Abstract
Temozolomide (TMZ) is the most effective chemotherapeutic agent for glioblastoma (GBM). Resistance to this methylating agent is linked to DNA repair enzyme O6-methylguanine-DNA methyltransferase (MGMT). However, in recent studies MGMT status was not completely accurate as a predictor of TMZ response in GBM, suggesting other mechanisms of resistance. As part of an effort aimed at discovery of genes involved in TMZ resistance in GBM, the expression of CD74 was evaluated in GBM patient samples and the influence of CD74 on TMZ response was evaluated in GBM tumor models. Reverse transcription-polymerase-chain reaction (RT-PCR) demonstrated differential expression of CD74 mRNA among the GBM xenografts; 8 of 20 (40%) expressed CD74 mRNA. In a preliminary evaluation of whether CD74 expression might influence TMZ response, CD74 mRNA expression levels were inversely associated with in vivo TMZ resistance in 20 GBM xenograft lines (median survival 122 vs. 62.5 days; r=−0.48 p = 0.032). In follow up to this observation, CD74 shRNA knock down in U87 cells significantly suppressed in vitro proliferation and increased TMZ sensitivity as compared to a non-specific control shRNA. Consistent with an effect on proliferation and survival, silencing of CD74 by shRNA was associated with reduced Akt and Erk1/2 activation in response to stimulation by CD74 ligand macrophage-migration inhibition factor (MIF). Lastly, expression of CD74 protein was assessed in patient samples (9 anaplastic astrocytoma [AA], and 62 GBM) by immunohistochemistry, and appreciable expression was observed in 28% of samples. Collectively, these findings suggest that CD74 is expressed in a subset of high grade gliomas and may contribute to TMZ resistance.
Keywords: CD74, glioblastoma xenografts, temozolomide, resistance
Introduction
The efficacy of standard temozolomide chemotherapy and radiation therapy for patients with newly diagnosed GBM often is compromised by inherent resistance to these therapies. Previous studies have demonstrated that high activity of the DNA repair protein O6-methylguanine methyltransferase (MGMT) can confer resistance to temozolomide in GBM patients, and molecular assays to evaluate MGMT status are being studied as predictive or prognostic indicators in GBM. However, the lack of perfect concordance between MGMT assays and TMZ resistance suggests that there are other factors that may influence TMZ responsiveness in GBM patients.
In the current study, we have identified CD74 as an additional potential modulator of TMZ responsiveness. CD74 was originally described as an MHC class II chaperone [1] and functions as a membrane receptor for the proinflammatory cytokine macrophage migration inhibitory factor (MIF) on immune cells [2]. MIF binding to CD74 activates downstream signaling through the MAPK and Akt pathways and promotes cell proliferation and survival [3, 4]. Beside expression by immune cells, CD74 overexpression has been observed in several non-CNS cancers, and CD74 expression in these tumors is associated with aggressive behavior and poor patient prognosis [5-9]. The current study provides the first report of CD74 overexpression in GBM, and links CD74 signaling to pro-survival effects that may contribute to TMZ resistance.
Materials and Methods
Experimental reagents
Monoclonal antibody specific for human CD74 (C-16) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and anti-phospho-AKT (Ser473), anti-total AKT, anti-phospho-(Thr202/Tyr204) ERK1/2, anti-total ERK1/2, anti-phospho Src (Tyr416) and anti-total Src antibodies were purchased from Cell Signaling Technologies, Inc. (Danvers, MA). Secondary biotinylated goat anti-rabbit and anti-mouse immunoglobulin G (IgG) were obtained from Cell Signaling Technologies,Inc. Avidin-biotin peroxidase complex was obtained from Vector Laboratories (Burlingame, CA), and horseradish-peroxidase-conjugated anti-rabbit and anti-mouse IgG were purchased from Amersham (Arlington Heights, IL). Recombinant MIF peptide was purchased from R&D systems (Minneapolis, MN).
Animal studies
Nude mice were purchased from National Institute of Health (NCI, Bethesda, MD). All experiments with animals were approved by the Mayo Institutional Animal Care and Use Committee. GBM xenografts were established as previously described [10]. Mice engrafted with intracranial tumors were randomized into groups of 8 to 10 mice each and treatment was initiated 2 weeks before mice were expected to become moribund. TMZ was purchased from the Mayo Clinic Pharmacy, suspended in Ora-plus (Paddock Laboratories, Minneapolis), and administered by oral gavage at 66 mg/kg/day for 5 days. This dosing regimen results in a drug exposure in mice that is equivalent to that obtained in humans with the routinely used adjuvant dosing regimen of 200 mg/m2/day x 5 days. Mice were observed daily and euthanized when they reached a moribund state.
Cell culture
GBM cells were plated on tissue culture flasks and cultured in DMEM medium (Gibco Life Technologies, Carlsbad, CA) supplemented with 10% fetal calf serum (Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin (100 U/ml; GIBCO) in a humidified incubator at 37°C and 5% CO2. To evaluate cell survival, cells were treated with graded concentrations of TMZ and survival was determined with the CyQuant cell proliferation kit purchased (Epicentre Biotechnologies, Madison, WI). Cell proliferation was assessed by direct cell counting using trypan blue or using CyQuant assay (Epicentre).
Western blot assay
Cells were lysed in a detergent-containing buffer (20 mM Tris HCl [pH. 7.4], 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 1% sodium deoxycholate, and 1 mM p-amidinophenyl methanesulfonyl fluoride hydrochloride) supplemented with a cocktail of protease inhibitors (Roche). The lysates were clarified by centrifugation, and protein concentration was determined by the Bio-Rad Protein Assay Reagent (Bio-Rad, Hercules, CA). Equal amounts of protein were diluted with SDS sample buffer (2% SDS, 25 mM Tris HCl [pH. 6.8], 5% 2-mercaptoethanol, and 10% glycerol), boiled for 5 minutes, and resolved by SDS-polyacrylamide gel electrophoresis. Separated proteins were electro-blotted onto a nitrocellulose membrane (Invitrogen). Nonspecific binding was blocked in 5% nonfat milk, 0.1% Tween-20, and 50 mM Tris [pH. 7.5]. All primary antibodies were incubated overnight at 4°C followed by 1 hour RT incubation with the secondary antibody, horseradish-peroxidase-conjugated anti-IgG. The membranes were developed according to the Pierce chemiluminescence protocol (Pierce, Rockford, IL). Membranes were stripped and reprobed using mouse anti-human β-Actin antibody to assess equivalent loading of samples.
Stable transfection of shRNA transfection
U87 glioma cells were plated on 6-well tissue culture plates at a density of 7 × 104 cells and incubated overnight. Cells were transfected with 4 μg of CD74-specific shRNA encoding pLKO-1 plasmid (Sigma Aldrich, St. Louis, MO) using Gene Porter® 2 transfection reagent according to the protocol supplied by the vendor (Genlantis, San Diego, CA). The transfection efficiency (75-80% - data not shown) was evaluated in a separate experiment using a green fluorescent protein (GFP) encoding plasmid. The transfected clones were selected using 2 μg/ml of puromycin (Invitrogen) and harvested by trypsinization within cloning rings. A non-specific targeting (NT) shRNA was transfected in parallel with CD74 shRNA for control experiments.
RNA isolation and Reverse transcription-Polymerase chain amplification (RT-PCR)
Total cellular RNA was isolated using RNeasy kit (Qiagen, Valencia, CA). RNA concentrations were determined by spectrophotometry at 260 nm and the quality of RNA was determined using Agilent Software (Agilent Technologies, Santa Clara, CA). RT-PCR was performed with 2 μg of isolated total RNA and synthesized to cDNA in a 20 μl reaction system using reverse transcriptase (Promega, Madison, WI). Reverse transcription conditions were 5 min denaturation at 70 °C, 60 min at 37°C and 5 min at 75 °C in a thermocycler (Perkin-Elmer). The following oligodeoxynucleotide primers used for PCR amplification were purchased from Integrated DNA Technologies (Coralville, IA); CD74: sense (5′-GACCTTATCTCCAACAATGAGCAAC-3′), and anti-sense (5′-AGCAGAGTCACCAGGATGGAA-3); MIF: sense (5′-GTGGACATCTTTGCTTTGGGCCTT-3′) and anti-sense (5′-TGTTCCTCCATTCAGCCAAGGTCT-3′) and β-actin: sense (CCAGAGATGGCCACGGCTGCT) and antisense (TCCTTCTGGATCCTGTCGGGA). PCR condition was 10 min denaturation at 95 °C, then 35 cycles of 30 sec at 94 °C, 30 sec at 55 °C and 1 min at 72 °C, and finally 10 min elongation at 72 °C in a thermocycler. PCR amplicons were analyzed by electrophoresis in a 2% agarose gel and visualized using UV fluorescence after staining with ethidium bromide. The β-actin mRNA was used as a loading control.
Quantitative RT -PCR (qRT-PCR)
The primers and probes used for quantitation of CD74 (catalogue no. 00269961_m1) and the GAPDH (catalogue no. 99999905_m1) endogenous control were purchased from ABI (assay-in-demand). The qRT-PCR was performed on the ABI prism 7900 (ABI) PCR and detection instrument. The qRT-PCR reactions were performed on plates using adhesive seals as covers. A master mix was prepared for target (CD74) and endogenous control (GAPDH) in a 20 μl reaction using a single-step RT-PCR reagents kit (ABI). The RT-PCR was programmed as follows: 42°C for 30 min, 95°C 10 min followed by 95 °C 15 s, 60 °C 1 min cycled 40 times. Each sample was amplified in triplicate. Control samples without template were included in all experiments. The relative levels of CD74 mRNA were determined using SDS RQ 1.3 software (ABI). For this analysis, GAPDH was used as endogenous control and CD74 mRNA expression level in U87 GBM cells was used as a calibrator.
Immunohistochemistry
Use of primary patient tissues was approved by the Mayo Institutional Review Board and patient consent to participate in research was obtained prior to staining and analysis. Tissue sections from 2 tissue micro-arrays (TMA) constructed from formalin fixed and paraffin embedded high grade glioma tissues were deparaffinized in xylene and dehydrated in alcohol. TMA sections were immersed in 0.01 mol/L citrate buffer (pH 6.0) and microwaved for 15 minutes for antigen retrieval followed by rinsing in phosphate-buffered saline (PBS). The endogenous peroxidase activity was quenched with methanol containing 0.3% hydrogen peroxide for 30 minutes at room temperature (RT). Nonspecific reactions were blocked by 3% bovine serum albumin (BSA) in PBS for 1 hour at RT. TMA sections were incubated with a primary CD74 antibody overnight at 4°C at a 1:100 dilution. Sections were incubated with biotinylated goat anti-mouse secondary antibodies for 1 hour at RT, followed by incubation for 30 minutes with the avidin-biotin peroxidase complex. The antigen-antibody binding was visualized with 3,3 diaminobenzidine/hydrogen peroxide followed by counterstaining with hematoxylin. Normal human tonsil was used as a positive control for CD74 expression. Analysis of the immunostaining for CD74 was performed by a neuropathologist, who was blinded to the samples. To assess the immunopositivity, all slides were viewed microscopically under low magnification and scored according to the intensity of immunoreaction as negative (−), mild (+), moderate (++) and strong (+++).
Statistical analysis
The differences in proliferation and TMZ sensitivity in relation to CD74 status was analyzed using a two-sample t-test (or two-sample rank sum, depending on the data distribution). Cumulative survival probabilities were estimated using the Kaplan-Meier method. The log rank test was used to compare survival of groups. Spearman’s correlation coefficient was used to assess the association of relative median survival prolongation in relation to CD74 expression. In all cases, two-sided p-values ≤0.05 were considered statistically significant.
Results
Preferential expression of CD74 by cells derived from TMZ resistance xenografts
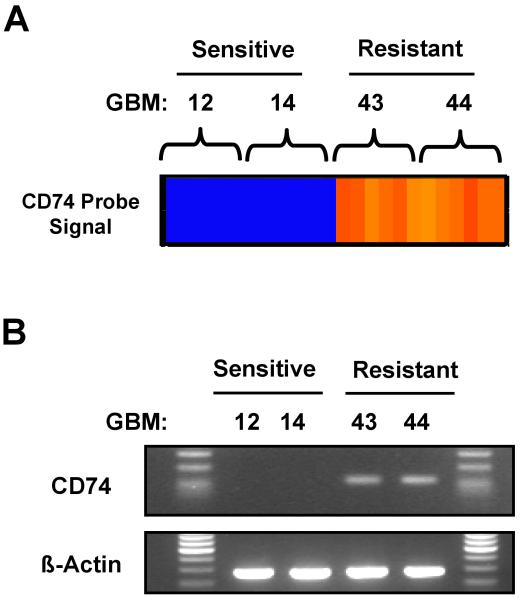
In previous studies, we have defined the TMZ sensitivity of a panel of primary GBM xenograft tumor lines [11], and as part of an effort aimed at discovery of genes associated with TMZ resistance, gene expression profiling was performed on short-term cell cultures derived from 2 TMZ sensitive xenograft lines (GBM12 and GBM14) and 2 TMZ resistant xenograft lines (GBM43 and GBM44). In a pooled analysis of gene expression levels for TMZ sensitive versus TMZ-resistant lines, a total of 120 genes were differentially expressed at a significance level of p < 0.001, of which the 50 genes with the greatest difference in expression are shown in Table 1. Interestingly, the most highly significant difference in expression was observed for the MHC II chaperone molecule CD74 (Fig. 1A). RT-PCR for CD-74 on these same 4 cell lines confirmed higher level CD74 mRNA expression in the TMZ resistant GBM43 and GBM44 lines as compared to the TMZ sensitive GBM12 and GBM14 tumor lines (Fig. 1B). While CD74 expression has not been described previously in GBM, these data suggested that CD74 may be an important modulator of TMZ response.
Table 1.
Top 50 genes preferentially overexpressed in short-term cultured cells from TMZ resistant GBM xenografts
| Rank | Cytoband | Gene Symbol |
Mean difference (log2) |
Ratio of means |
t-statistic | p-value |
|---|---|---|---|---|---|---|
| 1 | (5q32) | CD74 | 2.79663 | 6.94815 | 23.492 | < 0.0001 |
| 2 | (6p21.3) | HLA-DRB5 | 2.85072 | 7.21358 | 22.644 | < 0.0001 |
| 3 | (8p21) | NEFL | 1.17896 | 2.26413 | 20.054 | < 0.0001 |
| 4 | (15q13-q14) | SGNE1 | 1.27149 | 2.41411 | 19.233 | < 0.0001 |
| 5 | (6p21.2) | DAAM2 | 0.72751 | 1.65578 | 18.995 | < 0.0001 |
| 6 | (8p21) | NEFL | 1.13968 | 2.20333 | 17.16 | < 0.0001 |
| 7 | (6p21.3) | HLA-DRB1 | 3.52285 | 11.4943 | 16.573 | < 0.0001 |
| 8 | (7p12-p14) | STK17A | 0.9058 | 1.87358 | 16.047 | < 0.0001 |
| 9 | (6p21.3) | HLA-DRB1 | 3.01361 | 8.07584 | 15.383 | < 0.0001 |
| 10 | (Xq28) | L1CAM | 1.01344 | 2.01872 | 15.162 | < 0.0001 |
| 11 | (6p21.3) | IER3 | 2.28077 | 4.85938 | 13.452 | < 0.0001 |
| 12 | (6p21.3) | HLA-DQB1 | 0.9112 | 1.88061 | 13.054 | < 0.0001 |
| 13 | (1q24) | PRRX1 | 1.81618 | 3.52147 | 12.681 | < 0.0001 |
| 14 | (6p21.3) | HLA-DPB1 | 1.20809 | 2.31031 | 12.658 | < 0.0001 |
| 15 | (3p21-p14) | WNT5A | 0.73596 | 1.66551 | 12.586 | < 0.0001 |
| 16 | (6p21.3) | HLA-DRB4 | 1.98187 | 3.95004 | 12.402 | < 0.0001 |
| 17 | (6p21.3) | HLA-DRA | 3.08406 | 8.47999 | 12.29 | < 0.0001 |
| 18 | (6p21.3) | HLA-DRA | 3.54244 | 11.6515 | 12.252 | < 0.0001 |
| 19 | (2p14) | C2orf32 | 0.5908 | 1.50609 | 11.768 | < 0.0001 |
| 20 | (18p11.2) | PTPRM | 0.68664 | 1.60953 | 11.666 | < 0.0001 |
| 21 | (7q32-q35) | FLNC | 0.69961 | 1.62407 | 11.445 | < 0.0001 |
| 22 | (6p23) | RNF182 | 0.45141 | 1.36737 | 11.306 | < 0.0001 |
| 23 | (13q12.3) | LOC283537 | 0.3805 | 1.30179 | 11.24 | < 0.0001 |
| 24 | (6p21.3) | HLA-DPA1 | 1.9834 | 3.95425 | 11.131 | < 0.0001 |
| 25 | (5q34) | DUSP1 | 0.8521 | 1.80512 | 11.122 | < 0.0001 |
| 26 | (6p21.3) | HLA-DQB1 | 1.013 | 2.0181 | 10.946 | < 0.0001 |
| 27 | (5q32) | CD74 | 0.55819 | 1.47242 | 10.876 | < 0.0001 |
| 28 | (7q21-q31) | SEMA3C | 0.59447 | 1.50992 | 10.85 | < 0.0001 |
| 29 | (Xp22.1) | SAT | 2.11197 | 4.3228 | 10.75 | < 0.0001 |
| 30 | (1q24) | PRRX1 | 0.68108 | 1.60334 | 10.732 | < 0.0001 |
| 31 | (19q12) | ZNF537 | 0.45421 | 1.37003 | 10.653 | < 0.0001 |
| 32 | (2p22-p21) | LTBP1 | 0.58538 | 1.50044 | 10.433 | < 0.0001 |
| 33 | --- | NA | 0.95214 | 1.93474 | 10.408 | < 0.0001 |
| 34 | (6p21.3) | HLA-DQB1 | 0.70381 | 1.6288 | 10.23 | < 0.0001 |
| 35 | --- | NA | 0.9038 | 1.87099 | 10.136 | < 0.0001 |
| 36 | (16q11.2) | DNAJA2 | 0.37493 | 1.29678 | 10.07 | < 0.0001 |
| 37 | (6p21.3) | HLA-DPA1 | 3.05706 | 8.32275 | 10.028 | < 0.0001 |
| 38 | (7p12-p14) | STK17A | 1.04624 | 2.06514 | 9.91 | < 0.0001 |
| 39 | (6p21.3) | HLA-DQB1 | 0.32749 | 1.25482 | 9.839 | < 0.0001 |
| 40 | (6p21.3) | HLA-DQB1 | 0.61589 | 1.5325 | 9.794 | < 0.0001 |
| 41 | (17q25.3) | SYNGR2 | 0.46094 | 1.37643 | 9.712 | < 0.0001 |
| 42 | (11q13) | FOSL1 | 0.36663 | 1.28933 | 9.697 | < 0.0001 |
| 43 | (2q37.2) | SH3BP4 | 1.84982 | 3.60454 | 9.564 | < 0.0001 |
| 44 | (Xp22.1) | SAT | 1.39986 | 2.63876 | 9.539 | < 0.0001 |
| 45 | (4p16) | CTBP1 | 0.46968 | 1.38481 | 9.536 | < 0.0001 |
| 46 | (14q31) | NRXN3 | 0.81206 | 1.75572 | 9.394 | < 0.0001 |
| 47 | --- | ALS2CR4 | 0.33487 | 1.26127 | 9.351 | < 0.0001 |
| 48 | (10q26) | MGMT | 0.46494 | 1.38026 | 9.238 | < 0.0001 |
| 49 | (2p12) | POLE4 | 0.50419 | 1.41833 | 9.213 | < 0.0001 |
| 50 | (2p13) | TGFA | 0.80093 | 1.74222 | 9.193 | < 0.0001 |
Shown in bold are probes for CD74, which is further investigated in this study
Fig 1.
Expression of CD74 mRNA in 4 GBM xenograft lines relative to temozolomide sensitivity. A) The heat-map results of a microarray analysis of the CD74 probe set for the TMZ sensitive (GBM12 and GBM14) and TMZ resistant (GBM43 and GBM44) lines are shown. B) Total RNA from the same lines were reverse transcribed and PCR amplified using primers specific for human CD74 (upper panel). Beta-actin was used as internal control (lower panel)
Expression of CD74 in GBM xenografts and correlation with TMZ sensitivity
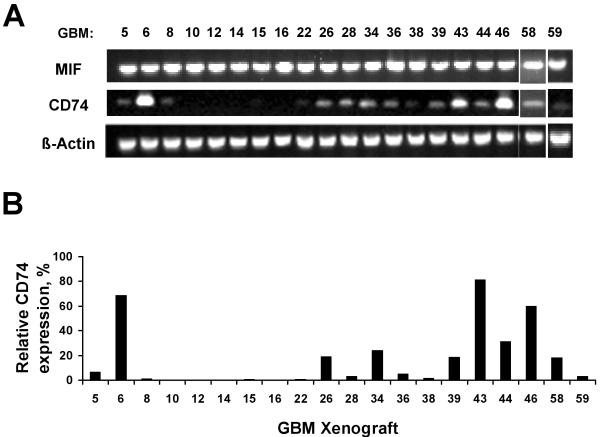
To follow up on the initial observation that CD74 expression may be related to TMZ sensitivity, the expression of CD74 and of MIF, the CD74 ligand, was evaluated in flank tumor tissue from 20 GBM xenograft lines in the Mayo GBM xenograft panel that have been characterized for TMZ response. These lines include the 4 xenograft lines from which the cell lines tested in our expression array study were derived. While robust MIF mRNA expression was observed in all xenograft lines, appreciable CD74 mRNA expression was observed only in a subset of tumors (8 of 20; 40%) (Fig.2A). CD74 mRNA levels were further quantitated by qRT-PCR, which showed similar distribution of CD74 expression in the GBM xenografts (Fig. 2B). Thus, CD74 expression was observed in approximately a third of the primary GBM xenografts.
Fig 2.
Expression of CD74 and MIF mRNA in a panel of GBM xenografts. A) Total RNA isolated for 20 different GBM xenograft lines was subject to RT-PCR using primers specific for CD74, MIF and beta-actin. B) Expression of CD74 mRNA in GBM xenografts was quantitated using qRT-PCR. The level of CD74 expression is calculated relative to control U87 GBM cells.
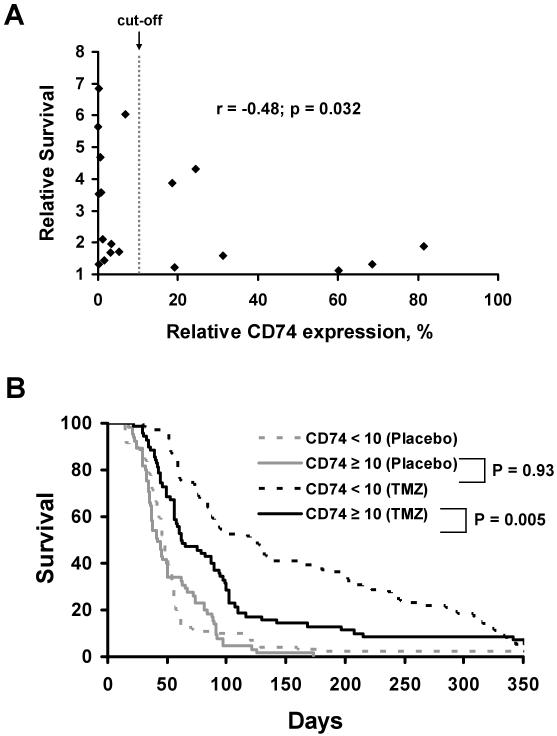
The in vivo TMZ sensitivity for each xenograft line has been previously evaluated in an orthotopic therapy evaluation model, and this allows for a direct comparison between CD74 expression levels and TMZ responsiveness in our panel of 20 GBM xenograft lines. In this previous study, mice with established intracranial tumors were randomized to therapy with placebo or TMZ (66 mg/kg/day x 5 days), and the ratio of median survival for TMZ-treated versus placebo-treated mice was used to define the survival benefit associated with TMZ treatment [11, 12]. These survival data were correlated with the qRT-PCR analysis of CD74 mRNA (Fig. 3A) and demonstrate an inverse relationship between CD74 mRNA expression levels and TMZ responsiveness (Spearman’s r = −0.48; p = 0.032); high CD74 expression was associated with a poor response to TMZ. Visual inspection of the qRT-PCR data suggests a cut-point for high versus low CD74 expression of approximately 10%, and based on this stratification, 12 of 20 (60%) xenografts had a CD74 score of < 10 and 8 (40%) with a CD74 expression score > 10. The survival benefit of TMZ treatment for xenografts with low CD74 expression was significantly greater (median: 4.68; range: 1.33 - 6.85) than those with high CD74 expression (median: 1.64; range: 1.11 - 4.31; p=0.03). TMZ survival determinations for each line were performed with approximately 10 mice each in the treatment and placebo groups for each tumor line. Therefore, in a second analysis, the survival data for all animals were pooled and the survival of mice implanted with tumor lines with high CD74 expression was compared to those with low CD74 expression in a Kaplan-Meier analysis (Fig. 3B). In this analysis, mice bearing xenograft lines with low CD74 expression (% expression <10; n = 105) had a significantly longer survival than those bearing tumor lines with high CD74 expression (% expression ≥ 10; n = 70) following TMZ therapy (median survival 122 vs. 62.5 days; Log rank test p = 0.005), while there was no association of CD74 expression level and survival of the placebo-treated mice (p=0.93). These findings suggest an inverse relationship between CD74 expression and TMZ sensitivity in GBM xenografts.
Fig 3.
CD74 expression relative to TMZ response in 20 GBM xenograft lines. A) Relative CD74 expression levels, determined from qRT-PCR, are plotted relative to the survival benefit for TMZ therapy in a panel of 20 GBM xenografts Survival benefit for each tumor line is defined as the ratio of median survival of TMZ treated versus placebo treated mice. B) Kaplan-Meier survival estimate curves associating CD74 mRNA expression with TMZ response for individual mice with intracranial GBM xenografts treated in the survival experiments.
Knock-down of CD74 expression by specific shRNA in U87 cells
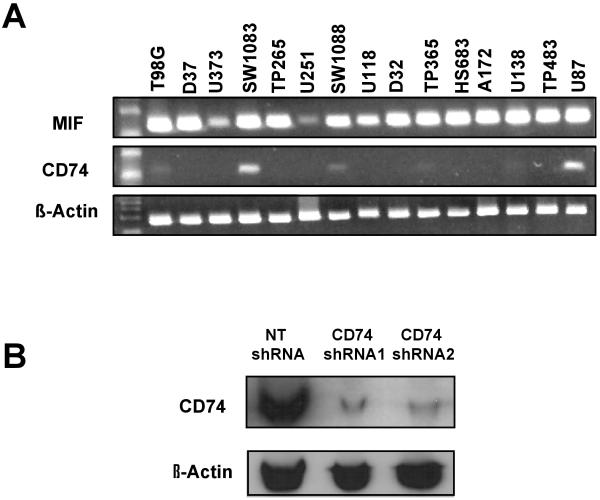
CD74 mRNA expression levels were assessed in a panel of conventional GBM cell lines using RT-PCR in order to identify GBM lines in which CD74 expression levels subsequently could be manipulated by shRNA. While the CD74 ligand MIF was overexpressed in all GBM cell lines tested, 6 of the 15 lines (40%) had detectable expression of CD74, and only U87 and SW1083 had robust CD74 expression (Fig. 4A). Based on this observation, U87 cells were selected to evaluate the influence of shRNA-mediated knock-down of CD74. Using a plasmid expression vector, stable clones expressing either a CD74 shRNA construct or a non-specific target (NT) shRNA were isolated and used for subsequent experiments. As seen in Fig. 4B, expression of CD74 protein, was significantly suppressed in 2 independent CD74 shRNA-expressing clones, while CD74 was unaffected in a NT shRNA-expressing clone. These 2 CD74 and 1 NT shRNA clones are used in the subsequent analyses described below.
Fig 4.
Expression of CD74 in established GBM cell lines and effective targeting of CD74 expression by shRNA. A) RT-PCR of CD74 and MIF expression in established GBM cell lines, relative to beta-actin control. B) Western blotting results for CD74 and beta-actin from U87 clones stably transfected with non-specific shRNA (NT shRNA) or CD74 shRNA (2 independent clones).
CD74-specific shRNA inhibits proliferation and enhances TMZ sensitivity
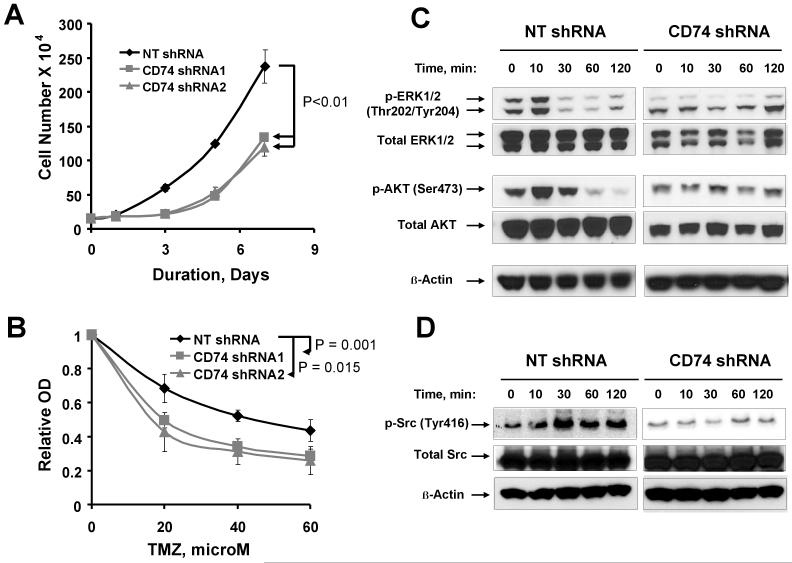
The effects of CD74 shRNA knockdown on cell growth and survival were assessed by counting using trypan blue. For both CD74 shRNA clones, cell proliferation was significantly blunted such that 7 days after plating the average number of cells was 133 × 104 and 119 × 104 as compared to 237 × 104 for NT-shRNA control (Fig. 5A; p < 0.01). Similar results were obtained using a calorimetric assay (data not shown). To determine whether CD74 expression promotes cell survival in GBM cells, the effect of CD74 silencing on the TMZ sensitivity was evaluated in U87 cells. Cells were treated with graded concentrations of TMZ and survival was determined using the CyQuant assay. As shown in Fig. 5B, U87 CD74 shRNA expressing cells were significantly more sensitive to TMZ than the NT shRNA control: 60 μM TMZ treatment of NT shRNA U87 was associated a 56 ± 5% reduction in absorbance (cell number) versus 72 ± 3% and 74 ± 4% for the two CD74 shRNA U87 clones, respectively (p<0.05 for either clone relative to NT shRNA clone). Together, these findings demonstrate that CD74 knockdown is associated with reduced proliferation and increased sensitivity to TMZ in U87 cells.
Fig 5.
Effect of silencing CD74 expression on proliferation and TMZ sensitivity in U87 GBM cells. U87 clones expressing CD74 shRNA or control non-specific targeting (NT) shRNA were evaluated by direct counting using trypan blue cell. A, Average cell number at each time-point is shown over the course of 7 day incubation. The error bars represent the standard error of the mean for two independent studies performed with triplicate samples. P-values shown are for comparing differences in proliferation at day 7. B, the effects of graded doses of TMZ on survival of the indicated U87 clones following 7 day incubation. Error bars represent the standard error of the mean for data generated from 4 independent experiments. P-values are shown for comparing control vs. CD74 shRNA transfected clones at a 60 μM concentration of TMZ. C - D, MIF induced activation of ERK1/2, AKT and Src in U87 cells is mediated by CD74. NT shRNA and CD74 shRNA U87 clones were serum deprived for 24 hours, stimulated with MIF (25 ng/ml) for the indicated duration prior to processing for western blotting. Membranes were probed for the indicated phospho-proteins and total proteins. Results are representative of 2 independent experiments.
CD74 is a mediator of MIF induced phosphorylation of MAPK and AKT
The effects of CD74 knock-down on MAPK and AKT signaling were evaluated in the U87 model to gain further insight into how CD74 shRNA might influence cell proliferation and survival. As shown in Fig. 5C, treatment of serum-starved cells with recombinant MIF induced robust phosphorylation of both ERK1/2 and AKT in U87 cells expressing the non-specific NT-shRNA control vector (left panel). In contrast, MIF treatment had no effect on the induction of ERK1/2 or AKT phosphorylation in U87 cells expressing CD74 shRNA (right panel). MIF treatment had no effect on the expression of total ERK1/2 or AKT in either cell line. In parallel with ERK1/2 and AKT activation, recombinant MIF induced phosphorylation of Src only in cells expressing the non-specific NT-shRNA (Fig. 5D). These findings suggest that CD74 signaling via MIF activation may provide important mitogenic and survival signals through the Src, MAPK and Akt signaling pathways.
Expression of CD74 in high grade glioma patient samples
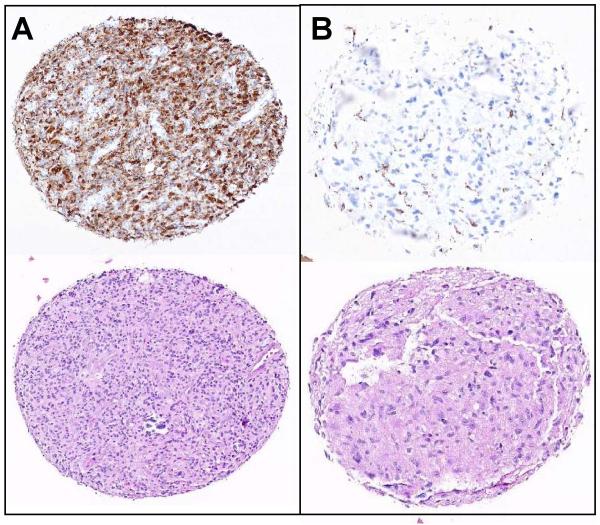
Because of our above xenograft results and since the expression of CD74 has not been previously characterized in GBM, the expression of CD74 first was evaluated by immunohistochemistry in 71 human samples of high grade gliomas. The studied cases included 44 males and 27 females; 9 cases were grade 3 (anaplastic astrocytoma, AA) and 62 cases were grade 4 (glioblastoma multiforme, GBM). Median patient age was 57 years (range 23-83 years). The cases were arrayed into a tissue micro-array, and representative CD74 staining results and corresponding H&E sections are shown in Fig. 6: CD74 positive (Fig. 6A, upper panel, left) and CD74 negative (Fig. 6B, upper panel, right). The scant CD74 staining in CD74-negative cases results from staining of intratumoral microglia and macrophages, which are known to express CD74. In contrast, CD74 was rarely expressed by the normal or reactive astrocytes. CD74 protein was expressed in 19 of 62 (31%) GBM cases, but only 1 of 9 (11%) AA cases (Table 2). This difference was not statistically significant (p=0.43). Of the positive GBM cases, 11 tumors had mild CD74 staining intensity, while the other 8 positive cases had moderate to strong staining. The survival data was available for 55 (77%) of 71 patient samples evaluated for CD74 expression. Because most these patients (38 (69%) out of 55) were not treated with TMZ, univariate analysis revealed no significant association between CD74 expression and overall survival of the studied cohort of high grade patients. In contrast, analysis of 17 patients who received TMZ therapy revealed a trend to shorter overall survival in patients expressing CD74 but the difference was not significantly (p=0.142; data not shown). Together, these results demonstrate that overexpression of CD74 protein is observed in a subset of high grade gliomas and establishment of its role as a predictor of TMZ response is an interesting subject for investigation in a larger cohort of TMZ treated GBM patients.
Fig 6.
Immunohistochemical expression of CD74 in GBM patient samples. A) Displays a representative case with strong CD74 expression with the corresponding H&E stain. B) Shows a representative CD74 negative case with the corresponding H&E stain. Specific CD74 staining of tumor cells is seen.
Table 2.
Extent of CD74 immunostaining in high grade gliomas
| CD74 Expression |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor grade | − | (%) | + | (%) | ++ | (%) | +++ | (%) | Total |
| Grade 3 | 8 | (89) | 0 | (0) | 1 | (11) | 0 | (0) | 9 |
| Grade 4 | 43 | (69) | 11 | (18) | 5 | (8) | 3 | (4) | 62 |
|
| |||||||||
| Total | 51 | (72) | 11 | (16) | 6 | (8) | 3 | (4) | 71 |
Discussion
Development of chemoresistance, particularly against TMZ is a common event in GBM that limits the efficacy of therapy. Several studies have demonstrated that MGMT expression is mechanistically linked to TMZ resistance [13, 14], and other studies suggest that suppression of MGMT transcription by promoter hypermethylation is associated with a favorable outcome following TMZ therapy [13, 15, 16]. However, not all tumors lacking MGMT protein or with MGMT promoter hypermethylation benefit from TMZ therapy [13], which supports the idea that TMZ resistance may be multi-factorial. The current study demonstrates over-expression of CD74 in a subset of GBM tumors and suggests that high-level CD74 expression may be an important factor that can contribute to TMZ resistance in addition to MGMT expression.
CD74 is an MHC class II associated invariant chain molecule expressed on the cell surface in a sub-set of immune cells, but expression in GBM tumors has not been previously described. Consistent with a role in malignant transformation of specific immune lineages, CD74 is highly overexpressed in the majority of B-cell neoplasms and multiple myeloma [17, 18], and function-inactivating CD74 antibodies have demonstrated anti-tumor activity in animal models for these indications [19-22]. CD74 overexpression also has been observed in a subset of thymic, gastric, renal, non-small cell lung and breast cancers [5, 7, 9, 23, 24]. Interestingly, expression of CD74 in some of these solid malignancies has been associated with more aggressive tumor behavior [7]. In the current study, CD74 expression was observed in 19 of 62 (31%) of GBM patient tumor samples but only 1 of 9 (11%) cases of grade 3 anaplastic astrocytoma (AA) (p-value = 0.429). While the difference in CD74 expression between AA and GBM was not statistically significant (perhaps due to low statistical power), the trend towards higher expression in GBM is consistent with the idea that CD74 expression may be linked to more aggressive behavior within the spectrum of malignant gliomas.
Our study raises the possibility that CD74 expression may contribute to TMZ resistance in GBM. In previous studies, we demonstrated that TMZ sensitivity, but not radiation sensitivity, in the Mayo GBM xenograft panel is significantly associated with MGMT promoter hypermethylation [11, 12], which is consistent with previous clinical data [13]. In the current study, analysis of this same in vivo response data from 20 primary GBM xenograft lines demonstrated a significant association between CD74 expression and TMZ resistance. Although CD74 did not remain a significant predictor of response in a multivariate analysis that included MGMT methylation status as an adjustable variable, shRNA knockdown of CD74 in the MGMT methylated U87 glioma cell line significantly sensitized the cells to TMZ. Collectively, these latter data suggest that CD74 signaling may be an additional factor that contributes to TMZ resistance in addition to the more dominant effects of MGMT activity.
CD74 signaling likely promotes cell survival through downstream signaling to pro-survival pathways. CD74 forms a complex with CD44 at the cell surface, and binding of the MIF ligand to this complex leads to recruitment of Src, which then can signal downstream to activate multiple signaling pathways including Ras/MAPK and Akt [2, 4, 25, 26]. These pathways are important modulators of cell proliferation and survival in GBM [27], and consistent with previous reports, MIF-induced phosphorylation of MAPK and AKT in U87 cells was selectively suppressed in those clones expressing CD74 shRNA (Fig. 5). MIF is highly expressed in the majority of GBM tumors (Fig. 2) [28, 29], and given the importance of MAPK and AKT signaling for cell survival, these data are consistent with the possibility that MIF-CD74 signaling through these pathways may contribute to the TMZ resistance associated with CD74 overexpression. On the basis of these data, future studies in our laboratory will evaluate whether CD74 confers resistance to other therapeutics with different mechanisms of action, and whether small molecule or monoclonal antibody inhibitors of this pathway can be used to potentiate the efficacy of TMZ in GBM.
Acknowledgments
This work was supported by NIH grant to J.N.S. (RO1 CA127716, P50 CA 108961) and Mayo Brain SPORE Career Development award to G.J.K. (P50 CA 108961).
References
- 1.Stumptner-Cuvelette PBP. Multiple roles of the invariant chain in MHC class II function. Biochim Biophys Acta. 2002;1542:1–13. doi: 10.1016/s0167-4889(01)00166-5. [DOI] [PubMed] [Google Scholar]
- 2.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starlets D, Gore Y, Binsky I, Haran M, Harpaz N, Shvidel L, Becker-Herman S, Berrebi A, Shachar I. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. 2006;107:4807–4816. doi: 10.1182/blood-2005-11-4334. [DOI] [PubMed] [Google Scholar]
- 4.Lue H, Thiele M, Franz J, Dahl E, Speckgens S, Leng L, Fingerle-Rowson G, Bucala R, Luscher B, Bernhagen J. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26:5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]
- 5.Datta MW, Shahsafaei A, Nadler LM, Freeman GJ, Dorfman DM. Expression of MHC class II-associated invariant chain (Ii;CD74) in thymic epithelial neoplasms. Appl Immunnohistochem Mol Morphol. 2000;8:210–215. doi: 10.1097/00129039-200009000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Siegler KL, Leifheit EC, Vera PL. Inhibition of macrophage migration inhibitory factor decreases proliferation and cytokine expression in bladder cancer cells. BMC Cancer. 2004;4:34. doi: 10.1186/1471-2407-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Iwashige H, Aridome K, Hokita S, Aikou T. Invariant chain expression in gastric cancer. Cancer Lett. 2001;168:87–97. doi: 10.1016/s0304-3835(01)00503-1. [DOI] [PubMed] [Google Scholar]
- 8.Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639–1651. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioachim HL, Pambuccian SE, Hekimgil M, Giancotti FR, Dorsett BH. Lymphoid monoclonal antibodies reactive with lung tumors. Diagnostic applications. Am J Surg Pathol. 1996;20:64–71. doi: 10.1097/00000478-199601000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, Schroeder MA, James CD. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-Oncology. 2005;7:164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitange GJ, Carlson BL, Mladek AC, Decker PA, Schroeder MA, Wu W, Grogan PT, Giannini C, Ballman KV, Buckner JC, James CD, Sarkaria JN. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009;92:23–31. doi: 10.1007/s11060-008-9737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson BL, Grogan PT, Mladek AC, Schroeder MA, Kitange GJ, Decker PA, Giannini C, Wu W, Ballman KA, James CD, Sarkaria JN. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int J Radiat Oncol Biol Phys. 2009;75:212–219. doi: 10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. see comment. [DOI] [PubMed] [Google Scholar]
- 14.Donson AM, Addo-Yobo SO, Handler MH, Gore L, Foreman NK. MGMT promoter methylation correlates with survival benefit and sensitivity to temozolomide in pediatric glioblastoma. Pediatric Blood & Cancer. 2007;48:403–407. doi: 10.1002/pbc.20803. [DOI] [PubMed] [Google Scholar]
- 15.Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, Bertorelle R, Bartolini S, Calbucci F, Andreoli A, Frezza G, Leonardi M, Spagnolli F, Ermani M. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26:2192–2197. doi: 10.1200/JCO.2007.14.8163. [DOI] [PubMed] [Google Scholar]
- 16.Ishii D, Natsume A, Wakabayashi T, Hatano H, Asano Y, Takeuchi H, Shimato S, Ito M, Fujii M, Yoshida J. Efficacy of temozolomide is correlated with 1p loss and methylation of the deoxyribonucleic acid repair gene MGMT in malignant gliomas. Neurol Med Chir (Tokyo) 2007;47:341–349. doi: 10.2176/nmc.47.341. discussion 350. [DOI] [PubMed] [Google Scholar]
- 17.Burton JD, Ely S, Reddy PK, Stein R, Gold DV, Cardillo TM, Goldenberg DM. CD74 is expressed by multiple myeloma and is a promising target for therapy. Clin Cancer Res. 2004;10:6606–6611. doi: 10.1158/1078-0432.CCR-04-0182. [DOI] [PubMed] [Google Scholar]
- 18.Shih L, Ong GL, Burton J, Mishina D, Goldenberg DM, Mattes MJ. Localization of an antibody to CD74 (MHC class II invariant chain) to human B cell lymphoma xenografts in nude mice. Cancer Immunol Immunother. 2000;49:208–216. doi: 10.1007/s002620000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mark T, Martin P, Leonard JP, Niesvizky R. Milatuzumab: a promising new agent for the treatment of lymphoid malignancies. Expert Opin Investig Drugs. 2009;18:99–104. doi: 10.1517/13543780802636162. [DOI] [PubMed] [Google Scholar]
- 20.Stein R, Mattes MJ, Cardillo TM, Hansen HJ, Chang CH, Burton J, Govindan S, Goldenberg DM. CD74: a new candidate target for the immunotherapy of B-cell neoplasms. Clin Cancer Res. 2007;13:5556s–5563s. doi: 10.1158/1078-0432.CCR-07-1167. [DOI] [PubMed] [Google Scholar]
- 21.Chang CH, Sapra P, Vanama SS, Hansen HJ, Horak ID, Goldenberg DM. Effective therapy of human lymphoma xenografts with a novel recombinant ribonuclease/anti-CD74 humanized IgG4 antibody immunotoxin. Blood. 2005;106:4308–4314. doi: 10.1182/blood-2005-03-1033. [DOI] [PubMed] [Google Scholar]
- 22.Stein R, Qu Z, Cardillo TM, Chen S, Rosario A, Horak ID, Hansen HJ, Goldenberg DM. Antiproliferative activity of a humanized anti-CD74 monoclonal antibody, hLL1, on B-cell malignancies. Blood. 2004;104:3705–3711. doi: 10.1182/blood-2004-03-0890. [DOI] [PubMed] [Google Scholar]
- 23.Mandal S, Curtis L, Pind M, Murphy LC, Watson PH. S100A7 (psoriasin) influences immune response genes in human breast cancer. Exp Cell Res. 2007;313:3016–3025. doi: 10.1016/j.yexcr.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, Schnitt S, Gabrielson E, Gelman R, Polyak K. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- 25.Lue H, Kapurniotu A, Fingerle-Rowson G, Roger T, Leng L, Thiele M, Calandra T, Bucala R, Bernhagen J. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal. 2006;18:688–703. doi: 10.1016/j.cellsig.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 28.Bacher M, Schrader J, Thompson N, Kuschela K, Gemsa D, Waeber G, Schlegel J. Up-regulation of macrophage migration inhibitory factor gene and protein expression in glial tumor cells during hypoxic and hypoglycemic stress indicates a critical role for angiogenesis in glioblastoma multiforme. Am J Pathol. 2003;162:11–17. doi: 10.1016/S0002-9440(10)63793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munaut CBJ, Foidart JM, Deprez M. Macrophage migration inhibitory factor (MIF) expression in human glioblastomas correlates with vascular endothelial growth factor (VEGF) expression. Neuropathol Appl Neurobiol. 2002;28:452–460. doi: 10.1046/j.1365-2990.2002.00416.x. [DOI] [PubMed] [Google Scholar]