Abstract
Cys-loop receptor binding sites characteristically possess an “aromatic box,” where several aromatic amino acid residues surround the bound ligand. A cation-π interaction between one of these residues and the natural agonist is common, although the residue type and location are not conserved. Even in the closely related vertebrate GABAA and GABAC receptors, residues in distinct locations perform this role: in GABAA receptors, a Tyr residue in loop A forms a cation-π interaction with GABA, while in GABAC receptors it is a loop B residue. GABA-activated Cys-loop receptors also exist in invertebrates, where they have distinct pharmacologies and are the target of a range of pesticides. Here we examine the location of GABA in an insect binding site by incorporating a series of fluorinated Phe derivatives into the receptor binding pocket using unnatural amino acid mutagenesis, and evaluating the resulting receptors when expressed in Xenopus oocytes. A homology model suggests that two aromatic residues (in loops B and C) are positioned such that they could contribute to a cation-π interaction with the primary ammonium of GABA, and the data reveal a clear correlation between the GABA EC50 and the cation-π binding ability both at Phe206 (loop B) and Tyr254 (loop C), demonstrating for the first time the contribution of two aromatic residues to a cation-π interaction in a Cys-loop receptor.
Introduction
Cys-loop receptors are comprised of five subunits arranged around a central ion-conducting pore. Each subunit consists of a large extracellular N-terminal domain and four transmembrane helices (Unwin, 1995; Miyazawa et al., 2003). Structural studies on acetylcholine binding proteins (AChBPs) (Brejc et al., 2001) and prokaryotic Cys-loop receptor homologs (Hilf and Dutzler, 2008, 2009; Bocquet et al., 2009) have confirmed the location of the agonist binding site at the interface of adjacent subunits. Three loops from one subunit (A–C) and three β strands from the adjacent subunit (“loops” D–F) constitute the binding pocket.
Homology models have been created for a range of Cys-loop receptors including nACh, 5-HT3, GABA, and glycine receptors, and support previous mutagenesis and labeling studies in showing that aromatic residues in the binding loops contribute to the formation of an “aromatic box” (Beene et al., 2002, 2004; Harrison and Lummis, 2006a; Padgett et al., 2007; Pless et al., 2007). However, the roles of specific residues are not necessarily equivalent for different receptors; even in the closely related GABAA and GABAC receptors, for example, GABA has different orientations in the binding pocket: In the former, there is a cation-π interaction between GABA and a Tyr residue in loop A, while in the latter this interaction is with a Tyr in loop B (Lummis et al., 2005; Padgett et al., 2007).
There is also a range of invertebrate Cys-loop receptors activated by GABA, but molecular details of insect GABA receptor binding sites are mostly poorly understood. The best studied insect GABA receptor is the RDL receptor, which is constituted from RDL subunits, products of the rdl (Resistance to DieLdrin) gene. These receptors, which can be expressed as homomers in Xenopus oocytes, are inhibited by picrotoxin, but not by bicuculline, both classic GABAA receptor antagonists, characteristics similar to GABA receptors of cultured Drosophila neurons (Zhang et al., 1994). A recent model we have proposed for this receptor indicates that two residues, Tyr206 in loop B and Phe 254 in loop C, both have the potential to form a cation-π interaction with the ammonium ion of GABA. In this study we used nonsense suppression methodology (Dougherty, 2000; Beene et al., 2003) to incorporate unnatural amino acids into the RDL binding site to determine which, if any, of the aromatic loop residues, form a cation-π interaction with GABA. The data suggest that both of the aromatic groups on these amino acids are involved.
Materials and Methods
Mutagenesis and preparation of mRNA and oocytes.
RDL subunit cDNA was subcloned from pRmHa3-RDL into pGEMHE for oocyte expression as previously described (Millar et al., 1994; McGonigle and Lummis, 2010). Site-directed mutagenesis was performed with the QuikChange Mutagenesis Kit (Stratagene). The mMessage mMachine Kit (Ambion) was used to generate capped mRNA for oocyte injection. Xenopus laevis (Nasco) oocytes from female frogs were prepared as previously described (Padgett et al., 2007) and injected with 5 ng of cRNA. After injection, oocytes were incubated for 24–48 h at 18°C.
Synthesis of tRNA and dCA amino acids.
This was performed as previously described (Padgett et al., 2007). In brief, unnatural amino acids (as shown in Fig. 1) were chemically synthesized as NVOC (nitroveratryloxycarbonyl)-protected cyanomethyl esters. They were coupled to the dinucleotide dCA that was enzymatically ligated to a 74-mer THG73 tRNACUA as described previously (Nowak et al., 1998). Directly before coinjection with the mRNA, the aminoacyl tRNA was deprotected by photolysis (Kearney et al., 1996). In a typical experiment, 10 ng of mRNA were injected with 25 ng of tRNA-aa in a total volume of 50 nl. In control experiments, mRNA was injected alone or together with THG74-dCA tRNA (with no unnatural amino acid attached).
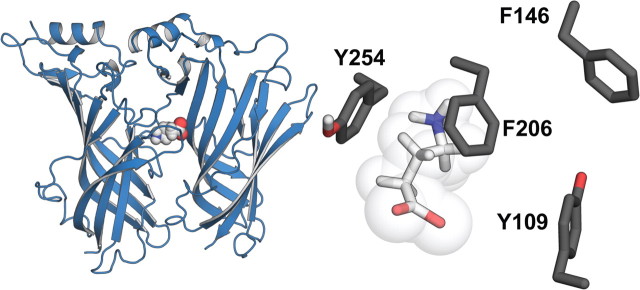
Figure 1.
Homology model of the extracellular domain of the RDL receptor based upon the crystal structure of AChBP with carbamylcholine bound (PDB ID; 1uv6). Only two of the five subunits have been shown for ease of viewing. The ligand binding site is located between the two subunits. Inset, Enlargement showing GABA docked into the binding site and surrounded by the “aromatic box” residues.
Characterization of mutant receptors.
GABA-induced currents were recorded from individual oocytes using the OpusXpress voltage-clamp system (Molecular Devices). All recordings were performed at 22–25°C. GABA (Sigma) was stored as 1 m aliquots at −20°C and diluted in ND96 (96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2, 5 mm HEPES, pH 7.4) before the experiment and delivered to the cells using the computer-controlled perfusion system of the OpusXpress. Glass microelectrodes were backfilled with 3 m KCl and had resistances between 0.5 and 3 MΩ. All recordings were performed at a holding potential of −60 mV. Analysis and curve fitting were performed using Prism V3.02 (GraphPad Software, www.graphpad.com). Concentration–response data for each oocyte were normalized to the maximum current for that oocyte. Responses were normalized to a maximal GABA evoked response. The mean response for a series of oocytes was iteratively fitted to the following equation: I = Imin + (Imax − Imin)/(1 + 10log(EC50−[A])nH), where Imax = maximal response, Imin = minimum response, [A] is the concentration of agonist, and nH is the Hill coefficient. Significance was calculated using a one-way ANOVA or an unpaired t test (Prism v3.02).
Homology modeling and docking.
Homology modeling and docking were as previously described (Harrison and Lummis, 2006). Briefly, the protein sequence of the RDL subunit was aligned with the sequence of AChBP (P58154) using FUGUE (Shi et al., 2001), and a three-dimensional homology model was generated using MODELLER 6v2 (Sali and Blundell, 1993) based on the crystal structure of AChBP (Brejc et al., 2001). The pentamer was generated by superimposing the RDL extracellular domain onto each protomer of AChBP and then was energy minimized using the force field implemented in MODELLER 6v2.
Docking of GABA into the RDL receptor homology model was performed using GOLD 3.0. The binding site was defined using the C atoms of conserved aromatics Phe206, Tyr254, and Tyr109, with a binding site radius of 10 Å. Hydrogen bonding interactions between ligands and binding site residues were identified using the hydrogen bond monitor function in ViewerLite.
Results
Functional responses
Application of GABA to Xenopus oocytes expressing RDL receptors elicited concentration-dependent inward currents. Plotting current amplitude against a range of GABA concentrations revealed an EC50 of 17 μm (Fig. 2), similar to previously published results (10–31 μm) (Belelli et al., 1996; Hosie and Sattelle, 1996a,b; McGurk et al., 1998; Hosie et al., 2001).
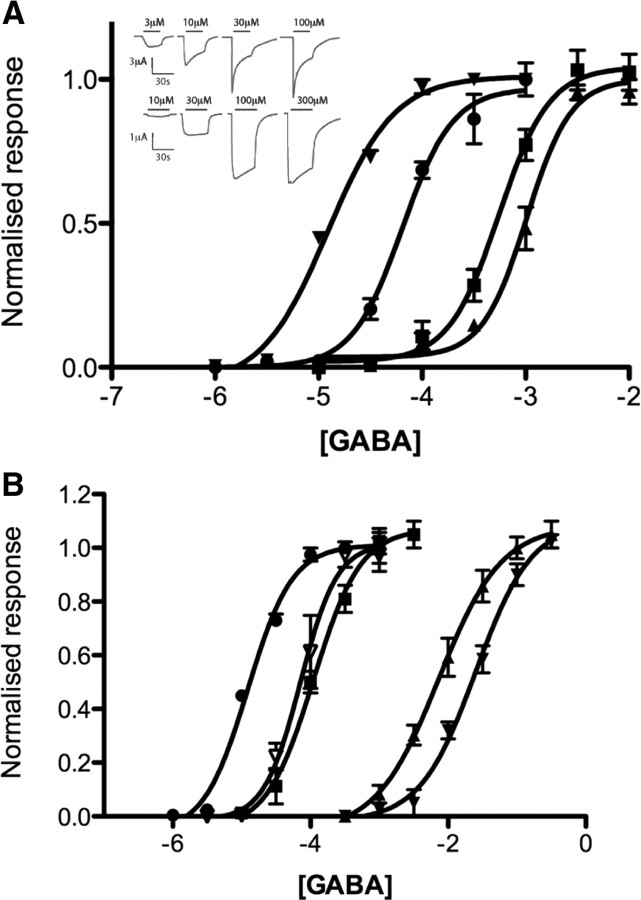
Figure 2.
GABA concentration response curves responses. A, GABA-induced current dose–response curves showing incremental increases in EC50 when RDL F206TAG mRNA was coinjected with Phe (▾), F-Phe (●), F2-Phe (■), and F3-Phe (▴) tRNA. Inset, Typical responses to a range of GABA concentrations in WT (upper) and FPhe-containing (lower) receptors. B, GABA-induced current dose–response curves showing incremental increases in EC50 when RDL Y254TAG mRNA was coinjected with Phe ▿, F-Phe ■, F2-Phe ▴, and F3-Phe ▾ tRNA. ●, WT.
Validation of the nonsense suppression method
Fluorinated Phe residues were incorporated using nonsense suppression. This method involves mutation the codon of the residue of interest to TAG (a nonsense codon), synthesizing mRNA, and then coinjecting this with a “suppressor” tRNA whose anticodon recognizes TAG. The acceptor site of this tRNA has been chemically ligated to the desired unnatural amino acid, which is incorporated into the peptide sequence in the same way as a “natural” amino acid. To validate the method, we coinjected the mutant mRNAs encoding Phe206TAG or Tyr254TAG receptors, and a suppressor tRNA chemically aminoacylated with Phe or Tyr, respectively. Mutant mRNAs produced functional receptors that responded to the application of GABA with EC50 and Hill coefficients similar to wild type. We thus conclude that the wild-type phenotype is successfully “rescued” by the nonsense suppression method.
No currents were recorded in response to high (5 mm) concentrations of GABA from oocytes injected with mutant mRNA alone or with mutant mRNA and tRNA-dCA with no amino acid attached (THG73 76-mer tRNA). This indicates that the stop codon at the position of the mutated residue, as expected, generates truncated and consequently nonfunctional receptors. Additionally, it shows that no endogenous tRNA-aa of the oocyte incorporates and functions at the mutated sites. Finally, endogenous synthetases do not seem to reacylate the tRNAs with naturally occurring amino acids (that function at this site in the channel) at a high rate, as we did not detect currents when mRNA and nonligated tRNA were coinjected.
Locating the residues of interest
In this study, we chose to focus on Phe206 (loop B) and Tyr254 (loop C), as aligning residues have been implicated in cation-π interactions in previous studies (Beene et al., 2002, 2004; Lummis et al., 2005; Harrison and Lummis, 2006a; Padgett et al., 2007; Pless et al., 2007). We initially also considered Phe146 (loop A), but substitution of this residue with Ala or with F3-Phe caused only a small increase in GABA EC50 (<4-fold), indicating that this residue did not contribute to a cation-π interaction. Substitution of Phe206 with Ala, however, resulted in an ∼50-fold increase in EC50, while Tyr254Ala receptors did not respond to GABA (up to 100 mm). We also examined substitution at these sites with an alternative aromatic: surprisingly, Phe206Tyr receptors had a large decrease (∼200-fold) in EC50 compared to WT, while substitution of Tyr 254 with Phe resulted in an ∼4-fold increase in EC50.
Phe206 forms a cation-π interaction with GABA
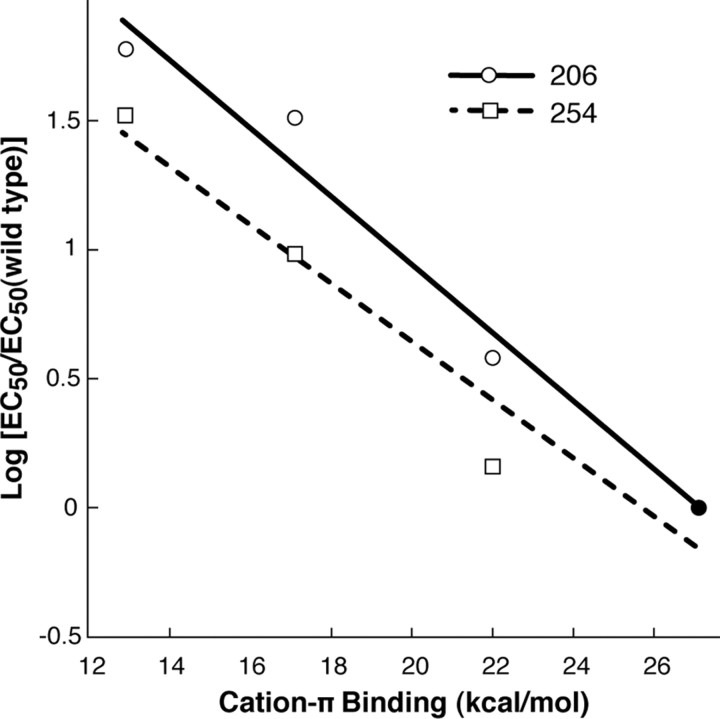
Successive addition of fluorine atoms to phenylalanine derivatives at position 206 in loop B led to a stepwise increase in the GABA EC50. Averaged dose–response data for all constructs are shown in Figure 2, with parameters of best fit summarized in Table 1. These results strongly suggest a cation-π interaction between GABA and Phe206. The resulting cation-π plot (Zhong et al., 1998) shows a strong linear correlation between the relative log EC50 value (scaled to WT) and the cation-π binding ability of the incorporated phenylalanine derivative (Fig. 3), and the consistency of this trend suggests that GABA forms a cation-π interaction with the phenylalanine at position 206 of the receptor.
Table 1.
Dose–response data for RDL with unnatural amino acids at positions 206 and 254
| Mutant | Log EC50 (m) | EC50 (μm) | nH | n |
|---|---|---|---|---|
| WT | −4.909 ± 0.34 | 17 | 1.4 ± 0.2 | 7 |
| F206A | >300 | 4 | ||
| F206Y | −6.941 ± 0.09 | 0.11 | 2.2 ± 0.3 | 6 |
| F206-FPhe | −4.190 ± 0.05 | 65 | 1.8 ± 0.3 | 5 |
| F206-F2Phe | −3.257 ± 0.05 | 554 | 1.8 ± 0.3 | 6 |
| F206-F3Phe | −2.990 ± 0.04 | 1022 | 2.0 ± 0.4 | 6 |
| Y254A | NF | 8 | ||
| Y254F | −4.050 ± 0.05 | 76 | 2.1 ± 0.4 | 4 |
| Y254-FPhe | −3.995 ± 0.07 | 110 | 1.3 ± 0.4 | 6 |
| Y254F2-Phe | −2.135 ± 0.08 | 733 | 1.1 ± 0.2 | 4 |
| Y254F3-Phe | −1.599 ± 0.07 | 2518 | 1.2 ± 0.3 | 7 |
| F206Y/Y254F | −5.754 ± 0.07 | 1.8 | 1.9 ± 0.5 | 7 |
| F146A | −4.140 ± 0.12 | 73 | 2.1 ± 0.2 | 5 |
| F146F3Phe | −4.084 ± 0.15 | 82 | 1.4 ± 0.4 | 4 |
All values are expressed as mean and SEM.
Figure 3.
Fluorination plots for residues 206 and 254. As discussed in text, both plots use Phe as the wild-type residue. For 206 and 254, respectively, the slopes of the fit lines are −0.13 and −0.11. Note that both plots share the point at y = 0, x = 27.1 (filled circle).
Tyr254 also forms a cation-π interaction with GABA
We have previously discussed the issues associated with successive incorporation of a tyrosine residue (Lummis et al., 2005). The pKa of the OH falls, possibly perturbing the analysis of the results. Therefore, as in previous studies, we have used Phe as the “wild-type” reference for fluorination studies, since the Y254F mutant is not strongly perturbed from wild type. Introduction of phenylalanine derivatives with a range of potential cation-π interaction energies revealed that Tyr 254 also forms such an interaction with GABA. Thus 4-F-Phe caused a small (∼1.5-fold) increase in EC50, and subsequent addition of 2 and 3 fluorines caused further stepwise increase of ∼10- and ∼33-fold, respectively, compared to Tyr254Phe receptors (Table 1). A plot of these data strongly suggests a cation-π interaction between GABA and Tyr 254 (Fig. 3).
An interaction between Phe206 and Tyr254
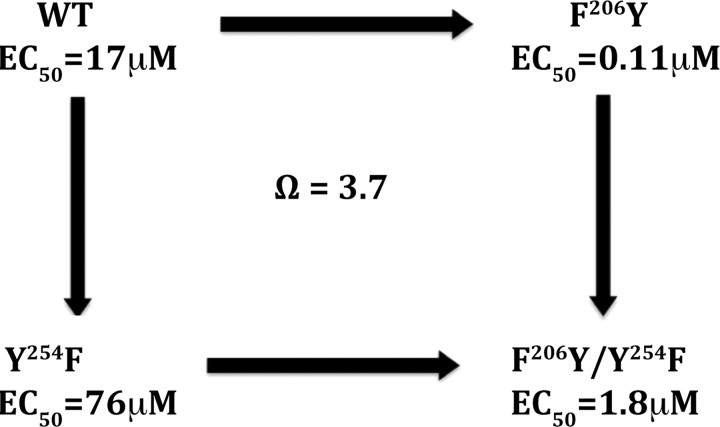
To probe whether there is a direct interaction of these two residues via GABA, we used double mutant cycle analysis. A coupling parameter, Ω, calculated from EC50 values, will be unity if a mutation to the potential interacting residue has no structural or energetic impact on the second potential interacting residue. In contrast, if the residues interact, the simultaneous change at both sites will lead to an effect that is either greater or less than the product of their individual effects, producing a Ω-value that significantly deviates from unity. As can be observed in Figure 4, the Ω-value was 3.7 for the subtle mutations interconverting Phe and Tyr, suggesting a significant interaction.
Figure 4.
Mutant cycle analysis. Determination of the EC50s of WT, single-substituted, and double-substituted receptors allows calculation of the coupling factor (Ω = EC50wt · EC50mutmut/EC50wtmut · EC50mutwt), which indicates there is a strong interaction between F206 and Y254 in RDL receptors.
Locating GABA in the binding pocket
We have previously created a model of the RDL receptor binding pocket, and docking of GABA revealed the potential for a strong cation-π interaction with Phe206 and/or Tyr254 (Harrison and Lummis, 2006b; McGonigle and Lummis, 2010). The current data therefore support the location of the positive amine of GABA between these two aromatic moieties in the RDL binding pocket (Fig. 5A).
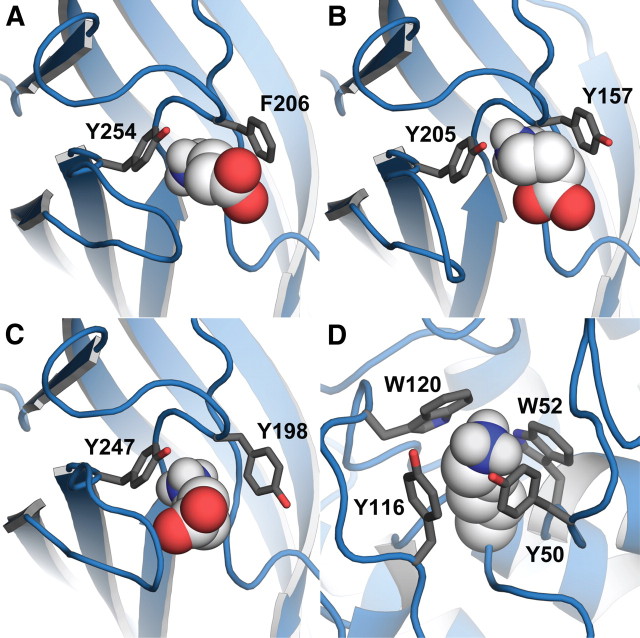
Figure 5.
A–C, GABA docked into GABA receptor models of RDL (A), GABAA (B), and GABAC (C) receptors, which have cation-π interaction with different residues in the binding site (loop B Tyr, loop A Tyr, and loops B and C Tyrs, respectively). D, Example of multiple aromatic residues contributing to a cation-π interaction in the enzyme gai 1 (Gallivan and Dougherty, 2000).
Discussion
Here we show that two aromatic residues in the binding site of the GABA-activated Cys-loop receptor RDL contribute to a cation-π interaction with GABA: Phe206 on loop B and Tyr 254 in loop C. Cation-π interactions that involve multiple residues are not uncommon; the enzyme glucoamylase, for example, has four aromatic residues around a single lysine (Gallivan and Dougherty, 2000). Previous studies on vertebrate Cys-loop receptors, however, have revealed only a single cation-π interaction, usually with the aromatic on loop B (Trp149 of several nAChRs; Trp183 of the 5-HT3 receptor, Tyr198 of the GABAC receptor, and Phe159 of the Gly receptor), although cation-π interactions with residues on loop A (Tyr97 in the GABAA receptor) and loop C (Trp226 on MOD-1 receptor) have also been observed, indicating that neither the location or the aromatic residue that participates in this interaction is conserved throughout this receptor family (Zhong et al., 1998; Beene et al., 2002, 2004; Lummis et al., 2005; Harrison and Lummis, 2006a; Padgett et al., 2007; Pless et al., 2007).
The fluorination plots of Phe206 and Tyr254 show convincing correlations and slopes that are in the typical range for a meaningful cation-π interaction. The slope for residue 206 (0.13) is slightly higher than for 254 (0.11), which could indicate that the cation-π interaction is stronger at the 206 site. Our recent study of cation-π interactions of different agonists in glycine receptors suggests that the exact orientation of the cationic moiety influences the strength of a cation-π interaction at a given site (Pless et al., 2011), consistent with a computational study that demonstrated that the orientation of the aromatic residue and the cation are crucial for a strong cation-π interaction (Gallivan and Dougherty, 2000).
The fluorination plots of Cys-loop receptors reveal variation in their slopes; ACh has the shallowest slope, probably because the quaternary ammonium of ACh has a less focused positive charge than the primary ammonium of 5-HT, GABA, and glycine; the strong electrostatic component of the cation-π interaction means that a more focused charge should produce a stronger effect (Zhong et al., 1998; Beene et al., 2002, 2004; Lummis et al., 2005; Harrison and Lummis, 2006a; Padgett et al., 2007; Pless et al., 2007). The slopes from the RDL data are, however, considerably less than those for vertebrate GABA-activated GABAA (0.29) and GABAC (0.21) receptors, which both form cation-π interactions with only a single residue. While the present data do not allow a quantitative analysis, we are tempted to speculate that residues 206 and 254 make comparable energetic contributions to binding that, individually, are weaker than seen in other Cys-loop receptors, but in combination produce a similar effect. As the cation-π interaction energy decreases only as 1/(rn), where n < 2 (described by Dougherty, 1996), it is quite feasible to propose a single cation making a significant cation-π interaction with two residues.
An interesting observation concerns the influence of a substituent at the 4 position for the two residues considered here. Surprisingly, replacement of Phe206 by Tyr caused a large decrease in EC50, showing that a Tyr here is more favorable for ligand binding and/or receptor activation. We speculate that this is because there is an interaction between a hydroxyl at the 4 position and either the ligand or the protein, resulting in a more efficient receptor. This could explain why the rightward shift in the concentration response curves is more pronounced between the monofluorinated and difluorinated derivatives, as the latter does not have a group at the 4 position; F3Phe again has an F at the 4 position and the rightward shift here is less pronounced. Tyr254 shows a more typical response, in that the Tyr254Phe mutant is loss of function, but the effect is not large. Again, the relatively modest impact of the first fluorine in the fluorination study, relative to Phe, could reflect a balancing of effects; i.e., we propose that adding a fluorine causes loss of function due to a diminution of the cation-π interaction, but the loss of function is less than expected due to a concomitant gain of function from the presence of a substituent at the 4 position.
The data strongly support the orientation of GABA in our model of the RDL binding site. GABA is a promiscuous neurotransmitter that binds to a range of receptors in vertebrates and invertebrates, but somewhat surprisingly, its exact location in the binding site and, in particular, the amino acid residue with which it forms a cation-π interaction, vary within different the binding pockets. All the receptors within which we have defined a cation-π interaction with GABA so far have different interacting residues: Tyr in loop A in GABAA receptors, Tyr in loop B in GABAC receptors, and now a joint interaction with a loop C Tyr and a loop B Phe in RDL receptors. Similarly, 5-HT binds in different locations in the 5-HT-activated 5-HT3 and MOD-1 receptors. In this instance, there is a Trp in loop B in the former that is equivalent to a Tyr in the latter, and vice versa in loop C. As the cation-π binding energy of Trp is considerably greater than Tyr, it is logical for the interaction with the primary amine of 5-HT to “move” from loops B to C. It is, however, less clear as to why this interaction should relocate in GABA receptors, as the cation-π binding abilities of Tyr and Phe are similar. Our models of GABA docked into the binding pockets of the three different receptors show distinct orientations of GABA in each of the receptors, but do not clarify why a cation-π interaction is favored at one aromatic rather than another, or at two aromatics compared to one. The overall results on Cys-loop receptors suggest that a cation-π interaction with a loop B residue is preferred over other locations (Table 2), and indeed the slopes of our fluorination plots indicate the loop B interaction is subtly stronger than the loop C interaction. We suggest that this may be because loop B remains static, while other regions, such as loop C, are more mobile (Thompson et al., 2008). It is also possible that other residues in the binding pocket, especially charged residues, play critical roles in the orientation of the agonist, e.g., Arg207 in the GABAA receptor binding site. We are tempted to speculate that the lack of charged residues in the RDL binding pocket may explain this apparently unique Cys-loop receptor double cation-π interaction.
Table 2.
Alignment of residues contributing to the aromatic box in the Cys-loop receptor family
| Receptor | A | B | C | D | |
|---|---|---|---|---|---|
| nACh | Y93 | W149 | Y190 | Y198 | W55 |
| 5-HT3 | E129 | W183 | F226 | Y234 | W90 |
| MOD-1 | C120 | Y180 | Y221 | W226 | F83 |
| GABAC | F138 | Y198 | Y241 | Y247 | Y102 |
| GABAA | Y97 | Y157 | F200 | Y205 | F65 |
| RDL | F146 | F206 | L249 | Y254 | Y109 |
| Gly | F99 | F159 | Y202 | F207 | F63 |
Appropriate binding segments are labeled accordingly. Residues in bold have been shown to be involved in cation-π interactions.
The homology model of GABA in the RDL receptor binding site supports the experimental evidence in suggesting that the B and C loop residues could contribute to a cation-π interaction. The binding pocket contains a range of aromatic residues that could potentially contribute to such an interaction, with Phe206 and Tyr254 the most likely candidates. Phe206 is on loop B, and is in a location equivalent to the Trp residues that have been identified as forming cation-π interactions in nACh (Trp147) and 5-HT3 (Trp183) receptors, and also to Tyr198, which forms a cation-π interaction with GABA in the GABAC receptor. Tyr254 on loop C is equivalent to Trp226 which form a cation-π interaction with 5-HT in the MOD-1 receptor. The loop A aromatic also can form such an interaction with GABA in the GABAA receptor. Here the equivalent residue is Phe146, but the relatively small increase in EC50 when this is substituted with a nonaromatic (Ala) or F3Phe indicates there is no cation π interaction with this residue.
The cationic moiety contributing to the cation-π interaction at Phe206 and/or Tyr254 could, in theory, originate from a positively charged Arg or Lys residue located in the vicinity. However our model indicates no Arg or Lys residues are located close enough to Phe206 or Tyr254 to interact with them; we therefore conclude that Phe206 and Tyr254 form cation-π interaction with the GABA amine group.
Thus, in conclusion, our data show a strong correlation between receptor function and the cation-π bonding abilities of Phe analogs when they are incorporated at two positions in the RDL receptor, Phe206 and Tyr254, indicating cation-π interactions between the primary amine of GABA and the aromatic side chains at these locations. Mutant cycle analysis reveals a significant interaction, providing further supporting evidence. Comparison with similar experiments in the GABAC receptor was not possible, as double mutants were nonfunctional, probably because both single mutants cause a significant increase in EC50; we would anticipate similar problems in the GABAA receptor. Nevertheless in these, and in a range of other Cys-loop receptors, the data suggest that only a single aromatic contributes to the cation-π interaction. Multiple aromatics, however, have been shown to contribute to cation-π interactions in a number of other proteins (see e.g., Fig. 5), but this is the first description of two aromatics contributing to such an interaction in Cys-loop receptors. These data will clarify our understanding of agonist binding in this family of receptors and will aid future efforts to design insecticidal agents that act at insect GABA receptors.
Footnotes
We thank The Wellcome Trust (WT 81925; S.C.R.L. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science), the Medical Research Council (a studentship to I.M.), the European Union (FP7 NeuroCypres; J.A.A., S.C.R.L.), and the U.S. National Institutes of Health (NS34407, D.A.D.).
The authors declare no competing financial interests.
References
- Beene DL, Brandt GS, Zhong W, Zacharias NM, Lester HA, Dougherty DA. Cation-pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry. 2002;41:10262–10269. doi: 10.1021/bi020266d. [DOI] [PubMed] [Google Scholar]
- Beene DL, Dougherty DA, Lester HA. Unnatural amino acid mutagenesis in mapping ion channel function. Curr Opin Neurobiol. 2003;13:264–270. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- Beene DL, Price KL, Lester HA, Dougherty DA, Lummis SCR. Tyrosine residues that control binding and gating in the 5-hydroxytryptamine3 receptor revealed by unnatural amino acid mutagenesis. J Neurosci. 2004;24:9097–9104. doi: 10.1523/JNEUROSCI.2429-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belelli D, Callachan H, Hill-Venning C, Peters JA, Lambert JJ. Interaction of positive allosteric modulators with human and Drosophila recombinant GABA receptors expressed in Xenopus laevis oocytes. Br J Pharmacol. 1996;118:563–576. doi: 10.1111/j.1476-5381.1996.tb15439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Dougherty DA. Cation-π interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- Dougherty DA. Unnatural amino acids as probes of protein structure and function. Curr Opin Chem Biol. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. A computational study of cation-pi interactions vs. salt bridges in aqueous media: implications for protein engineering. J Am Chem Soc. 2000;122:870–874. [Google Scholar]
- Harrison NJ, Lummis SCR. Locating the carboxylate group of GABA in the homomeric rho GABA(A) receptor ligand-binding pocket. J Biol Chem. 2006a;281:24455–24461. doi: 10.1074/jbc.M601775200. [DOI] [PubMed] [Google Scholar]
- Harrison NJ, Lummis SCR. Molecular modeling of the GABAC receptor ligand-binding domain. J Mol Model. 2006b;12:317–324. doi: 10.1007/s00894-005-0034-6. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- Hosie AM, Sattelle DB. Allosteric modulation of an expressed homo-oligomeric GABA-gated chloride channel of Drosophila melanogaster. Br J Pharmacol. 1996a;117:1229–1237. doi: 10.1111/j.1476-5381.1996.tb16720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie AM, Sattelle DB. Agonist pharmacology of two Drosophila GABA receptor splice variants. Br J Pharmacol. 1996b;119:1577–1585. doi: 10.1111/j.1476-5381.1996.tb16075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie AM, Buckingham SD, Presnail JK, Sattelle DB. Alternative splicing of a Drosophila GABA receptor subunit gene identifies determinants of agonist potency. Neuroscience. 2001;102:709–714. doi: 10.1016/s0306-4522(00)00483-8. [DOI] [PubMed] [Google Scholar]
- Kearney PC, Nowak MW, Zhong W, Silverman SK, Lester HA, Dougherty DA. Dose-response relations for unnatural amino acids at the agonist binding site of the nicotinic acetylcholine receptor: tests with novel side chains and with several agonists. Mol Pharmacol. 1996;50:1401–1412. [PubMed] [Google Scholar]
- Lummis SC, Beene DL, Harrison NJ, Lester HA, Dougherty DA. A cation-pi binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem Biol. 2005;12:993–997. doi: 10.1016/j.chembiol.2005.06.012. [DOI] [PubMed] [Google Scholar]
- McGonigle I, Lummis SC. Molecular characterization of agonists that bind to an insect GABA receptor. Biochemistry. 2010;49:2897–2902. doi: 10.1021/bi901698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk KA, Pistis M, Belelli D, Hope AG, Lambert JJ. The effect of a transmembrane amino acid on etomidate sensitivity of an invertebrate GABA receptor. Br J Pharmacol. 1998;124:13–20. doi: 10.1038/sj.bjp.0701787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar NS, Buckingham SD, Sattelle DB. Stable expression of a functional homo-oligomeric Drosophila GABA receptor in a Drosophila cell line. Proc Biol Sci. 1994;258:307–314. doi: 10.1098/rspb.1994.0178. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Nowak MW, Gallivan JP, Silverman SK, Labarca CG, Dougherty DA, Lester HA. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- Padgett CL, Hanek AP, Lester HA, Dougherty DA, Lummis SCR. Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation-pi interaction between GABA and beta 2Tyr97. J Neurosci. 2007;27:886–892. doi: 10.1523/JNEUROSCI.4791-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Dibas MI, Lester HA, Lynch JW. Conformational variability of the glycine receptor M2 domain in response to activation by different agonists. J Biol Chem. 2007;282:36057–36067. doi: 10.1074/jbc.M706468200. [DOI] [PubMed] [Google Scholar]
- Pless SA, Hanek AP, Price KL, Lynch JW, Lester HA, Dougherty DA, Lummis SCR. A cation-π interaction at a phenylalanine residue in the glycine receptor binding site is conserved for different agonists. Mol Pharmacol. 2011;79:742–748. doi: 10.1124/mol.110.069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Lochner M, Lummis SCR. Loop B is a major structural component of the 5-HT3 receptor. Biophys J. 2008;95:5728–5736. doi: 10.1529/biophysj.108.135624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- Zhang HG, ffrench-Constant RH, Jackson MB. A unique amino acid of the Drosophila GABA receptor with influence on drug sensitivity by two mechanisms. J Physiol. 1994;479:65–75. doi: 10.1113/jphysiol.1994.sp020278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci U S A. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]