Abstract
The nature, size and distribution of the genomic regions underlying divergence and promoting reproductive isolation remain largely unknown. Here, we summarize ongoing efforts using young (12 000 yr BP) species pairs of lake whitefish (Coregonus clupeaformis) to expand our understanding of the initial genomic patterns of divergence observed during speciation. Our results confirmed the predictions that: (i) on average, phenotypic quantitative trait loci (pQTL) show higher FST values and are more likely to be outliers (and therefore candidates for being targets of divergent selection) than non-pQTL markers; (ii) large islands of divergence rather than small independent regions under selection characterize the early stages of adaptive divergence of lake whitefish; and (iii) there is a general trend towards an increase in terms of numbers and size of genomic regions of divergence from the least (East L.) to the most differentiated species pair (Cliff L.). This is consistent with previous estimates of reproductive isolation between these species pairs being driven by the same selective forces responsible for environment specialization. Altogether, dwarf and normal whitefish species pairs represent a continuum of both morphological and genomic differentiation contributing to ecological speciation. Admittedly, much progress is still required to more finely map and circumscribe genomic islands of speciation. This will be achieved through the use of next generation sequencing data but also through a better quantification of phenotypic traits moulded by selection as organisms adapt to new environmental conditions.
Keywords: divergent selection, gene expression, quantitative trait loci, reproductive isolation, speciation islands
1. Introduction
Dobzhansky [1] initially proposed that the genotype of a species is an integrated system adapted to the environment or ecological niche in which the species lives, whereby recombination in hybrid offspring may lead to the formation of discordant gene patterns. Understanding how speciation can take place in the presence of the homogenizing effects of gene flow remains a major challenge in evolutionary biology. The ecological theory of adaptive radiation hypothesizes that shifts of organisms into novel habitats will be adaptive, whereby populations will diverge for specific phenotypes and genotypes influencing survival and reproduction when exposed to different environments [2,3]. Under the genic view of speciation, divergent selection should create heterogeneous genomic differentiation by causing adaptive loci (and those physically linked to them) to flow between populations less readily than others. This will result in accentuated genetic divergence of regions affected by selection while the homogenizing effects of gene flow should preclude divergence in other regions [4–6].
2. The nature of genomic islands of divergence
The nature, size and distribution of these genomic regions underlying divergence and promoting reproductive isolation are still a contentious and debated area of research. While extensive theoretical and empirical research has been conducted, the precise genetic processes that regulate and favour regions of genomic divergence promoting speciation remain largely unknown [5,7–10]. How populations go from little evidence of phenotypic and genotypic divergence to fully incompatible and reproductively isolated entities continues to be one of the key unanswered questions in evolutionary biology.
There are two conceptually different ideas to explain how genetic differences between populations and species arise and are maintained in the presence of gene flow. First, genetic differences and thus isolation are predicted to accumulate in a few, but large genomic ‘islands’ of reduced gene flow [5,10–12]. Conversely, selection can also act simultaneously on many physically unlinked genomic regions. Under this view, genomes are highly porous and islands of speciation, as small as a single gene, are scattered throughout the genome [7,9,13,14]. Both scenarios may act concurrently, as large regions of differentiation are created while, simultaneously, selection isolates single genes or mutations from the homogenizing effects of gene flow. If the process of speciation takes its course, these regions under the effect of divergent selection, originally expected to be rare, will tend to grow in both size and number until eventually genomic islands merge and the whole genome becomes fully incompatible [5,11]. Given that, with time, islands of speciation will become drowned in a sea of divergence, this process should also be studied in the early step of a speciation event.
Chromosomal rearrangements are one genetic mechanism that may promote the initial appearance and further spread of genomic islands of divergence [8,14,15]. These rearrangements are expected to impede gene flow through the suppression of recombination around chromosomal breakpoints, thus promoting the growth of diverging regions and ultimately facilitating speciation. While there is theoretically support for this idea, empirical evidence has been mixed [16–20]. In a manner analogous to the effect of chromosomal rearrangements, strong divergent selection at specific loci can also promote the accumulation and spread of divergent genomic regions. This view has materialized recently into the verbal ‘divergence hitchhiking’ model proposed by Via & West [21]. Indeed, Via & West [21] empirically showed that quantitative trait loci (QTL) for adaptive traits between pea aphid populations were linked to regions of higher genetic differentiation and that this effect extended far (greater than 10 cM) from the putative target of selection. Via [10] recently highlighted a similar scenario in three-spined sticklebacks [22] where divergence hitchhiking protected and isolated large genomic regions surrounding genes responsible for adaptation to freshwater. While attractive, this idea is again supported by mixed empirical evidence [16,23–27]. Moreover, theoretical modelling has shown that divergence hitchhiking will only create large regions of differentiation around selected loci under restrictive conditions [9]. Thus, locally reduced effective recombination, low migration rates (m < 0.001), small effective population sizes (Ne < 1000) and strong selective pressures (s > 0.10) should promote the maintenance and subsequent growth of genomic islands of divergence.
3. The role of gene expression in the study of speciation
Initial attempts to identify regions of genomic divergence underlying adaptive traits has necessarily focused on easily measured morphological, and to a lesser degree, behavioural traits [28]. However, focusing only on easily identifiable traits may bias our view about the number and nature of phenotypic traits involved in the process of ecological speciation. One way to circumvent these limits is to use gene expression data as a way to reveal otherwise hidden phenotypes of potential ecological relevance [29]. Moreover, in an analogous manner to traditional phenotypic QTL (pQTL) studies, expression QTL (eQTL) studies allow the identification of specific genomic regions responsible for gene expression differences [30]. These studies have revealed patterns of significant clustering of genes in regulation ‘hotspots’ and hinted at the existence of localized genomic islands of expression divergence [29]. If these hotspots are studied in the context of a recent divergence event, they may be representative of regions involved in ecological speciation [29]. Expression QTL studies can potentially inform about the mechanisms of gene regulation, and provide insights into the process of speciation [31]. Thus, studying gene expression may improve our understanding of speciation, particularly when integrative approaches are applied in the context of recent divergence.
4. The lake whitefish study system
Here, we investigated key issues pertaining to the nature, number and size of genomic regions underlying species divergence in lake whitefish by conducting novel analyses on previously published data [32–35]. The lake whitefish species complex includes several sympatric populations inhabiting northern temperate lakes in Canada and Maine [36]. These species pairs are characterized by two post-glacial derived forms living in sympatry. A ‘dwarf’ form, typically growing slower, maturing at a much earlier age and size, and living in the limnetic zone of lakes, and a larger ‘normal’ form, which grows faster, reaches a larger size, matures at a later age and lives within the benthic zone of lakes. Their phenotypic divergence is recent (less than 12 000 yr BP) and involves both a phase of allopatry (geographical isolation) and sympatry (secondary contact) [31,37,38]. This system is especially well suited to study the genetics of species boundaries for several reasons. First, lake whitefish represents a rare illustration of a continuum of both morphological and genetic differentiation within a given taxon. This differentiation spans from complete introgression to near complete reproductive isolation, depending on the history [39,40] and the potential for competitive interactions imposed by unique ecological characteristics of each lake [41–43]. Second, previous work has identified several genomic regions responsible for the control of adaptive phenotypic traits (pQTL mapping) [32,33] and gene expression differences (eQTL mapping) [34,35]. Third, these genetic markers have also been characterized in natural populations, thus permitting inference about the effect of selection acting on phenotypic traits, including gene expression. Fourth, the repeated independent parallel evolution of normal and dwarf phenotypes implies that populations evolved under similar selective pressures [31]. Lastly, these lake whitefish populations have small effective population sizes (Ne < 1000), low migration rates (m varying between 0.0004 and 0.0019) [41,44], as well as strong selection against hybrids [45–47]. Under these conditions, divergence hitchhiking is theoretically expected to facilitate the formation of large genomic islands of divergence [9].
In this study, we pursued three objectives pertaining to the genomic patterns of divergence observed during speciation: (i) to test the prediction that genetic markers associated with potentially adaptive traits (identified through pQTL and eQTL mapping) will show stronger evidence of divergent selection in natural populations than non-QTL linked markers; (ii) to assess the prevalence of a few large islands of divergence along the genome rather than small independent regions; and (iii) to test whether sympatric species pairs showing more pronounced ecological, phenotypic and genetic divergence are also characterized by sharper speciation boundaries in terms of number and sizes of genomic regions of divergence.
5. Methods
(a). Genetic mapping
We used genetic linkage maps previously generated from two backcross hybrid families (F1 × limnetic dwarf: BCD map, F1 × benthic normal: BCN map). Briefly, hybrids were produced between parents representing two allopatric whitefish populations belonging to two different glacial races and permitted to genotype and position about 900 amplified fragment length polymorphism (AFLP) and microsatellite loci among 336 progeny (for details see [32]). These particular populations were chosen because they overlap in reproductive schedule and showed strong parallel phenotypic differentiation with sympatric populations studied here. Nine different phenotypes characterizing normal and dwarf populations were measured on these progeny and 34 pQTL linked to eight of these traits were mapped over 13 linkage groups using interval mapping [33]. While using a single mapping population is a common practice, it must be noted that it leads to the possibility that pQTL from the crosses may not always correspond to pQTL in the populations under study here.
(b). Gene expression and genome scan in natural populations
We used gene expression data obtained from the analysis of white muscle [35] and whole brain [34] tissues. Briefly, these studies used microarrays and the linkage map of Rogers et al. [32] to localize eQTL of genes expressed in white muscle (262 transcripts localized) and brain (249 transcripts localized) for one of the backcross family (BCD map). The markers (AFLP) used for genetic mapping were also genotyped in four lakes containing dwarf and normal sympatric populations [33]. Combining both QTL and genome scan information revealed that a total of six eQTL regions co-localized with pQTL and were also outliers in a genome scan study [31,33]. Moreover, these genomic regions were non-randomly distributed across the genome and hinted at the role of major regulatory hotspots controlling the expression of numerous genes [34,35].
(c). Association between FST and phenotypic quantitative trait loci, expression quantitative trait loci and expression quantitative trait loci hotspots
t-Tests were performed for each lake individually to test whether mean FST was greater for genetic markers associated with pQTL, eQTL or eQTL hotspots (QTL associated with the expression of five or more (up to 52) genes; see details in Rogers & Bernatchez [33]). In addition, χ2-tests were performed for each lake individually to test whether there were significantly more FST outliers for markers associated with pQTL, eQTL or eQTL hotspots.
(d). Spatial autocorrelation
Spatial autocorrelation analyses were run and Moran's I was quantified for each lake individually in order to assess whether FST outlier loci were clustered together more than expected by chance, thus representing evidence of relatively large speciation islands. Moran's I is a measure of spatial autocorrelation where −1 indicates perfect dispersion and +1 perfect correlation among samples [48].
(e). Distance from nearest outlier phenotypic quantitative trait loci
First, we identified outlier markers (markers showing FST value > 95% quantile of FST distribution in natural populations) associated with an adaptive trait (hereafter referred as outlier pQTL). Then, we tested the correlation between genetic distance (in centiMorgans) from an outlier pQTL and FST values for all other mapped markers on the same linkage group. This was done separately for all four lakes. A linear regression between FST and genetic distance was fitted in order to test whether the effect of divergence extended far from the region under selection itself, thus suggesting the existence of relatively large speciation islands. This relationship could be tested only for pQTL given that neither eQTL nor eQTL hotspots were significantly associated with higher FST values compared to non-eQTL markers (table 1). A logistic regression was also fitted, however, relationships were not significant in any of the lakes. In addition, linear models always had lower Akaike Information Criterion (AIC) and thus represented a better fit for the data (i.e. the differences in AIC between the logistic and linear regressions were 47.4, 50.9, 24.4 and 23.5 for Indian, East, Webster and Cliff lakes, respectively).
Table 1.
Genetic divergence (mean FST and number of outliers observed and expected) for markers linked to pQTL, eQTL and eQTL hotspots for all four lakes containing sympatric dwarf and normal populations. BCD (an F1 hybrid parent backcrossed with a pure dwarf parent) and BCN (an F1 hybrid parent backcrossed with a pure normal parent) represent the two backcross families analysed here. T-tests and χ2-tests were performed to assess if, respectively, mean FST and number of outliers deviated for markers associated with a phenotype. (bold, p-value < 0.05).
| lake | markers | genetic map | mean FST | p-value (t-test) | observed outliers | expected outliers | p-value (χ2-test) |
|---|---|---|---|---|---|---|---|
| East L. | pQTL | BCD | 0.07 | 0.05 | 3 | 0.94 | 0.02 |
| no pQTL | 0.04 | — | 1 | 3.06 | — | ||
| pQTL | BCN | 0.03 | 0.69 | 0 | 0 | — | |
| no pQTL | 0.03 | — | 0 | 0 | — | ||
| eQTL | BCD | 0.05 | 0.43 | 1 | 1.35 | 0.71 | |
| no eQTL | 0.04 | — | 3 | 2.65 | — | ||
| eQTL hotspot | BCD | 0.07 | 0.63 | 0 | 0.09 | 0.76 | |
| no eQTL hotspot | 0.05 | — | 4 | 3.91 | — | ||
| Indian L. | pQTL | BCD | 0.04 | 0.96 | 4 | 1.89 | 0.08 |
| no pQTL | 0.04 | — | 4 | 6.11 | — | ||
| pQTL | BCN | 0.01 | 0.0003 | 0 | 0.11 | 0.73 | |
| no pQTL | 0.03 | — | 1 | 0.89 | — | ||
| eQTL | BCD | 0.04 | 0.73 | 2 | 2.70 | 0.60 | |
| no eQTL | 0.05 | — | 6 | 5.30 | — | ||
| eQTL hotspot | BCD | 0.05 | 0.81 | 0 | 0.18 | 0.67 | |
| no eQTL hotspot | 0.04 | — | 8 | 7.82 | — | ||
| Webster L. | pQTL | BCD | 0.12 | 0.26 | 1 | 0.94 | 0.95 |
| no pQTL | 0.10 | — | 3 | 3.06 | — | ||
| pQTL | BCN | 0.07 | 0.45 | 0 | 0.11 | 0.73 | |
| no pQTL | 0.09 | — | 1 | 0.89 | — | ||
| eQTL | BCD | 0.10 | 0.60 | 0 | 1.35 | 0.15 | |
| no eQTL | 0.10 | — | 4 | 2.65 | — | ||
| eQTL hotspot | BCD | 0.07 | 0.51 | 0 | 0.09 | 0.76 | |
| no eQTL hotspot | 0.10 | — | 4 | 3.91 | — | ||
| Cliff L. | pQTL | BCD | 0.19 | 0.05 | 4 | 2.12 | 0.14 |
| no pQTL | 0.11 | — | 5 | 6.88 | — | ||
| pQTL | BCN | 0.25 | 0.07 | 3 | 0.53 | 0.0004 | |
| no pQTL | 0.11 | — | 2 | 4.47 | — | ||
| eQTL | BCD | 0.14 | 0.31 | 1 | 3.03 | 0.15 | |
| no eQTL | 0.11 | — | 8 | 5.97 | — | ||
| eQTL hotspot | BCD | 0.21 | 0.72 | 1 | 0.20 | 0.07 | |
| no eQTL hotspot | 0.13 | — | 8 | 8.80 | — | ||
| all lakes combined | pQTL | BCD | 0.11 | 0.01 | 12 | 4.76 | 0.0002 |
| no pQTL | 0.07 | — | 13 | 20.24 | — | ||
| pQTL | BCN | 0.09 | 0.26 | 3 | 0.23 | 4.3e-9 | |
| no pQTL | 0.06 | — | 4 | 6.77 | — | ||
| eQTL | BCD | 0.08 | 0.27 | 4 | 7.86 | 0.10 | |
| no eQTL | 0.07 | — | 21 | 17.14 | — | ||
| eQTL hotspot | BCD | 0.10 | 0.68 | 1 | 0.62 | 0.62 | |
| no eQTL hotspot | 0.08 | — | 24 | 24.38 | — |
(f). Among lake differences characterizing genomic islands of divergence
In order to provide a relative basis to compare the extent of differences in the characteristics of genomic islands of divergence among lakes, the following parameters were quantified: (i) the number of islands: defined as the number of unlinked outliers detected in each sympatric whitefish species pairs; (ii) the relative size of islands: defined by the x-intercept at the lake-specific mean FST value for the relationship depicted in figure 1 (FST values versus chromosomal distance to the nearest outlier pQTL). These values (in centiMorgans) were then divided by the value for the least divergent lake (East L.) in order to get relative estimates among lakes; (iii) the sea level was defined for each lake separately as the FST outlier threshold value (95% quantile) above which markers were considered outliers; and (iv) the mean island height was defined in each lake as the difference between the mean FST value of all outliers minus the FST outlier threshold value. Note that none of these parameters should be taken as an absolute quantification of the actual physical or genetic size of a region of divergence, but instead used purely as comparative measures.
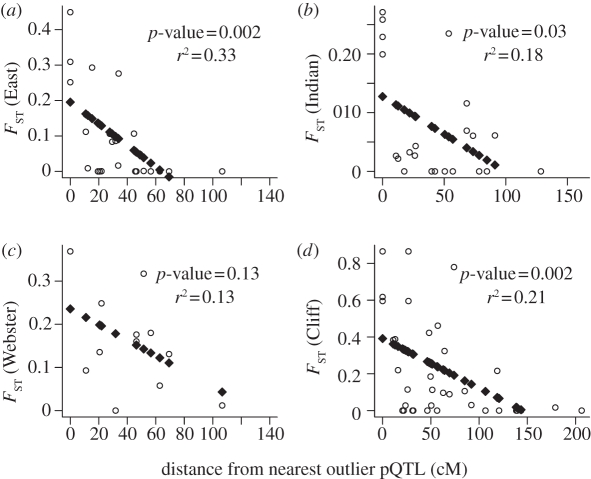
Figure 1.
(a–d) The relationship (linear regression) between distance from the nearest outlier pQTL and genetic divergence (FST) for all four lakes.
6. Results
(a). Phenotypic quantitative trait loci, expression quantitative trait loci and expression quantitative trait loci hotspot
Results in table 1 indicate that pQTL are characterized on average both by higher FST values (t-tests) and an outlier status (χ2-tests) relative to non-pQTL markers. In contrast, neither eQTL nor eQTL hotspots were significantly associated with either higher FST or an outlier status in all four lakes individually or combined. However, the fact that all relationships were stronger and tended to be more significant when combining the data from the four lakes for pQTL suggests that the absence of significant correlation may be partly due to a lack of statistical power caused by the limited number of QTL in each category.
(b). Spatial autocorrelation
In the BCD map, we found weak, yet significant autocorrelation between outliers in three of the four lakes studied: Indian (Moran's I = 0.044, p-value = 0.02), East (Moran's I = 0.09, p-value < 0.01) and Cliff (Moran's I = 0.051, p-value = 0.01). The significant autocorrelation values were mainly due to three regions where two linked markers were outliers (two Indian outliers in linkage group 7 (LG7) which were 16.6 cM apart; two East outliers in LG4, 12.1 cM apart and two Cliff outliers on LG8, 26.8 cM apart). No significant association was found in the BCN family (table 2). This is probably due to the much lower number of pQTL segregating in that family (10 pQTL for BCN versus 24 for BCD) possibly because the phenotypic differences between hybrid and normal individuals were less marked than between hybrid and dwarf [33].
Table 2.
Spatial autocorrelation statistics for all four lakes. Moran's I ranges from −1 (perfect dispersion) to +1 (perfect correlation) among FST outliers within each lake.
| Moran's I statistics |
|||
|---|---|---|---|
| genetic map | observed | expected | |
| East L. | |||
| BCD | +0.09 | 0 | |
| BCN | no mapped outliers | ||
| Indian L. | |||
| BCD | +0.04 | 0 | |
| BCN | 0 | 0 | |
| Webster L. | |||
| BCD | −0.01 | 0 | |
| BCN | 0 | 0 | |
| Cliff L. | |||
| BCD | +0.05 | 0 | |
| BCN | −0.01 | 0 | |
(c). Distance from nearest outlier phenotypic quantitative trait loci
Figure 1 illustrates the relationship between the distance (in centiMorgan) from an outlier pQTL and FST value for all markers on the same linkage group, in each lake separately. All outlier pQTL were found on different linkage groups. A significant negative linear relationship (p-value < 0.05) was detected for three of the four lakes studied (East L. r2 = 0.33, Indian L. r2 = 0.18 and Cliff L. r2 = 0.21), whereas a similar, albeit not significant trend was observed in Webster L. (r2 = 0.14, p-value = 0.13). In the BCN map, the relationship could only be tested in Cliff L., given that the other lakes did not have any outlier QTL. In this case, the relationship was significant (p-value = 0.004, results not shown).
(d). Among lakes differences in characteristics of genomic islands of divergence
Table 3 illustrates that the four lakes differed in their parameters characterizing genomic islands of divergence. Lakes were ordered according to a gradient of the expected intensity of divergent natural selection [41–43]. We observed a general trend towards an increase in number, size and height of genomic islands of divergence from the least genetically and phenotypically differentiated sympatric pair (East L.) towards the most differentiated whitefish pair (Cliff L.). Roughly speaking, the three genomic island characteristics doubled from the least to the most differentiated lake. In particular, it is noteworthy that the relative size of islands of divergence increased from 1 (East L.) to 1.43 (Indian L.), 1.57 (Webster L.) and 1.95 (Cliff L.). Given that the threshold was much higher in the most differentiated lake, it should become increasingly difficult to observe islands of divergence above the global sea level. Despite this, the mean height of the islands above sea level (defined as the FST outlier threshold) increased from the least to the most differentiated lake (table 3).
Table 3.
Characteristics of genomic islands of divergence for each of the four whitefish species pairs. See §5 for detailed explanations of how each parameter was defined.
| mean FST | AFLP outliers | QTL outliersa (number of islands) | relative size of islandsb | outlier threshold (sea level) | height of island | |
|---|---|---|---|---|---|---|
| East L. | 0.05 | 4 | 4 | 1.00 | 0.25 | 0.08 |
| Indian L. | 0.04 | 9 | 3 | 1.43 | 0.20 | 0.08 |
| Webster L. | 0.10 | 5 | 1 | 1.57 | 0.37 | 0.18 |
| Cliff L. | 0.14 | 14 | 7 | 1.95 | 0.46 | 0.29 |
aNote that QTL outliers are the sum of both BCD and BCN pQTL and also outliers in natural populations.
bSizes are based only on BCD map. They represent the x value where y is equal to the mean FST value for a given species pair in figure 1. Sizes are then divided by the value for the least divergent lake (East) in order to get among lake relative estimates.
7. Discussion
(a). Association between FST and quantitative trait loci
The general goal of this study was to investigate key predictions pertaining to the nature of regions of genomic divergence among lake whitefish species pairs. Our results partly confirmed our first prediction given that in general, pQTL showed higher FST values and were more likely to be outliers (and therefore candidates for being targets of divergent selection) than non-pQTL markers. The detection of a significant association between genetic divergence and pQTL would imply that the genetic bases of traits conferring adaptability are also under divergent natural selection. While this is perhaps a simplistic or self-evident assumption of ecological speciation, it has seldom been explicitly demonstrated [49]. This is in part because, even in an optimal experimental system, one may not expect this relationship to always hold true, such as in a situation where adaptation results from incomplete selective sweeps [50] or if traits under selection are highly polygenic [51]. In the end, pQTL will always allow a limited view of an organism's genetic complexity; some phenotypes will never be measured, and therefore, QTL underlying these traits never be detected. This is in part why gene expression and eQTL studies could have such great potential to survey all relevant molecular phenotypes in an unbiased manner.
In contrast to pQTL, neither eQTL nor eQTL hotspots were associated with high (outlier) divergence values. There are biological reasons explaining why gene expression differences (eQTL) are less prone to be associated with signs of divergent natural selection. For one, given the numerous levels of control that exist between the transcription of mRNA and the expression of a phenotype, it is plausible that gene expression differences do not always translate into meaningful phenotypic differences. Segregating genetic variation affecting expression level may also evolve neutrally, owing to the redundancy of developmental and physiological systems, and therefore, remain hidden from selection acting at the level of the phenotype [52]. In addition, compensatory mutations evolving in a neutral fashion can result in transgressive patterns of expression when recombined in hybrids. For these reasons, gene expression differences in hybrids are not necessarily correlated with difference in parental species and a significant proportion of gene regulation is expected to behave in a non-additive fashion [53–55].
(b). The size of genomic islands of divergence
To this day, there is no consensus into what should be the best method to measure speciation islands, and what are the patterns of genomic divergence expected under varying conditions of gene flow [10,49,56]. Further theoretical work will be needed to objectively characterize speciation islands given that different approaches can lead to substantially variable conclusions. Here, we must clarify that our analyses and conclusions differ from the ones suggested by Via & West [21] and Via [49] who concluded that FST outliers were separated from their closest pQTL on average by 10.6 cM in pea aphids (Acyrthosiphon pisum pisum) and by 16.5 cM in the lake whitefish species pairs discussed in the present study [33]. They suggested that divergence hitchhiking created surprisingly large genomic regions (greater than 10 cM) around divergently selected pQTL. Given that, as a rough rule of thumb, 1 cM corresponds to a physical distance of one megabase [57] this would indeed imply extremely large islands of genomic divergence. Because of the relatively low marker density of the studies by Rogers & Bernatchez [33] and Via & West [21], the hypothesis of linkage between a given FST outlier and the closest pQTL has to be interpreted carefully. Indeed, an FST outlier could also be in linkage disequilibrium with other loci targeted by natural selection and be independent of the closest measured pQTL.
In the present analysis, the data were treated differently. Analyses were performed separately for each lake, because each environment represents an independent speciation event [40,58]. Moreover, all QTL cannot be viewed as being implicitly under divergent natural selection and thus, should not necessarily be expected to show high (outlier) divergence values [59]. Therefore, we restricted our analysis to outlier QTL, the genetic regions most likely to be directly responsible for adaptive traits and evolving under divergent natural selection. The linear regressions in figure 1 imply that selection influences relatively large regions of differentiation. The spatial autocorrelation analyses are also indicative that some outlier loci are clustered even if they are genetically far apart (table 3). As such, our results provided support for the second prediction: the occurrence of large islands of divergence rather than small independent regions under selection. This is also in concordance with the recent work of Hohenlohe et al. [22] and the interpretation of Via [10], which argue for large chromosomal regions around selected QTL. Yet, it does not rule out the existence of much smaller and localized regions of divergence. Namely, our small autocorrelation values suggest that outlier loci show limited clustering and mostly represent independent regions of divergence.
While the occurrence of both small regions under selection and large speciation islands may seem contradictory, we re-emphasize that the two opposing views on how genomic divergence spread throughout the genome are merely the end of a continuum. It then seems that, while strong selection can create relatively few and large islands of divergence, it may also act in a much more localized sense on several independent regions. This scenario is probably closer to reality than either extreme views of speciation under the presence of gene flow. Consider, for instance, the case of Anopheles gambiae regarding the recent divergence of several reproductively isolated populations. In fact, the expression ‘islands of speciation’ itself comes from Turner et al. [12], who first identified three large regions of divergence among a sea of neutral divergence between M and S forms in A. gambiae. Recent whole genome sequencing and genotyping have revealed a more complex scenario with pervasive genomic divergence inconsistent with contemporary inter-form gene flow [60,61]. In another example, strongly selected Drosophila populations harbouring standing genetic variation did not reveal large islands of divergence or complete selective sweep after 600 generations of selection, a scenario inconsistent with a large speciation island model [50]. If, early in divergence, selection acts on standing genetic variation through soft or incomplete sweeps and on traits which are highly polygenic, then identifying genes critical to initiating the speciation process may become much more difficult than initially perceived.
Another confounding factor which could explain the observed wide spread of highly divergent loci is the effect of selective sweeps on mutations that are beneficial in all populations. In the traditional interpretation, loci with higher levels of population differentiation than the expected neutral level are seen as evidence of divergent natural selection acting between populations. However, unconditionally beneficial mutations are expected to appear often in nature. Therefore, depending on how far the selective sweep has progressed when populations are sampled, such mutations may appear highly divergent, even though they are merely incidental in explaining the maintenance of species boundaries [62]. This alternative explanation has been largely overlooked and emphasizes that genetic data should not be interpreted without an understanding of the genotype–phenotype association.
(c). Among environment differences in characteristics of genomic islands of divergence
Cases of closely related species, such as dwarf and normal whitefish, permit evaluations of the effect of divergent selection according to a gradient of ongoing ecological speciation. Here, our third prediction of a general trend towards an increase in terms of numbers and sizes of genomic regions of divergence from the least differentiated (East L.) to the most diverged species pair (Cliff L.) was also confirmed.
Previously, Landry et al. [42] and Landry & Bernatchez [43] showed that sympatric lakes with the most divergent sympatric populations (Cliff, Webster and Indian Lakes) were characterized by less habitat availability, less zooplanktonic prey biomass, smaller prey size range and larger gap in prey size distribution between the limnetic and benthic niches compared with the least divergent populations (East, Témiscouata and Crescent Lakes). The authors concluded that resource limitation resulted in increased potential for competition and selective pressure towards optimal normal and dwarf adaptive peaks. In addition, Lu & Bernatchez [47] and Renaut & Bernatchez [46] also showed that there was a continuum of differentiation among lakes both from a phenotypic and genetic standpoint, from East Lake being the least differentiated to Indian, Webster and Cliff L., being the most differentiated. Here, East L. generally differed from the three more differentiated species pairs, being characterized by a smaller number of putative islands of divergence. These were also smaller in relative size, both in terms of the size of chromosomal regions being hitchhiked, as well as the height of the islands (table 3). On the contrary, the Cliff Lake species were the most distinct, showing the highest number of islands, which were also on average the largest. As such, our results confirm, in whitefish, one of the basic premises of the genic view of speciation [5], whereby populations that are more representative of the early steps of ecological speciation are also those characterized by a lower and generally smaller number of islands.
Admittedly, the criteria we used to define genomic island characteristics were arbitrary and should be interpreted as qualitative and relative measures of divergence. Yet, they still provided a useful means to visualize the main differences observed between whitefish species pairs from different lakes. More rigorous confirmation of the nature and extent of genomic divergence among different species pairs will require re-analysis of this system with a denser genomic coverage, both for the genetic map and the genomic scan of wild populations. Such endeavour is currently underway.
8. Outlook
As recently suggested, detection of widespread genomic divergence may support a very different speciation model where the identification of genetic changes responsible for ecological and behavioural divergence will be more difficult than initially hoped [60,61]. Certainly, from a technological standpoint, next generation sequencing technologies are rapidly revolutionizing the field of evolutionary biology, genetics and ecology [63]. However, there is also a marked lag in the development of analytical tools to take full advantage of this new type and sheer quantity of data [64,65]. Moreover, one has to keep in mind that merely identifying the genetic basis of phenotypic traits does not fulfil the conditions necessary to define a phenotype-environment association [3]. Ultimately, phenotypes are what matter for organisms. Therefore, we indisputably maintain the importance of knowing the genotype–phenotype–environment association [66]. One of the great advantages of gene expression studies lies in the fact that they allow simultaneous measurement of thousands of different phenotypic traits and thus get a phenome-wide view of the extent of divergence [67]. Unfortunately, while high-throughput sequencing, genotyping and gene expression studies have matured into widely available procedures, high-throughput phenotyping remains in its infancy, and will be required to better quantify the phenotypic space occupied by diverging populations. The integrative approach developed in the lake whitefish study system certainly brings insights into the multi-dimensional nature of the genomic patterns of divergence observed during speciation.
Acknowledgements
We would like to thank the organizers Patrik Nosil and Jeffrey L. Feder for kindly inviting us to contribute to this special issue. In addition, we thank Loren H. Rieseberg for fruitful discussions on the genetics of speciation and are grateful to P. Nosil, S. Via and one anonymous referee for their constructive comments. This research programme on whitefish ecological genomics was funded by a Natural Science and Engineering Research Council of Canada (NSERC) and Canadian Research Chair in Genomics and Conservation of Aquatic Resources to L.B., an NSERC postgraduate scholarship to S.R. and NSERC discovery grant to S.M.R.
References
- 1.Dobzhansky T. 1951. Genetics and the origin of species, 3rd edn. New York, NY: Columbia University Press [Google Scholar]
- 2.Mayr E. 1963. Animal species and evolution. Cambridge, MA: Harvard University Press [Google Scholar]
- 3.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press [Google Scholar]
- 4.Lewontin R. C., Krakauer J. 1973. Distribution of gene frequency as a test of the theory of selective neutrality of polymorphisms. Genetics 74, 175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C. I. 2001. The genic view of the process of speciation. J. Evol. Biol. 14, 851–865 10.1046/j.1420-9101.2001.00335.x (doi:10.1046/j.1420-9101.2001.00335.x) [DOI] [Google Scholar]
- 6.Nosil P., Funk D. J., Ortiz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18, 375–402 10.1111/j.1365-294X.2008.03946.x (doi:10.1111/j.1365-294X.2008.03946.x) [DOI] [PubMed] [Google Scholar]
- 7.Rice W. R., Hostert E. E. 1993. Laboratory experiments on speciation: what have we learned in 40 years? Evolution 47, 1637–1653 10.2307/2410209 (doi:10.2307/2410209) [DOI] [PubMed] [Google Scholar]
- 8.Rieseberg L. H. 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358 10.1016/S0169-5347(01)02187-5 (doi:10.1016/S0169-5347(01)02187-5) [DOI] [PubMed] [Google Scholar]
- 9.Feder J. L., Nosil P. 2010. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64, 1729–1747 10.1111/j.1558-5646.2009.00943.x (doi:10.1111/j.1558-5646.2009.00943.x) [DOI] [PubMed] [Google Scholar]
- 10.Via S. 2011. Divergence hitchhiking and the spread of genomic isolation during ecological speciation with gene flow. Phil. Trans. R. Soc. B 367, 451–460 10.1098/rstb.2011.0260 (doi:10.1098/rstb.2011.0260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C. I., Ting C. T. 2004. Genes and speciation. Nat. Rev. Genet. 5, 114–122 10.1038/nrg1269 (doi:10.1038/nrg1269) [DOI] [PubMed] [Google Scholar]
- 12.Turner T. L., Hahn M. W., Nuzhdin S. V. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3, e285. 10.1371/journal.pbio.0030285 (doi:10.1371/journal.pbio.0030285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane N. C., King M. G., Barker M. S., Raduski A., Karrenberg S., Yatabe Y., Knapp S. J., Rieseberg L. H. 2009. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution 63, 2061–2075 10.1111/j.1558-5646.2009.00703.x (doi:10.1111/j.1558-5646.2009.00703.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noor M. A., Bennett S. M. 2009. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103, 439–444 10.1038/hdy.2009.151 (doi:10.1038/hdy.2009.151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faria R., Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 11, 660–669 10.1016/j.tree.2010.07.008 (doi:10.1016/j.tree.2010.07.008) [DOI] [PubMed] [Google Scholar]
- 16.Strasburg J. L., Sherman N. A., Wright K. M., Moyle L. C., Willis J. H., Rieseberg L. H. 2012. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Phil. Trans. R. Soc. B 367, 364–373 10.1098/rstb.2011.0199 (doi:10.1098/rstb.2011.0199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rieseberg L. H., et al. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216 10.1126/science.1086949 (doi:10.1126/science.1086949) [DOI] [PubMed] [Google Scholar]
- 18.Noor M. A., Garfield D. A., Schaeffer S. W., Machado C. A. 2007. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics 177, 1417–1428 10.1534/genetics.107.070672 (doi:10.1534/genetics.107.070672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry D. B., Willis J. H. 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8, e1000500. 10.1371/journal.pbio.1000500 (doi:10.1371/journal.pbio.1000500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stemshorn K. C., Reed F. A., Nolte A. W., Tautz D. 2011. Rapid formation of distinct hybrid lineages after secondary contact of two fish species (Cottus sp.). Mol. Ecol. 20, 1475–1491 10.1111/j.1365-294X.2010.04997.x (doi:10.1111/j.1365-294X.2010.04997.x) [DOI] [PubMed] [Google Scholar]
- 21.Via S., West J. 2008. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol. Ecol. 17, 4334–4345 10.1111/j.1365-294X.2008.03921.x (doi:10.1111/j.1365-294X.2008.03921.x) [DOI] [PubMed] [Google Scholar]
- 22.Hohenlohe P. A., Bassham S., Etter P. D., Stiffler N., Johnson E. A., Cresko W. A. 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6, e1000862. 10.1371/journal.pgen.1000862 (doi:10.1371/journal.pgen.1000862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawthorne D. J., Via S. 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412, 904–907 10.1038/35091062 (doi:10.1038/35091062) [DOI] [PubMed] [Google Scholar]
- 24.Emelianov I., Marec F., Mallet J. 2004. Genomic evidence for divergence with gene flow in host races of the larch budmoth. Proc. R. Soc. Lond. B 271, 97–105 10.1098/rspb.2003.2574 (doi:10.1098/rspb.2003.2574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harr B. 2006. Genomic islands of differentiation between house mouse subspecies. Genome Res. 16, 730–737 10.1101/gr.5045006 (doi:10.1101/gr.5045006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Counterman B. A., et al. 2010. Genomic hotspots for adaptation: the population genetics of Mullerian mimicry in Heliconius erato. PLoS Genet. 6, e1000796. 10.1371/journal.pgen.1000796 (doi:10.1371/journal.pgen.1000796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadeau N. J., et al. 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Phil. Trans. R. Soc. B 367, 343–353 10.1098/rstb.2011.0198 (doi:10.1098/rstb.2011.0198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schluter D., Conte G. L. 2009. Genetics and ecological speciation. Proc. Natl Acad. Sci. USA 106, 9955–9962 10.1073/pnas.0901264106 (doi:10.1073/pnas.0901264106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavey S. A., Collin H., Nosil P., Rogers S. M. 2010. The role of gene expression in ecological speciation. Ann. NY Acad. Sci. 1206, 110–129 10.1111/j.1749-6632.2010.05765.x (doi:10.1111/j.1749-6632.2010.05765.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kammenga J. E., Herman M. A., Ouborg N. J., Johnson L., Breitling R. 2007. Microarray challenges in ecology. Trends Ecol. Evol. 22, 273–279 10.1016/j.tree.2007.01.013 (doi:10.1016/j.tree.2007.01.013) [DOI] [PubMed] [Google Scholar]
- 31.Bernatchez L., et al. 2010. On the origin of species: insights from the ecological genomics of lake whitefish. Phil. Trans. R. Soc. B 365, 1783–1800 10.1098/rstb.2009.0274 (doi:10.1098/rstb.2009.0274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers S. M., Isabel N., Bernatchez L. 2007. Linkage maps of the dwarf and normal lake whitefish (Coregonus clupeaformis) species complex and their hybrids reveal the genetic architecture of population divergence. Genetics 175, 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers S. M., Bernatchez L. 2007. The genetic architecture of ecological speciation and the association with signatures of selection in natural lake whitefish (Coregonus sp. Salmonidae) species pairs. Mol. Biol. Evol. 24, 1423–1438 10.1093/molbev/msm066 (doi:10.1093/molbev/msm066) [DOI] [PubMed] [Google Scholar]
- 34.Whiteley A. R., et al. 2008. The phenomics and expression quantitative trait locus mapping of brain transcriptomes regulating adaptive divergence in lake whitefish species pairs (Coregonus sp.). Genetics 180, 147–164 10.1534/genetics.108.089938 (doi:10.1534/genetics.108.089938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derome N., Bougas B., Rogers S. M., Whiteley A. R., Labbe A., Laroche J., Bernatchez L. 2008. Pervasive sex-linked effects on transcription regulation as revealed by eQTL mapping in lake whitefish species pairs (Coregonus sp. Salmonidae). Genetics 179, 1903–1917 10.1534/genetics.107.086306 (doi:10.1534/genetics.107.086306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenderson O. C. 1964. Evidence of subpopulations of lake whitefish, Coregonus clupeaformis, involving a dwarf form. Trans. Am. Fish. Soc. 93, 77–94 10.1577/1548-8659(1964)93[77:EOSOLW]2.0.CO;2 (doi:10.1577/1548-8659(1964)93[77:EOSOLW]2.0.CO;2) [DOI] [Google Scholar]
- 37.Bernatchez L., Dodson J. J. 1990. Allopatric origin of sympatric populations of lake whitefish (Coregonus clupeaformis) as revealed by mitochondrial DNA restriction analysis. Evolution 44, 1263–1271 10.2307/2409287 (doi:10.2307/2409287) [DOI] [PubMed] [Google Scholar]
- 38.Bernatchez L. 2004. Ecological theory of adaptive radiation: an empirical assessment from coregonine fishes (Salmoniformes). In Evolution illuminated: salmon and their relatives (eds Hendry A. P., Stearns S. C.), pp. 175–207 Oxford, UK: Oxford University Press [Google Scholar]
- 39.Lu G., Basley D. J., Bernatchez L. 2001. Contrasting patterns of mitochondrial DNA and microsatellite introgressive hybridization between lineages of lake whitefish (Coregonus clupeaformis): relevance for speciation. Mol. Ecol. 10, 965–985 10.1046/j.1365-294X.2001.01252.x (doi:10.1046/j.1365-294X.2001.01252.x) [DOI] [PubMed] [Google Scholar]
- 40.Renaut S., Nolte A. W., Rogers S. M., Derome N., Bernatchez L. 2011. SNP signatures of selection on standing genetic variation and their association with adaptive phenotypes along gradients of ecological speciation in lake whitefish species pairs (Coregonus spp.). Mol. Ecol. 20, 545–559 10.1111/j.1365-294X.2010.04952.x (doi:10.1111/j.1365-294X.2010.04952.x) [DOI] [PubMed] [Google Scholar]
- 41.Lu G., Bernatchez L. 1999. Correlated trophic specialization and genetic divergence in sympatric lake whitefish ecotypes (Coregonus clupeaformis): support for the ecological speciation hypothesis. Evolution 53, 1491–1505 10.2307/2640895 (doi:10.2307/2640895) [DOI] [PubMed] [Google Scholar]
- 42.Landry L., Vincent W. F., Bernatchez L. 2007. Parallelism between limnological features and phenotypic evolution of lake whitefish dwarf ecotypes. J. Evol. Biol. 20, 971–984 10.1111/j.1420-9101.2007.01304.x (doi:10.1111/j.1420-9101.2007.01304.x) [DOI] [PubMed] [Google Scholar]
- 43.Landry L., Bernatchez L. 2010. Role of epibenthic resource opportunities in the parallel evolution of lake whitefish species pairs (Coregonus sp.). J. Evol. Biol. 23, 2602–2613 10.1111/j.1420-9101.2010.02121.x (doi:10.1111/j.1420-9101.2010.02121.x) [DOI] [PubMed] [Google Scholar]
- 44.Campbell D., Bernatchez L. 2004. Generic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Mol. Biol. Evol. 21, 945–956 10.1093/molbev/msh101 (doi:10.1093/molbev/msh101) [DOI] [PubMed] [Google Scholar]
- 45.Rogers S. M., Bernatchez L. 2006. The genetic basis of intrinsic and extrinsic postzygotic reproductive isolation jointly promoting speciation in the lake whitefish species complex (Coregonus clupeaformis). J. Evol. Biol. 19, 1979–1994 10.1111/j.1420-9101.2006.01150.x (doi:10.1111/j.1420-9101.2006.01150.x) [DOI] [PubMed] [Google Scholar]
- 46.Renaut S., Bernatchez L. 2011. Transcriptome-wide signature of hybrid breakdown associated with intrinsic reproductive isolation in lake whitefish species pairs (Coregonus spp. Salmonidae). Heredity 106, 1003–1011 10.1038/hdy.2010.149 (doi:10.1038/hdy.2010.149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu G., Bernatchez L. 1998. Experimental evidence for reduced hybrid viability between dwarf and normal ecotypes of lake whitefish (Coregonus clupeaformis Mitchill). Proc. R. Soc. Lond. B 265, 1025–1030 10.1098/rspb.1998.0394 (doi:10.1098/rspb.1998.0394) [DOI] [Google Scholar]
- 48.Moran P. A. P. 1950. Notes on continuous stochastic phenomena. Biometrika 37, 17–33 [PubMed] [Google Scholar]
- 49.Via S. 2009. Natural selection in action during speciation. Proc. Natl Acad. Sci. USA 106, 9939–9946 10.1073/pnas.0901397106 (doi:10.1073/pnas.0901397106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R., Long A. D. 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 40, 587–590 10.1038/nature09352 (doi:10.1038/nature09352) [DOI] [PubMed] [Google Scholar]
- 51.Storz J. F., Wheat C. W. 2010. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution 64, 2489–2509 10.1111/j.1558-5646.2010.01044.x (doi:10.1111/j.1558-5646.2010.01044.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson G., Dworkin I. 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5, 681–690 10.1038/nrg1426 (doi:10.1038/nrg1426) [DOI] [PubMed] [Google Scholar]
- 53.Ranz J. M., Namgyal K., Gibson G., Hartl D. 2004. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 14, 373–379 10.1101/gr.2019804 (doi:10.1101/gr.2019804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bougas B., Granier S., Audet C., Bernatchez L. 2010. The transcriptional landscape of cross-specific hybrids and its possible link with growth in brook charr (Salvelinus fontinalis Mitchill). Genetics 186, 97–107 10.1534/genetics.110.118158 (doi:10.1534/genetics.110.118158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renaut S., Nolte A. W., Bernatchez L. 2009. Gene expression divergence and hybrid misexpression between lake whitefish species pairs (Coregonus spp. Salmonidae). Mol. Biol. Evol. 26, 925–936 10.1093/molbev/msp017 (doi:10.1093/molbev/msp017) [DOI] [PubMed] [Google Scholar]
- 56.Feder J. L., Gejji R., Yeaman S., Nosil P. 2012. Establishment of new mutations under divergence and genomic hitchhiking. Phil. Trans. R. Soc. B 367, 461–474 10.1098/rstb.2011.0256 (doi:10.1098/rstb.2011.0256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allen-Brady K., Camp N. J. 2011. Genetic distance and markers used in linkage mapping. Methods Mol. Biol. 713, 43–53 10.1007/978-1-60327-416-6_4 (doi:10.1007/978-1-60327-416-6_4) [DOI] [PubMed] [Google Scholar]
- 58.Pigeon D., Chouinard A., Bernatchez L. 1997. Multiple modes of speciation involved in the parallel evolution of sympatric morphotypes of lake whitefish (Coregonus clupeaformis, Salmonidae). Evolution 51, 196–205 10.2307/2410973 (doi:10.2307/2410973) [DOI] [PubMed] [Google Scholar]
- 59.Rogers S. M., Bernatchez L. 2005. Integrating QTL mapping and genomic scans towards the characterization of candidate loci under parallel directional selection in the lake whitefish (Coregonus clupeaformis). Mol. Ecol. 14, 351–361 10.1111/j.1365-294X.2004.02396.x (doi:10.1111/j.1365-294X.2004.02396.x) [DOI] [PubMed] [Google Scholar]
- 60.Neafsey D. E., et al. 2010. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330, 514–517 10.1126/science.1193036 (doi:10.1126/science.1193036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawniczak M. K., et al. 2010. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330, 512–514 10.1126/science.1195755 (doi:10.1126/science.1195755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bierne N. 2010. The distinctive footprints of local hitchhiking in a varied environment and global hitchhiking in a subdivided population. Evolution 64, 3254–3272 10.1111/j.1558-5646.2010.01050.x (doi:10.1111/j.1558-5646.2010.01050.x) [DOI] [PubMed] [Google Scholar]
- 63.Tautz D., Ellegren H., Weigel D. 2010. Next generation molecular ecology. Mol. Ecol. 19(Suppl. 1), 1–3 10.1111/j.1365-294X.2009.04489.x (doi:10.1111/j.1365-294X.2009.04489.x) [DOI] [PubMed] [Google Scholar]
- 64.McPherson J. D. 2009. Next-generation gap. Nat. Methods 6, S2–S5 10.1038/nmeth.f.268 (doi:10.1038/nmeth.f.268) [DOI] [PubMed] [Google Scholar]
- 65.Gompert Z., Buerkle C. A. 2011. A hierarchical Bayesian model for next-generation population genomics. Genetics 187, 903–917 10.1534/genetics.110.124693 (doi:10.1534/genetics.110.124693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Houle D., Govindaraju D. R., Omholt S. 2010. Phenomics: the next challenge. Nat. Rev. Genet. 11, 855–866 10.1038/nrg2897 (doi:10.1038/nrg2897) [DOI] [PubMed] [Google Scholar]
- 67.Ranz J. M., Machado C. A. 2006. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol. Evol. 21, 29–37 10.1016/j.tree.2005.09.002 (doi:10.1016/j.tree.2005.09.002) [DOI] [PubMed] [Google Scholar]