Abstract
The molecular mechanisms contributing to the development and progression of gingivobuccal complex (GBC) cancers–a sub-site of oral cancer, comprising the buccal mucosa, the gingivobuccal sulcus, the lower gingival region and the retromolar trigone-remain poorly understood. Identifying the GBC cancer-related gene expression signature and the driver genes residing on the altered chromosomal regions is critical for understanding the molecular basis of its pathogenesis. Genome-wide expression profiling of 27 GBC cancers with known chromosomal alterations was performed to reveal differentially expressed genes. Putative driver genes were identified by integrating copy number and gene expression data. A total of 315 genes were found differentially expressed (P≤0.05, logFC>2.0) of which eleven genes were validated by real-time quantitative reverse transcriptase-PCR (qRT-PCR) in tumors (n=57) and normal GBC tissues (n=18). Overexpression of LY6K, in chromosome band 8q24.3, was validated by immunohistochemical (IHC) analysis. We found that 78.5% (2,417/3,079) of the genes located in regions of recurrent chromosomal alterations show copy number dependent expression indicating that copy number alteration has a direct effect on global gene expression. The integrative analysis revealed BIRC3 in 11q22.2 as a candidate driver gene associated with poor clinical outcome. Our study identified previously unreported differentially expressed genes in a homogeneous subtype of oral cancer and the candidate driver genes that may contribute to the development and progression of the disease.
INTRODUCTION
Copy number alterations (CNAs) of the genome have a strong effect on gene expression and determine cancer cell survival and progression (Hyman et al., 2002; Albertson et al., 2003; Järvinen et al., 2006; Järvinen et al., 2008). CNAs observed in tumors often involve a large group of genes located consecutively along the same chromosome. Only a minority of these genes are likely to be the true “driver” genes contributing to tumorigenesis. The other genes, called “passenger” genes, may be altered simply because of their chromosomal location and proximity to the target gene(s) (Leary et al., 2008). Driver genes activate the neoplastic process and mutations in these genes contribute to the transformation of normal cells to proliferating cancer cells (Akavia et al., 2010). Distinguishing driver genes from passenger genes is an intriguing question in cancer genetics. One approach to answer this question is to integrate copy number, gene expression and clinical data to distinguish passenger genes, of which the expression mainly reflects the genomic copy number, from driver genes, which promote the disease and have clinical relevance as biomarkers (Santarius et al., 2010).
This study presents a gene expression analysis of oral cancer, focusing on squamous cell carcinomas of the “gingivobuccal complex” (GBC). The GBC is composed of the buccal mucosa, the gingivobuccal sulcus, the lower gingiva and the retromolar trigone. It is the most common sub-site of oral cancer, closely linked to the habit of chewing betel quid containing tobacco in the Indian population (Pathak et al., 2005). Oral cancers from different sites within the entire oral cavity differ in their biological and clinical behavior, leading to discordance in genomic and transcriptomic profiling studies (Estilo et al., 2009). Most reports on gene expression profiling in oral cancer have either used tumors from multiple, heterogeneous sites in the oral cavity (Roepman et al., 2006; Chen et al., 2008; Sticht et al., 2008; Méndez et al., 2009) or have focused on tongue cancers (Carinci et al., 2005; Shimada et al., 2005; Zhou et al., 2006; Ye et al., 2008; Estilo et al., 2009). There are no previous studies on gene expression profiling of GBC cancers.
Our array comparative genome hybridization (aCGH) study revealed clinically relevant CNAs in oral cancers (Ambatipudi et al., 2011), but the relative importance of individual genes in these regions remained elusive. To our knowledge, only a single study has utilized the potential of integrating copy number and gene expression data for identifying genes associated with poor survival using tumor cells from metastatic lymph nodes in heterogeneous oral sites (Xu et al., 2010). Therefore, we performed an integrative analysis of primary tumors from a homogeneous site to understand the influence of CNAs on the gene expression signature in GBC cancer.
MATERIALS AND METHODS
Tissue Specimen Collection and Tumor Microdissection
This project was approved by the Institutional Review Board and the Local Ethics Committee of Tata Memorial Hospital (Approval number HEC No.559 of 2008). Written informed consents were obtained from all study participants. Neo-primary oral tumor samples were collected from 57 patients undergoing surgery at the Head and Neck Unit and from the ICMR National Tumour Tissue Repository, Tata Memorial Centre, Mumbai. Patients received neither radiation nor chemotherapy before surgery. Tissues with more than 70% tumor content were subjected to RNA extraction. A possible presence of Human Papilloma Virus (HPV) DNA in tumor specimens was analyzed by polymerase chain reaction (PCR) using GP5+/GP6+ (L1 region) generic primers (Ambatipudi et al., 2011). Non-inflamed GBC mucosa was collected from 18 healthy individuals to serve as reference tissues. All the samples were snap frozen in liquid nitrogen and later kept at −80°C until they were used. For the microarray gene expression analysis, a test set consisting of 27 GBC cancers (with known copy number alterations) and five normal GBC samples (4 individual normal and 1 pool of RNA from 9 other normal samples) was used. The differentially expressed genes were validated by real-time quantitative reverse transcriptase-PCR (qRT-PCR) on an independent set of GBC samples from 30 patients and 9 additional normal subjects. Immunohistochemical (IHC) analysis was performed on 55 formalin fixed paraffin-embedded tumors and 12 unrelated GBC tissues.
RNA Isolation and Gene Expression Analysis
RNA was isolated from the tumor and normal tissues using the RNeasy mini kit according to the manufacturer’s instructions (Qiagen GmbH, Hilden), as described in the Supporting Information Text. The Whole Human Genome Microarray Kit, 4×44K (Agilent Technologies, Santa Clara, CA) with 43,376 oligonucleotide reporters was used for the gene expression study. Single color labeling, hybridization and data extraction were done according to the manufacturer’s protocol. Briefly, 1.65 μg of RNA was labeled with Cy3 dye using the Quick Amp one color labeling kit (Agilent Technologies, Santa Clara, CA). Slides were washed, dried and scanned on an Agilent high resolution scanner (Agilent Technologies, Santa Clara, CA) with default parameters followed by feature extraction using the FE software version 10.5 (Agilent Technologies). All data generated was MIAME compliant and the raw data is deposited in the GEO database (accession number GSE23558) following the instructions on the MGED Society website http://www.mged.org/Workgroups/MIAME/miame.html.
Data Pre-processing
The Agilent 44K array has multiple oligonucleotide reporters for some genes and many reporters that cannot be unambiguously interpreted as representing the expression level of a single gene (Gertz et al., 2009). Hence, in this study, we restricted the analysis to 25,505 reporters annotated “fully valid”. Raw intensity values were first normalized and quality filtered with the Agi 4×44 PreProcess Bioconductor package (Gentleman et al., 2004). After initial quality filtering, 15,946 reporters mapping to 11,718 genes were used for the subsequent expression analysis.
Analysis of Gene Expression Data
We identified differentially expressed genes with fully valid reporters using the Bioconductor package limma (Smyth, 2004). Each reporter was tested for a difference in the mean logarithmic intensity between the tumor and the normal samples by a moderated t-test. When there was more than one fully valid reporter for a gene, p-values for the significance of the differential expression were combined using the method of R. A. Fisher (Elston, 1991). Gene-specific p-values were then corrected for testing across multiple genes by the method of Benjamini and Hochberg (Benjamini and Hochberg, 1995). A corrected p-value below 0.05 was considered statistically significant. The genes so obtained were considered differentially expressed if the absolute logarithmic expression fold-change to the base 2 (logFC) between the groups was above 2, corresponding to a 4-fold expression change.
Integrative Gene Expression and Copy Number Analysis
For 93 recurrent copy number alterations (Ambatipudi et al., 2011), the set of genes covered by each CNA was determined. Then, for each CNA, the expression Yi of reporter i was modeled according to the linear model E[Yi] = Xαi, where αi = (αiN, αiOCαiCNA) describes the average expression values for normal, tumors, and tumors with a CNA at the locus of reporter i, respectively. Ambiguous genes with spatially overlapping CNAs of opposing type were excluded. Tests on the averages were done by means of the moderated t-test and combined p-values for genes were then computed from the p-values of reporters with Fisher’s method, as described above. To test the overall gene dosage effect of a given CNA and each of 14 peak regions of maximal recurrence (Ambatipudi et al., 2011) within a CNA, we first computed the logFC value for each tumor sample and compared it with the averaged value in normal samples of all valid reporters of the genes located in the CNA under consideration. Then we tested with a single-sided t-test if the logFCs in cases with the alteration were, on average, different from those without the alteration. The tail of the t-test was chosen to test for a higher fold-change for gains, and a lower fold-change for losses.
Survival Analysis
The survival analysis was based on the Cox proportional hazard model. The risk was modeled by the function exp(−λ0(t)βTX), where λ 0(t) denotes the baseline risk, β represents a vector of risk coefficients and X denotes the expression of genes. The coefficients β were estimated from the data. Additional backward model selection was performed based on the Bayesian Information Criterion (BIC) to avoid overfitting. The R package survfit was used for these analyses.
Validation of Copy Number Alteration of the 8q24.3 Locus by Interphase Fluorescence In Situ Hybridization (I-FISH)
The copy number status of the 8q24.3 locus was evaluated by dual color I-FISH. The BAC clones RP11-642A1 (8q24.3) and RP11-73M19 (8 centromere), used for FISH, were obtained from the Children’s Hospital Oakland Research Institute (CHORI) BACPAC resources center. A centromeric probe served as hybridization control and was used as a reference to determine the copy number status of the 8q24.3 locus. FISH images were captured under a fluorescence microscope (Axioskop II, Carl Zeiss, Germany) and analyzed using the ISIS imaging software (Metasystems, Altlussheim, Germany).
Literature Mining
A literature search was done to find genes that were previously suggested to have either differential expression or copy number alterations in oral cancer using PubMed, PubMedCentral, and the Science Citation Index and was completed in May 2010. We noted whether the changes in copy number or expression were associated with prognosis. Although there were numerous reviews of genomics and oral cancer, we could not find a comprehensive list of implicated genes in any review.
Real-time qRT-PCR Analysis of Differentially Expressed Genes
The qRT-PCR assays were carried out for selected candidate genes found significantly upregulated (SPP1, CA9, HOXC9, TNFRSF12A, LY6K, INHBA, FST, MFAP5 and DHRS2) or downregulated (MAL and GPX3). Two genes (TSN and SLC4A1AP), which showed no significant differential expression, were used to validate the findings of gene expression data. The fluorescent TaqMan probes were obtained from Applied Biosystems. The assay IDs and the protocol details are provided in Supporting Information Table 1 and Supporting Information Text, respectively.
Statistical Analysis of qRT-PCR Data
The Shapiro-Wilk test was used to assess the normal distribution of differentially expressed genes in normal and tumor samples. A p-value below 0.05 was considered statistically significant. The genes that followed a normal distribution were compared using an independent samples t-test, while a Mann-Whitney test was used for comparison of the genes which did not follow a normal distribution. For eleven selected genes, the Pearson correlation between logFC values of the microarray and qRT-PCR was computed. The analysis was done in SPSS 15.0.
Immunohistochemical Analysis
The immunohistochemical analysis of formalin fixed paraffin embedded tissues was performed using the Vectastain® universal elite ABC kit (PK-6200, Vector Laboratories, Inc, Burlingame, CA). Tissue sections of 5 μm thickness were deparaffinised, rehydrated and treated with peroxidase block. After heat based antigen retrieval, sections were incubated with normal horse serum. The sections were incubated overnight with rabbit polyclonal anti-human LY6K antibody (HPA017770, Prestige Antibodies, Sigma-Aldrich St. Louis, MO) at a dilution of 1:250. These sections were next kept for 30min with biotinylated universal secondary antibody solution followed by incubation with Vectastain® elite ABC reagent for 30min. The immunoreaction in tissue sections was visualized us i n g 3, 3′ –Diaminobenzidine tetrahydrochloridehydrate (D5637, Sigma-Aldrich St. Louis, MO). The slides were finally counterstained with haematoxylin. Sections treated with normal rabbit serum were used as negative controls.
RESULTS
Patient Characteristics
The clinicopathological and demographic characteristics of all tumor samples are summarized in Table 1. The patients in this study cohort were predominantly male tobacco habitués. Tumors were of moderate or poor grade, mainly of pTNM stages III or IV and approximately half the number of cases showed lymph node invasion. Thirty additional GBC cancers with similar characteristics and nine unrelated normal samples from GBC sites were used for validating the findings by qRT-PCR. All samples were unrelated to human papillomavirus infection (Ambatipudi et al., 2011). Case-wise details for all GBC cancer patient samples used for gene expression, qRT-PCR and IHC analysis are summarized in Supporting Information Table 2.
Table 1.
Clinicopathological and demographic details of study cohort
| Characteristics | Test Set (n=27) (GE* and qRT-PCR)† | Validation Set (n=30) (qRT-PCR)† |
|---|---|---|
| Gender | ||
| Males | 20 (74%) | 20 (67%) |
| Females | 7 (26%) | 10 (33%) |
| Age | ||
| Median (IQR)# | 53 (44–60) | 51 (42–56) |
| Habit profile | ||
| Exclusive Chewers | 21 (78%) | 22 (73%) |
| Exclusive Smokers | 1 (4%) | 2 (7%) |
| Chewing and Smoking | 5 (18%) | 6 (20%) |
| Grade | ||
| Well | 0 (0%) | 2 (6%) |
| Moderate | 19 (70%) | 20 (67%) |
| Poor | 8 (30%) | 8 (27%) |
| Nodal involvement | ||
| Negative (N0) | 14 (52%) | 15 (50%) |
| Positive (N+) | 13 (48%) | 15 (50%) |
| Stage (pTNM) | ||
| I & II | 3 (11%) | 0 (0%) |
| III & IV | 24 (89%) | 30 (100%) |
Shown is the number of cases, except for Age
GE: Gene Expression analysis;
IQR: Interquartile range
Differentially Expressed Genes in GBC Cancers
We found 315 genes differentially expressed (80 upregulated and 235 downregulated) in GBC cancers, as shown in Supporting Information Table 3. The 40 genes with the strongest fold-change in either direction are listed in Supporting Information Table 4.
Validation of Gene Expression Results by qRT-PCR
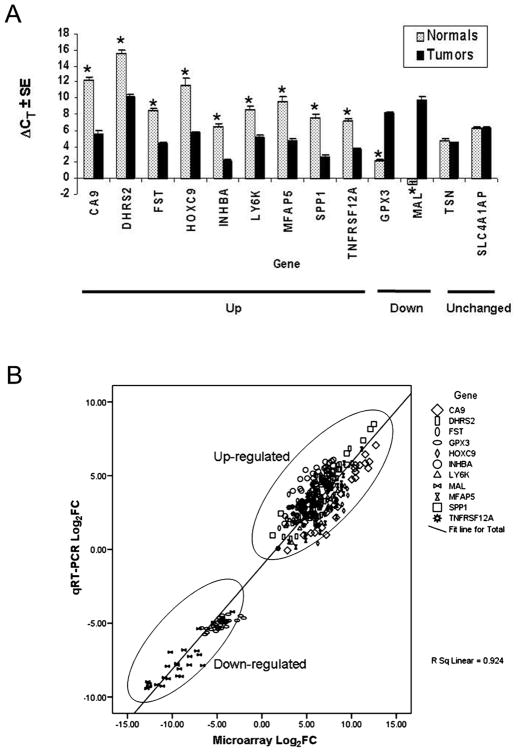
From the 80 genes with the most extreme logFC values (40 upregulated and 40 downregulated), we selected 11 genes for individual validation either due to the absence of prior reports (see literature search below) or their potential as putative serum biomarkers. Real-time qRT-PCR was done for the genes SPP1, CA9, HOXC9, TNFRSF12A, LY6K, INHBA, FST, MFAP5, DHRS2, MAL, and GPX3 in both the test and the validation sets, to confirm the results obtained by genome-wide differential gene expression analysis in the test set only. We found statistically significant differences in the ΔCT values between normal and tumor specimens. Overexpressed genes showed lower ΔCT values in tumors as compared to normal tissue while the opposite was observed for underexpressed genes. TSN and SLC4A1AP showed no significant change in the ΔCT values (Figure 1A) validating the gene expression findings. The correlation of the microarray and the qRT-PCR data for the test set was determined for all validated differentially expressed genes. Our qRT-PCR results correlated strongly with those from microarray analyses (R2=0.92, P<0.0001; Pearson correlation test) (Figure 1B). Real-time qRT-PCR confirmed the direction of the change in the validation set of tumors (n=30) and in normal samples (n=9).
Figure 1.
A: Validation of eleven genes found differentially expressed in the test set by qRT-PCR (tumors n = 57 and Normals n = 14). Each tumor-normal pair of vertical bars represents a gene grouped according to the type of differential expression. The height of a bar is the average difference in PCR cycles compared to 18S RNA, ΔCT. Error bars represent the standard error. On the right, TSN and SLC4A1AP represent genes without any change in expression between tumors and normals. Statistically significant differences (see Methods in Supporting Information Text for details) in the ΔCT-values are indicated by stars (*).
B: Correlation analysis between the microarray and qRT-PCR fold-change values for eleven differentially expressed genes in test set of 27 GBC samples. Plot Shows the Log2FC values for microarray data on the x-axis versus Log2FC values for qRT-PCR data on the y-axis.
Integrative Analysis of Gene Expression and Copy Number Alterations
We found that 26.3% (3,079/11,718) of the analyzed genes were located in regions of recurrent CNAs found in a previous study (Ambatipudi et al., 2011), as shown in Supporting Information Table 5. Of these genes, 78.5% (2,417/3,079) showed copy number dependent expression effects, meaning higher average expression in tumors with a copy number gain, or lower average expression in tumors with genomic losses. The frequency of a copy number dosage effect on gene expression did not differ significantly between gains (1,751/2,203 genes with increased expression in gains) and losses (673/890 with decreased expression in losses).
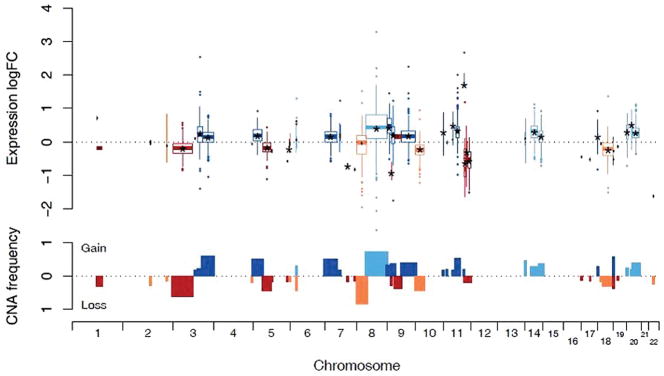
The recurrent CNAs showing a high concordance (mean |logFC|>0.5, P<0.05) with gene expression results were the losses of 7q31.1, 9p21.3, 11q22.3-q23.1, 11q23.3-q25 and 22q11.23 as well as the gains of 9p21.3-p24.3, 11q22.1-q22.2, and 20p11.21 (Figure 2 and Supporting Information Table 6). The amplification of 11q22.1-q22.2 spanning genes MMP7, TMEM123, BIRC2, YAP1, BIRC3 and MMP10 showed the highest mean fold-change in copy number dependent overexpression (logFC=1.71). Conversely, the most extreme underexpression (logFC=−0.91) was observed in the region of 9p21 containing CDKN2B, CDKN2A, and MTAP.
Figure 2.
Effects of gene dosage on the underlying genes in GBC tumors (n = 27). In the upper panel, each of the 93 recurring CNAs is represented by a boxplot describing the distribution of the logarithmic expression fold-change of all genes located in the CNA compared to those cancer cases without the aberration. The position on the x-axis and the width of each box denotes the genomic position and spatial extent of the alteration respectively. Blue colors represent gains and show a positive expression fold-change while red color representing losses show a negative fold-change. Any significant average fold-changes (P < 0.05, Benjamini-Hochberg adjusted t-test) are indicated by star marks (*). In the lower panel, bars indicate the observed frequencies of CNAs; gains are plotted upwards and losses downwards.
Survival Analysis
A univariate survival analysis of differentially expressed genes revealed no significant association with survival after correction for multiple testing. Multivariate Cox proportional hazards models were used to assess the contribution of genes residing in focal CNAs that contained fewer than ten genes with valid reporters and were significantly correlated with expression (P < 0.05; t-test). These restrictions were made to keep the model complexity tractable and to preserve power otherwise lost due to multiple testing. The best subsets of genes for prediction, which may also contain no genes, were selected by the BIC.
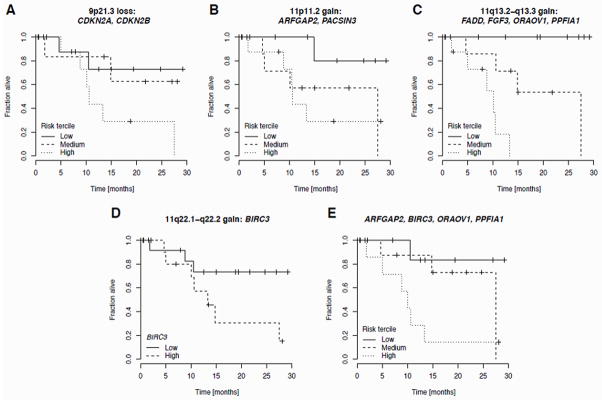
For the losses of 5q35.3 and 7q31.1, the BIC method did not select any significant genes. For the deletion of 9p21.3, a combination of CDKN2A (λ=0.287, risk coefficient) and CDKN2B (λ=−0.472) was selected (P = 0.07, likelihood ratio test). For one of two focal gains on 11p11.2, the BIC selected the ARFGAP2 (λ=1.62) and PACSIN3 (λ=−1.53) genes (P = 0.054). For the gain of 11q13.2-q13.3, the best subset consists of the genes FADD (λ=2.47), FGF3 (λ=−1.84), ORAOV1 (λ=1.9), and PPFIA1 (λ=−2.74; P = 0.006). For the gain of 11q22.1-q22.2, the covariates in the Cox model could be reduced to only BIRC3 (λ=0.65; P=0.04). Kaplan-Meier plots with risk terciles for all multivariate cases are shown in Figure 3A–C. A Kaplan-Meier plot demonstrating association of high BIRC3 expression with poor survival is shown in Figure 3D. Finally, we computed the best combination of genetic predictors from the combined set of genes selected for each CNA. BIC model selection yielded the combination of ARFGAP2 (λ=3.51), BIRC3 (λ=0.97), ORAOV1 (λ=2.90), and PPFIA1 (λ=−1.32), as the optimal multivariate predictor (P = 0.002, Figure 3E).
Figure 3.
Kaplan-Meier curves for the survival of GBC cancer patients. A–D: BIC-optimal multivariate Cox proportional hazard model for genes located in focal gains. Patients were split into three risk groups according to the terciles of the observed hazard function (A–C); in the univariate case (D) high and low expression groups are defined by BIRC3 expression above and below the median, respectively. E: Combined optimal model based on all genes analyzed in A–D. Multiple gene symbols are listed alphabetically in a panel, so the order implies nothing about their relative importance.
FISH Analysis for Validation of 8q24.3 Gain
Of all genes on chromosomal region 8q24, LY6K showed the highest overexpression (logFC=2.3) irrespective of the copy number change which was apparent from the homogeneous overexpression in the samples with or without 8q24 gain (logFC=0.54, P=0.19). The copy number increase of the 8q24.3 locus was validated using interphase FISH (Supporting Information Figure 1).
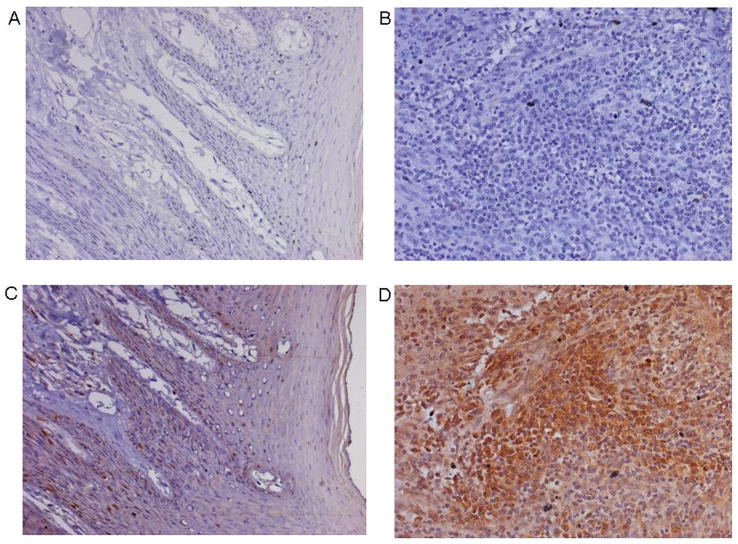
IHC Analysis of LY6K
The immunohistochemical analysis showed higher protein abundance at the plasma membrane and cytoplasm in cancer specimens than in normal tissues (Figure 4), thereby validating the findings of the expression analysis.
Figure 4.
Immunohistochemical analysis of LY6K in normal and tumor tissues of GBC. A – B: represent serum controls for normal and tumor tissues, C: unrelated normal GBC sample showing weak staining; D: GBC cancer showing strong staining with cytoplasmic and membranous localization (200X Magnification).
Literature Search
The literature search identified 277 genes previously suggested to have either copy number variations or differential expression in oral cancer. The full list is shown as Supporting Information Table 7 and is based on 73 references. Of these 277 genes, 214 genes that have both a valid reporter on the microarray and an adequate signal in the experiments are marked with a “#”. Only 39/214 genes (Supporting Information Table 8A) were mentioned in multiple papers, even though we explicitly searched for replication papers, and 28/214 genes (Supporting Information Table 8B) had evidence for both copy number changes and differential expression. Among these 28 genes, 23 genes had evidence for gain and overexpression, 4 had evidence for loss and underexpression, and the gene FAT1 had conflicting evidence on expression. There were 129 genes with evidence of only differential expression, and 57 genes had evidence only of copy number changes (Supporting Information Tables 8C and 8D). Surprisingly, these 57 genes are divided into 24 genes with gains and 33 genes with losses, significantly different from the gain/loss pattern for the 28 genes with both types of evidence (P < 0.0005, two-sided Fisher’s exact test).
DISCUSSION
We performed gene expression profiling of 27 GBC tumors to delineate differentially expressed genes and did an integrative analysis to identify candidate driver genes. Three hundred and fifteen genes were found differentially expressed in GBC cancers and selected genes were validated by qRT-PCR and IHC on an independent set of tumors and unrelated normal GBC tissues. The integrative copy number and gene expression analysis revealed a large number of genes in which the two types of changes were associated. Our study identified novel genes with a potential role in tumorigenesis and demonstrated the utility of an integrative approach in identifying candidate driver genes.
To our knowledge, this is the first study that provides a comprehensive genome-wide gene expression profile and integrative analysis of GBC cancers. A recent integrative study identified genes associated with poor survival in metastatic OSCC (Xu et al., 2010), but the use of microdissection in isolating tumor cells from nodes did not account for tumor-stromal interactions at the primary site which play an important role in the development and progression of OSCC (Chen et al., 2008). Hence, we used primary GBC tissues and avoided microdissecting tumor cells. Field cancerization has been reported to alter the genetic profile of histologically normal tissues surrounding primary tumors (Braakhuis et al., 2003). To avoid any such influence, unrelated, non-inflamed normal tissues were accrued from the GBC site.
A number of genes found differentially expressed in our study cohort have been previously reported in oral or head and neck cancers including INHBA, SERPINE1, GBP5, MMP10, MMP3, PTHLH, KRT4, and MAL (Chung et al., 2006; Chen et al., 2008; Ye et al., 2008; Méndez et al., 2009). In addition, several previously unreported genes such as LY6K, MFAP5, HOXC9, TNFRSF12A, DHRS2, FST, and GPX3 were also found to be differentially expressed and warrant further investigation. A majority of these differentially expressed genes (MMPs, PTHLH, INHBA, LY6K, MFAP5) are secretory proteins and may be utilized as potential serum biomarkers to assess disease aggressiveness.
The literature search revealed 214 genes with valid reporters in our study that had been previously implicated in oral cancer. Among these, 42 genes showed differential expression in our study (Supporting Information Table 8E); for all but one gene, the direction of expression change agreed with previous reports. Forty-one of the 42 genes were among the 129 genes for which the prior evidence was only about differential expression; only one gene, FSTL3, had prior evidence for both differential expression and copy number alterations. Interestingly, 32 of the 42 genes with prior evidence were previously reported in at least one of two studies (Nagata et al., 2003; Kornberg et al., 2005), while other studies in our literature search had few if any genes replicated in our GBC cohort. The concentration of the replicated prior evidence in two studies suggests that the failure to replicate genes across multiple studies of oral cancer may be in part due to heterogeneity between tumor sites which was avoided in our study.
The gain of chromosome band 8q24 is a frequent event in head and neck squamous cell carcinomas, including oral cancers (Huang et al., 2002; Pathare et al., 2009). Previous reports have suggested the overexpression of the known oncogene MYC as the underlying event in the gain of 8q24 (Bhattacharya et al., 2009; Grisanzio and Freedman, 2010). We did not find overexpression of MYC, within this region, indicating that MYC is not primarily regulated via copy number gain; a similar negative finding about MYC has been reported in cervical cancers (Lando et al., 2009). From our study, it appears that LY6K is constitutively overexpressed either by copy number changes, or by yet unknown mechanisms. LY6K belongs to the LY6 family and by sequence analysis, is predicted to encode a protein anchored to the cell membrane via glycosyl-phosphatidyl-inositol (GPI). LY6K is also predicted to be secreted and involved in cell signaling because these functions have been proven for other members of the LY6 protein family (de Nooij-van Dalen et al., 2003), although the precise role of LY6K in carcinogenesis is unknown. Ishikawa et al. have suggested the use of LY6K as a potential serologic and histochemical biomarker for lung and esophageal cancers and its potential activation in a range of tumor samples, including cervical cancers (Ishikawa et al., 2007). We, for the first time, show the overexpression of LY6K in GBC cancers, which adds to the tumor types possibly benefiting from drugs targeting LY6K.
The mystery of how LY6K could be both GPI-anchored and secreted was resolved in a recent study of the orthologous mouse Ly6k in which it is shown that there are two protein isoforms, one anchored and one secreted (Maruyama et al., 2010). The two Ly6k isoforms differ by a post-translational modification that changes the molecular weight, and hence they cannot be distinguished by RT-PCR or any other technique that considers the cDNA/mRNA (Maruyama et al., 2010).
MFAP5 (MAGP2) located in 12p13.1-p12.3 is one of the genes showing over- expression independently of copy number status in GBC cancers that has not been previously reported as differentially expressed in oral cancer. MFAP5 is a secreted factor and an important component in the assembly of microfibrils. It has recently been shown to mediate ovarian tumor cell survival and endothelial cell motility via the alpha(V)beta(3) integrin receptor, and it was associated with chemoresistance and poor clinical outcome (Penner et al., 2002; Mok et al., 2009). The novel finding of increased expression of MFAP5 may be due to specific relevance of this gene to the GBC site or to preponderance (in our cohort) of locally advanced lesions that show higher tendency to migrate and attract blood vessel formation. Considering the secretory nature of MFAP5 and its role in cell motility and survival, we hypothesize that it may be utilized as a serum biomarker to assess disease aggressiveness.
An analysis of affected signaling pathways using the DAVID database (Supporting Information Text and Supporting Information Table 9) indicated an overrepresentation of differentially expressed genes in the KEGG pathways related to extracellular matrix receptor interaction, cytokine-cytokine receptor interaction, focal adhesion, arachidonic acid metabolism, chemokine signalling, cell adhesion molecules and complement and coagulation cascades. Yet, the power of this approach is challenged by the large number of tests performed, and the significance is weak after correcting for multiple testing. We selected a few genes from these pathways based on their very high (SPP1, CA9, FST, INHBA, TNFRSF12A) or low (GPX3) expression levels in tumors. We also validated genes previously unexplored in oral cancers, including LY6K, HOXC9, DHRS2 and MFAP5. We selected MAL because it was reported to be associated with disease progression and metastasis in head and neck tumors (Beder et al., 2009), which may share some risk factors with GBC tumors.
Cytokine-cytokine receptor interactions in the tumor milieu play an important role in cancer pathogenesis. Cancer cells can respond to host-derived cytokines that promote growth, attenuate apoptosis and facilitate invasion and metastasis (Dranoff, 2004). Our analysis revealed overexpression of INHBA, TNFRSF12A and CXCL chemokines (CXCL10, CXCL12, CXCL13). One function of cytokines is to trigger inflammatory responses that play decisive roles at different stages of tumor development (Grivennikov et al., 2010). Arachidonic acid and its inflammation-related metabolites play a role in tumor biology (Hyde and Missailidis, 2009). We report the downregulation of many genes in this pathway including ALOX12, EPHX2, PTGDS and GPX3. Reactive oxygen species (ROS) generated during tobacco usage are reported to cause increased genomic instability, growth and invasion (Nair et al., 2004; Yu et al., 2007). As most of the patients in our study are tobacco users, we speculate that the reduced levels of the GPX3 enzyme may make the oral epithelial cells unable to quench ROS, contributing to tumor progression.
Integration of gene expression data with array CGH data revealed that gene expression and copy number alterations were coherently linked for the majority of genes located in recurring CNAs. Because the pattern of genomic alterations varies substantially between patients, the correlation between expression and copy number implies a similar heterogeneity among patients, at the level of gene expression. As a consequence, it is a challenge to identify subsets of patients with differential prognosis or who respond to different treatments based on either genomic alterations or gene expression alone. Another challenge is the identification of candidate driver genes and their distinction from passengers (Akavia et al., 2010). Based on the wide-spread gene-dosage effect on mRNA expression observed here, it appears that a correlation of gene expression and DNA copy number is, on its own, a rather unspecific criterion. On the contrary, one may argue that driver genes essential for carcinogenesis should be constitutively differentially expressed also in the absence of a CNA, and may be distinguished from passenger genes whose expression mostly reflects the copy number status. An additional criterion to identify a driver gene could be the correlation of expression with clinicopathological parameters such as lymph node invasion or survival (Santarius et al., 2010).
To get further insights into the role of genes located in regions of recurring genomic alterations, we assessed their prognostic relevance. As the identification of genes underlying large CNAs remains difficult, we restricted the analysis to genes in focal alterations that were found to correlate with copy number status. We found that the expression of genes located in 2 distinct regions, 11q13–2 q13.3 and 11q22.1-q22.2 serve as the strongest survival predictors.
A gain of the 11q13.2–q13.3 region containing 12 genes was observed in 37.7% of all the GBC cancers analyzed. Our results identified the expression of ORAOV1, FGF3, FADD, and PPFIA1 as strong predictors of patient survival. A systematic analysis of gene expression and amplification of the same region by Huang et al. (2006) found that, among others, the overexpression of ORAOV1, FADD, PPFIA1 was copy number dependent, while the overexpression of ANO1 (Anoctamin 1, formerly ORAOV2, oral cancer overexpressed 2) was not (Huang et al., 2006). These findings are in agreement with a recent report by Ayoub et al., where a copy number dependent expression of PPFIA1, OROAV1, FADD, and ANO1 was observed. Further, the study identified ANO1 and ORAOV1 as candidate genes for predicting metastasis in head and neck squamous cell carcinoma (Ayoub et al., 2010).
The consistent overexpression of ORAOV1 (oral cancer overexpressed 1) in all three studies, including ours, underlines its functional importance in head and neck, and oral squamous cell carcinomas. ORAOV1 regulates angiogenesis and tumor growth in OSCC (Jiang et al., 2008) and a recent report identified its role in regulating cell cycle and apoptosis (Jiang et al., 2010). Moreover, we found a tendency for overexpression of ANO1 in tumors (logFC = 0.7, P = 0.2) in addition to a strong copy number dependent effect (logFC = 1.61, P = 10−5), in line with recent reports (Huang et al., 2006; Kashyap et al., 2009; Ayoub et al., 2010). In our study, however, ANO1 was not found as a survival predictor. FADD (Fas (TNFRSF6)-associated via death domain) mediates apoptotic signaling by binding to different cell-surface receptors of the tumor necrosis factor superfamily. Recently, FADD was found to be overexpressed in another study of oral cancer (Prapinjumrune et al., 2010). FGF3 (fibroblast growth factor 3) expression was found to be variable in this study, in agreement with the findings of Huang et al. (2006), and a lower expression was associated with worse prognosis. PPFIA1 (protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1) regulates cell-matrix interactions and displayed a copy number dependent overexpression (logFC = 1.29, P = 3 · 10−6). Interestingly, higher PPFIA1 expression was found to coincide with a better prognosis, in line with recent findings in HNSCC (Tan et al., 2008). It thus appears that the complex process of tumor progression is a consequence of the combined effect of multiple driver genes in 11q13..
We previously reported a gain of 11q22.1-q22.2 in 14.8% of the GBC cases and found it to be associated with poor prognosis in oral cancers (Ambatipudi et al., 2011). This amplicon caused the highest average fold-change in gene expression and it includes the genes BIRC3, BIRC2, TMEM123, YAP1, MMP7, and MMP10 (Supporting Information Table 6). The expression of the first five increases approximately by a factor of two in the presence of the CNA. MMP10, however, is consistently overexpressed (logFC=5.1, P=0.002), irrespective of the copy number status. Snijders et al. reported the importance of this amplicon in oral cancers suggesting that MMP7, YAP1, BIRC2, and BIRC3 could be driver genes (Snijders et al., 2005). However, they observed very little overexpression of BIRC3 (baculoviral IAP repeat-containing 3) when amplified (Snijders et al., 2005). Overexpression of BIRC2 and BIRC3 which encode IAP1 and IAP2 respectively, is consistent with other evidence that these apoptosis-related genes are coordinately regulated (Conze et al., 2005). Overexpression of BIRC3, but not BIRC2, was associated with poor clinical outcome in our study cohort thus suggesting that BIRC3 is the driver gene.
BIRC3 encodes a protein (IAP2) that inhibits apoptosis by binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2. Tumors with 11q22.1-q22.2 amplification could thus gain aggressiveness through copy number dependent BIRC3 overexpression which helps tumor cells to evade hypoxia induced apoptosis (Kilic et al., 2007; Lando et al., 2009). Overexpression of BIRC3/cIAP2 has been implicated in the progression of both solid and haematological malignancies (Friboulet et al., 2008; Karasawa et al., 2009; Grzybowska-Izydorczyk et al., 2010; Wu et al., 2010). When cellular inhibitor of apoptosis proteins (c-IAPs) are overexpressed or overly active, as in many cancers, cells are no longer able to die in a physiologically programmed fashion and become increasingly resistant to standard radiation and chemotherapies (Hunter et al., 2007). Higher levels of cIAP2 inhibit the effectiveness of commonly used cancer drugs 5-Flurouracil (Karasawa et al., 2009) and cisplatin (Wu et al., 2010). Both cIAP1 and cIAP2, and also other proteins with IAP domains can be inhibited pharmacologically and are the molecular targets of various cancer clinical trials (Vucic and Fairbrother, 2007).
Cancer is a complex multigenic and multifactorial disease (Hanahan and Weinberg, 2011) and the presence of a single driver gene on a chromosomal location may not be the sole requirement for GBC cancer progression. It is important to identify the gene(s) in a region of copy number change that provide a selective advantage during the process of clonal evolution and tumor progression. Based on our integrative analysis, we hypothesize that the driver genes ORAOV1 and BIRC3 may interact with each other or with other genes in their 11q amplicons to evade apoptosis in GBC cancer cells. The possible interactions between genes on 11q and the underlying molecular mechanisms need further investigation.
In summary, our study analyzed the gene expression patterns in GBC cancers. We reported several novel and previously reported genes contributing to various pathways deregulated in GBC cancers. The integrative analysis revealed a widespread dosage effect of CNAs on global gene expression. The study could identify the driver gene BIRC3 amongst several individually expressed genes located in the focal gain of 11q22.1-q22.2 previously shown associated with poor clinical outcome in oral cancers. Further, the genes so identified, including some of the secretory factors we validated, may serve as potential biomarkers with prognostic value to assess clinical outcome of GBC cancer patients.
Supplementary Material
Acknowledgments
Supported by: The Lady Tata Memorial Trust, Mumbai and the Council of Scientific and Industrial Research [CSIR Grant No: 27(0207)/09/EMR-II]. This research was supported in part by the Intramural Research program of the National Institutes of Health, NLM. MG and NB received funding from SystemsX.ch, the Swiss initiative in systems biology, under grant No. 2009/024, evaluated by the Swiss National Science Foundation. SA was supported by a fellowship from CSIR during his tenure as a PhD student.
The authors thank all participants of the study. ICMR National Tumour Tissue Repository, Tata Memorial Centre, Mumbai is acknowledged for providing tumor tissues. They also thank two anonymous reviewers for their thoughtful suggestions that helped to improve the manuscript.
References
- Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Ambatipudi S, Gerstung M, Gowda R, Pai P, Borges A, Schäffer AA, Beerenwinkel N, Mahimkar MB. Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome. PLoS One. 2011;6:e17250. doi: 10.1371/journal.pone.0017250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub C, Wasylyk C, Li Y, Thomas E, Marisa L, Robe A, Roux M, Abecassis J, de Reynies A, Wasylyk B. ANO1 amplification and expression in HNSCC with a high propensity for future distant metastasis and its functions in HNSCC cell lines. Br J Cancer. 2010;103:715–726. doi: 10.1038/sj.bjc.6605823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beder LB, Gunduz M, Hotomi M, Fujihara K, Shimada J, Tamura S, Gunduz E, Fukushima K, Yaykasli K, Grenman R, Shimizu K, Yamanaka N. T-lymphocyte maturation-associated protein gene as a candidate metastasis suppressor for head and neck squamous cell carcinomas. Cancer Sci. 2009;100:873–880. doi: 10.1111/j.1349-7006.2009.01132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Soc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bhattacharya N, Roy A, Roy B, Roychoudhury S, Panda CK. MYC gene amplification reveals clinical association with head and neck squamous cell carcinoma in Indian patients. J Oral Pathol Med. 2009;38:759–763. doi: 10.1111/j.1600-0714.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- Carinci F, Lo Muzio L, Piattelli A, Rubini C, Chiesa F, Ionna F, Palmieri A, Maiorano E, Pastore A, Laino G, Dolci M, Pezzetti F. Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunopathol Pharmacol. 2005;18:513–524. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]
- Chen C, Méndez E, Houck J, Fan W, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG, Schwartz SM, Zhao LP. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2152–2162. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CH, Parker JS, Ely K, Carter J, Yi Y, Murphy BA, Ang KK, El-Naggar AK, Zanation AM, Cmelak AJ, Levy S, Slebos RJ, Yarbrough WG. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-κB signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res. 2006;66:8210–8218. doi: 10.1158/0008-5472.CAN-06-1213. [DOI] [PubMed] [Google Scholar]
- Conze DB, Albert L, Ferrick DA, Goeddel DV, Yeh WC, Mak T, Ashwell JD. Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol Cell Biol. 2005;25:3348–3356. doi: 10.1128/MCB.25.8.3348-3356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nooij-van Dalen AG, van Dongen GAMS, Smeets SJ, Nieuwenhuis EJC, Stigter-van Walsum M, Snow GB, Brakenhoff RH. Characterization of the human Ly-6 antigens, the newly annotated member Ly-6K included, as molecular markers for head-and-neck squamous cell carcinoma. Int J Cancer. 2003;103:768–774. doi: 10.1002/ijc.10903. [DOI] [PubMed] [Google Scholar]
- Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- Elston RC. On Fisher’s method of combining p-values. Biom. J. 1991;33:339–345. [Google Scholar]
- Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, Kraus DH, Patel S, Shaha AR, Wong RJ, Huryn JM, Shah JP, Singh B. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friboulet L, Pioche-Durieu C, Rodriguez S, Valent A, Souquere S, Ripoche H, Khabir A, Tsao SW, Bosq J, Lo KW, Busson P. Recurrent overexpression of c-IAP2 in EBV-associated nasopharyngeal carcinomas: critical role in resistance to Toll-like receptor 3-mediated apoptosis. Neoplasia. 2008;10:1183–1194. doi: 10.1593/neo.08590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz EM, Sengupta K, Difilippantonio MJ, Ried T, Schaffer AA. Evaluating annotations of an Agilent expression chip suggests that many features cannot be interpreted. BMC Genomics. 2009;10:566. doi: 10.1186/1471-2164-10-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisanzio C, Freedman ML. Chromosome 8q24-Associated Cancers and MYC. Genes Cancer. 2010;1:555–559. doi: 10.1177/1947601910381380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowska-Izydorczyk O, Cebula B, Robak T, Smolewski P. Expression and prognostic significance of the inhibitor of apoptosis protein (IAP) family and its antagonists in chronic lymphocytic leukaemia. Eur J Cancer. 2010;46:800–810. doi: 10.1016/j.ejca.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Huang Q, Yu GP, McCormick SA, Mo J, Datta B, Mahimkar M, Lazarus P, Schäffer AA, Desper R, Schantz SP. Genetic differences detected by comparative genomic hybridization in head and neck squamous cell carcinomas from different tumor sites: construction of oncogenetic trees for tumor progression. Genes Chromosomes Cancer. 2002;34:224–233. doi: 10.1002/gcc.10062. [DOI] [PubMed] [Google Scholar]
- Huang X, Godfrey TE, Gooding WE, McCarty KS, Jr, Gollin SM. Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosomes Cancer. 2006;45:1058–1069. doi: 10.1002/gcc.20371. [DOI] [PubMed] [Google Scholar]
- Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- Hyde CAC, Missailidis S. Inhibition of arachidonic acid metabolism and its implication on cell proliferation and tumour-angiogenesis. Int Immunopharmacol. 2009;9:701–715. doi: 10.1016/j.intimp.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringnér M, Sauter G, Monni O, Elkahloun A, Kallioniemi O-P, Kallioniemi A. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- Ishikawa N, Takano A, Yasui W, Inai K, Nishimura H, Ito H, Miyagi Y, Nakayama H, Fujita M, Hosokawa M, Tsuchiya E, Kohno N, Nakamura Y, Daigo Y. Cancer-testis antigen lymphocyte antigen 6 complex locus K is a serologic biomarker and a therapeutic target for lung and esophageal carcinomas. Cancer Res. 2007;67:11601–11611. doi: 10.1158/0008-5472.CAN-07-3243. [DOI] [PubMed] [Google Scholar]
- Järvinen A-K, Autio R, Haapa-Paananen S, Wolf M, Saarela M, Grénman R, Leivo I, Kallioniemi O, Mäkitie AA, Monni O. Identification of target genes in laryngeal squamous cell carcinoma by high-resolution copy number and gene expression microarray analyses. Oncogene. 2006;25:6997–7008. doi: 10.1038/sj.onc.1209690. [DOI] [PubMed] [Google Scholar]
- Järvinen A-K, Autio R, Kilpinen S, Saarela M, Leivo I, Grénman R, Mäkitie AA, Monni O. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer. 2008;47:500–509. doi: 10.1002/gcc.20551. [DOI] [PubMed] [Google Scholar]
- Jiang L, Zeng X, Wang Z, Ji N, Zhou Y, Liu X, Chen Q. Oral cancer overexpressed 1 (ORAOV1) regulates cell cycle and apoptosis in cervical cancer HeLa cells. Mol Cancer. 2010;9:20. doi: 10.1186/1476-4598-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zeng X, Yang H, Wang Z, Shen J, Bai J, Zhang Y, Gao F, Zhou M, Chen Q. Oral cancer overexpressed 1 (ORAOV1): a regulator for the cell growth and tumor angiogenesis in oral squamous cell carcinoma. Int J Cancer. 2008;123:1779–1786. doi: 10.1002/ijc.23734. [DOI] [PubMed] [Google Scholar]
- Karasawa H, Miura K, Fujibuchi W, Ishida K, Kaneko N, Kinouchi M, Okabe M, Ando T, Murata Y, Sasaki H, Takami K, Yamamura A, Shibata C, Sasaki I. Down-regulation of cIAP2 enhances 5-FU sensitivity through the apoptotic pathway in human colon cancer cells. Cancer Sci. 2009;100:903–913. doi: 10.1111/j.1349-7006.2009.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap MK, Marimuthu A, Kishore CJH, Peri S, Keerthikumar S, Prasad TSK, Mahmood R, Rao S, Ranganathan P, Sanjeeviah RC, Vijayakumar M, Kumar KV, Montgomery EA, Kumar RVV, Pandey A. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol Ther. 2009;8:36–46. doi: 10.4161/cbt.8.1.7090. [DOI] [PubMed] [Google Scholar]
- Kilic M, Kasperczyk H, Fulda S, Debatin KM. Role of hypoxia inducible factor-1 alpha in modulation of apoptosis resistance. Oncogene. 2007;26:2027–2038. doi: 10.1038/sj.onc.1210008. [DOI] [PubMed] [Google Scholar]
- Kornberg LJ, Villaret D, Popp M, Lui L, McLaren R, Brown H, Cohen D, Yun J, McFadden M. Gene expression profiling in squamous cell carcinoma of the oral cavity shows abnormalities in several signaling pathways. Laryngoscope. 2005;115:690–698. doi: 10.1097/01.mlg.0000161333.67977.93. [DOI] [PubMed] [Google Scholar]
- Lando M, Holden M, Bergersen LC, Svendsrud DH, Stokke T, Sundfor K, Glad IK, Kristensen GB, Lyng H. Gene dosage, expression, and ontology analysis identifies driver genes in the carcinogenesis and chemoradioresistance of cervical cancer. PLoS Genet. 2009;5:e1000719. doi: 10.1371/journal.pgen.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, Jones S, Sjöblom T, Park B-H, Parsons R, Willis J, Dawson D, Willson JKV, Nikolskaya T, Nikolsky Y, Kopelovich L, Papadopoulos N, Pennacchio LA, Wang T-L, Markowitz SD, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M, Yoshitake H, Tsukamoto H, Takamori K, Araki Y. Molecular expression of Ly6k, a putative glycosylphosphatidyl-inositol-anchored membrane protein on the mouse testicular germ cells. Biochem Biophys Res Commun. 2010;402:75–81. doi: 10.1016/j.bbrc.2010.09.117. [DOI] [PubMed] [Google Scholar]
- Méndez E, Houck JR, Doody DR, Fan W, Lohavanichbutr P, Rue TC, Yueh B, Futran ND, Upton MP, Farwell DG, Heagerty PJ, Zhao LP, Schwartz SM, Chen C. A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin Cancer Res. 2009;15:1353–1361. doi: 10.1158/1078-0432.CCR-08-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong K-K, Park D-C, Hao K, Yip DKP, Donninger H, Ozbun L, Samimi G, Brady J, Randonovich M, Pise-Masison CA, Barrett JC, Wong WH, Welch WR, Berkowitz RS, Birrer MJ. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata M, Fujita H, Ida H, Hoshina H, Inoue T, Seki Y, Ohnishi M, Ohyama T, Shingaki S, Kaji M, Saku T, Takagi R. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer. 2003;106:683–689. doi: 10.1002/ijc.11283. [DOI] [PubMed] [Google Scholar]
- Nair U, Bartsch H, Nair J. Alert for an epidemic of oral cancer due to use of the betel quid substitutes gutkha and pan masala: a review of agents and causative mechanisms. Mutagenesis. 2004;19:251–262. doi: 10.1093/mutage/geh036. [DOI] [PubMed] [Google Scholar]
- Pathak KA, Gupta S, Talole S, Khanna V, Chaturvedi P, Deshpande MS, Pai PS, Chaukar DA, D’Cruz AK. Advanced squamous cell carcinoma of lower gingivobuccal complex: patterns of spread and failure. Head Neck. 2005;27:597–602. doi: 10.1002/hed.20195. [DOI] [PubMed] [Google Scholar]
- Pathare S, Schäffer AA, Beerenwinkel N, Mahimkar M. Construction of oncogenetic tree models reveals multiple pathways of oral cancer progression. Int J Cancer. 2009;124:2864–2871. doi: 10.1002/ijc.24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner AS, Rock MJ, Kielty CM, Shipley JM. Microfibril-associated glycoprotein-2 interacts with fibrillin-1 and fibrillin-2 suggesting a role for MAGP-2 in elastic fiber assembly. J Biol Chem. 2002;277:35044–35049. doi: 10.1074/jbc.M206363200. [DOI] [PubMed] [Google Scholar]
- Prapinjumrune C, Morita K, Kuribayashi Y, Hanabata Y, Shi Q, Nakajima Y, Inazawa J, Omura K. DNA amplification and expression of FADD in oral squamous cell carcinoma. J Oral Pathol Med. 2010;39:525–532. doi: 10.1111/j.1600-0714.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- Roepman P, de Jager A, Groot Koerkamp MJA, Kummer JA, Slootweg PJ, Holstege FCP. Maintenance of head and neck tumor gene expression profiles upon lymph node metastasis. Cancer Res. 2006;66:11110–11114. doi: 10.1158/0008-5472.CAN-06-3161. [DOI] [PubMed] [Google Scholar]
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- Shimada K, Uzawa K, Kato M, Endo Y, Shiiba M, Bukawa H, Yokoe H, Seki N, Tanzawa H. Aberrant expression of RAB1A in human tongue cancer. Br J Cancer. 2005;92:1915–1921. doi: 10.1038/sj.bjc.6602594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3.3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RCK, Albertson DG. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Sticht C, Freier K, Knöpfle K, Flechtenmacher C, Pungs S, Hofele C, Hahn M, Joos S, Lichter P. Activation of MAP kinase signaling through ERK5 but not ERK1 expression is associated with lymph node metastases in oral squamous cell carcinoma (OSCC) Neoplasia. 2008;10:462–470. doi: 10.1593/neo.08164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KD, Zhu Y, Tan HK, Rajasegaran V, Aggarwal A, Wu J, Wu HY, Hwang J, Lim DT, Soo KC, Tan P. Amplification and overexpression of PPFIA1, a putative 11q13 invasion suppressor gene, in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2008;47:353–362. doi: 10.1002/gcc.20539. [DOI] [PubMed] [Google Scholar]
- Vucic D, Fairbrother WJ. The inhibitor of apoptosis proteins as therapeutic targets in cancer. Clin Cancer Res. 2007;13:5995–6000. doi: 10.1158/1078-0432.CCR-07-0729. [DOI] [PubMed] [Google Scholar]
- Wu HH, Wu JY, Cheng YW, Chen CY, Lee MC, Goan YG, Lee H. cIAP2 upregulated by E6 oncoprotein via epidermal growth factor receptor/phosphatidylinositol 3-kinase/AKT pathway confers resistance to cisplatin in human papillomavirus 16/18-infected lung cancer. Clin Cancer Res. 2010;16:5200–5210. doi: 10.1158/1078-0432.CCR-10-0020. [DOI] [PubMed] [Google Scholar]
- Xu C, Liu Y, Wang P, Fan W, Rue TC, Upton MP, Houck JR, Lohavanichbutr P, Doody DR, Futran ND, Zhao LP, Schwartz SM, Chen C, Méndez E. Integrative analysis of DNA copy number and gene expression in metastatic oral squamous cell carcinoma identifies genes associated with poor survival. Mol Cancer. 2010;9:143. doi: 10.1186/1476-4598-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YP, Yu G, Tseng G, Cieply K, Nelson J, Defrances M, Zarnegar R, Michalopoulos G, Luo J-H. Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 2007;67:8043–8050. doi: 10.1158/0008-5472.CAN-07-0648. [DOI] [PubMed] [Google Scholar]
- Zhou X, Temam S, Oh M, Pungpravat N, Huang B-L, Mao L, Wong DT. Global expression-based classification of lymph node metastasis and extracapsular spread of oral tongue squamous cell carcinoma. Neoplasia. 2006;8:925–932. doi: 10.1593/neo.06430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.