Abstract
Chromosomal inversions impact genetic variation and facilitate speciation in part by reducing recombination in heterokaryotypes. We generated multiple whole-genome shotgun sequences of the parapatric species pair Drosophila pseudoobscura and Drosophila persimilis and their sympatric outgroup (Drosophila miranda) and compared the average pairwise differences for neutral sites within, just outside and far outside of the three large inversions. Divergence between D. pseudoobscura and D. persimilis is high inside the inversions and in the suppressed recombination regions extending 2.5 Mb outside of inversions, but significantly lower in collinear regions further from the inversions. We observe little evidence of decreased divergence predicted to exist in the centre of inversions, suggesting that gene flow through double crossovers or gene conversion is limited within the inversion, or selection is acting within the inversion to maintain divergence in the face of gene flow. In combination with past studies, we provide evidence that inversions in this system maintain areas of high divergence in the face of hybridization, and have done so for a substantial period of time. The left arm of the X chromosome and chromosome 2 inversions appear to have arisen in the lineage leading to D. persimilis approximately 2 Ma, near the time of the split of D. persimilis–D. pseudoobscura–D. miranda, but likely fixed within D. persimilis much more recently, as diversity within D. persimilis is substantially reduced inside and near these two inversions. We also hypothesize that the inversions in D. persimilis may provide an empirical example of the ‘mixed geographical mode’ theory of inversion origin and fixation, whereby allopatry and secondary contact both play a role.
Keywords: genome resequencing, divergence, diversity, Drosophila pseudoobscura, Drosophila persimilis
1. Introduction
Examining the genomic landscape between closely related species can reveal areas of high divergence that are often targeted as potential signatures of speciation. Between some species, regions of high divergence appear to have been established and maintained purely by selection (reviewed in [1]). In others, establishment and maintenance of regions of high divergence appears facilitated by structural differences between species [2,3].
Chromosomal rearrangements such as inversions (an orientation reversal of a piece of DNA within a single chromosome) impact species divergence by reducing recombination in heterokaryotypes [2,4–6]. For paracentric inversions, which do not span a centromere, a single meiotic recombination event between heterokaryotypes results in acentric and dicentric products—both of which do not produce viable gametes (electronic supplementary material, figure S1). In Drosophila, acentric and dicentric products remain in the polar bodies in females (males, generally, do not recombine); therefore, paracentric inversions (i) do not affect fertility, (ii) are not structurally underdominant, (iii) effectively prevent recombination within the inverted region and (iv) are the most common type of inversion observed in Drosophila [7,8]. This suppressed effective recombination between heterokaryotypes of paracentric inversions may facilitate speciation by protecting locally adapted alleles inside the inversions from gene flow with other populations, allowing further divergence between two karyotypes (reviewed in earlier studies [9,10]). While theoretical [11–14] and empirical [15–21] support for this hypothesis exists, the maintenance of genomic divergence-within inversions over long time periods (many hundreds of thousands of generations), and the shape of elevated divergence-within inversions have been less explored empirically (but see [2–5]).
The diverging sister taxa of Drosophila pseudoobscura and Drosophila persimilis offer a suitable system for evaluating the impacts of inversions on genome-wide diversity and divergence. Drosophila pseudoobscura is a widespread species, ranging across western Canada, the United States and part of Central America, with a disjunct, allopatric subspecies in Colombia (D. pseudoobscura bogotana). Its sister species, D. persimilis, is restricted to the far west of Canada and the United States. The two D. pseudoobscura subspecies shared a common ancestor with D. persimilis approximately 500 000 years ago [22,23] and the two subspecies diverged from each other approximately 200 000 years ago [22,24]. With one exception, hybrid males from all possible crosses of the three species/subspecies are sterile, and a very small number of D. pseudoobscura–D. persimilis hybrids have been found in the wild [25,26]. In total, these species have six chromosome arms (three telocentric autosomes (chromosomes 2–4), one ‘dot’ autosome (chromosome 5) and a metacentric X chromosome with a left and right arm, XL and XR, respectively), and two inversions—one on the XL and one on the second chromosome—are fixed between D. pseudoobscura and D. persimilis [27]. The inversions on the second chromosome and the XL are slightly greater than 7 Mb each (23% and 34% of each chromosome arm, respectively), and another inversion on the XR, which is polymorphic within D. persimilis, is 13 Mb (45% of the XR chromosome arm) [5]. These three inversions are derived in D. persimilis [27,28], and D. persimilis that do not harbour the XR inversion exhibit sex-ratio distortion [28]. Chromosome 3 segregates for more than 30 different inversion arrangements within D. pseudoobscura [29].
The three inversions differentiate between D. persimilis and D. pseudoobscura and encompass important alleles for maintaining reproduction isolation in the face of gene flow [15,16]. For hybrids of D. persimilis and D. pseudoobscura, reproductive traits such as male sterility, male mating success, female species preferences and hybrid inviability map to the XL and chromosome 2 inversions [15,16], and male courtship dysfunction localizes to the XR inversion as well as the fixed inversions [16]. In the allopatric species pair, D. ps. bogotana and D. persimilis, no gene flow occurs, reproductive isolation alleles do not mix in collinear regions and alleles for reproductive isolation have accumulated outside of inversions [30–33]. In contrast, in the feebly hybridizing, parapatric D. pseudoobscura and D. persimilis, sterility of hybrid males localizes predominantly to inversions [30]. Thus, it appears that inversions play an important role in maintaining species differences in sympatry [15].
Based on sequence analyses and genetic mapping data, we inferred that gene flow has homogenized the collinear regions of D. pseudoobscura and D. persimilis [2,4,5,34], but sequence divergence is maintained in and approximately 2.5 Mb outside of the chromosome 2, XL and XR inversions owing to reduced recombination [2,4–6,34]. Little reduction of divergence toward the centre of these inversions is observed [2,5,6]. However, these inferences are based on sparse sequence coverage (either in number of genomes or in coverage of genomes) rather than high-sequence depth comparisons.
Gene flow may erode divergence within inversions through the action of gene conversions and double crossovers. Small gene conversion events (approx. 200 bp [35]) are expected to act homogeneously across the inversion [36], except at regions extremely close to the inversion breakpoints (e.g. less than 17 kb [35]). Double crossovers between the karyotypes rarely encompass areas near breakpoints, and breakpoints remain diverged even when the centre of the inversion experiences gene flow [3,37–39].
Recent theoretical work has precipitated a need to re-examine divergence patterns across fixed inversions between D. persimilis and D. pseudoobscura. First, in some cases, theoretical simulations indicate that long-term maintenance of divergence in inversion-localized genes may wear away upon secondary contact through the action of gene conversion and double crossovers [40]. Second, coalescent models suggest that linked neutral divergence between karyotypes is expected to be higher at breakpoints and near locally adapted alleles in the inversion, even for relatively old inversions [14]. We examine the diversity and divergence patterns across fixed inversions for D. persimilis and D. pseudoobscura, in the light of this recent theoretical work and with more extensive sequence data than past studies, to evaluate divergence in inversion centres relative to breakpoints.
We also compare the divergence patterns of D. persimilis and D. pseudoobscura with divergence patterns of the two species to their closest outgroup, D. miranda. Drosophila miranda diverged from D. pseudoobscura and D. persimilis approximately 2 Ma [22,41,42]. This outgroup does not harbour the three inversions and cannot successfully hybridize with any of the other species in our study. By comparing the D. persimilis–D. pseudoobscura divergence with the divergence patterns to their closest outgroup, we draw inferences regarding the coalescence of the arrangements for each inversion.
We leverage new whole-genome shotgun sequences, originally sequenced for a companion study, to estimate pairwise differences in intergenic regions across collinear and inverted regions on chromosome 2, XL and XR. Specifically, our comparisons include two resequenced lines of D. persimilis, 10 resequenced lines of D. pseudoobscura and three resequenced lines of D. miranda. We measured nucleotide differences outside and within the inversions with special attention to the distance from the inversion breakpoints. Recombination suppression extending outside of the inversions has been more finely pinpointed [6] compared with past work [2]; therefore, the effect of inversion-associated recombination suppression can be more accurately measured.
2. Material and methods
For each line, DNA from 15 to 20 virgin female inbred flies was extracted using the GentraSystems PureGene DNA preparation (Minneapolis, MN, USA) and sequenced with Illumina Genome Analyzer IIx at the University of North Carolina-Chapel Hill High-Throughput Sequencing Facility (see electronic supplementary material, table S1 for type of read and amount of data obtained for each line). Illumina reads were aligned to the D. pseudoobscura reference genome v2.9 using bwa-0.5.5 (default alignment settings were used except the maximum number of gap extensions was set to 4) [43]. Consensus assemblies for each resequenced line were generated using the bwa alignments and samtools 0.1.6 pileup [44]. Default pileup settings were used except the theta parameter (error-dependency coefficient) in the maq consensus calling model was set to 0.9, and the number of haplotypes in the sample was set to 1 because each resequenced line was inbred. Each consensus sequence was filtered with custom python scripts. We excluded bases with read coverage less than four or with Phred-scale consensus mapping score of less than 30. We also excluded insertions and deletions that were supported by less than 70 per cent of reads and excluded 5 bp flanking the indel.
We estimated average pairwise differences for intergenic regions across collinear and inverted regions on chromosome 2 and the XL chromosome arm using two resequenced lines of D. persimilis and the reference D. persimilis genome, 10 resequenced lines of D. pseudoobscura and the D. pseudoobscura reference genome, and three resequenced lines of D. miranda. For the XR, we included all D. pseudoobscura and D. miranda lines, but only one resequenced line of D. persimilis and the reference D. persimilis genome. The other resequenced line of D. persimilis was a sex-ratio distortion line and does not contain the XR inversion. This sequencing effort was initially motivated by a companion study, which did not require even sampling across species. Intergenic regions are best suited for this assay, as opposed to intron or synonymous sites, because the distance between the inversion breakpoint and the nearest predicted gene is often very large [5]. Bases were considered intergenic if no coding or intron regions were annotated in Flybase for D. pseudoobscura. Results from intron bases and fourfold degenerate sites are consistent with intergenic regions (see electronic supplementary material, figures S2–S7). Inversion breakpoints were defined as in Noor et al. [5].
All analyses were performed in jmp v. 8.0.2. We used Mann–Whitney U-test and Wilcoxon signed-rank matched pairs tests to examine whether average pairwise divergence between regions differed, and corrected for multiple-testing using sequential Bonferroni [45,46]. All statistics were performed on average pairwise divergence across (i) 10 and (ii) 100 kb non-overlapping windows. For all tests, chromosomes were divided into four regions: inside the inversion (IN), regions of recombination suppression outside but near the inversion (2.45–2.84 Mb; see [5,6]; NEAR), regions outside of the inversion and the region of recombination suppression (FAR) and breakpoints. The number of bases included in the breakpoint region was dependent on the window size. For the 10 kb window, we included two 10 kb windows on each side of the inversion. For the 100 kb window, we used one 100 kb window spanning the breakpoint. We explored various numbers of windows and performed the analysis with (i) ten 10 kb windows spanning each inversion breakpoint or (ii) five 100 kb windows spanning each inversion breakpoint. These added windows served only to obscure the divergence difference between the breakpoint and the inverted region and were not used in further analysis. We excluded 3 Mb proximal to the centromere and 1 Mb closest to the telomere; centromeric effects on diversity and divergence appear to extend farther into the genome. For the XL chromosome arm, the FAR region was only on the centromeric side of the inversion because any bases that qualified on the telomeric side were within 1 Mb of the end of the chromosome.
3. Results and discussion
For chromosomal speciation by suppressed recombination to be supported: (i) sister taxa must maintain fixed inversion differences, (ii) recombination must be suppressed within/near the inversion, and (iii) reproductive isolation alleles must localize to the inverted region [9]. The D. pseudoobscura–D. persimilis system satisfies all requirements [6,15,16,47], and our study quantifies the genomic impacts of chromosomal speciation between D. pseudoobscura and D. persimilis. Our results suggest that within and near the inversions (especially, chromosome 2 and XL) divergence between heterokaryotypes has been maintained for approximately 2 Myr.
(a). Divergence patterns in and near inversions
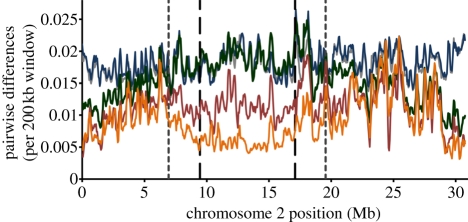
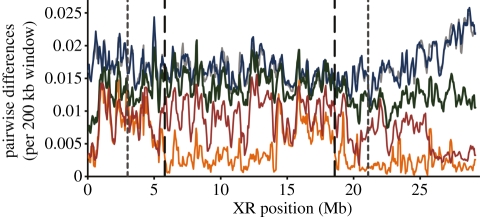
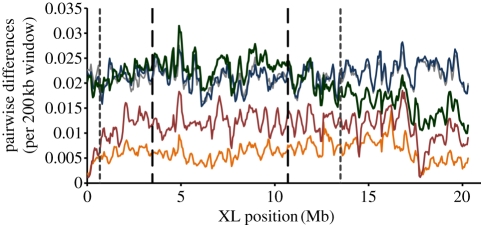
Evidence from other systems, where chromosomal rearrangements have played a role in speciation, suggests that divergence is maintained predominantly very close to the inversion breakpoints [3,39]. Between D. pseudoobscura and D. persimilis for intergenic regions, breakpoints for the XL chromosome did not exhibit higher divergence relative to that observed inside the inversions. However, for both of the scales measured (10 or 100 kb windows; electronic supplementary material, tables S2 and S3), chromosome 2 and XR breakpoints exhibited qualitatively higher divergence in intergenic regions relative to that observed inside the inversions. The largest observed difference was in the 100 kb window analysis of chromosome 2; the median average divergence for breakpoint windows (one 100 kb window at each breakpoint, 0.032) was approximately 1.7 times the median average divergence inside the inversion (0.019). Likewise, breakpoints of the XR inversion harbour approximately 1.1–1.4 times the amount of divergence seen inside the inversion (100 and 10 kb windows, respectively). Power to assess significance was low in all cases, because the number of intervals which spanned the breakpoint was small, but overall, we do not observe a striking difference in divergence between the regions immediately spanning the breakpoints and regions further inside the inversion.
In some cases, gene flow during secondary contact between diverged karyotypes may erode divergence in the centre of inversions [36,48] if low levels of gene flux occur in heterokaryotypes. In D. pseudoobscura–D. persimilis heterokaryotypes, the rate of double crossovers within the inverted region is approximately 1 × 10−4 (measured over the XR inversion [6])—four orders of magnitude higher than values that could theoretically erode allele frequency differences between karyotypes under certain scenarios [40]. However, for all three inverted regions between D. pseudoobscura and D. persimilis, average pairwise divergence in intergenic regions is significantly higher in rearranged versus FAR collinear regions (figures 1–3; electronic supplementary material, tables S2 and S3; p < 0.0001, Z > 5.748 in all cases for both 10 and 100 kb windows) even though these two species diverged several millions of generations ago.
Figure 1.
Sliding window analysis of the fraction of bases in intergenic regions differentiating two genome sequences along a 200 kbp interval, iterated every 50 kbp for chromosome 2. Average pairwise differences were used for comparisons where a total of more than two genome sequences were available. Eleven Drosophila pseudoobscura genomes, three Drosophila persimilis and three Drosophila miranda were used in generating the graphs. Dashed lines represent the boundaries of the inversions and dotted lines are the boundaries of recombination suppression as defined in Stevison et al. [6]. Chromosomes are presented from telomere to centromere. Orange, D. persimilis–D. persimilis; pink, D. pseudoobscura–D. pseudoobscura; green, D. pseuodoobscura–D. persimilis; blue, D. miranda–D. seudoobscura; grey, D. miranda–D. persimilis.
Figure 3.
Sliding window analysis of the fraction of bases in intergenic regions differentiating two genome sequences along a 200 kbp interval, iterated every 50 kbp for the XR chromosome arm. All legends and data used same as in figure 1, except that one resequenced line of Drosophila persimilis was not included.
Divergence also extends beyond the inversion boundaries into collinear regions [4,5], likely because recombination is suppressed in these regions as well [2,6]. For the XL and chromosome 2, we confirmed that divergence for NEAR regions was significantly higher than FAR regions (figures 1 and 2; electronic supplementary material, tables S2 and S3; p < 0.0001, Z > 5.428 in all cases for both 10 and 100 kb windows). For the XR inversion, NEAR regions were significantly more divergent than FAR regions only for the 10 kb window analysis before Bonferroni adjustment (p = 0.0331 before Bonferroni, Z = 2.131; figure 3; electronic supplementary material, tables S2 and S3). The putatively ancestral XR arrangement is still segregating in some populations of D. persimilis (homosequential to D. pseudoobscura); thus differences between IN, FAR, and NEAR genomic regions in the XR are expected to be less defined than the inversions on the XL and chromosome 2.
Figure 2.
Sliding window analysis of the fraction of bases in intergenic regions differentiating two genome sequences along a 200 kbp interval, iterated every 50 kbp for the XL chromosome arm. All legends and data used same as in figure 1.
(b). Heterogeneity between inversions in magnitude of D. persimilis–D. pseudoobscura divergence
The D. pseudoobscura–D. persimilis divergence between the inversions follows the pattern: XL > 2 > XR. This pattern was suggested previously to indicate that the inversions arose and fixed sequentially—each capturing a different coalescence point and preventing further gene flow within the bounds of the inversion. In this scenario, gene flow continued for the remainder of the genome until a subsequent inversion arose and became established [5]. Alternatively, the heterogeneous divergence pattern (XL > 2 > XR) seen across the inversions was posited the result of differential mutation pressures across the genome [2], as a similar divergence pattern across chromosomes was observed for D. pseudoobscura and D. miranda which are not separated by inversion. The working model for this system from Kulathinal et al. [2] is that the three inversions arose and the inversions on XL and chromosome 2 became fixed in the nascent D. persimilis lineage shortly after the separation of D. miranda–D. pseudoobscura–D. persimilis [2]. Our data refine this hypothesis in several ways.
First, we found that divergence in inverted regions for both the XL and chromosome 2 between D. pseudoobscura and D. persimilis was greater than the divergence of either with putative outgroup species D. miranda (matched pairs for all sequential Bonferroni-corrected comparisons for both 10 and 100 kb windows: XL: p < 0.001 median average pairwise divergence across all windows in inversion ps–per = 0.0231, ps–mir = 0.0208, per–mir = 0.0212; chromosome 2: p < 0.042, ps–per = 0.0185, ps–mir = 0.0178 and per–mir = 0.0178). These results suggest that, within the XL and chromosome 2 inversions, D. persimilis and D. pseudoobscura share a deeper ancestor than they do with D. miranda. However, the D. pseudoobscura–D. miranda divergence is equal to the D. persimilis–D. miranda divergence (sequential Bonferroni-corrected p > 0.084 in all cases) which would argue that the three arose at nearly the same time. To account for this discrepancy, D. persimilis–D. pseudoobscura divergence would have had to increase within the inversions, perhaps at and near sites under divergent selection between D. persimilis and D. pseudoobscura (reviewed in [49]). While very speculative, this hypothesis is indirectly supported in that sites thought to be under less selective constraint (intron and fourfold degenerate sites; electronic supplementary material, figures S2–S7) exhibit this ‘overly elevated’ D. persimilis–D. pseudoobscura divergence relative to D. miranda to a lesser extent than putatively less-neutral intergenic regions.
The inversions also may coalesce deeper in evolutionary history than the remainder of the genome. Using the collinear FAR region on the centromeric side (FAR-C) for the XL and chromosome 2, we observed that the D. pseudoobscura–D. persimilis divergence is less than the divergence of either from D. miranda, indicating that gene flow has homogenized collinear regions relative to inverted regions (XL: ps–per = 0.0166, ps–mir = 0.0227, per–mir = 0.0219; chromosome 2: ps–per = 0.0150, ps–mir = 0.0191, per–mir = 0.0185, match pairs p < 0.0001, for all sequential Bonferroni-corrected comparisons for both 10 and 100 kb windows).
Second, the hypothesis that the inversions arose sequentially, and captured different time points during divergence of the species, can be examined with our data. While the D. pseudoobscura–D. persimilis divergence is heterogeneous among inversions, the D. pseudoobscura–D. persimilis divergence relative to the D. pseudoobscura–D. miranda divergence (which are not differentiated by these inversions) is also heterogeneous among inversions, and such a comparison accounts for heterogeneous mutation rates across chromosomes. The D. pseudoobscura–D. persimilis divergence relative to the D. pseudoobscura–D. miranda divergence is approximately 1.10 times higher within the XL inversion than within the chromosome 2 inversion and approximately 1.27 times higher within the XL inversion than within the XR inversion (all pairwise comparisons, p < 0.0001). Likewise, the D. pseudoobscura–D. persimilis divergence relative to the D. pseudoobscura–D. miranda divergence is approximately 1.17 times higher for the chromosome 2 inversion than the XR inversion. This result tentatively suggests a mechanism of a sequential origin of the inversions, with the inversions on the XL and chromosome 2 arising before XR, and possibly with the XL inversion arising before the chromosome 2 inversion. However, the XR inversion in D. persimilis remains polymorphic. Within-D. persimilis gene flow between arrangements on the XR is relatively substantial [6], and may be why the XR inversion appears much younger than the XL and chromosome 2 inversions. Thus, any accurate estimation of the timing of the XR inversion's origin is difficult. Likewise, it is possible that the apparent relative age of the inversions (XL > 2 > XR) is actually more a reflection of the relative time that each inversion fixed, not necessarily the timing of their origin; so, definitive conclusions regarding the sequential origin and fixation of these inversions are nearly impossible to make at this time.
Finally, because the XR inversion remains polymorphic and is not fixed within D. persimilis, we can assert that the timing of fixation of the XR inversion is secondary to the fixation of the chromosome 2 and XL inversions. The strong reduction in nucleotide diversity in the XL and chromosome 2 inversions of D. persimilis suggests that, while the inversions may have arisen long ago, the fixation of the XL and chromosome 2 inversions was more recent, probably near or since the D. persimilis–D. pseudoobscura–D. miranda split (sensu [14,48]). Within D. persimilis, nucleotide diversity is low over the regions encompassed in and near the XL (median diversity: FAR = 0.008, NEAR = 0.006 and IN = 0.005; Z > 3.174, Bonferroni-corrected p = 0.024; figures 1 and 2) and chromosome 2 inversions (median diversity: FAR = 0.010, NEAR = 0.008 and IN = 0.005; Z > 4.487, p < 0.001, in all cases), whereas a similar reduction in diversity is much weaker or non-existent for D. pseudoobscura (XL median diversity: FAR = 0.012, NEAR = 0.012 and IN = 0.012; chromosome 2 median diversity: FAR = 0.012, NEAR = 0.012 and IN = 0.011, all comparisons not significant; figures 1 and 2). Interestingly, the D. persimilis nucleotide diversity in and near the XR inversion is greater than the diversity in the region outside of the inversion (FAR = 0.001, NEAR = 0.003 and IN = 0.003; significant for 10 kb windows only, Bonferroni-corrected p < 0.044 both comparisons; figure 3), consistent with this inversion being polymorphic within D. persimilis and the inferred exchange between the two arrangements within that species [6].
Our data appear consistent with a model that treats the origin and fixation of an inversion as two separate steps. Theoretically, the inversions may have persisted as low-frequency variants within the nascent D. persimilis lineage for a long period of time (accounting for the deep coalescence) and more recently the inversions were driven to fixation (accounting for the reduced diversity within D. persimilis). Theoretical underpinnings of such a mechanism were recently presented (‘mixed mode geographic model’) [50] and rely on periods of allopatry (for an inversion to arise in a population) and secondary contact (to drive the inversion to fixation). This theory appears to describe our system well. The inversions on the XL and chromosome 2 may have arisen in allopatric lineages leading to D. persimilis. More recently, allopatric populations of D. persimilis and D. pseudoobscura came into contact and hybridized, favouring the spread of the low-frequency D. persimilis inversions owing to their recombination-reducing effect. Other predictions of the mixed mode model are also consistent with our data. For example, when populations re-establish secondary contact and population sizes differ, inversions will more often elevate in frequency in the smaller of the two populations [50] (in this case D. persimilis).
4. Future directions
First, while we have qualitatively described patterns of diversity and divergence with respect to inversions, formal model testing would better elucidate the processes responsible for these patterns. With formal model testing, we could evaluate a range of plausible parameter values for migration, gene flux, and selection to develop a more quantitative understanding of the forces influencing the D. pseudoobscura–D. persimilis inversions. Recent theoretical works [14,40,50] offer promising steps to enable such parametrization, and such analysis may be plausible in the future.
Second, inversions may have a greater role in shaping genomic diversity than the effects explored in our current study. When present in a heterozygous state, inversions stall the pachytene checkpoint and likely allow more time during meiosis for double strand breaks to be resolved as crossovers; thus, heterozygous inversions increase crossing over across the genome (‘interchromosomal effect’) [51–54] and this effect has been observed in the D. pseudoobscura system in particular [6,55]. Broadly, crossing over is positively associated with genomic diversity [56,57]; thus, inversions fixed between parapatric species may inadvertently increase genomic diversity in collinear regions when these species hybridize through the interchromosomal effect. Investigating this impact of inversions in creating and maintaining genomic diversity through the interchromosomal effects may reveal in part how hybrid zones contribute to within species diversity and the evolution of new adaptations.
In conclusion, while inversions play an important role in reproductive isolation in the D. pseudoobscura–D. persimilis system, they are just one avenue in which accentuated divergence may accumulate between the genomes of diverging, but occasionally hybridizing taxa [1,40]. Reduced recombination, in general, can contribute to divergence during the speciation process (reviewed in Nachman & Payseur [58]). Further, genomic scans and experimental approaches have identified regions with greater differentiation than expected under neutrality that are clustered but not associated with inversions (reviewed in earlier studies [1,59,60]). We look forward to a more comprehensive understanding of how often and when chromosomal rearrangements and these other processes facilitate the speciation process.
Acknowledgements
We thank L. Stevison, R. Guerrero, M. Kirkpatrick and three additional anonymous reviewers for critical comments on the manuscript. C. Jones provided useful discussion, and P. Nosil and J. Feder provided valuable insights. Funding for this research was provided by NIH grants GM076051, GM086445 and GM092501.
References
- 1.Nosil P., Funk D. J., Ortiz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18, 375–402 10.1111/j.1365-294X.2008.03946.x (doi:10.1111/j.1365-294X.2008.03946.x) [DOI] [PubMed] [Google Scholar]
- 2.Kulathinal R. J., Stevison L. S., Noor M. A. F. 2009. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5, e1000550. 10.1371/journal.pgen.1000550 (doi:10.1371/journal.pgen.1000550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasburg J. L., Scotti-Saintagne C., Scotti I., Lai Z., Rieseberg L. H. 2009. Genomic patterns of adaptive divergence between chromosomally differentiated sunflower species. Mol. Biol. Evol. 26, 1341–1355 10.1093/molbev/msp043 (doi:10.1093/molbev/msp043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machado C. A., Haselkorn T. S., Noor M. A. F. 2007. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics 175, 1289–1306 10.1534/genetics.106.064758 (doi:10.1534/genetics.106.064758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noor M. A., Garfield D. A., Schaeffer S. W., Machado C. A. 2007. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics 177, 1417–1428 10.1534/genetics.107.070672 (doi:10.1534/genetics.107.070672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevison L. S., Hoehn K., Noor M. A. F. 2011. The effect of chromosomal inversions on recombination in hybrid between Drosophila pseudoobscura and D. persimilis. Genome Biol. Evol. 3, 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturtevant A. H., Beadle G. W. 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21, 554–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts P. A. 1967. A positive correlation between crossing over within heterozygous pericentric inversions and reduced egg hatch of Drosophila females. Genetics 56, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faria R., Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol. Evol. 25, 660–669 10.1016/j.tree.2010.07.008 (doi:10.1016/j.tree.2010.07.008) [DOI] [PubMed] [Google Scholar]
- 10.Jackson B. C. 2011. Recombination-suppression: how many mechanisms for chromosomal speciation? Genetica 139, 393–402 10.1007/s10709-011-9558-0 (doi:10.1007/s10709-011-9558-0) [DOI] [PubMed] [Google Scholar]
- 11.Trickett A. J., Butlin R. K. 1994. Recombination suppressors and the evolution of new species. Heredity 73, 339–345 10.1038/hdy.1994.180 (doi:10.1038/hdy.1994.180) [DOI] [PubMed] [Google Scholar]
- 12.Navarro A., Barton N. H. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57, 447–459 10.1111/j.0014-3820.2003.tb01537.x (doi:10.1111/j.0014-3820.2003.tb01537.x) [DOI] [PubMed] [Google Scholar]
- 13.Kirkpatrick M., Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434 10.1534/genetics.105.047985 (doi:10.1534/genetics.105.047985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerrero R. F., Rousset F., Kirkpatrick M. 2012. Coalescent patterns for chromosomal inversions in divergent populations. Phil. Trans. R. Soc. B 367, 430–438 10.1098/rstb.2011.0246 (doi:10.1098/rstb.2011.0246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noor M. A., Grams K. L., Bertucci L. A., Reiland J. 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl Acad. Sci. USA 98, 12 084–12 088 10.1073/pnas.221274498 (doi:10.1073/pnas.221274498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noor M. A., Grams K. L., Bertucci L. A., Almendarez Y., Reiland J., Smith K. R. 2001. The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution 55, 512–521 10.1111/j.0014-3820.2001.tb00785.x (doi:10.1111/j.0014-3820.2001.tb00785.x) [DOI] [PubMed] [Google Scholar]
- 17.Rieseberg L. H. 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358 10.1016/S0169-5347(01)02187-5 (doi:10.1016/S0169-5347(01)02187-5) [DOI] [PubMed] [Google Scholar]
- 18.Feder J. L., et al. 2003. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl Acad. Sci. USA 100, 10 314–10 319 10.1073/pnas.1730757100 (doi:10.1073/pnas.1730757100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feder J. L., Roethele J. B., Filchak K., Niedbalski J., Romero-Severson J. 2003. Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics 163, 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayala F. J., Coluzzi M. 2005. Chromosome speciation: humans. Drosophila, and mosquitoes. Proc. Natl Acad. Sci. USA 102, 6535–6542 10.1073/pnas.0501847102 (doi:10.1073/pnas.0501847102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandul N. P., Lukhtanov V. A., Pierce N. E. 2007. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 61, 546–559 10.1111/j.1558-5646.2007.00046.x (doi:10.1111/j.1558-5646.2007.00046.x) [DOI] [PubMed] [Google Scholar]
- 22.Wang R. L., Wakeley J., Hey J. 1997. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics 147, 1091–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hey J., Nielsen R. 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167, 747–760 10.1534/genetics.103.024182 (doi:10.1534/genetics.103.024182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaeffer S. W., Miller E. L. 1991. Nucleotide sequence analysis of Adh genes estimates the time of geographic isolation of the Bogota population of Drosophila pseudoobscura. Proc. Natl Acad. Sci. USA 88, 6097–6101 10.1073/pnas.88.14.6097 (doi:10.1073/pnas.88.14.6097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobzhansky T. 1973. Is there gene exchange between Drosophila pseudoobsura and Drosophila persimilis in their natural habitats? Am. Nat. 107, 312–314 10.1086/282833 (doi:10.1086/282833) [DOI] [Google Scholar]
- 26.Powell J. R. 1983. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: evidence from Drosophila. Proc. Natl Acad. Sci. USA 80, 492–495 10.1073/pnas.80.2.492 (doi:10.1073/pnas.80.2.492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobzhansky T., Tan C. C. 1936. Studies on hybrid sterility III. A comparison of the gene arrangement in two species, Drosophila pseudoobscura and Drosophila miranda. Z. Indukt. Abstamm. Vererbungsl 72, 88–114 10.1007/BF01850144 (doi:10.1007/BF01850144) [DOI] [Google Scholar]
- 28.Dobzhansky T., Epling C. 1944. Contributions to the genetics, taxonomy, and ecology of Drosophila pseudoobscura and its relatives. Washington, DC: Carnegie Institute of Washington [Google Scholar]
- 29.Dobzhansky T., Powell J. R. 1975. Drosophila pseudoobscura and its American relatives, Drosophila persimilis and Drosophila miranda. Handb. Genet. 3, 537–587 [Google Scholar]
- 30.Brown K. M., Burk L. M., Henagan L. M., Noor M. A. 2004. A test of the chromosomal rearrangement model of speciation in Drosophila pseudoobscura. Evolution 58, 1856–1860 10.1111/j.0014-3820.2004.tb00469.x (doi:10.1111/j.0014-3820.2004.tb00469.x) [DOI] [PubMed] [Google Scholar]
- 31.Chang A. S., Noor M. A. F. 2007. The genetics of hybrid male sterility between the allopatric species pair Drosophila persimilis and D. pseudoobscura bogotana: dominant sterility alleles in collinear autosomal regions. Genetics 176, 343–349 10.1534/genetics.106.067314 (doi:10.1534/genetics.106.067314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang A. S., Noor M. A. 2009. Epistasis modifies the dominance of loci causing hybrid male sterility in the Drosophila pseudoobscura species group. Evolution 64, 253–260 10.1111/j.1558-5646.2009.00823.x (doi:10.1111/j.1558-5646.2009.00823.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang A. S., Bennett S. M., Noor M. A. F. 2010. Epistasis among Drosophila persimilis factors conferring hybrid male sterility with D. pseudoobscura bogotana. PLoS ONE 5, e15377. 10.1371/journal.pone.0015377 (doi:10.1371/journal.pone.0015377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machado C. A., Kliman R. M., Markert J. A., Hey J. 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19, 472–488 [DOI] [PubMed] [Google Scholar]
- 35.Schaeffer S. W., Anderson W. W. 2005. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics 171, 1729–1739 10.1534/genetics.105.041947 (doi:10.1534/genetics.105.041947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro A., Ruiz A. 1997. On the fertility effects of pericentric inversions. Genetics 147, 931–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andolfatto P., Depaulis F., Navarro A. 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77, 1–8 10.1017/S0016672301004955 (doi:10.1017/S0016672301004955) [DOI] [PubMed] [Google Scholar]
- 38.Laayouni H., Hasson E., Santos M., Fontdevila A. 2003. The evolutionary history of Drosophila buzzatii. XXXV. Inversion polymorphism and nucleotide variability in different regions of the second chromosome. Mol. Biol. Evol. 20, 931–944 10.1093/molbev/msg099 (doi:10.1093/molbev/msg099) [DOI] [PubMed] [Google Scholar]
- 39.Yatabe Y., Kane N. C., Scotti-Saintagne C., Rieseberg L. H. 2007. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 175, 1883–1893 10.1534/genetics.106.064469 (doi:10.1534/genetics.106.064469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feder J. L., Nosil P. 2009. Chromosomal inversions and species differences: when are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution 63, 3061–3075 10.1111/j.1558-5646.2009.00786.x (doi:10.1111/j.1558-5646.2009.00786.x) [DOI] [PubMed] [Google Scholar]
- 41.Beckenbach A. T., Wei Y. W., Liu H. 1993. Relationships in the Drosophila obscura species group, inferred from mitochondrial cytochrome oxidase II sequences. Mol. Biol. Evol. 10, 619–634 [DOI] [PubMed] [Google Scholar]
- 42.Machado C. A., Hey J. 2003. The causes of phylogenetic conflict in a classic Drosophila species group. Proc. R. Soc. Lond. B 270, 1193–1202 10.1098/rspb.2003.2333 (doi:10.1098/rspb.2003.2333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 10.1093/bioinformatics/btp324 (doi:10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. & 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079 10.1093/bioinformatics/btp352 (doi:10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm S. 1979. A simple sequentially rejective Bonferroni test procedure. Scand. J. Stat. 6, 65–70 [Google Scholar]
- 46.Rice W. R. 1989. Analyzing tables of statistical tests. Evolution 43, 223–225 10.2307/2409177 (doi:10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- 47.Dobzhansky T. 1936. Studies of hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21, 113–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro A., Barbadilla A., Ruiz A. 2000. Effect of inversion polymorphism on the neutral nucleotide variability of linked chromosomal regions in Drosophila. Genetics 155, 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faria R., Neto S., Noor M. A. F., Navarro A. 2011. Role of natural selection in chromosomal speciation. ELS. Chichester, UK: John Wiley & Sons, Ltd; (doi:10.1002/9780470015902.a0022850) [Google Scholar]
- 50.Feder J. L., Gejji R., Powell T. H. Q., Nosil P. 2011. Adaptive chromosomal divergence driven by mixed geographic mode of evolution. Evolution 65, 2157–2170 10.1111/j.1558-5646.2011.01321.x (doi:10.1111/j.1558-5646.2011.01321.x) [DOI] [PubMed] [Google Scholar]
- 51.Sturtevant A. H. 1919. Contributions to the genetics of Drosophila melanogaster. III. Inherited linkage variation in the second chromosome. Carnegie Inst. Wash. Publ. 278, 305–341 [Google Scholar]
- 52.Schultz J., Redfield H. 1951. Interchromosomal effects on crossing over in Drosophila. Cold Spring Harbor Symp. Quant. Biol. 16, 175–197 [DOI] [PubMed] [Google Scholar]
- 53.Joyce E. F., McKim K. S. 2010. Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS Genet. 6, e1001059. 10.1371/journal.pgen.1001059 (doi:10.1371/journal.pgen.1001059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pegueroles C., Ordónez V., Mestres F., Pascual M. 2010. Recombination and selection in the maintenance of the adaptive value of inversions. J. Evol. Biol. 23, 2709–2717 10.1111/j.1420-9101.2010.02136.x (doi:10.1111/j.1420-9101.2010.02136.x) [DOI] [PubMed] [Google Scholar]
- 55.Ortiz-Barrientos D., Chang A. S., Noor M. A. 2006. A recombinational portrait of the Drosophila pseudoobscura genome. Genet. Res. 87, 23–31 10.1017/S0016672306007932 (doi:10.1017/S0016672306007932) [DOI] [PubMed] [Google Scholar]
- 56.Begun D. J., Aquadro C. F. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356, 519–520 10.1038/356519a0 (doi:10.1038/356519a0) [DOI] [PubMed] [Google Scholar]
- 57.Nachman M. 2002. Variation in recombination rate across the genome: evidence and implications. Curr. Opin. Genet. Dev. 12, 657–663 10.1016/S0959-437X(02)00358-1 (doi:10.1016/S0959-437X(02)00358-1) [DOI] [PubMed] [Google Scholar]
- 58.Nachman M. W., Payseur B. A. 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Phil. Trans. R. Soc. B 367, 409–421 10.1098/rstb.2011.0249 (doi:10.1098/rstb.2011.0249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nosil P., Feder J. L. 2012. Genomic divergence during speciation: causes and consequences. Phil. Trans. R. Soc. B 367, 332–342 10.1098/rstb.2011.0263 (doi:10.1098/rstb.2011.0263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feder J. L., Gejji R., Yeaman S., Nosil P. 2012. Establishment of new mutations under divergence and genome hitchhiking. Phil. Trans. R. Soc. B 367, 461–474 10.1098/rstb.2011.0256 (doi:10.1098/rstb.2011.0256) [DOI] [PMC free article] [PubMed] [Google Scholar]