Abstract
In cultured pulmonary artery endothelial cells and other cell types, overexpression of mt-targeted DNA repair enzymes protects against oxidant-induced mitochondrial DNA (mtDNA) damage and cell death. Whether mtDNA integrity governs functional properties of the endothelium in the intact pulmonary circulation is unknown. Accordingly, the present study used isolated, buffer-perfused rat lungs to determine whether fusion proteins targeting 8-oxoguanine DNA glycosylase 1 (Ogg1) or endonuclease III (Endo III) to mitochondria attenuated mtDNA damage and vascular barrier dysfunction evoked by glucose oxidase (GOX)-generated hydrogen peroxide. We found that both Endo III and Ogg1 fusion proteins accumulated in lung cell mitochondria within 30 min of addition to the perfusion medium. Both constructs prevented GOX-induced increases in the vascular filtration coefficient. Although GOX-induced nuclear DNA damage could not be detected, quantitative Southern blot analysis revealed substantial GOX-induced oxidative mtDNA damage that was prevented by pretreatment with both fusion proteins. The Ogg1 construct also reversed preexisting GOX-induced vascular barrier dysfunction and oxidative mtDNA damage. Collectively, these findings support the ideas that mtDNA is a sentinel molecule governing lung vascular barrier responses to oxidant stress in the intact lung and that the mtDNA repair pathway could be a target for pharmacological intervention in oxidant lung injury.
Keywords: Ogg1, reactive oxygen species, pulmonary edema
reactive oxygen species (ROS) play a pathogenic role in acute lung injury (ALI) and its more severe sequel, the acute respiratory distress syndrome (ARDS) (4). Although numerous studies have explored antioxidants as therapeutic agents in ALI/ARDS, their outcome in clinical trials has been disappointing (27). One explanation for this failure is that the key molecule(s) integrating ROS actions on the pulmonary vasculature have yet to be identified; antioxidants may fail to protect against damage to these putative sentinel molecule(s) governing activation of cell death and dysfunction pathways.
Provocative evidence suggests that mtDNA may serve as such a molecular sentinel. In cultured pulmonary vascular endothelial cells (EC) and other cell types, mtDNA is highly sensitive to oxidative damage (1, 11, 29) and there is a conspicuous association between mtDNA damage and oxidant-induced cell death. For example, in pulmonary vascular EC phenotypes the propensity for apoptosis is closely linked to the efficiency of mtDNA repair (10). Genetic modulation of the first and rate-limiting step in mtDNA repair, mediated by Ogg1, a DNA glycosylase responsible for detecting and excising oxidatively damaged purines, exerts coordinate effects on ROS-induced mtDNA damage and cell death in all cell populations so far examined (8, 22, 23, 25, 26). Recent observations in intact mice with Staphylococcus aureus peritonitis and in critically ill human subjects indicate that mitochondrial accrual of Ogg1 and mitochondrial biogenesis are closely associated with survival (2, 5).
Collectively, these observations raise the interesting idea that mtDNA repair may be an isolated pharmacological target to modulate ROS-mediated endothelial barrier dysfunction in the intact pulmonary circulation. As an initial test of this concept, we used perfused rat lungs challenged with glucose oxidase (GOX)-generated H2O2 to determine whether novel fusion protein constructs targeting DNA glycosylases to mitochondria suppressed ROS-induced mtDNA damage and whether protection against oxidative mtDNA damage was linked to inhibition of ROS-induced lung vascular barrier dysfunction. Our results support the idea that oxidative mtDNA damage may initiate oxidant-mediated EC barrier dysfunction and in addition raise the prospect that the link between oxidative mtDNA damage and repair and cell dysfunction may be more dynamic than previously appreciated.
MATERIALS AND METHODS
Fusion proteins and other reagents.
Codon-optimized constructs were placed in plasmids for expression in Escherichia coli of fusion proteins containing either Endo III or Ogg1 coupled to a TAT sequence to facilitate cellular uptake, the mitochondrial targeting sequence from MnSOD, a hemagglutinin (HA) tag for immunological localization and a histidine tail as previously described for the preparation of fusion proteins containing exonuclease III or green fluorescent protein (14). Liquid cultures of bacterial cells transfected with plasmids containing the constructs were grown to an OD60 = 0.6 and induced with IPTG for 3 h. Bacteria were pelleted by centrifugation and resuspended in buffer A [20 mM Tris·HCl pH 8.0, 500 mM NaCl, 1× protein inhibitor cocktail EDTA-free (Calbiochem), 100 mM PMSF, and 5 mM imidazole]. Bacteria were lysed by sonication with a Branson Sonifier 250. After sonication, bacterial lysates were spun in a Beckman ultracentrifuge for 20 min at 105 g. After centrifugation, cleared lysates were incubated with Ni-NTA-agarose. The Ni-NTA-agarose was placed in a column and washed with several volumes of wash buffer (buffer A containing 30 mM imidazole). The bound protein was eluted from the column with elution buffer (buffer A containing 500 mM imidazole). The purity of the eluted protein was assessed by SDS-PAGE. All reagents for fusion protein production were obtained from Sigma unless otherwise indicated.
Colorimetric H2O2 assay.
A standard colorimetric assay was used to determine whether mt-targeted DNA repair proteins directly inhibited GOX-induced accumulation of hydrogen peroxide in aqueous solution (9).
Western immunoblot analysis of subcellular fusion protein localization.
Subcellular fractions were prepared from lung homogenates as described previously (16) with certain modifications. Lung tissue (1 g) was homogenized in a glass homogenizer with Teflon pestle eight times with 6 ml of homogenization buffer (0.25 M sucrose, 20 mM HEPES-NaOH pH 7.4, and 1 mM EDTA). Protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) was added to all isolation buffers. The homogenate was filtered through 70-μm mesh (BD Biosciences, Bedford, MA) and centrifuged on a cushion (5 ml) containing 0.35 M sucrose, 20 mM HEPES-NaOH pH 7.4, and 1 mM EDTA at 700 g for 10 min at 4°C. The fraction around and above the interphase was collected as crude mitochondria and reserved for mitochondrial isolation. The nuclear pellet was suspended in 3 ml of nuclear isolation buffer (0.25 M sucrose, 20 mM HEPES-NaOH pH 7.4, 25 mM KCl, and 5 mM MgCl2) and purified on a 3-ml cushion containing 0.8 M sucrose, 20 mM HEPES-NaOH pH 7.4, 25 mM KCl, and 5 mM MgCl2 at 3,000 g for 15 min at 4°C. The nuclear pellet so obtained was washed with nuclear isolation buffer and centrifuged at 1,000 g for 10 min. The pellet containing purified nuclei was suspended in 300 μl of RIPA buffer (Cell Signaling Technology, Danvers, MA), incubated for 30 min on ice, and centrifuged at 18,000 g for 15 min. The supernatant was designated as the “nuclear fraction.” The crude mitochondrial fraction, collected as described above, was centrifuged at 18,000 g for 20 min to pellet mitochondria, which were suspended in 2 ml of mitochondrial isolation buffer (0.2 M mannitol, 50 mM sucrose, 20 mM HEPES-NaOH pH 7.4, and 1 mM EDTA) and centrifuged under the same conditions. This supernatant was designated as the cytosolic fraction, and the pellet containing mitochondria was suspended in 300 μl of RIPA buffer (Cell Signaling Technology, Danvers, MA), incubated for 30 min on ice, and centrifuged at 18,000 g for 15 min. This latter supernatant was designated as the mitochondrial fraction. Cytosolic, nuclear, and mitochondrial fractions were subjected to Western immunoblot analysis for specific markers and for HA-tagged fusion protein constructs.
Western blot analyses were performed as described earlier (25) using antibodies against the HA tag (Sigma-Aldrich) to determine subcellular distribution of the fusion proteins. Antibodies against ATP synthase (complex V) subunit alpha (Invitrogen, Camarillo, CA) and against lamin B (Santa Cruz Biotechnology, Santa Cruz, CA) were used as mitochondrial and nuclear fraction markers respectively. Antibody against β-actin was used as a loading control for total lysate and cytosolic fractions.
Analysis of mtDNA content and damage and nuclear DNA damage.
Immediately after perfusion, lungs were snap frozen in liquid nitrogen and saved for determination of mtDNA content and oxidative mtDNA damage, global nuclear DNA damage, as well as oxidative damage to the indicated sequences of selected nuclear genes. Total DNA was isolated from lung samples powdered with a mortar and pestle, by previously described methods (19, 26). Purified DNA samples were digested with PpuMI and AhdI restriction enzymes (New England Biolabs, Beverly, MA) and used for further analyses.
To search for oxidative damage to the mitochondrial genome, Southern blot analysis was performed as published previously with minor modifications (26). In brief, digested DNA samples were precipitated, dissolved in TE buffer, and precisely quantified on the Hoefer DyNA Quant 200 fluorometer (Hoefer, San Francisco, CA) with use of Hoechst 33258 dye. To reveal oxidative base modifications, DNA was treated with formamidopyrimidine glycosylase (Fpg; New England Biolabs), a bacterial DNA repair enzyme that cleaves DNA at sites of oxidized purines, thereby creating single-strand breaks. Subsequently, Fpg-treated and untreated samples were incubated with 0.1 N NaOH for 15 min at 37°C, mixed with loading dye, and resolved in 0.6% agarose alkaline gel. After electrophoresis, DNA was vacuum transferred to a nylon membrane (Roche Diagnostics, Mannheim, Germany) and hybridized with a PCR-generated probe to the corresponding region of mtDNA. The mtDNA probe, labeled with a DIG-labeling kit (Roche Diagnostics), was generated with rat mtDNA sequence used as template and the following primers: 5′-CCCTACTTACTGGCTTCAATCTAC-3′ for the sense strand and 5′-CATACCATACCTATATATCCGAAGG-3′ for the antisense strand. The 1,016-bp product was hybridized with a 13.6-kb fragment of rat mtDNA obtained after PpuMI and AhdI digestion. Hybridization bands were detected with Amersham Hyperfilm ECL (GE Healthcare, Piscataway, NJ) and a Gel Logic 1500 Imaging System (Kodak, Rochester, NY). The hybridization band intensity depends on the relative amount and integrity of mtDNA. Changes in the equilibrium lesion density were calculated as negative ln of the quotient of hybridization intensities in treated and control bands and normalized to 10 kb (3).
The content of mtDNA in rat lung explants was determined by slot blot analysis (20). In brief, isolated and digested with restriction enzymes DNA was precisely quantified, adjusted to the same concentration with H2O, and treated with 0.4 N NaOH for 10 min at room temperature to denature the DNA. The indicated amounts of total DNA were then blotted onto a nylon membrane (Roche Diagnostics) using a slot blot apparatus (Hoefer), and membranes were hybridized with a DIG-labeled mtDNA-specific probe, after which they were washed and processed according to the manufacturer's suggestions. Mitochondrial DNA (mtDNA) content was normalized to total DNA.
Two different strategies were used to detect damage to the nuclear genome. First, qualitative alkaline gel electrophoresis was used to determine the mean fragment length of ethidium-stained DNA, as previously described (13). In this analysis, line scans of pixel densities of ethidium-stained alkaline gels were used to identify the position of the mean DNA fragments, whose lengths were then determined by relating the mean fragment position to the position of size standards on the same gels. Second, we used a PCR-based assay to detect base modifications in short sequences of selected nuclear genes previously demonstrated to be sensitive to oxidative base modifications in response to hypoxia and other physiological signals (19, 31). The sequences examined in each gene and primers used to amplify the sequences of interest are listed in Table 1. The basis of the assay is similar to that described above for the Southern blot technique; treatment of DNA with Fpg results in strand cleavage at sites of oxidized purines, thereby creating single-strand breaks that block PCR amplification. Differences in PCR amplification between Fpg-treated and untreated DNA are thus a specific indicator of the presence of oxidative base damage. The Fpg cleavage reaction was performed by incubating 250 ng of genomic DNA with 8 units of Fpg in 1× NEBuffer 1 (10 mM Bis-Tris propane-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.0) and 100 μg/ml BSA in a volume of 50 μl. Incubations were carried out at 37°C for 16 h. Fpg was then inactivated by heating at 60°C for 5 min. An aliquot containing 10 ng genomic DNA was then used for the PCR assay to detect Fpg-sensitive cleavage sites. Data are presented as the fraction intact DNA, calculated as the quotient of band intensities in Fpg-treated and untreated DNA.
Table 1.
Primer sequences for PCR assessment of Fpg-sensitive oxidative base damage in nuclear DNA sequences
| Gene | Forward | Reverse |
|---|---|---|
| Vegf promoter: (+ hypoxia response element) | 5′-GCTCTGCCAGACTCCACAGT-3′ | 5′-ACCTCGGAAACAGAAACTCTAGG-3′ |
| Vegf intron | 5′-GAAGAGTACGCATATCCATAGC-3′ | 5′-CACTCCAGGGCTTCATCATTGC-3′ |
| HO-1 | 5′-TTCCTCTTCCTCTTCTTC-3′ | 5′-AACCAACTATACACTATACTC-3′ |
| ODC | 5′-TTAATTGGCTTGTATCTC-3′ | 5′-TTATGCTGAATGACTTAG-3′ |
| 28 S | 5′-CTCAACCTATTCTCAAAC-3′ | 5′-GTCTATATCAACCAACAC-3′ |
Fpg, formamide pyrimidine glycosylase.
Assessment of the vascular filtration coefficient and histopathology in perfused rat lungs.
Adolescent male Sprague-Dawley rats (250–350 g, Charles Rivers Laboratories, Wilmington, MA) were anesthetized with pentobarbital and a tracheotomy catheter was inserted. Lungs were ventilated (Harvard rodent ventilator model 683) with a humidified gas mixture of 21% O2, 5% CO2, and N2 at an airway pressure of 6 cmH2O, a positive end-expiratory pressure of 2.5 cmH2O, and a respiratory rate of 60 breaths per min. After median sternotomy and anticoagulation with heparin (100 units), pulmonary arterial and left ventricular catheters were inserted, and the lungs were removed en bloc and suspended from a force displacement transducer to measure real-time changes in lung weight (Grass FT03, Grass Instruments, Quincy, MA) in a humidified chamber. Pulmonary artery and venous pressures were continuously monitored with Cope pressure transducers (model 041-500-503) and recorded on a Grass polygraph (model 7F). Zone 3 conditions were maintained in all experiments. All procedures involving animals were approved by the Institutional Animal Care and Use Committee.
The perfusate consisted of 4% bovine albumin in physiological salt solution (in mM: 3.2 CaCl2, 119 NaCl, 4.7 KCl, 1.17 MgSO4, 1.18 KH2PO4, 22.6 NaHCO3, and 5.5 d-glucose). The lungs were perfused in a recirculating circuit by use of a peristaltic pump (Gilson Minipuls 2) at a constant flow of 0.04 ml·g body wt−1·min−1. Prior to continuous perfusate recirculation, 50 ml of perfusate was used to wash the lungs free of blood and then discarded. Study agents were administered directly into the perfusate reservoir.
The vascular filtration coefficient (Kf), an index of hydraulic conductivity, was determined by a previously described method (18). In experiments to determine whether mt-targeted DNA repair proteins prevented oxidant-induced lung injury, isolated lungs were allowed to equilibrate and attain isogravimetric conditions for 30 min, after which baseline Kf was determined, normalized to lung dry weight, and expressed as milliliters per minute per cmH2O per 100 g of lung tissue. Fusion proteins or their vehicle were then added to the perfusion reservoir to attain final concentrations of 220 and 100 nM for the Ogg1 and Endo III constructs, respectively, and Kf was determined a second time 60 min thereafter. Subsequently, GOX (0.5 units/50 ml) was added to the reservoir, and Kf was measured a third and final time after 60 min of additional perfusion.
To corroborate the physiological analysis of endothelial barrier properties, qualitative histopathological analyses were performed on control and GOX-treated isolated rat lungs in the absence and presence of treatment with mt-targeted Ogg1. These experiments utilized lungs that were not subjected to Kf measurements. In brief, lungs were fixed by airways instillation of neutral buffered formalin at a constant pressure of 20 cmH2O, embedded in paraffin, stained with hematoxylin and eosin (H&E), and evaluated by light microscopy at a magnification of ×10 for the presence of perivascular cuffs indicative of endothelial barrier degradation.
We also determined whether mt-targeted Ogg1 reversed ongoing oxidant-induced endothelial barrier dysfunction. In this instance, after determination of baseline Kf, GOX was added to the reservoir and a second Kf was determined after 60 min of perfusion. The Ogg1 construct or its vehicle were then added to the reservoir, and a final Kf value was ascertained 30 min later. In separate experiments, lung tissue was harvested for determination of mtDNA integrity by quantitative Southern blot analyses as described above.
Statistical analysis.
Results are expressed as means ± SE. Comparisons were made with one-way analysis of variance. Tukey's post hoc tests were applied as appropriate. Differences in mean values were considered statistically significant when P < 0.05.
RESULTS
Mt-targeted DNA repair enzymes do not directly inhibit GOX-induced accumulation of hydrogen peroxide.
To ascribe pharmacological effects of the fusion proteins to repair of GOX-induced mtDNA damage, it is important to determine whether they directly inhibit GOX-induced accumulation of hydrogen peroxide. Accordingly, we used a standard colorimetric assay for hydrogen peroxide in aqueous solution and found, not surprisingly, that GOX-induced accumulation of the reactive species was unaffected by either mt-targeted fusion protein (data not shown).
Disposition of mt-targeted Ogg1 after vascular administration to perfused rat lungs.
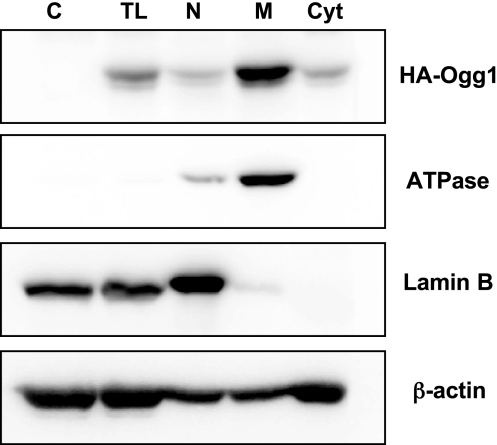
As shown in Fig. 1, HA immunoreactivity associated with the Ogg1 fusion protein construct was detected most prominently in the mitochondrial fraction in lung tissue harvested after 30 min perfusion. Minimal deposition was detected in the nuclear fraction.
Fig. 1.
Distribution of hemagglutinin (HA)-tagged 8-oxoguanine DNA glycosylase 1 (Ogg1) fusion protein in subcellular fractions derived from perfused rat lung. Isolated rat lungs were perfused for 30 min with 220 nM Ogg1-HA fusion protein construct, after which lung tissue was homogenized and subcellular fractions were prepared as described in materials and methods. Western analysis of HA immunoreactivity was applied to total lysate (TL) derived from control lung tissue not treated with the fusion protein (C), as well as TL, nuclear (N), mitochondrial (M), and cytosolic (Cyt) fractions derived from lungs perfused with the Ogg1 fusion protein. Representative of 3 experiments.
Impact of mt-targeted DNA repair enzymes on GOX-induced mtDNA content and integrity.
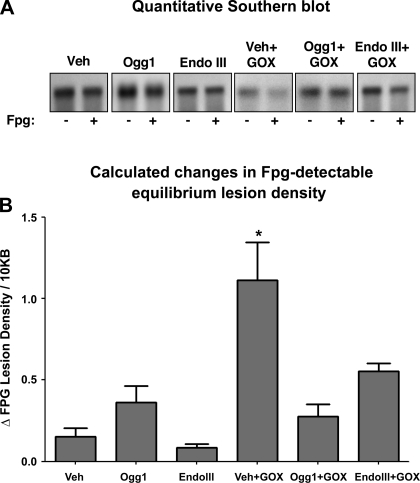
Slot blot analysis indicated that neither the fusion proteins nor GOX altered mtDNA content relative to untreated lung tissue (N = 4–5/group; data not shown). To measure oxidative mtDNA damage, we restricted total lung DNA into fragments of interest, treated the DNA fragments with alkali or alkali plus Fpg, and then hybridized to a genomic probe for a 13.6-kb sequence of the mitochondrial genome. There are two important considerations in this analysis: First, since lung mtDNA content did not differ between the treatment groups, differences in mtDNA probe hybridization to alkali-treated DNA likely reflect differences in the density of abasic sites or deoxyribose backbone damage. A second consideration is that further decreases in hybridization intensities caused by Fpg treatment reflect the relative density of oxidized purine base products. Inspection of the quantitative Southern blot analysis shown in Fig. 2A indicates that GOX treatment alone reduced hybridization intensity in alkali-treated DNA and that this decrease was prevented by pretreatment with either of the mt-targeted fusion proteins. The magnitude of the Fpg-related decrease in hybridization intensity was more prominent in GOX-exposed lungs than in controls, and this too was blunted by pretreatment with the mt-targeted fusion proteins. Assuming a Poisson distribution of the damaged bases, it is possible to calculate the relative increase in purine lesion density revealed by Fpg treatment for each experimental group using a simple equation (3). These quantitative data, depicted in Fig. 2B, show that GOX caused a large increase in the equilibrium density of purine base oxidation products in the mitochondrial genome, which was attenuated by pretreatment with the mt-targeted DNA repair proteins (N = 4–6/group).
Fig. 2.
Impact of Ogg1 and endonuclease III (Endo III) fusion proteins on glucose oxidase (GOX)-induced oxidative mtDNA damage in perfused rat lungs. A: representative quantitative Southern blot analyses of oxidative mitochondrial DNA (mtDNA) damage detected by alkali without (−) and with (+) formamidopyrimidine DNA glycosylase (Fpg) in isolated lungs perfused with the fusion protein vehicle (Veh), the Ogg1 or Endo III fusion proteins in the absence of an oxidant stress, or the 2 fusion proteins administered prior to challenging the lungs with GOX-generated H2O2. Note that diminished hybridization intensity relative to PSS or Veh controls is indicative. B: calculated changes in Fpg-detectable oxidative base lesion for the above-mentioned experimental groups. N = 4–6 per group. *Significantly different from Veh controls.
Impact of GOX on nuclear DNA damage.
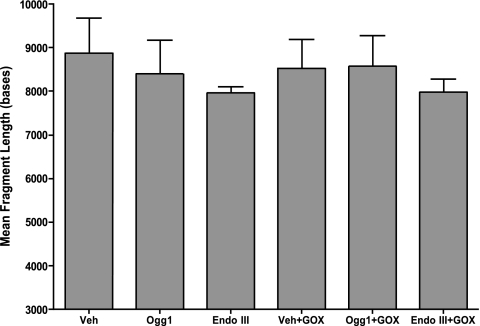
Initial experiments determined the mean DNA fragment length from ethidium-stained gels as an estimate of nuclear DNA damage. As shown in Fig. 3, mean fragment length in alkali-treated DNA did not differ between control and GOX-challenged lungs nor did it differ as a result of treatment with either fusion protein construct (N = 4–6/group). Mean fragment lengths detected by alkali plus Fpg also did not differ between experimental groups (data not shown). Whereas mean fragment length analysis is well suited for examination of total genomic damage in intact tissue, it may not be sufficiently sensitive to detect damage restricted to a few sensitive sequences. Accordingly, we applied a PCR-based assay to search for damage in short DNA sequences previously shown to be prone to oxidative base modifications evoked hypoxia and other physiological signals (19, 31). As shown in Table 2, challenge with GOX failed to cause oxidative base damage to any of the nuclear gene sequences examined, and, not surprisingly, neither of the mt-targeted DNA repair enzymes altered the density of oxidative base modifications in total genomic DNA or in the selected sequences from control or GOX-treated lung DNA (N = 4–7/group).
Fig. 3.
Impact of GOX and of Ogg1 and Endo III fusion proteins on mean DNA fragment length determined from intact perfused rat lung tissue. Mean fragment length of alkali-treated DNA was determined in isolated rat lungs perfused with the fusion protein vehicle (Veh), the Ogg1 or Endo III fusion proteins in the absence of an oxidant stress, or the 2 fusion proteins administered prior to challenging the lungs with GOX-generated H2O2. There were no detectable changes in mean DNA fragment length between the experimental groups. N = 4–6 per group.
Table 2.
Glucose oxidase-generated hydrogen peroxide fails to cause Fpg-sensitive oxidative base damage in selected sequences of nuclear genes
| Fraction Intact DNA |
|||||
|---|---|---|---|---|---|
| Vegf promoter (HRE) | Vegf intron | HO-1 | ODC | 28S | |
| Veh | 1.03 ± 0.04 | 0.98 ± 0.11 | 0.97 ± 0.06 | 0.92 ± 0.12 | 1.05 ± 0.04 |
| Ogg1 | 0.97 ± 0.07 | 0.94 ± 0.05 | 0.99 ± 0.13 | 1.06 ± 0.04 | 1.03 ± 0.12 |
| Endo III | 0.94 ± 0.04 | 0.98 ± 0.03 | 0.94 ± 0.10 | 0.97 ± 0.09 | 0.97 ± 0.04 |
| Veh+GOX | 0.97 ± 0.04 | 1.02 ± 0.06 | 1.03 ± 0.05 | 0.98 ± 0.08 | 1.04 ± 0.02 |
| Ogg1+GOX | 0.98 ± 0.08 | 0.99 ± 0.05 | 0.92 ± 0.13 | 1.02 ± 0.14 | 1.04 ± 0.06 |
| Endo III+GOX | 1.00 ± 0.04 | 0.99 ± 0.03 | 0.96 ± 0.04 | 0.98 ± 0.07 | 1.00 ± 0.03 |
Values are mean ± SE (n = 4–7). Veh, vehicle; Ogg1, 8-oxoguanine DNA glycosylase 1; Endo III, endonuclease III; GOX, glucose oxidase.
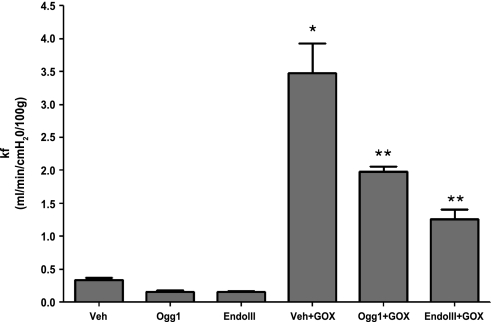
Impact of mt-targeted DNA repair enzymes on GOX-induced increases in lung vascular permeability.
Sixty-minute exposure to the Ogg1 or Endo III fusion proteins failed to alter baseline Kf compared with controls. GOX was then added to the perfusate reservoir and allowed to recirculate for an additional 60 min, after which Kf was again determined. As displayed in Fig. 4, GOX caused a large increase in vascular filtration in control lungs. Pretreatment for 60 min with Ogg1 or Endo III fusion proteins significantly attenuated increases in vascular filtration evoked by GOX. The vehicle control and fusion proteins had no effect in the absence of GOX (N = 4–6/group).
Fig. 4.
Impact of Ogg1 and Endo III fusion proteins on GOX-induced changes the vascular filtration coefficient (Kf) in perfused rat lungs. Vascular filtration coefficient was determined in rat lungs perfused with Veh, the Ogg1 or Endo III fusion proteins in the absence of an oxidant stress, or the 2 fusion proteins administered prior to challenging the lungs with GOX-generated H2O2. Note that GOX caused large increases in Kf that were attenuated by either the Ogg1 or the Endo III fusion proteins. N = 4–6 per group. *Significantly increased compared with Veh control at P < 0.05. **Significantly decreased compared with Veh + GOX at P < 0.05.
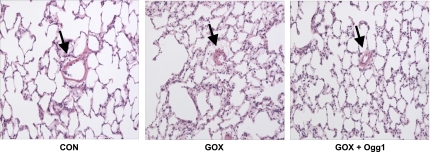
To corroborate the above physiological assessment of endothelial barrier integrity, histopathological analyses were performed on H&E-stained sections of control lungs, lungs treated with the mt-targeted Ogg1 fusion protein, GOX-treated lung preparations, and lungs pretreated with mt-targeted Ogg1 and then challenged with GOX. As depicted by the representative photomicrographs shown in Fig. 5, whereas GOX-challenged lungs displayed prominent perivascular cuffs indicative of endothelial barrier dysfunction, lungs pretreated with mt-targeted Ogg1 were largely devoid of such cuffs, thus supporting the contention that enhanced mtDNA repair suppressed GOX-mediated loss of endothelial integrity.
Fig. 5.
Photomicrographs (magnification: ×10) of hematoxylin and eosin-stained lung tissue from control (Con) lungs, lungs challenged with GOX alone and in the presence of mt-targeted Ogg1. Arrow denotes presence of perivascular cuffs indicative of endothelial barrier dysfunction. Lungs treated with mt-targeted Ogg1 alone were indistinguishable from controls (data not shown). Photomicrographs representative of 3 lung preparations per experimental group.
Mitochondrially targeted Ogg1 reverses GOX-induced mtDNA damage and vascular barrier dysfunction.
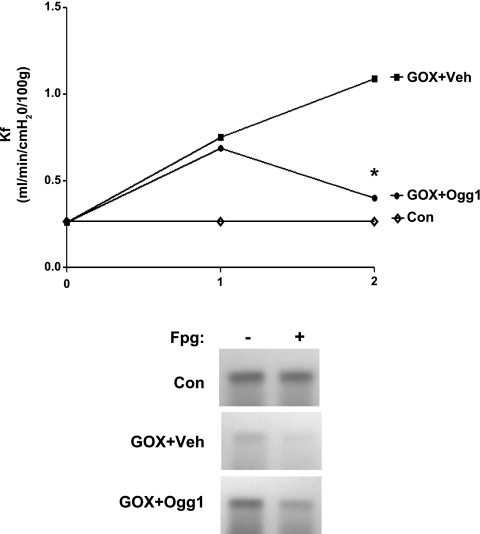
After an initial determination of Kf, lungs were perfused for 60 min with GOX and Kf was measured again. Lungs were then reserved for determination of mtDNA damage or were treated with mt-targeted Ogg1 or its vehicle. Forty-five minutes later, Kf was measured a final time, and lung tissues were prepared for mtDNA damage assessment. The impact of the DNA repair enzyme on Kf, shown in Fig. 6, indicates that Ogg1 restored Kf to near baseline values. Complementing these observations, a representative Southern blot analyses of mtDNA damage also is shown in Fig. 6 and reveals that, within 45 min of addition to the perfusion medium, mt-targeted Ogg1 engendered partial recovery of mtDNA integrity.
Fig. 6.
Mitochondria-targeted Ogg1 reverses GOX-induced vascular barrier dysfunction and mtDNA damage in perfused rat lungs. Top: after baseline Kf measurement (0), either GOX or vehicle was added to the reservoir and Kf was assessed 60 min later (1). Mitochondria-targeted Ogg1 fusion protein or vehicle was then added to the reservoir and a final Kf was measured 45 min later (2). Control lungs were perfused with no additions for 105 min, with Kf determined at 90 min and termination of perfusion. N = 4–6 for each group. *Different from Veh at P < 0.05. Bottom: in separate experiments, lungs were perfused and treated as just described, and at the end of perfusion lung tissue DNA was isolated for determination of alkali and alkali + Fpg-detectable mtDNA damage by quantitative Southern blot analysis. In the representative Southern blot shown, note that hybridization intensities are reduced in GOX + Veh compared with Veh alone, indicating the persistence of mtDNA damage in GOX-treated lungs, and that addition of mitochondria-targeted Ogg1 partially restores hybridization intensity measured 45 min thereafter, indicating incomplete repair of mtDNA lesions. Representative of 3 experiments.
DISCUSSION
This study shows that intravascular delivery of fusion protein constructs consisting of TAT and MTS sequences coupled to purine- and pyrimidine-specific DNA glycosylases suppress mtDNA damage and vascular barrier dysfunction induced by GOX-generated ROS in buffer-perfused rat lungs. Critical to interpreting these findings within the framework of our concept that mtDNA repair serves as an isolated target for pharmacological intervention in oxidant-mediated lung injury is whether the fusion proteins exert effects unrelated to their impact on mtDNA repair.
We offer several lines of evidence that the constructs are in fact acting at the level of the mitochondrial genome. First, the present data exclude the possibility that the mt-targeted DNA repair proteins inhibit GOX-mediated H2O2 accumulation in aqueous solution. Second, the fusion proteins were prominently localized in lung cell mitochondria, with relatively minor deposition in cytosolic and nuclear fractions. It should be noted that although the present study defined the subcellular distribution of the fusion proteins, the cellular disposition of the mt-targeted DNA repair enzymes within lung tissue is currently unknown. Based on the results of the physiological and histopathological assessments discussed below; however, it would seem that endothelial cells are a key cellular target of fusion protein effects. Third, and consistent with earlier reports attesting to the relative sensitivity of the mitochondrial genome to oxidant stress (1, 11, 29), we found in intact lung tissue that GOX-generated reactive species selectively damaged the mitochondrial genome, characterized by increased densities of strand breaks and apurinic/pyrimidinic sites and of purine base oxidation products. GOX-induced nuclear DNA damage could not be detected by use of two complementary strategies. In light of these observations, and since the extensively studied Ogg1 and Endo III are highly specific for oxidized purine and pyrimidine base products, respectively (21, 28), we believe that the protective actions of the fusion proteins described subsequently are most likely ascribed to their effects on the mitochondrial genome in lung vascular endothelial cells.
mtDNA damage induced by GOX was accompanied by increases in lung vascular permeability as detected by increases in Kf and histopathologically. Importantly, development of mtDNA lesions and barrier dysfunction were attenuated by pretreatment with mt-targeted Ogg1 and Endo III fusion proteins. Treatment of isolated lungs with mt-targeted Ogg1 after GOX-mtDNA damage and vascular barrier dysfunction were established also was associated with partial repair of the mitochondrial genome and complete restoration of normal barrier properties. Because of the specificity for Ogg1 and Endo III for purine and pyrimidine base oxidation products, respectively, these findings imply that damage to either class of mtDNA base can provoke loss of endothelial barrier integrity in the intact pulmonary circulation. These observations also extend previous reports in multiple cultured cell populations demonstrating that modulation of Ogg1-dependent mtDNA repair exerts coordinate effects on ROS-mediated cell death (8, 22, 23, 25, 26). Although beyond the scope of the present study, it is interesting to speculate that oxidative mtDNA damage is a controlling event in a range of ROS-mediated effects on pulmonary vascular endothelial cells, including an early phase of barrier dysfunction linked to alterations in cell-cell and cell-matrix adhesions followed in some instances by loss of cell viability. Additional studies will be needed to resolve the functional link between mtDNA integrity-dependent alterations in EC barrier properties and EC fate in the setting of oxidant stress.
A traditional model linking mtDNA damage to cell death holds that oxidative lesions impair mtDNA transcription, thereby reducing mtDNA-encoded subunits of electron transport and leading to enhanced mitochondrial generation of proapoptotic ROS (12, 15, 24, 30). In this model, overexpression of mt-targeted DNA glycosylases prevents accumulation of oxidative mtDNA damage and thereby suppresses its downstream consequences. Some of our data do not easily fit with this traditional concept. In particular, even though reversal of H2O2-induced EC barrier dysfunction in perfused lungs by mt-targeted Ogg1 was accompanied by partial mtDNA damage repair, the time course of this response seems too rapid to be explained by normalization of mtDNA transcription and mRNA translation and subsequent reversal of mitochondrially driven proapoptotic signaling. It should be considered that the reported turnover rate for most of the mtDNA-encoded electron transport chain components is on the order of 2 h (7), but substantial reversal of GOX-induced EC barrier dysfunction in perfused lungs occurred within only 45 min of Ogg1 fusion protein administration.
These unexpectedly rapid actions of mt-targeted Ogg1 raise questions about the mechanism(s) by which enhanced mtDNA glycosylases protects against ROS-mediated barrier dysfunction. Indeed, in the specific case of Ogg1, whereas one recent report demonstrates than a mutant lacking both lyase and glycosylase activities fails to suppress ROS-mediated mtDNA damage and cell death (6), another shows that a different mutant Ogg1 devoid of glycosylase activity but with unknown lyase activity exerts a normal level of protection against ROS stress and cytotoxicity (17). Importantly, the latter report demonstrated that both mutant and wild-type Ogg1 bound the proapoptotic aconitase, and on this basis it was proposed that stabilization of aconitase by Ogg1, rather than mtDNA repair per se, was the mechanism of protection. Against this background, the possibility might be considered that mt-targeted DNA repair enzymes protect against the adverse effects of oxidant stress not solely by repairing mtDNA, but also by stabilizing mtDNA-protein interactions and the functions so governed, including interactions governing EC barrier maintenance. Future studies on the ability of mt-targeted Endo III, an enzyme structurally and functionally different than Ogg1, to rapidly reverse GOX-mediated endothelial barrier dysfunction will be useful in exploring this complicated issue.
In summary, the data presented herein support the possibility that the mitochondrial genome, and specifically the base excision mtDNA repair pathway, could be an isolated targeted for intervention in ROS-mediated lung endothelial barrier dysfunction. Obviously, experiments in pathologically relevant intact animals will be required as a next step in the translation of this idea into practical utility.
GRANTS
This work was supported in part by grants from the National Heart, Lung, and Blood Institute (HL058234, HL073244, and Project 3 in PO1 HL66299).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Ballinger SW, Patterson C, Yan CN, Doan R, Burow DL, Young CG, Yakes FM, Van Houten B, Ballinger CA, Freeman BA, Runge MS. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res 86: 960–966, 2000 [DOI] [PubMed] [Google Scholar]
- 2. Bartz RR, Suliman HB, Fu P, Welty-Wolf K, Carraway MS, MacGarvey NC, Withers CM, Sweeney TE, Piantadosi CA. Staphylococcus aureus sepsis and mitochondrial accrual of the 8-oxoguanine DNA glycosylase DNA repair enzyme in mice. Am J Respir Crit Care Med 183: 226–233, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40: 359–369, 1985 [DOI] [PubMed] [Google Scholar]
- 4. Brigham KL. Oxidant stress and adult respiratory distress syndrome. Eur Respir J Suppl 11: 482s–484s, 1990 [PubMed] [Google Scholar]
- 5. Carre JE, Orban JC, Re L, Felsmann K, Iffert W, Bauer M, Suliman HB, Piantadosi CA, Mayhew TM, Breen P, Stotz M, Singer M. Survival in critical illness is associated with early activation of mitochondrial biogenesis. Am J Respir Crit Care Med 182: 745–751, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterjee A, Mambo E, Zhang Y, Deweese T, Sidransky D. Targeting of mutant hogg1 in mammalian mitochondria and nucleus: effect on cellular survival upon oxidative stress. BMC Cancer 6: 235, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol 7: 453–478, 1991 [DOI] [PubMed] [Google Scholar]
- 8. Dobson AW, Grishko V, LeDoux SP, Kelley MR, Wilson GL, Gillespie MN. Enhanced mtDNA repair capacity protects pulmonary artery endothelial cells from oxidant-mediated death. Am J Physiol Lung Cell Mol Physiol 283: L205–L210, 2002 [DOI] [PubMed] [Google Scholar]
- 9. Gillespie MN, Moore CG, Wright CE, O'Connor WN. Salutary effects of prostaglandin E1 in perfused rat lungs injured with hydrogen peroxide. J Pharmacol Exp Ther 241: 1–5, 1987 [PubMed] [Google Scholar]
- 10. Grishko V, Solomon M, Breit JF, Killilea DW, Ledoux SP, Wilson GL, Gillespie MN. Hypoxia promotes oxidative base modifications in the pulmonary artery endothelial cell VEGF gene. FASEB J 15: 1267–1269, 2001 [DOI] [PubMed] [Google Scholar]
- 11. Grishko V, Solomon M, Wilson GL, LeDoux SP, Gillespie MN. Oxygen radical-induced mitochondrial DNA damage and repair in pulmonary vascular endothelial cell phenotypes. Am J Physiol Lung Cell Mol Physiol 280: L1300–L1308, 2001 [DOI] [PubMed] [Google Scholar]
- 12. Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res 88: 529–535, 2001 [DOI] [PubMed] [Google Scholar]
- 13. Koczor CA, Shokolenko IN, Boyd AK, Balk SP, Wilson GL, Ledoux SP. Mitochondrial DNA damage initiates a cell cycle arrest by a Chk2-associated mechanism in mammalian cells. J Biol Chem 284: 36191–36201, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koczor CA, Snyder JW, Shokolenko IN, Dobson AW, Wilson GL, Ledoux SP. Targeting repair proteins to the mitochondria of mammalian cells through stable transfection, transient transfection, viral transduction, and TAT-mediated protein transduction. Methods Mol Biol 554: 233–249, 2009 [DOI] [PubMed] [Google Scholar]
- 15. Mayer B, Oberbauer R. Mitochondrial regulation of apoptosis. News Physiol Sci 18: 89–94, 2003 [DOI] [PubMed] [Google Scholar]
- 16. Nakamoto H, Kaneko T, Tahara S, Hayashi E, Naito H, Radak Z, Goto S. Regular exercise reduces 8-oxodG in the nuclear and mitochondrial DNA and modulates the DNA repair activity in the liver of old rats. Exp Gerontol 42: 287–295, 2007 [DOI] [PubMed] [Google Scholar]
- 17. Panduri V, Liu G, Surapureddi S, Kondapalli J, Soberanes S, de Souza-Pinto NC, Bohr VA, Budinger GR, Schumacker PT, Weitzman SA, Kamp DW. Role of mitochondrial hOGG1 and aconitase in oxidant-induced lung epithelial cell apoptosis. Free Radic Biol Med 47: 750–759, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parker JC, Gillespie MN, Taylor AE, Martin SL. Capillary filtration coefficient, vascular resistance, and compliance in isolated mouse lungs. J Appl Physiol 87: 1421–1427, 1999 [DOI] [PubMed] [Google Scholar]
- 19. Pastukh V, Ruchko M, Gorodnya O, Wilson GL, Gillespie MN. Sequence-specific oxidative base modifications in hypoxia-inducible genes. Free Radic Biol Med 43: 1616–1626, 2007 [DOI] [PubMed] [Google Scholar]
- 20. Pastukh VM, Zhang L, Ruchko MV, Gorodnya O, Bardwell GC, Tuder RM, Gillespie MN. Oxidative DNA damage in lung tissue from patients with COPD is clustered in functionally significant sequences. Int J Chron Obstruct Pulmon Dis 6: 209–217, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rachek LI, Grishko VI, Alexeyev MF, Pastukh VV, LeDoux SP, Wilson GL. Endonuclease III and endonuclease VIII conditionally targeted into mitochondria enhance mitochondrial DNA repair and cell survival following oxidative stress. Nucleic Acids Res 32: 3240–3247, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rachek LI, Grishko VI, Musiyenko SI, Kelley MR, LeDoux SP, Wilson GL. Conditional targeting of the DNA repair enzyme hOGG1 into mitochondria. J Biol Chem 277: 44932–44937, 2002 [DOI] [PubMed] [Google Scholar]
- 23. Rachek LI, Thornley NP, Grishko VI, LeDoux SP, Wilson GL. Protection of INS-1 cells from free fatty acid-induced apoptosis by targeting hOGG1 to mitochondria. Diabetes 55: 1022–1028, 2006 [DOI] [PubMed] [Google Scholar]
- 24. Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol 294: C413–C422, 2008 [DOI] [PubMed] [Google Scholar]
- 25. Ruchko M, Gorodnya O, LeDoux SP, Alexeyev MF, Al-Mehdi AB, Gillespie MN. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. Am J Physiol Lung Cell Mol Physiol 288: L530–L535, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Ruchko MV, Gorodnya OM, Zuleta A, Pastukh VM, Gillespie MN. The DNA glycosylase Ogg1 defends against oxidant-induced mtDNA damage and apoptosis in pulmonary artery endothelial cells. Free Radic Biol Med 50: 1107–1113, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsushima K, King LS, Aggarwal NR, De Gorordo A, D'Alessio FR, Kubo K. Acute lung injury review. Intern Med 48: 621–630, 2009 [DOI] [PubMed] [Google Scholar]
- 28. van der Kemp PA, Charbonnier JB, Audebert M, Boiteux S. Catalytic and DNA-binding properties of the human Ogg1 DNA N-glycosylase/AP lyase: biochemical exploration of H270, Q315 and F319, three amino acids of the 8-oxoguanine-binding pocket. Nucleic Acids Res 32: 570–578, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA 94: 514–519, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang D, Mott JL, Chang SW, Stevens M, Mikolajczak P, Zassenhaus HP. Mitochondrial DNA mutations activate programmed cell survival in the mouse heart. Am J Physiol Heart Circ Physiol 288: H2476–H2483, 2005 [DOI] [PubMed] [Google Scholar]
- 31. Ziel KA, Grishko V, Campbell CC, Breit JF, Wilson GL, Gillespie MN. Oxidants in signal transduction: impact on DNA integrity and gene expression. FASEB J 19: 387–394, 2005 [DOI] [PubMed] [Google Scholar]