Manduca sexta. Insect Biochem Molec Biol. 2011 doi: 10.1016/j.ibmb.2011.06.002. In press. [DOI] [PubMed] [Google Scholar]
- 21.Kataoka H, Toschi A, Li JP, Carney RL, Schooley DA, Kramer SJ. Identification of an allatotropin from adult Manduca sexta. Science. 1989;243:1481–3. doi: 10.1126/science.243.4897.1481. [DOI] [PubMed] [Google Scholar]
- 22.Koladich PM, Cusson M, Bendena WG, Tobe SS, McNeil JN. Cardioacceleratory effects of Manduca sexta allatotropin in the true armyworm moth, Pseudaletia unipuncta. Peptides. 2002;23:645–51. doi: 10.1016/s0196-9781(01)00658-1. [DOI] [PubMed] [Google Scholar]
- 23.Kreienkamp HJ, Larusson HJ, Witte I, Roeder T, Birgül N, Hönck HH, Harder S, Ellinghausen G, Buck F, Richter D. Functional annotation of two orphan G-protein-coupled receptors, Drostar1 and -2, from Drosophila melanogaster and their ligands by reverse pharmacology. J Biol Chem. 2002;42:39937–43. doi: 10.1074/jbc.M206931200. [DOI] [PubMed] [Google Scholar]
- 24.Lawson D, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Research. 2009;37:D583–587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K-Y, Horodyski FM, Chamberlin ME. Inhibition of midgut ion transport by allatotropin (Mas-AT) and Manduca FLRFamide peptides in the tobacco hornworm, Manduca sexta. J Exp Biol. 1998;201:3067–74. doi: 10.1242/jeb.201.22.3067. [DOI] [PubMed] [Google Scholar]
- 26.Lee K-Y, Chamberlin ME, Horodyski FM. Biological activity of Manduca sexta allatotropin-like peptides, predicted products of tissue-specific and developmentally-regulated alternatively spliced mRNAs. Peptides. 2002;23:1933–41. doi: 10.1016/s0196-9781(02)00181-x. [DOI] [PubMed] [Google Scholar]
- 27.Lee KY, Horodyski FM. Effects of starvation and mating on corpora allata activity and allatotropin (Manse-AT) gene expression in Manduca sexta. Peptides. 2006;27:567–74. doi: 10.1016/j.peptides.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Hernández-Martínez S, Unnithan GC, Feyereisen R, Noriega FG. Activity of the corpora allata of adult female Aedes aegypti: effects of mating and feeding. Ins Biochem Mol Biol. 2003;33:1307–15. doi: 10.1016/j.ibmb.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Unnithan GC, Veenstra JA, Feyereisen R, Noriega FG. Stimulation of JH biosynthesis by the corpora allata of adult female Aedes aegypti in vitro: effect of farnesoic acid and Aedes allatotropin. J Exp Biol. 2003;206:1825–32. doi: 10.1242/jeb.00371. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Hernandez-Martinez S, Fernandez F, Mayoral JG, Topalis P, Priestap H, Perez M, Navare A, Noriega FG. Biochemical, molecular and functional characterization of PISCF-allatostatin, a regulator of juvenile hormone biosynthesis in the mosquito Aedes aegypti. J Biol Chem. 2006;245:34048–55. doi: 10.1074/jbc.M606341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lwalaba D, Hoffmann KH, Woodring J. Control of the release of digestive enzymes in the larvae of the fall armyworm, Spodoptera frugiperda. Arch Insect Biochem Physiol. 2010;73:14–29. doi: 10.1002/arch.20332. [DOI] [PubMed] [Google Scholar]
- 32.Mayoral JG, Nouzova M, Yoshiyama M, Shinoda’ T, Hernandez-Martinez S, Dolghih E, Turjanski AG, Roitberg AR, Priestap H, Perez M, Mackenzie L, Li Y, Noriega FG. Molecular and functional characterization of a juvenile hormone acid methyltransferase expressed in the corpora allata of mosquitoes. Insect Bioch Mol Biol. 2009;39:31–7. doi: 10.1016/j.ibmb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chemical Senses. 2010;35:157–70. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 34.Michineau S, Muller L, Pizard A, Alhenc-Gelas F, Rajerison RM. N-linked glycosylation of the human bradykinin B2 receptor is required for optimal cell-surface expression and coupling. Biol Chem. 2004;385:49–57. doi: 10.1515/BC.2004.007. [DOI] [PubMed] [Google Scholar]
- 35.Nene, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noriega FG, Wells MA. A comparison of three methods for isolating RNA from mosquitoes. Insect Molec Biol. 1993;2:21–24. doi: 10.1111/j.1365-2583.1993.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 37.Noriega FG, Shaa D, Wells MA. Juvenile Hormone controls early trypsin gene expression in the midgut of Aedes aegypti. Insect Molec Biol. 1997;6:63–6. doi: 10.1046/j.1365-2583.1997.00154.x. [DOI] [PubMed] [Google Scholar]
- 38.Noriega FG, Colonna AE, Wells MA. Increase in the size of the amino acid pool is sufficient to activate translation of early trypsin mRNA in Aedes aegypti midgut. Insect Biochem Mol Biol. 1999;29:243–247. doi: 10.1016/s0965-1748(98)00132-5. [DOI] [PubMed] [Google Scholar]
- 39.Nouzova M, Edwards M, Mayoral JG, Noriega FG. A coordinated expression of biosynthetic enzymes controls the flux of juvenile hormone precursors in the corpora allata of mosquitoes. Insect Biochem Molec Biol. 2011 doi: 10.1016/j.ibmb.2011.04.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Offermanns S, Simon MI. G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J Biol Chem. 1995;270:15175–80. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- 41.Petri B, Homberg U, Loesel R, Stengl M. Evidence for a role of GABA and Mas-allatotropin in photic entrainment of the circadian clock of the cockroach Leucophaea maderae. J Exp Biol. 2002;205:1459–69. doi: 10.1242/jeb.205.10.1459. [DOI] [PubMed] [Google Scholar]
- 42.Rachinsky A, Tobe SS. Role of second messengers in the regulation of juvenile hormone production in insects, with particular emphasis on calcium and phosphoinositide signaling. Arch Ins Biochem Physiol. 1996;33:259–82. [Google Scholar]
- 43.Rachinsky A, Srinivasan A, Ramaswamy SB. Regulation of juvenile hormone biosynthesis in Heliothis virescens by Manduca sexta allatotropin. Arch Ins Biochem Physiol. 2003;54:121–33. doi: 10.1002/arch.10107. [DOI] [PubMed] [Google Scholar]
- 44.Rands E, Candelore MR, Cheung AH, Hill WS, Strader CD, Dixon RA. Mutational analysis of beta-adrenergic receptor glycosylation. J Biol Chem. 1990;65:10759–10764. [PubMed] [Google Scholar]
- 45.Reichling C, Meyerhof W, Behrens M. Functions of human bitter taste receptors depend on N-glycosylation. J Neurochem. 2008;106:1138–1148. doi: 10.1111/j.1471-4159.2008.05453.x. [DOI] [PubMed] [Google Scholar]
- 46.Rudwall AJ, Sliwowska J, Nässel DR. Allatotropin-like neuropeptide in the cockroach abdominal nervous system: Myotropic actions, sexually dimorphic distribution and colocalization with serotonin. J Comp Neurol. 2000;428:159–73. doi: 10.1002/1096-9861(20001204)428:1<159::aid-cne11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 47.Sheng Z, Ma L, Cao M-X, Li S, Jiang R-J. Biochemical and molecular characterization of allatotropin and allatostatin from the Eri silkworm, Samia cynthia ricini. Insect Biochem. Molec Biol. 2007;37:90–6. doi: 10.1016/j.ibmb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Skrzypski MT, Le T, Kaczmarek P, Pruszynska-Oszmalek E, Pietrzak P, Szczepankiewicz D, Kolodziejski PA, Sassek M, Arafat A, Wiedenmann B, Nowak KW, Strowski MZ. Orexin A stimulates glucose uptake, lipid accumulation and adiponectin secretion from 3T3-L1 adipocytes and isolated primary rat adipocytes. Diabetologia. 2011 doi: 10.1007/s00125-011-2152-2. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 49.Swart I, Overton JM, Houpt TA. The effect of food deprivation and experimental diabetes on orexin and NPY mRNA levels. Peptides. 2001;12:2175–9. doi: 10.1016/s0196-9781(01)00552-6. [DOI] [PubMed] [Google Scholar]
- 50.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2 MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molec Biol Evolution. 2011 doi: 10.1093/molbev/msr121. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda T, Ugawa S, Yamamura H, Imaizumi Y, Shimada S. Functional interaction between T2R taste receptors and G-protein α subunits expressed in taste receptor cells. J Neuorsci. 2003;23:7376–80. doi: 10.1523/JNEUROSCI.23-19-07376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueda H, Shinoda T, Hiruma K. Spatial expression of the mevalonate enzymes involved in juvenile hormone biosynthesis in the corpora allata in Bombyx mori. J Insect Physiol. 2009;55:798–9804. doi: 10.1016/j.jinsphys.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Veenstra JA, Costes L. Isolation and identification of a peptide and its cDNA from the mosquito Aedes aegypti related to Manduca sexta allatotropin. Peptides. 1999;20:1145–51. doi: 10.1016/s0196-9781(99)00117-5. [DOI] [PubMed] [Google Scholar]
- 54.Veenstra JA, Lehman HK, Davis NT. Allatotropin is a cardioacceleratory peptide in Manduca sexta. J Exp Biol. 1994;188:347–54. doi: 10.1242/jeb.188.1.347. [DOI] [PubMed] [Google Scholar]
- 55.Voisin T, Rouet-Benzineb P, Reuter N, Laburthe M. Orexins and their receptors:structural aspects and role in peripheral tissues. Cell Mol Life Sci. 2003;60:72–87. doi: 10.1007/s000180300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vuerinckx K, Verlinder H, Lindermans M, Vanden Broeck J, Huybrechts R. Characterization of an allatotropin-like peptide receptor in the red flour beetle, Tribolium castaneum. Insect Biochem Molec Biol. 2011 doi: 10.1016/j.ibmb.2011.06.003. In press. [DOI] [PubMed] [Google Scholar]
- 57.Weaver RJ, Audsley N. Neuropeptide regulators of juvenile hormone synthesis. Ann NY Acad Sci. 2009;1163:316–329. doi: 10.1111/j.1749-6632.2009.04459.x. [DOI] [PubMed] [Google Scholar]
- 58.Yamanaka N, Yamamoto S, Žitňan D, Watanabe K, Kawada T, Satake H, Kaneko Y, Hiruma K, Tanaka Y, Shinoda T, Kataoka H. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One. 2008;3:e3048. doi: 10.1371/journal.pone.0003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin H, Zhang T-Y, Xu W-H. Structural organization and expression analysis of the cDNA encoding allatotropin in the cotton bollworm, Helicoverpa armigera. Arch Ins Biochem Physiol. 2005;60:71–83. doi: 10.1002/arch.20082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.