Abstract
The function of the tumor suppressor promyelocytic leukemia (PML) protein is disrupted in promyelocytic leukemia. PML has been reported to function as a negative regulator of mTOR (mammalian target of rapamycin) and nuclear Akt under some conditions. mTOR and Akt pathways regulate a diverse array of pathways, including those that control insulin signaling, energy metabolism, growth, cellular survival, and lifespan. Although the PML-mTOR/Akt link suggests that PML may have metabolic functions in the whole organism, very little is known about the metabolic functions of PML. Here we report that PML−/− mice did not show any significant metabolic defects. There was no impairment in the mTOR/Akt or AMPK signaling in white adipose tissue, liver, or muscle. However, despite having normal food intake and activity levels, PML−/− mice gained body weight faster and had more fat mass, particularly subcutaneous fat mass, in the diet-induced obesity model. Using in vitro adipogenesis models, we discovered that PML is a suppressor of adipogenesis. PML expression decreased during adipogenesis and was undetectable in fully differentiated adipocytes. Loss of PML increased expression of the adipogenic transcription factors CCAAT/enhancer binding protein-α and peroxisome proliferator-activated receptor-γ. We found that the Sirt1-NCor-SMRT corepressor complex, which represses pparg transcription, does not bind to the pparg promoter efficiently upon PML depletion. On the basis of these findings, we propose that PML is a negative regulator of the adipogenic transcription factors and that, in times of energy excess, PML may limit fat accumulation by suppressing the differentiation of preadipocytes into adipocytes.
Keywords: adipogenesis
the promyelocytic leukemia (pml) gene, which is translocated in most acute promyelocytic leukemias, encodes a tumor suppressor protein (59). The loss of PML is associated with the pathogenesis of a variety of hematopoietic malignancies and solid tumors. PML suppresses proliferation and promotes apoptosis and senescence. PML is localized to nuclear foci termed PML nuclear bodies (NB). It is also found diffusely throughout the nucleus and in the cytoplasm. Despite the fact that a variety of biological functions have been connected to PML, PML-deficient mice exhibit only subtle defects and develop normally (71, 72).
Recent studies reported that PML may exert its tumor-suppressive role, at least in part, by suppressing mTOR and nuclear Akt activities (7, 30, 64). These studies suggested that PML colocalizes with mammalian target of rapamycin (mTOR) and Akt at PML NBs, inhibiting mTOR and nuclear phospho-Akt function in some conditions. Under hypoxic condition, PML represses mTOR through sequestration of mTOR into PML NBs, inhibiting the interaction of mTOR with its negative regulator Rheb (Ras homolog enriched in brain; a small GTPase) (7). In mouse cancer models, PML inactivates Akt by recruiting nuclear phospho-Akt together with its cognate phosphatase PP2aC into PML NBs (64).
Although the mTOR/Akt signaling network is critically important for oncogenesis, it also plays a key role in obesity and insulin resistance (41, 68). mTOR exists in two complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Growth factors and hormones such as insulin regulate mTORC1 through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway (60). Activated Akt transduces signals to mTORC1 through tuberous sclerosis protein (TSC) 2 of the tumor suppressor TSC1/TSC2 complex by modulating Rheb (29). The activated GTP-bound Rheb then enables mTORC1 signaling to its downstream effectors such as S6K1 (S6 kinase 1) and 4E-BPs (eukaryotic initiation factor 4E-binding proteins), enhancing mRNA translation. Nutrients activate S6K1 and 4E-BP1 via mTORC1.
mTORC1 and its effectors S6K1 and 4E-BP1/2 act as critical mediators for the development of obesity and insulin resistance (15, 41, 68). S6K1−/− mice are resistant to age- and diet-induced obesity and insulin resistance, have reduced adipose tissue mass, and have increased energy expenditure and lipolysis (12, 25, 66, 68). Mice lacking 4E-BP1 and 4E-BP2 have exhibited elevated sensitivity to diet-induced obesity and insulin resistance (41). Insulin and mTOR/Akt signaling have also been shown to be involved in the control of adipogenesis (20, 36, 41, 54, 77, 79).
The proposed connection of PML to mTOR and Akt in tumorigenesis raises a question of whether PML deficiency would cause metabolic defects in vivo. Here, we tested the metabolic functions of PML by characterizing the metabolic phenotypes of PML−/− mice. We found that the mTOR and Akt signaling was not affected in the white adipose tissue (WAT), liver, or muscle in PML−/− mice. Although PML deficiency did not lead to any significant metabolic deregulation, it did result in accelerated fat accumulation on high-fat diet (HFD) but not on breeder diet (BD). Fat accumulation in the subcutaneous fat depot was substantially greater than that in epididymal fat. The increased adiposity in vivo was associated with increased adipogenesis in the PML-depleted cells in vitro. PML depletion inhibited sirtuin 1 (Sirt1)/nuclear corepressor (NCoR)/silencing mediator for retinoic and thyroid hormone receptor (SMRT) recruitment to the pparγ promoter, which likely contributes in part to the transcriptional repression of peroxisome proliferator-activated receptor-γ (PPARγ) and to the subsequent adipogenesis. Therefore, we have uncovered a novel role of PML in adipogenesis regulation.
MATERIALS AND METHODS
Mice and metabolic studies.
Animal care was in accordance with institutional guidelines, and all experiments were approved by the National Heart, Lung, and Blood Institute Animal Care and Use Committee. Wild-type C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were housed with a 12:12-h light-dark cycle; 129Sv PML−/− mice (72) were crossed to C57BL/6J for at least six generations before the study. PML+/+ littermates from these matings were used as a wild-type control. PML+/+ and PML−/− male and female mice were fed either BD or HFD beginning at weaning (4 wk) for indicated times. For some experiments, HFD feeding continued ≤40 mo. The HFD consisted of 45% calories from fat, 35% from carbohydrate, and 20% from protein (D12451, 4.73 kcal/gm; Research Diets, New Brunswick, NJ). The BD (National Institutes of Health standard facility diet) consisted of 21% calories from fat, 55% from carbohydrate, and 24% from protein (LabDiets, Richmond, IN). Metabolic measurements such as body weight, fat mass, blood glucose, insulin level, glucose tolerance test, insulin tolerance test, locomoter activity, indirect calorimetry, mitochondrial DNA quantitation, and serum analysis were performed as described in our previous studies (67, 76). Food consumption was measured by subtracting the amount of food left on the grid and the amount of spilled food from the initial weight of food supplied. Food intake was measured over 14 days. Circulating free fatty acid level was measured using kits from Wako chemicals (Richmond, VA). Because both male and female mice showed similar trends in metabolic studies, data obtained from male or female mice were presented interchangeably in this study.
Body fat index calculation.
Fat mass was first measured by nuclear magnetic resonance spectroscopy using Minispec (Bruker Biospin, Houston, TX). Body fat index was calculated by dividing the fat mass by total body weight.
RNA isolation and RT-PCR.
Total cellular RNA was prepared using TRIzol (Invitrogen) according to the manufacturer's instructions. Ten micrograms of total RNA was used for making cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). Real-time PCR was performed using a 7900HT sequence detection system (Applied Biosystems). All of the RT-PCR reagents and primers were purchased from Applied Biosystems. Mouse ribosome 18S rRNA was used as an internal control.
Cell culture and lentiviral infection.
3T3-L1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% bovine serum and antibiotics. For PML shRNA expression, packaged lentivirus was produced from 293T cells (69). 3T3-L1 cells were infected with the control or mouse PML shRNA lentivirus (catalog no. TRCN0000040483, sequence: CCGGGCCCAGCATATCTACTCCTTTCTCGAGAAAGGAGTAGATATGCTGGGCTTTTTG; Sigma, St. Louis, MO) for 24 h. Cells were then selected with puromycin (2 μg/ml) for 2 wk and induced for adipogenesis. PML knockdown efficiency was confirmed by Western blotting with anti-(mouse) PML antibody (Supplemental Fig. S3C; Supplemental Material for this article is available online at the AJP-Endocrinology and Metabolism web site). PML shRNA cells or PML sh indicated in the figure legends and text refer to the 3T3-L1 cells infected with PML shRNA lentivirus. Control shRNA cells or control sh refer to 3T3-L1 cells infected with control shRNA lentivirus. Primary mouse embroyonic fibroblasts (MEFs) were isolated at day 12.5 postcoitum from PML+/+ and PML−/− mice. Each embryo was sheared with an 18-gauge needle and cultured in DMEM with 15% FBS for three passages. Primary littermate PML+/+ and PML−/− MEFs were used in this study.
Adipocyte differentiation.
3T3-L1 preadipocytes or MEFs were grown in DMEM containing 10% fetal calf serum until they were confluent and then maintained in the same medium for an additional 2 days. To induce differentiation, 2 days after confluence (day 0) cells were treated with fresh DMEM containing 10% fetal calf serum, 0.5 mM isobutylmethylxanthine, and 1 μM dexamethasone for 2 days. The media were then removed and cells incubated with DMEM containing 10% fetal calf serum and 10 μg/ml insulin for 2 days. After that, the media were removed and cells maintained with DMEM containing 10% fetal calf serum. Cells were refed every 2 days with DMEM containing 10% fetal calf serum.
Oil Red O staining.
3T3-L1 cells or MEFs were induced with adipogenesis for indicated times. Cells were rinsed with PBS and fixed with 10% formalin in PBS for 15 min. After two washes in PBS, cells were stained for 1 h in Oil Red O solution (0.55% in isopropyl alcohol solution-distilled water, 60:40). The stain was then removed, and cells were washed with water, and an optical density of 564 nm was measured.
Confocal microscopy. Confocal microscopy was performed as described (75) using anti-(mouse) PML antibody. For Fig. 3C and Supplemental Fig. S3B, cells were first stained with anti-(mouse) PML antibody and then stained with Oil Red O.
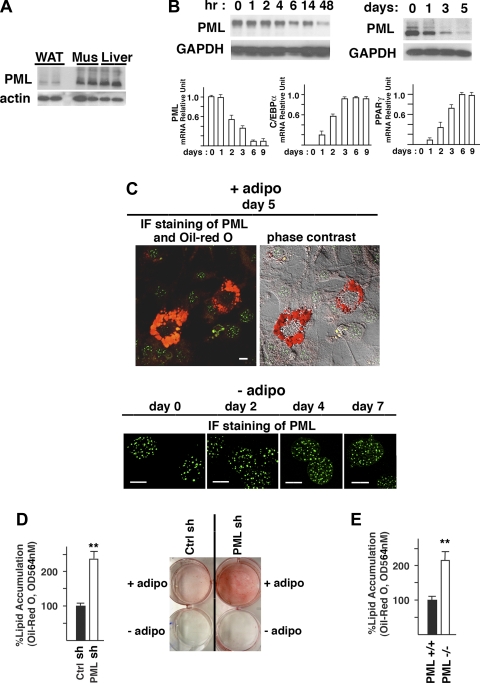
Fig. 3.
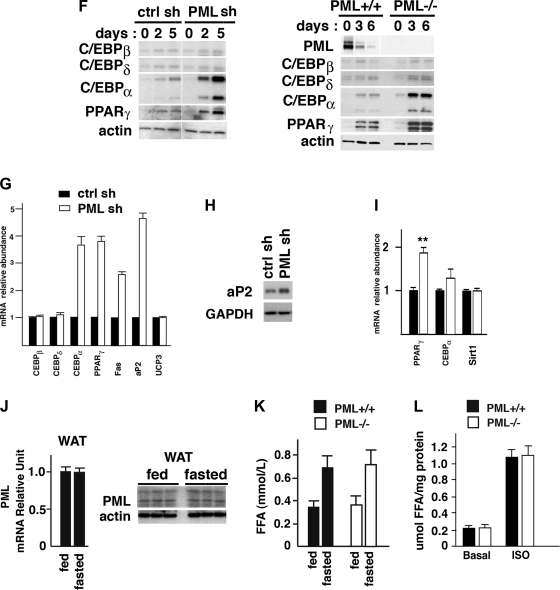
PML regulates adipogenesis in culture. A: PML protein in WAT, skeletal muscle (Mus), and liver from male BD-fed PML+/+ mice (BD for 12 wk). B: decrease in PML expression during adipogenesis (0–5 days) in 3T3-L1 cells shown by Western blotting analysis (upper panel). RT-PCR analyses of PML, CCAAT/enhancer-binding protein (C/EBP)α and peroxisome proliferator-activated receptor-γ (PPARγ) during adipogenesis (0–9 days) in 3T3-L1 cells (bottom). C: PML staining and Oil Red O staining. Top: confocal microscopy of 3T3-L1 cells on day 5 after adipogenic induction (+adipo). Bottom: representative pictures of the control 3T3-L1 cells cultured in the absence of adipogenic stimuli (0–7 days) (−adipo). Scale bars, 10 μm. D: %lipid accumulation in ctrl shRNA and PML shRNA 3T3-L1 cells on day 5 after adipogenic induction (left) and a picture of the Oil Red O staining of control (ctrl) shRNA and PML shRNA 3T3-L1 cells cultured in the presence (+adipo) or absence (−adipo) of the adipogenic stimuli for 5 days (right). E. %lipid accumulation in PML+/+ and PML−/− MEFs on day 5 after adipogenic induction. F: expressions of the C/EBP proteins and PPARγ during adipogenesis in ctrl shRNA and PML shRNA cells (left) and in PML+/+ and PML−/− MEFs (right). G: mRNA expression levels (RT-PCR) of various genes on day 5 after adipogenic induction in ctrl shRNA and PML shRNA-3T3-L1 cells. H: Western blotting analysis of the aP2 protein level in ctrl shRNA and PML shRNA-3T3-L1 cells (on day 5 after adipogenic induction). I: mRNA expression levels (RT-PCR) of PPARγ, C/EBPα, and sirtuin 1 (Sirt1) in subcutaneous fat of male HFD-fed PML+/+ and PML−/− mice (HFD for 16 wk); n = 3. J: RT-PCR and Western blotting analyses of the PML expression level in WAT from fed or fasted male BD-fed PML+/+ mice. Mice were either fed ad libitum or fasted (24 h). Each lane in Western blot represents an individual mouse. K: circulating free fatty acid (FFA) levels in PML+/+ and PML−/− mice under fed or fasted conditions. Male BD-fed PML+/+ and PML−/− mice were used (BD for 14–16 wk; n = 5–6 of each group). L: FFA release in the culture medium in the absence (basal) or presence of 10 μM isoproterenol (ISO; 2 h) analyzed using epididymal white adipose samples obtained from female HFD-fed PML+/+ and PML−/− mice (HFD for 4–6 wk; n = 3 of each group). **P < 0.01.
Western blotting analysis.
Whole cell lysates from mouse tissue, 3T3-L1, and primary MEFs were prepared by adding SDS sample buffer and denatured for 5 min at 100°C. For tissue extraction, samples were pulverized in liquid nitrogen and homogenized in a lysis buffer (20 mM HEPES, 250 mM sucrose, 4 mM EDTA, 1% Triton, 0.5% NP40 and protease inhibitors). Proteins were separated by electrophoresis on 4–20% gradient Tris-glycine polyacrylamide gels (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Millipore). Enhanced chemiluminescence (ECL) was performed per the manufacturer's instructions (ECL-PLUS; GE Biosciences). Antibodies were from Cell Signaling Technology [anti-Akt, anti-phospho-Akt Ser473, anti-S6K, anti-phospho-S6K Thr389, anti-S6, anti-phospho-S6 Ser240/244 antibodies, anti-4E-BP1, anti-phospho-4EBP1 Thr37/46, anti-AMP-activated protein kinase-α (AMPKα), anti-phospho-AMPK Thr172], Santa Cruz Biotechnology [anti-human PML, anti-CCAAT/enhancer-binding protein-α (C/EBP)α, anti-C/EBPβ, anti-C/EBPδ, anti-PPARγ, anti-aP2, anti-actin], BD Bioscience (anti-GAPDH), and Upstate Biotechnology (anti-mouse PML, anti-Sirt1, anti-NCoR, anti-SMRT). All secondary antibodies were purchased from Amersham Biosciences.
Coimmunoprecipitation assay.
Coimmunoprecipitation assays were performed as described previously (75).
Adipose tissue sample preparation and lipolysis assay.
Epididymal adipose tissue was excised from indicated animals. Adipose samples were minced in DMEM containing 2% fatty acid-free bovine serum albumin (Sigma). Aliquots of minced adipose samples were transferred to 12-well plates and incubated with isoproterenol (10 μM) for 2 h. Free fatty acid (FFA) levels present in the medium were measured by using commercial kits (Wako Chemicals) and normalized for protein content.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were carried out as described (33) using control shRNA or PML shRNA 3T3-L1 cells. Cells were selected with puromycin for 2 wk and induced adipogenesis. Cells at various stages of differentiation were treated with 1% formaldehyde for 10 min at room temperature, rinsed with PBS, and lysed in buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris·HCl (pH 8.1), and protease inhibitors. Samples were centrifuged at 11,500 rpm for 5 min to remove non-cross-linked protein with chromatin, and the resulting pellets were resuspended in 1 ml of the same lysis buffer and sonicated with sonicator for 2 min. Chromatin solution was precleared with protein A/G beads for 2–4 h. DNA-protein complexes were then immunoprecipitated with anti-Sirt1, anti-PPARγ, anti-NCoR, or anti-SMRT antibodies for 4 h and then collected with protein A-agarose for 3 h. The beads were washed, and chromatin complexes were eluted from the beads. After heating at 66°C for 8 h to release cross-linked DNA, the samples were digested with proteinase K, and the DNA was purified with QIAquick PCR Kit (Qiagen, Valencia, CA). The samples were subjected to PCR. The following primers were used: pparγ promoter, −297 5′-CAGATTTGTGGCTCACTTCGTG-3′(forward) and +8 5′-GACTTCGGCAGCTGCTCACACC-3′(reverse); aP2 promoter, −5,436 5′-CACTATGTTAGCCAGGATGGTCTC-3′ (forward) and −5,234 5′-CTCTTCCTCCTGATAGCTCCATGA-3′(reverse). These promoter regions contain PPARγ binding site. Twenty percent of the ChIP DNA was subjected to PCR amplification using Hotstar Taq polymerase (Qiagen).
Bromodeoxyuridine incorporation.
Cells were grown on coverslips until 2 days postconfluence and induced to differentiation. Because insulin has been shown to be important for induction of clonal expansion, cell number counts, and bromodeoxyuridine (BrdU) incorporation in Supplemental Fig. S3, E and F, cells were differentiated as described by Qiu et al. (56), except that 10 μg/ml insulin was used. Briefly, 2 days after confluence, cells were treated with DMEM-10% fetal calf serum-isobutylmethylxanthine-dexamethasone-10 μg/ml insulin. After 48 h, the culture medium was replaced with DMEM with 10% fetal calf serum and 10 μg/ml insulin, and the cells were then fed every other day with DMEM containing 10% fetal calf serum. For BrdU incorporation, BrdU (10 μM) was pulsed to the cells from the 16th to the 18th hour after the adipogenic induction. Cells were changed to normal medium. On day 3, coverslips were washed with PBS and fixed with ethanol [50 mM glycine-HCl (pH 2.0) in 70% ethanol] for 1 h, blocked with 2% bovine serum albumin in PBS, and incubated with anti-BrdU antibody for 1 h. After washing, fluorescein isothiocyanate-conjugated secondary antibody containing 0.1 μg/ml 4,6-diamidino-2-phenylindole was added to the coverslips for 1 h at room temperature, and a percentage of labeled cells was measured under the immunofluorescence microscope.
Luciferase assay.
The murine pparγ promoter was cloned by PCR from genomic DNA using oligonucleotide primers based on the published sequence (5) with an additional 5′ Kpn1 site and 3′ Xho1 site for subcloning purposes: −609/−588, 5′-CGGGGTACCGAATTTGGATAGCAGTAACATT-3′; +52/+31, 5′-CCGCTCGAGCAGAGATTTGCTGTAATTCACA-3′. The PCR fragments were cloned into pGL3-basic vector (Promega, Madison, WI). pGL3-pparγ luciferase construct was transfected into 293T cells along with expression vectors for PPARγ or human PML4 plus Renilla luciferase plasmid using Lipofectamine 2000 (Invitrogen). After 48 h, cells were harvested and lysed in the lysis buffer. Luciferase activity was measured using the Dual Luciferase assay kit (Promega). Experiments were performed in triplicate and repeated three times.
Statistics.
Comparisons between the treatment groups were analyzed by unpaired Student's t-test. Results are expressed as means ± SE. Comparisons involving repeated measurements were analyzed by repeated-measures ANOVA followed by Bonferroni posttest. P < 0.05 was considered as statistically significant.
RESULTS
Loss of PML results in fat accumulation after HFD in mice.
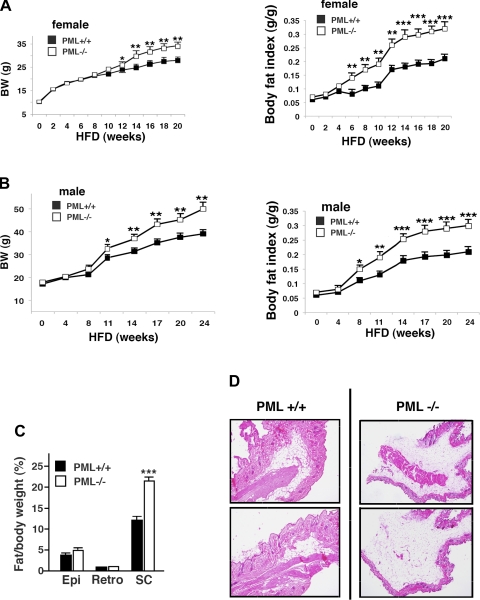
To understand the role of PML in metabolism, we fed PML+/+ and PML−/− mice either our standard facility diet (BD; 21% calories by fat) or HFD (45% calories by fat) starting at age 4 wk. We first compared weight gains of the BD-fed PML+/+ and PML−/− mice in both sexes. On BD, the growth curves of PML+/+ and PML−/− mice were identical (Supplemental Fig. S1A). However, on the HFD both female (Fig. 1A, left) and male PML−/− mice (Fig. 1B, left, and Supplemental Fig. S1B) had accelerated weight gain starting at 12 wk or at 11 wk of the feeding, respectively. For example, after 20 wk on HFD, female HFD-fed PML−/− mice gained 24.1 g, whereas female PML+/+ mice gained 18.0 g. In male mice, HFD-fed PML−/− mice gained 32 g, whereas PML+/+ mice gained 21.9 g. In both sexes of HFD-fed PML−/− mice, the increased body weight was associated with increased fat accumulation (Fig. 1, A and B, right, and Supplemental Fig. S1B). The lean body mass of HFD-fed PML+/+ and PML−/− mice was similar in both sexes (not shown). The body fat index (total fat mass corrected for body weight) was significantly higher in PML−/− mice on the HFD in both sexes, starting from 6 wk in females and 8 wk in males.
Fig. 1.
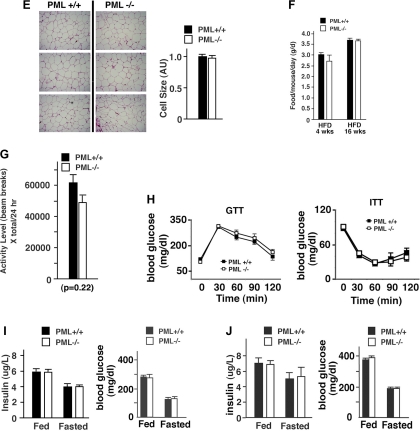
Loss of promyelocytic luekemia (PML) results in fat accumulation after high-fat diet (HFD) in mice. A: body weight (BW) and body fat index of female HFD-fed PML+/+ and PML−/− mice (n = 10–15/genotype). Body fat index was calculated by dividing the fat mass by total BW. B: BW and body fat index of male HFD-fed PML+/+ and PML−/− mice (n = 8–10/genotype). C: weights of epididymal (Epi) and retroperitoneal (Retro) fat normalized by BW. The weight of subcutaneous (sc) fat was estimated roughly by subtracting the sum of Epi and Retro fat from the total body fat and normalized by BW. Fat pads were obtained from HFD-fed male PML+/+ and PML−/− mice (HFD for 16 wk; n = 3–4/genotype). D: skin sections of the male HFD-fed PML+/+ and PML−/− mice (HFD for 24 wk) were stained with hematoxylin and eosin (H & E). Two different individual mice/genotype were used. E: histological sections of sc fat pads of male HFD-fed PML+/+ and PML−/− mice (HFD for 24 wk; left). E, right: adipocyte cell size in sc fat [arbitrary units (AU)]. For fat cell size histogram, digital images of H & E-stained adipose tissue slides were analyzed using image scope software (Aperio). Three different individual mice/genotype were used. F: food intake per mouse per day measured over 14 days (n = 5–6/genotype). Female HFD-fed PML+/+ and PML−/− mice were used (HFD for 4 and 16 wk). G: locomoter activity as measured by beam breaks/24 h using female HFD-fed PML+/+ and PML−/− mice (HFD for 16 wk; n = 8/genotype). X total indicates X total laser beam interruptions. H: glucose (GTT) and insulin tolerance tests (ITT) of female HFD-fed PML+/+ and PML−/− mice (HFD for 16–18 wk). For GTT, mice were fasted for 16 h, and 1 mg/g glucose was injected intraperitoneally. Blood glucose was measured at indicated times after injection. For the ITT, mice were fasted for 4 h before intraperitoneal injection with 0.4 U/kg of human insulin (Sigma) (n = 8–12/genotype). I: blood insulin (left) and glucose levels (right) in female HFD-fed PML+/+ and PML−/− mice (HFD for 16–18 wk; n = 8–12 of each group). Mice were either fed ad libitum or fasted (16 h). J: blood insulin (left) and glucose levels (right) in male HFD-fed PML+/+ and PML−/− mice (HFD for 40 wk; n = 8–10 of each group). Mice were either fed ad libitum or fasted (16 h). Weeks indicate weeks on HFD. *P < 0.05; **P < 0.01; ***P < 0.001.
Fat depots can be divided roughly into two compartments, subcutaneous and visceral. By examining the histological sections of the skin (subcutaneous; Fig. 1D), as well as by measuring the mass of epididymal fat (visceral; Fig. 1C) using HFD-fed PML+/+ and PML−/− mice (HFD for 16 wk), we compared the accumulation of fat in the two depots. The weight of subcutaneous fat was estimated roughly by subtracting the sum of epididymal and retroperitoneal fat from the total body fat, as shown by other studies (74). PML−/− mice gained 9.2 g in subcutaneous fat and 2.04 g in epididymal fat over 16 wk on HFD. PML+/+ gained 4.3 g in subcutaneous fat and 1.43 g in epididymal fat. In Fig. 1C, when normalized for body weight, subcutaneous fat and epididymal fat were increased 74 and 17%, respectively, in HFD-fed PML−/− mice (HFD for 16 wk) compared with PML+/+ mice (Fig. 1C). The subcutaneous/epididymal fat weight ratio was 50% higher in PML−/− mice (the ratios in PML+/+ and PML−/− mice were 3.01 and 4.51, respectively), indicating a redistribution of adipose depots in the HFD-fed PML−/− mice (Fig. 1C). Consistent with these results, histological examination of the skin sections revealed substantial increases in subcutaneous fat mass in PML−/− mice (Fig. 1D). Increased fat mass may be a result of increased fat cell number, fat cell size, or both. Analyses of the fat cell size demonstrated that there was no difference in fat cell size in PML−/− mice (Fig. 1E), suggesting that PML−/− mice have increased fat mass largely as a result of increased fat cell number.
Obesity is frequently associated with lipid accumulation in the liver, leading to nonalcoholic fatty liver disease (6, 49). To examine whether increased fat accumulation in PML−/− mice is leading to increased lipid accumulation in the liver, we measured the mass and triglyceride content in the liver of HFD-fed PML+/+ and PML−/− mice (HFD for 16 wk). The liver mass was 1.42 g for PML+/+ and 1.41 g for PML−/−, and the liver triglyceride content was 14.4 mg/g tissue in both PML+/+ and PML−/− mice. Consistent with these results, hematoxylin and eosin staining of the liver sections showed that PML+/+ and PML−/− mice had similar levels of lipid droplets (Supplemental Fig. S1C). Fatty liver disease is frequently associated with inflammation. Since there was no hepatic steatosis in PML−/− mice, we expected to find no difference in inflammatory markers. Indeed, the mRNA levels of macrophage markers F4/80, TNFα, and monocyte chemoattractant protein-1 were not increased in PML−/− liver (data not shown). These findings indicate that although PML−/− mice accumulated more fat, it was stored mostly in the subcutaneous depot, sparing visceral organs such as the liver.
Loss of PML increases fat accumulation without causing metabolic dysregulation.
To understand why PML−/− mice accumulated more fat on the HFD, we measured their food intake for 3 days after 4 and 12 wk on the HFD (not shown) or for 14 days after 4 and 16 wk on the HFD in female PML+/+ and PML−/− mice (Fig. 1F). As shown in Fig. 1F, PML−/− mice consumed a similar amount of food as PML+/+ mice. Food intake measured for 3 days also did not show any significant difference (not shown). The physical activity levels as measured by beam breaks were slightly decreased in female PML−/− mice, although the difference was not statistically significant (P = 0.22; Fig. 1G). The metabolic rates of PML+/+ and PML−/− mice, as measured by indirect calorimetry, were nearly identical when normalized to the body weight (Supplemental Fig. S1D, left). When oxygen uptake (V̇o2) was normalized to lean body mass, PML−/− mice actually had higher, not lower, V̇o2 than PML+/+ mice, a conclusion that does not correlate with the fat mass gain in PML−/− mice (Supplemental Fig. S1D, right). In the discussion, we have provided our reasoning as to why indirect calorimetry data would not provide precise assessment in energy expenditure in the case of PML+/+ and PML−/− mice.
PML+/+ and PML−/− mice had similar body temperatures in both fed and fasted conditions (Supplemental Fig. S1E) and similar mitochondrial content in the major thermogenic tissues, i.e., skeletal muscle and brown adipose tissue (BAT) (Supplemental Fig. S1F). Consistent with this, mRNA expressions of the genes that are important for mitochondrial biogenesis and function, such as PPARγ coactivator-1α (PGC-1α), PGC-1β, medium-chain acyl-CoA dehydrogenase, estrogen-related receptor-α, carnitine palmitoyltransferase-1b, uncoupling protein (UCP) 1, UCP2, and UCP3, were also similar in WAT, BAT, or liver from HFD-fed PML+/+ and PML−/− mice (not shown).
Increased adiposity is usually associated with decreased glucose tolerance and insulin sensitivity. To investigate whether this is also the case with PML−/− mice, we performed glucose tolerance test and insulin tolerance test after 16–18 wk on HFD in PML+/+ and PML−/− mice. Surprisingly, we found that PML−/− mice, despite having more fat accumulation, had similar glucose tolerance and insulin sensitivity as PML+/+ mice (Fig. 1H). Consistent with this, the levels of insulin and blood glucose in both the fed and fasted states were very similar between PML+/+ and PML−/− mice (Fig. 1I).
We then extended the HFD feeding to 40 wk to increase fat accumulation. After 40 wk on HFD, male PML−/− mice gained 40.9 g of body weight and 16.2 g of fat, whereas male PML+/+ mice had gained 29.8 g of body weight and 9.1 g of fat. Even after 40 wk of HFD, the levels of serum insulin and glucose in fed (P = 0.9) and fasted (P = 0.8) conditions were similar (Fig. 1J). Circulating triglyceride, cholesterol, and FFA levels were also similar in PML+/+ and PML−/− mice at all time points tested (not shown).
Taken together, these findings indicate that the increased fat mass in PML−/− mice did not lead to the metabolic problems normally associated with obesity.
PML depletion does not affect Akt, mTOR/S6K1, or AMPK activities in WAT, liver, or muscle.
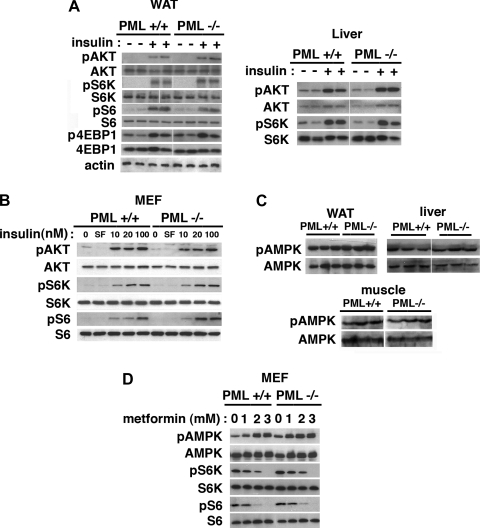
PML is thought to be involved in the regulation of Akt/mTOR under some conditions. To investigate whether PML regulates the Akt-mTOR pathway in metabolically relevant tissues in vivo, we visualized phosphorylation of Akt, S6K1, S6, or 4E-BP1 in WAT, liver (for HFD; Fig. 2A), and muscle (not shown) after insulin treatment. We found that they were not altered significantly in either BD-fed (not shown) or HFD-fed PML−/− mice (Fig. 2A) or in primary PML+/+ and PML−/− MEFs (Fig. 2B).
Fig. 2.
PML depletion does not affect Akt, mTOR/S6K1, or AMP-activated protein kinase (AMPK) signaling. A: Western blotting analysis of phospho-Akt (p-Akt; phospho-Ser473), p-S6 kinase 1 (p-S6K; phospho-Thr389), p-S6 (phospho-Ser240/244), and phospho-eukaryotic initiation factor 4E-binding protein-1 (p-4E-BP1; phospho-Thr37/46). White adipose tissue (WAT) and liver proteins from 2 different male HFD-fed PML+/+ and PML−/− mice (HFD for 20 wk) were loaded, electrophoresed, blotted, and probed with various antibodies. For insulin treatment, mice were fasted for 6 h, injected intraperitoneally with 0.75 U/kg insulin. Tissues were collected 10 min after injection. Some images were obtained from spliced blots. Spaces were inserted at the splice sites. B: Western blotting analysis of the primary PML+/+ and PML−/− MEFs treated with insulin (0–100 nM, 15 min). SF, serum-free condition, cells were serum-starved overnight. C: AMPK activation in vivo measured by p-AMPK (phospho-Thr172) level in WAT, liver, and muscle from female HFD-fed PML+/+ and PML−/− mice (HFD for 20 wk). Each lane represents an individual mouse. Spaces were inserted at the splice sites on the blots. D: Western blotting analysis of the primary PML+/+ and PML−/− myocyte enhancer factors (MEFs) after metformin treatment (0–3 mM, 4 h).
The fuel-sensing kinase AMPK is activated by ATP-depleting conditions such as nutrient deprivation or pathological stresses (31). AMPK increases ATP production by inhibiting energy-consuming anabolic pathways such as mTOR (29, 37) and is involved in metabolic control (4, 45). Activation of AMPK has been shown to reduce fat accumulation and increase glucose tolerance, insulin sensitivity, mitochondrial biogenesis, and physical endurance. We investigated whether accelerated fat accumulation in PML−/− mice may be a result of altered AMPK activity. In Fig. 2C, the levels of phospho (Thr172)-AMPK, the active form of AMPK, in WAT, liver, and muscle were similar in both the PML+/+ and PML−/− mice after HFD or BD (Fig. 2C for HFD, BD not shown). AMPK activation (Supplemental Fig. S2A) as well as the decrease in blood glucose level (Supplemental Fig. S2B) induced by AMPK activator metformin were similar between PML+/+ and PML−/− mice. PML−/− MEFs also showed no defect in AMPK signaling after metformin treatment compared with the PML+/+ MEFs (Fig. 2D). Taken together, these results suggest that PML−/− mice and MEFs, at least in C57BL/6J background, have normal Akt, mTOR, and AMPK signaling.
Fat mass gain in PML−/− mice is associated with enhanced adipogenesis in PML-depleted cells.
Adipogenesis, differentiation of immature preadipocytes into mature fat-accumulating adipocytes, requires the sequential expressions of transcription factors, including the C/EBP family proteins and PPARγ. PPARγ is a transcription factor from the nuclear receptor superfamily, and the expression of PPARγ is both necessary and sufficient for adipogenesis (57). The induction of adipogenesis in vitro involves transitions in cell cycle regulation from confluence arrest to mitotic clonal expansion and then terminal cell cycle exit. During clonal expansion, growth-arrested preadipocytes reenter the cell cycle and undergo one or two rounds of cell division (52). The gene expression program leading to terminal adipocyte differentiation is initiated during and after the mitotic clonal expansion period. Among the transcription factors, C/EBPβ and C/EBPδ are expressed during the early phases of differentiation (39, 43) and are required for the subsequent expression of C/EBPα and PPARγ that is central to adipogenic differentiation in the later phases. PPARγ and C/EBPα cross-regulate expressions of each other (58).
Despite the normal food intake and only a slightly decreased physical activity in PML−/− mice, the data shown in Fig. 1 suggested the possibility that the accelerated fat accumulation in HFD-fed PML−/− mice may be due to a cell-autonomous defect in regulation of adipogenesis. To test this possibility, we first examined PML expression levels in WAT, skeletal muscle, and liver using an anti-(mouse) PML antibody that is highly specific for PML (Supplemental Fig. S3A). Although PML is considered to be a ubiquitously expressed protein, we found that the PML expression in WAT is very low compared with that in skeletal muscle and liver (Fig. 3A).
We then monitored the PML expression level during the differentiation of 3T3-L1 cells into adipocytes. PML expression, as measured by Western blotting and RT-PCR analyses, was highest in growing (not shown) and confluent 3T3-L1 cells but decreased after adipogenic induction (Fig. 3B). The PML level declined progressively to very low levels by days 5 and 6. In contrast, PPARγ and C/EBPα expressions increased progressively, as shown by RT-PCR analyses. PML level also decreased during the differentiation of MEFs into adipocytes (Fig. 3F). Consistent with these results, confocal microscopy showed that PML NBs are strongly visible in undifferentiated 3T3-L1 cells cultured without adipogenic stimuli (−adipo; Fig. 3C, bottom) but are nearly undetectable in fully differentiated adipocytes with intense Oil Red O staining in the cytoplasm (+adipo; Fig. 3C, top, and Supplemental Fig. S3B).
Accelerated fat accumulation in PML−/− mice on the HFD in conjunction with the disappearance of PML during adipogenesis suggested to us that PML may play a role in adipogenesis. To test this hypothesis, we used shRNA to knock down PML in 3T3-L1 preadipocytes (the efficacy of PML knockdown at the protein level was assessed by Western blotting analysis; Supplemental Fig. S3C). As shown in Fig. 3D, PML shRNA more than doubled the accumulation of lipid droplets in differentiated adipocytes. We also induced adipogenesis in PML+/+ and PML−/− MEFs and measured lipid accumulation. We found that PML−/− MEFs also accumulated more than twice as many lipids as PML+/+ MEFs after adipogenesis (Fig. 3E and Supplemental Fig. S3D).
Genes that regulate cell cycle are activated or inactivated during the clonal expansion phase (47, 61). The levels of the key cell cycle inhibitors, the cyclin-dependent kinase inhibitors p21 and p27, decline during mitotic clonal expansion. It is well known that PML is required for the retinoic acid-dependent transactivation of the p21 gene (71). PML is also known to modulate p21 through a tumor suppressor, p53, for apoptosis and cell senescence (23, 53, 59). PML overexpression in cultured cells results in growth inhibition (42), and PML−/− MEFs proliferate faster than the wild-type MEFs (71, 72). Since PML is related to the cell cycle regulation, we asked whether loss of PML enhanced adipogenesis by inducing clonal expansion rather than regulating the subsequent differentiation phase. Figure S3, E and F, shows that PML depletion did not affect the increase in cell number or DNA replication during adipogenesis, indicating that PML depletion most likely affected adipogenesis not by affecting cell division but by enhancing terminal differentiation.
Since adipogenesis requires the induction of the expression of several critical transcription factors, we investigated whether PML regulates their expression. In PML shRNA 3T3-L1 cells and in PML−/− MEFs, the expression levels of the early regulators C/EBPβ and C/EBPδ were not changed, but the expression levels of PPARγ and C/EBPα, which is in part activated by PPARγ, were elevated (Fig. 3, F and G). In RT-PCR analyses (not shown), the mRNA levels of C/EBPβ and C/EBPδ in control siRNA and PML siRNA 3T3-L1 cells were similar at any time points tested (6, 12, 24, 48, and 72 h after adipogenic induction). In both control siRNA and PML siRNA 3T3-L1 cells, the transcription levels of C/EBPβ and C/EBPδ were transiently upregulated 2.8- and 3.2-fold, respectively, relative to the levels on day 0 adipogenic induction, and declined afterward. PPARγ also induces expression of target genes that play important roles in lipogenesis such as the fatty acid-binding protein aP2, a marker of differentiated adipocytes, and fatty acid synthase (FAS) (22). Fas and aP2 levels were increased in PML shRNA 3T3-L1 cells (Fig. 3, G and H).
To confirm the increased expression of PPARγ in vivo upon PML loss, we measured mRNA levels of PPARγ, C/EBPα, and Sirt1 in subcutaneous fat obtained from HFD-fed PML+/+ and PML−/− mice (Fig. 3I). We found that PML−/− fat expressed significantly higher levels of PPARγ mRNA but slightly increased levels of C/EBPα mRNA and similar levels of Sirt1 mRNA.
Increased lipid accumulation with the loss of PML may be a consequence of decreased lipolysis. Food deprivation promotes lipolysis in WAT, releasing FFA into the blood. Although the level of PML expression is very low in WAT, as shown in Fig. 3A, it is possible that PML is induced in WAT by fasting or caloric restriction, thereby playing a role in lipolysis. As shown in Fig. 3J, fasting did not induce PML expression in WAT. Furthermore, the increase in circulating FFA levels induced by fasting was similar in PML+/+ and PML−/− mice (Fig. 3K).
Catecholamine-induced lipolysis can reduce fat mass (38). To examine the role of PML in catecholamine-induced lipolysis, we treated PML+/+ and PML−/− (male BD-fed) mice with a β-adrenergic agonist, isoproterenol (ISO; 10 mg/kg for 6 h), and measured circulating FFA levels (70). The ISO-stimulated increase in circulating FFA levels was also similar between PML+/+ and PML−/− mice (not shown). We also analyzed the ISO-stimulated FFA release in the culture medium using the epididymal adipose tissue samples obtained from HFD-fed PML+/+ and PML−/− mice. As shown in Fig. 3L, both the basal and ISO-stimulated release of FFA from PML−/− WAT were similar to those from PML+/+ WAT. These results suggested that PML is not involved in the regulation of lipolysis, making it unlikely that the increased fat accumulation in PML−/− mice is a result of decreased lipolysis.
PML promotes Sirt1-PPARγ interaction and Sirt1-NCoR-SMRT recruitment to the pparγ promoter during adipogenesis in 3T3-L1 cells.
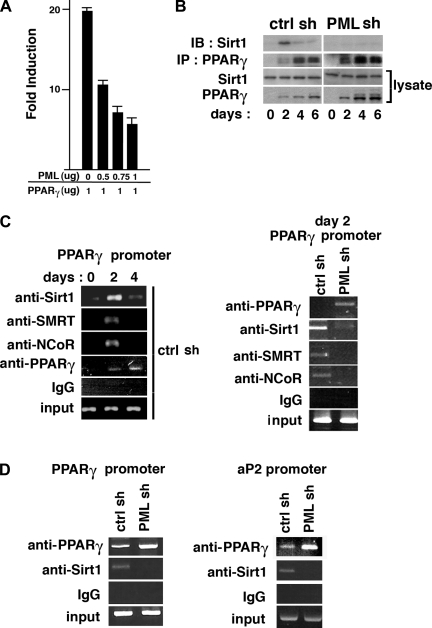
The results shown above indicate that PML may suppress adipogenesis by inhibiting the transcription of PPARγ and the genes that are regulated by PPARγ. To test this possibility, we cotransfected a pparg reporter construct and an expression vector for PPARγ, which regulates its own promoter (62), with increasing amounts of PML expression vector. As shown in Fig. 4A, PML repressed PPARγ-mediated transactivation of the pparγ promoter in a dose-dependent manner. PML does not bind DNA directly (1, 18), but it can regulate transcription by interacting with transcriptional coactivators and corepressors in the PML NBs (3, 73). For example, PML interacts directly with transcriptional coactivator CBP/p300 and is required for the ability of CBP to act as a transcriptional coactivator of the retinoic acid receptor (80). PML also interacts with multiple corepressors such as NCoR-SMRT and both class I and class II histone deacetylases to repress transcription (35, 65, 73).
Fig. 4.
PML loss attenuates Sirt1-PPARγ interaction and inhibits Sirt1-nuclear corepressor(NCoR)-silencing mediator for retinoic and thyroid hormone receptor (SMRT) recruitment to the pparγ promoter during adipogenesis in 3T3-L1 cells. A: pGL3-pparγ-luc reporter plasmid was cotransfected into 293T cells with expression vectors for human PML4 and PPARγ together with renilla luciferase vector. Results are expressed as firefly luciferase activity normalized to renilla luciferase activity. B: coimmunoprecipitation of Sirt1 with PPARγ. Cell lysates of ctrl shRNA and PML shRNA-3T3-L1 cells were obtained during 0–6 days of adipogenesis and subjected to immunoprecipitation using anti-PPARγ (Santa Cruz Biotechnology) antibody. The immunoprecipitated complex was immunoblotted with anti-Sirt1 (Upstate Biotechnology) antibody. C, left: chromatin immunoprecipitation (ChIP) analysis on the pparγ promoter in ctrl shRNA 3T3-L1 cells using IgG and antibodies against Sirt1 (Upstate Biotechnology), SMRT (Santa Cruz Biotechnology), NCoR (Santa Cruz Biotechnology) and PPARγ (H-100; Santa Cruz Biotechnology) at indicated days after adipogenic induction. C, right: ChIP analyses on the pparγ promoter in ctrl shRNA and PML shRNA 3T3-L1 cells on day 2 after adipogenic induction. Twenty-five to 35 cycles of PCR reaction were performed. D: ChIP analysis on the pparγ or aP2 promoter. The chromatin-associated DNA samples were obtained in PML shRNA and ctrl shRNA cells on day 6 after differentiation and immunoprecipitated with IgG or antibodies against Sirt1 and PPARγ.
PPARγ represses transcription via the corepressors NCoR-SMRT and activates transcription by recruitment of coactivators (14, 26, 28). The NAD-dependent histone deacetylase Sirt1 represses PPARγ and its target genes and inhibits lipid accumulation in adipocytes by forming the Sirt1-NCoR-SMRT complex at the PPARγ-binding sites (55, 63). Sirt1 has also been reported to interact with PML when PML is overexpressed (40). Sirt1 has a diffuse nuclear localization pattern but is recruited to the PML NBs upon overexpression of PML, where Sirt1 is thought to stabilize PML, promoting its sumoylation (11, 40).
To understand the possible role of PML in the formation of the Sirt1-NCoR-SMRT-PPARγ complex, we first examined how PML affects the Sirt1-PPARγ interaction (55) during the course of adipogenesis in 3T3-L1 cells. The Sirt1-PPARγ interaction was highest on day 2 after adipogenic induction and decreased afterward, although the level of PPARγ expression on day 2 was lower than that in the later stages (Fig. 4B). The rise in Sirt1-PPARγ interaction on day 2 was suppressed by PML shRNA (Fig. 4B). The Sirt1 level was reported to be increased (55) during the early phase of 3T3-L1 adipogenesis. However, we found that Sirt1 levels did not change significantly either during adipogenesis or by PML shRNA.
To visualize the formation of the Sirt1-NCoR-SMRT-PPARγ complex on the pparγ promoter, we performed ChIP assays. As shown in Fig. 4C, left, recruitment of the Sirt1-NCoR-SMRT complex to the pparγ promoter was also highest on day 2 and declined afterward in control shRNA 3T3-L1 cells. As expected, PPARγ binding to the promoter increased during adipogenesis in control shRNA 3T3-L1 cells (Fig. 4C, left). In Fig. 4C, right, the increase in the recruitment of Sirt1-NCoR-SMRT on day 2 was inhibited by PML shRNA. Even on day 6, the recruitment of Sirt1 to the pparγ promoter was blocked by PML shRNA, but the recruitment of PPARγ was stimulated by PML shRNA (Fig. 4D, left). The same pattern of recruitment also occurred on the aP2 promoter (Fig. 4D, right). Expression of NCoR-SMRT did not change much over the course of adipogenesis (not shown). We were not able to visualize PML recruitment with the ChIP assay because the anti-(mouse) PML antibody that is currently available does not immunoprecipitate the mouse PML protein. Taken together, these results suggest that PML contributes to recruitment of the corepressor complex containing Sirt1-NCoR-SMRT to the pparγ promoter and PPARγ-dependent promoters such as aP2 promoter during adipogenesis.
To understand how PML enhances the formation of the Sirt1-NCoR-SMRT-PPARγ complex, we examined whether PML interacts directly with either PPARγ or Sirt1 endogenously during the course of 3T3-L1 adipogenesis. We found that endogenous PML did not coimmunoprecipitate with PPARγ or Sirt1 during 3T3-L1 adipogenesis. However, exogenously expressed PML coimmunoprecipitated with endogenous Sirt1 or PPARγ upon overexpression of PPARγ in 293T cells (not shown). The failure to coimmunoprecipitate PML with Sirt1 or PPARγ without overexpression suggests that PML may regulate them without directly contacting them during adipogenesis. Because PML is required to form a macromolecular structure, PML NBs, it is possible that their recruitment into PML NBs, rather than a direct contact with PML, may regulate the activities of Sirt1 and PPARγ, at least in part.
Although many studies have demonstrated a role of PML in many diverse pathways important for growth, apoptosis, and senescence, the metabolic functions of PML have not been examined until now. Our discovery that PML−/− mice are prone to accelerated fat accumulation in the presence of excess caloric intake reveals an entirely unanticipated role of PML in vivo. Although a number of questions regarding the function of PML in vivo remains to be further elucidated, our work provides insights and establishes a framework for future studies to better understand PML function.
DISCUSSION
Visceral fat is associated with an increased risk for metabolic syndrome, but subcutaneous fat has protective effects. Excess visceral fat content is more closely related to insulin resistance than total fat mass. In old age, fat is redistributed from subcutaneous to intra-abdominal visceral depots (17, 21, 78). In this study, we have shown that PML-deficient mice gained body weight faster and had more fat mass in a diet-induced obesity model. Despite the weight gain, PML−/− mice did not have insulin resistance, glucose intolerance, or inflammation, which are normally associated with obesity. The fact that fat accumulation in PML−/− mice occurred largely in the subcutaneous fat depot (Fig. 1, C and D) may explain why PML−/− mice are relatively healthy.
PML−/− mice, despite having more fat mass, had similar fat cell size, suggesting that PML deficiency increased fat mass by increasing fat cell number (i.e., adipogenesis). Using in vitro adipogenesis models, we discovered that PML, the expression of which decreased during adipogenesis, is a suppressor of adipogenesis and that PML deficiency enhanced adipogenesis.
Since PML deficiency increases PPARγ activity, it is not surprising that the changes in fat metabolism seen in PML−/− mice resemble those produced by treatment with PPARγ agonist thiazolidinediones (TZDs), which increases adipogenesis and reduces fat cell size (13, 19, 48). TZDs also shift the distribution of fat from the visceral fat depot to the subcutaneous fat depot (2, 16, 46). It is believed that these properties of TZDs contribute to the improvement of insulin sensitivity and reduction in glucose and lipid levels in humans and rodents (8, 50).
If PML−/− mice have normal food intake but accumulate more fat, it is predicted that the metabolic rate of PML−/− mice is lower than that of PML+/+ mice. However, we were not able to detect a lower metabolic rate in PML−/− mice with indirect calorimetry (Fig. S1D). We believe this discrepancy is due to the inherent imprecision associated with indirect calorimetry. The weight gain of 10.1 g over 24 wk in male PML−/− mice and of 6.1 g over 20 wk in female PML−/− mice reflects a decrease in the energy expenditure of 0.54 and 0.39 kcal/day, respectively. Assuming the daily energy expenditure is 50–60 KJ/day, as indicated in the study by Butler and Kozak (9), these values, under similar caloric intake conditions, fall below the lower limit of the discriminatory capacity of indirect calorimetry (2.7–3.3% in females and 3.8–4.5% in males), which is thought to be ∼5% of the daily total energy expenditure. Therefore, although PML−/− mice may have a lower metabolic rate than PML+/+ mice, we were not be able to detect it by using this method. In Fig. S1D (right), we also normalized V̇o2 measurement by the lean body mass, as suggested by Butler and Kozak (9). It has been reported that adipose tissue contributes relatively little to the whole body energy expenditure compared with lean mass (9, 27, 32). Whereas some studies suggested that energy expenditure may be normalized by lean body mass, other studies showed that normalization by lean body mass alone could be problematic as well (32). In our case, PML−/− mice actually had higher, not lower, V̇o2 than PML+/+ mice when V̇o2 was normalized to lean body mass (Fig. S1D, right). In the studies reported by Butler and Kozak (9), mouse obesity models carrying a mutation or a deletion in the leptin (Lepob/Lepob) or melanocortin-4 receptor gene (Mc4r−/−) also had substantially higher energy expenditure than control mice when energy expenditure was normalized to lean body mass, a conclusion that does not correlate with large fat mass gain in these mice. Future analyses that utilize a cohort size large enough for regression analysis would be helpful for precise assessment of energy expenditure in PML+/+ and PML−/− mice.
Obesity is a risk factor for a broad spectrum of cancers, including colorectal, pancreatic, postmenopausal breast, and endometrial cancers (34, 44). PML loss has been also shown to increase colon cancer and prostate cancer incidence in mouse cancer models. The mechanism by which abdominal obesity may increase cancer risk includes the chronic inflammation (51) and insulin resistance seen in obesity (10). Given that PML is a pleiotropic tumor suppressor and that PML loss is highly associated with tumor progression and metastatic status in human cancers (24), PML deficiency and the resulting obesity may increase the risks for cancer. However, alternatively, by decreasing abdominal obesity and reducing obesity-induced inflammation, PML deficiency might provide some protection against certain types of cancers.
Fat-specific depletion of PML would provide a better understanding of the role of PML in adipogenesis and metabolic regulation in vivo. However, if PML is indeed not required for the maintenance of WAT, as suggested by the very low level expression in the fully differentiated adipocytes, fat-specific depletion of PML using differentiated markers such as aP2 to drive Cre expression would not provide information on the role of PML in vivo.
Our study indicates that deficiency of PML increases adipogenesis and fat accumulation without causing metabolic dysregulation. For unknown reasons, aging is associated with reduced subcutaneous fat depot but with increased abdominal fat depot. Although much more study is needed, it is possible to speculate that targeting PML-dependent pathways in subcutaneous preadipocytes may increase the subcutaneous fat depot and divert excess fat away from the abdominal depot, thus reducing the risk for diseases associated with abdominal obesity. One possibility is that, in old animals, adipogenesis in the subcutaneous depot may be decreased with increased function of PML in preadipocytes. PML deficiency may provide insights into therapeutic strategies for increasing adipogenesis without causing metabolic disease.
GRANTS
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
DISCLOSURES
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
We are grateful to Dalton Saunders, Yiying Tsai, and Adam Weidenhammer for technical support.
K. H. Lee is currently affiliated with the Department of Physiology, Seoul National University College of Medicine, Seoul, South Korea. M. Liu is currently affiliated with Trinity School of Arts and Sciences, Duke University, Durham, NC.
REFERENCES
- 1. Ahn JH, Brignole EJ, 3rd, Hayward GS. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol 18: 4899–4913, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akazawa S, Sun F, Ito M, Kawasaki E, Eguchi K. Efficacy of troglitazone on body fat distribution in type 2 diabetes. Diabetes Care 23: 1067–1071, 2000 [DOI] [PubMed] [Google Scholar]
- 3. Alcalay M, Tomassoni L, Colombo E, Stoldt S, Grignani F, Fagioli M, Szekely L, Helin K, Pelicci PG. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol Cell Biol 18: 1084–1093, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson U, Filipsson K, Abbott CR, Woods A, Smith K, Bloom SR, Carling D, Small CJ. AMP-activated protein kinase plays a role in the control of food intake. J Biol Chem 279: 12005–12008, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Batchvarova N, Wang XZ, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153). EMBO J 14: 4654–4661, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellentani S, Saccoccio G, Masutti F, Croce LS, Brandi G, Sasso F, Cristanini G, Tiribelli C. Prevalence of and risk factors for hepatic steatosis in Northern Italy. Ann Intern Med 132: 112–117, 2000 [DOI] [PubMed] [Google Scholar]
- 7. Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Simon MC, Rafii S, Pandolfi PP. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature 442: 779–785, 2006 [DOI] [PubMed] [Google Scholar]
- 8. Boden G, Cheung P, Mozzoli M, Fried SK. Effect of thiazolidinediones on glucose and fatty acid metabolism in patients with type 2 diabetes. Metabolism 52: 753–759, 2003 [DOI] [PubMed] [Google Scholar]
- 9. Butler AA, Kozak LP. A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes 59: 323–329, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer 4: 579–591, 2004 [DOI] [PubMed] [Google Scholar]
- 11. Campagna M, Herranz D, Garcia MA, Marcos-Villar L, González-Santamaría J, Gallego P, Gutierrez S, Collado M, Serrano M, Esteban M, Rivas C. SIRT1 stabilizes PML promoting its sumoylation. Cell Death Differ 18: 72–79, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carnevalli LS, Masuda K, Frigerio F, Le Bacquer O, Um SH, Gandin V, Topisirovic I, Sonenberg N, Thomas G, Kozma SC. S6K1 plays a critical role in early adipocyte differentiation. Dev Cell 18: 763–774, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang AY, Wyse BM, Gilchrist BJ. Ciglitazone, a new hypoglycemic agent. II. Effect on glucose and lipid metabolisms and insulin binding in the adipose tissue of C57BL/6J-ob/ob and −+/? mice. Diabetes 32: 839–845, 1983 [DOI] [PubMed] [Google Scholar]
- 14. Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377: 454–457, 1995 [DOI] [PubMed] [Google Scholar]
- 15. Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 13: 252–259, 2007 [DOI] [PubMed] [Google Scholar]
- 16. de Souza CJ, Eckhardt M, Gagen K, Dong M, Chen W, Laurent D, Burkey BF. Effects of pioglitazone on adipose tissue remodeling within the setting of obesity and insulin resistance. Diabetes 50: 1863–1871, 2001 [DOI] [PubMed] [Google Scholar]
- 17. DeNino WF, Tchernof A, Dionne IJ, Toth MJ, Ades PA, Sites CK, Poehlman ET. Contribution of abdominal adiposity to age-related differences in insulin sensitivity and plasma lipids in healthy nonobese women. Diabetes Care 24: 925–932, 2001 [DOI] [PubMed] [Google Scholar]
- 18. Doucas V, Tini M, Egan DA, Evans RM. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc Natl Acad Sci USA 96: 2627–2632, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujiwara T, Yoshioka S, Yoshioka T, Ushiyama I, Horikoshi H. Characterization of new oral antidiabetic agent CS-045. Studies in KK and ob/ob mice and Zucker fatty rats. Diabetes 37: 1549–1558, 1988 [DOI] [PubMed] [Google Scholar]
- 20. Gagnon A, Lau S, Sorisky A. Rapamycin-sensitive phase of 3T3-L1 preadipocyte differentiation after clonal expansion. J Cell Physiol 189: 14–22, 2001 [DOI] [PubMed] [Google Scholar]
- 21. Goodpaster BH, Krishnaswami S, Resnick H, Kelley DE, Haggerty C, Harris TB, Schwartz AV, Kritchevsky S, Newman AB. Association between regional adipose tissue distribution and both type 2 diabetes and impaired glucose tolerance in elderly men and women. Diabetes Care 26: 372–379, 2003 [DOI] [PubMed] [Google Scholar]
- 22. Graves RA, Tontonoz P, Spiegelman BM. Analysis of a tissue-specific enhancer: ARF6 regulates adipogenic gene expression. Mol Cell Biol 12: 1202–1208, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. The function of PML in p53-dependent apoptosis. Nat Cell Biol 2: 730–736, 2000 [DOI] [PubMed] [Google Scholar]
- 24. Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst 96: 269–279, 2004 [DOI] [PubMed] [Google Scholar]
- 25. Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 166: 213–223, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387: 43–48, 1997 [DOI] [PubMed] [Google Scholar]
- 27. Himms-Hagen J. On raising energy expenditure in ob/ob mice. Science 276: 1132–1133, 1997 [DOI] [PubMed] [Google Scholar]
- 28. Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377: 397–404, 1995 [DOI] [PubMed] [Google Scholar]
- 29. Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Ito K, Bernardi R, Pandolfi PP. A novel signaling network as a critical rheostat for the biology and maintenance of the normal stem cell and the cancer-initiating cell. Curr Opin Genet Dev 19: 51–59, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1: 15–25, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Kaiyala KJ, Schwartz MW. Toward a more complete (and less controversial) understanding of energy expenditure and its role in obesity pathogenesis. Diabetes 60: 17–23, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol 24: 1188–1199, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karin M. Nuclear factor-kappaB in cancer development and progression. Nature 441: 431–436, 2006 [DOI] [PubMed] [Google Scholar]
- 35. Khan MM, Nomura T, Kim H, Kaul SC, Wadhwa R, Shinagawa T, Ichikawa-Iwata E, Zhong S, Pandolfi PP, Ishii S. Role of PML and PML-RARalpha in Mad-mediated transcriptional repression. Mol Cell 7: 1233–1243, 2001 [DOI] [PubMed] [Google Scholar]
- 36. Kim JE, Chen J. Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes 53: 2748–2756, 2004 [DOI] [PubMed] [Google Scholar]
- 37. Krause U, Bertrand L, Hue L. Control of p70 ribosomal protein S6 kinase and acetyl-CoA carboxylase by AMP-activated protein kinase and protein phosphatases in isolated hepatocytes. Eur J Biochem 269: 3751–3759, 2002 [DOI] [PubMed] [Google Scholar]
- 38. Lafontan M, Berlan M, Carpene C. Fat cell adrenoceptors: inter- and intraspecific differences and hormone regulation. Int J Obes 9, Suppl 1: 117–127, 1985 [PubMed] [Google Scholar]
- 39. Lane MD, Tang QQ, Jiang MS. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem Biophys Res Commun 266: 677–683, 1999 [DOI] [PubMed] [Google Scholar]
- 40. Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J 21: 2383–2396, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 117: 387–396, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Le XF, Vallian S, Mu ZM, Hung MC, Chang KS. Recombinant PML adenovirus suppresses growth and tumorigenicity of human breast cancer cells by inducing G1 cell cycle arrest and apoptosis. Oncogene 16: 1839–1849, 1998 [DOI] [PubMed] [Google Scholar]
- 43. MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem 64: 345–373, 1995 [DOI] [PubMed] [Google Scholar]
- 44. Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature 454: 436–444, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Minokoshi Y, Alquier T, Furukawa N, Kim YB, Lee A, Xue B, Mu J, Foufelle F, Ferre P, Birnbaum MJ, Stuck BJ, Kahn BB. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428: 569–574, 2004 [DOI] [PubMed] [Google Scholar]
- 46. Miyazaki Y, Mahankali A, Matsuda M, Mahankali S, Hardies J, Cusi K, Mandarino LJ, DeFronzo RA. Effect of pioglitazone on abdominal fat distribution and insulin sensitivity in type 2 diabetic patients. J Clin Endocrinol Metab 87: 2784–2791, 2002 [DOI] [PubMed] [Google Scholar]
- 47. Morrison RF, Farmer SR. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J Biol Chem 274: 17088–17097, 1999 [DOI] [PubMed] [Google Scholar]
- 48. Nolan JJ, Ludvik B, Beerdsen P, Joyce M, Olefsky J. Improvement in glucose tolerance and insulin resistance in obese subjects treated with troglitazone. N Engl J Med 331: 1188–1193, 1994 [DOI] [PubMed] [Google Scholar]
- 49. Nomura H, Kashiwagi S, Hayashi J, Kajiyama W, Tani S, Goto M. Prevalence of fatty liver in a general population of Okinawa, Japan. Jpn J Med 27: 142–149, 1988 [DOI] [PubMed] [Google Scholar]
- 50. Okuno A, Tamemoto H, Tobe K, Ueki K, Mori Y, Iwamoto K, Umesono K, Akanuma Y, Fujiwara T, Horikoshi H, Yazaki Y, Kadowaki T. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J Clin Invest 101: 1354–1361, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140: 197–208, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Patel YM, Lane MD. Mitotic clonal expansion during preadipocyte differentiation: calpain-mediated turnover of p27. J Biol Chem 275: 17653–17660, 2000 [DOI] [PubMed] [Google Scholar]
- 53. Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406: 207–210, 2000 [DOI] [PubMed] [Google Scholar]
- 54. Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev 17: 1352–1365, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429: 771–776, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qiu Z, Wei Y, Chen N, Jiang M, Wu J, Liao K. DNA synthesis and mitotic clonal expansion is not a required step for 3T3-L1 preadipocyte differentiation into adipocytes. J Biol Chem 276: 11988–11995, 2001 [DOI] [PubMed] [Google Scholar]
- 57. Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7: 885–896, 2006 [DOI] [PubMed] [Google Scholar]
- 58. Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev 14: 1293–1307, 2000 [PubMed] [Google Scholar]
- 59. Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell 108: 165–170, 2002 [DOI] [PubMed] [Google Scholar]
- 60. Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101, 2005 [DOI] [PubMed] [Google Scholar]
- 61. Timchenko NA, Wilde M, Nakanishi M, Smith JR, Darlington GJ. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev 10: 804–815, 1996 [DOI] [PubMed] [Google Scholar]
- 62. Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 8: 1224–1234, 1994 [DOI] [PubMed] [Google Scholar]
- 63. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 77: 289–312, 2008 [DOI] [PubMed] [Google Scholar]
- 64. Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP. Identification of a tumour suppressor network opposing nuclear Akt function. Nature 441: 523–527, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tussie-Luna MI, Bayarsaihan D, Seto E, Ruddle FH, Roy AL. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc Natl Acad Sci USA 99: 12807–12812, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tzatsos A, Kandror KV. Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol Cell Biol 26: 63–76, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes 59: 554–563, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431: 200–205, 2004 [DOI] [PubMed] [Google Scholar]
- 69. Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 6: 505–514, 2007 [DOI] [PubMed] [Google Scholar]
- 70. Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol Biol Cell 20: 801–808, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science 279: 1547–1551, 1998 [DOI] [PubMed] [Google Scholar]
- 72. Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP. PML is essential for multiple apoptotic pathways. Nat Genet 20: 266–272, 1998 [DOI] [PubMed] [Google Scholar]
- 73. Wu WS, Vallian S, Seto E, Yang WM, Edmondson D, Roth S, Chang KS. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol Cell Biol 21: 2259–2268, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology 151: 2504–2514, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang S, Jeong JH, Brown AL, Lee CH, Pandolfi PP, Chung JH, Kim MK. PML activates CHK2 by mediating CHK2 autophosphorylation. J Biol Chem 281: 26645–26654, 2006 [DOI] [PubMed] [Google Scholar]
- 76. Yang S, Liu A, Weidenhammer A, Cooksey RC, McClain D, Kim MK, Aguilera G, Abel ED, Chung JH. The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology 150: 2153–2160, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yeh WC, Bierer BE, McKnight SL. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci USA 92: 11086–11090, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zamboni M, Armellini F, Harris T, Turcato E, Micciolo R, Bergamo-Andreis IA, Bosello O. Effects of age on body fat distribution and cardiovascular risk factors in women. Am J Clin Nutr 66: 111–115, 1997 [DOI] [PubMed] [Google Scholar]
- 79. Zhang HH, Huang J, Duvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One 4: e6189, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene 18: 7941–7947, 1999 [DOI] [PubMed] [Google Scholar]