Summary
Background
Medicine and biomedical sciences have become data-intensive fields, which, at the same time, enable the application of data-driven approaches and require sophisticated data analysis and data mining methods. Biomedical informatics provides a proper interdisciplinary context to integrate data and knowledge when processing available information, with the aim of giving effective decision-making support in clinics and translational research.
Objectives
To reflect on different perspectives related to the role of data analysis and data mining in biomedical informatics.
Methods
On the occasion of the 50th year of Methods of Information in Medicine a symposium was organized, that reflected on opportunities, challenges and priorities of organizing, representing and analysing data, information and knowledge in biomedicine and health care. The contributions of experts with a variety of backgrounds in the area of biomedical data analysis have been collected as one outcome of this symposium, in order to provide a broad, though coherent, overview of some of the most interesting aspects of the field.
Results
The paper presents sections on data accumulation and data-driven approaches in medical informatics, data and knowledge integration, statistical issues for the evaluation of data mining models, translational bioinformatics and bioinformatics aspects of genetic epidemiology.
Conclusions
Biomedical informatics represents a natural framework to properly and effectively apply data analysis and data mining methods in a decision-making context. In the future, it will be necessary to preserve the inclusive nature of the field and to foster an increasing sharing of data and methods between researchers.
Keywords: Biomedical informatics, data mining, data analysis, data-driven methods, translational bioinformatics
Introduction
The current era can be considered as a golden age for Biomedical Informatics (BI) [1,2,3]. After the early days of enthusiasm followed by a period of disillusion, [4,5], BI has now matured to the level of being an essential component of health care activities and biomedical research [6,7]. On the one hand, health care institutions are leveraging hospital information systems, which are a crucial asset of these complex organizations, and increasing investments of the public and private sectors show that medical and health informatics have become a solid field [7]. On the other hand, the ‘–omics’ data explosion and the need for translating research results in clinical practice have boosted the activities of BI, which is now an irreplaceable component of molecular medicine [8].
On the occasion of the 50th year of Methods of Information in Medicine a scientific symposium was organized, which took place in Heidelberg, Germany from June 9 to 11, 2011. A select number of distinguished colleagues from around the world gathered in Heidelberg to participate in the symposium which had as its theme: ‘Biomedical Informatics: Confluence of Multiple Disciplines’, reflecting on opportunities, challenges and priorities of organizing, representing and analysing data, information and knowledge in biomedicine and health care.
As one outcome of this symposium, the contributions of experts with different backgrounds in the area of biomedical data analysis have been collected in this paper, in order to provide a broad, though coherent, overview of some of the most interesting aspects of the field.
Looking at the current scenario of biomedical sciences, it is easy to notice that, now more than ever, data analysis and information processing have become basic and crucial components of the day-to-day activities of researchers, scientists, clinicians, nurses and decision-makers. It is not surprising, therefore, that on the occasion of its 50th anniversary, Methods of Information in Medicine proposes a careful reflection on the current perspectives of the role of data analysis and data mining in BI.
Rather interestingly, since its beginnings, data-driven approaches and data mining methods have been sources of controversies. First of all, the transformation of biology and medicine into a “data-intensive” field has provided validity to experiments, in which the goal was to gather data to generate new unbiased knowledge [9]. The risk of false discoveries, however, has raised skepticism and several studies have partially lower the initial enthusiasms [10]. Second, there still exists an unresolved tension between data miners, who agnostically use methods from computer science, signal processing, optimization and statistics, and data analysts, who mainly ground their approaches in well-established statistical theory and tools.
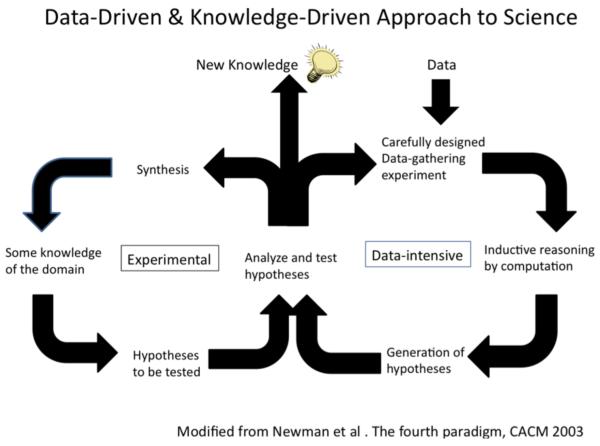
Being at the intersection of many disciplines, BI represents the natural space for reconciling in a coherent scenario different paradigms and perspectives for the benefit of scientific progress (See Figure 1). The basic dilemma between empiricism and rationalism, underlying many of the above mentioned disputes, is resolved in BI following a pragmatic strategy, aimed at solving problems in the best possible way, given the current status of knowledge and taking into account technological constraints and limitations [11,12]. Moreover, the availability of knowledge repositories in electronic format so strongly empowers biomedical research that data analysis and knowledge generation steps are now part of a unique, continuous cycle [13].
Figure 1.
Data-driven and knowledge-driven approaches will co-operate and make rapid progress in biomedical science [8].
The first two sections of the paper deepen the insight on this crucial theme. The role of data-driven approaches and the integration of data and knowledge in BI are analyzed and future challenges outlined.
As science is progressing, data mining and statistical approaches are no longer seen as alternative ways of dealing with data analysis problems. On the contrary, they are beginning to be seen as fully complementary. One of the aspects of such relationships is the ability to evaluate predictive models, such as classification or regression, on the basis of sound strategies. The third section of the paper describes a number of suitable methods to be applied for assessing the performance of learning “machines” grounded in confidence intervals theory.
Certainly, one of the main engines of the data-driven revolution in biomedicine has been high-throughput –omics biotechnology and the related bioinformatics needs. The natural conjunction of genomic medicine, bioinformatics and medical informatics is represented by translational bioinformatics, which bridges the different fields into a unique, purposive, discipline aimed at exploiting research results in clinical practice. The fourth and fifth sections of the paper describe the data analysis aspects of this field, with a particular focus on information integration and genetic epidemiology.
The paper ends stressing two main issues: i) the potential enabling role that BI may have to provide open-access information to clinical data; ii) the need for keeping the BI field open to diverse methodological contributions that will strengthen its innovation capabilities.
Data Accumulation and Data-Driven Approaches across Biomedical Informatics
One of the most outstanding features of all electronic information, especially in biomedicine, is its integrity and expandability. In the last two decades, data and knowledge have been rapidly accumulated in each subdiscipline of biomedical informatics. This wealth of information is now about to change the circumstances and methods of investigation in every field. In clinical medicine, not only the rapid progress of the technology of modern medicine, but also the progress of computational science has contributed to dramatic medical advances. For example, electronic medical literature can be easily searched from PubMed on the Internet. Thus PubMed enables scientists and medical doctors to obtain the most up-to-date knowledge relevant to their work quickly and easily, thereby expediting the progress of medicine. Also, electronic documentation systems have made it possible to collect a very high volume of active patient data relevant to what was impossible in the paper-based era.
Once these data have been collected, we can create a data warehouse and retrieve special case data or apply data mining to find hidden facts and rules [14] Electronic discharge summaries are now being collected and can be reused to retrieve similar cases by using text mining [15]. Laboratory data have been recorded for as long as one’s entire life and can be integrated from several facilities and made ready to be analyzed for disease management. In addition, as the volume of data increases beyond medical facilities, it becomes more important for databases to be re-utilized. Regional healthcare information systems can provide more data than one medical facility and electronic health records (EHRs) can further be expanded to a national or global scale [16]. For example, all billing data for every month, which includes main disease names, types and times of laboratory tests, and names and doses of drugs administrated or injected in hospitals, can be electronically collected from all medical facilities in Korea and Japan. Even only having this information, we can analyze the nationwide tendency of clinical practices for a disease [17]. Now, because EHRs have evolved, all the events affecting a person’s health can be collected electronically throughout one’s lifetime, which can be considered as a personal health record (PHR), or a personal life record (PLR). In a PHR, not only medical data but also health data are included, such as blood pressure and body weight measurements taken at a health club, or the list of lunch items consumed in a company cafeteria. Thus, a PHR can include a complete personal history about health. The same or even a more extreme phenomenon of huge data accumulation is occurring in genetic research. This research includes genome and sequence analysis, microarray data or genetic expression data analysis, high-throughput genome mapping, gene regulation, protein structure prediction and classification, and disease classification, for which informatics plays a very important role [18]. In this rapidly emerging field, an extensive amount of knowledge has also been accumulated in many genomic databases for reuse by many researchers seeking to discover new relations by using the techniques of informatics.
Going forward, active research will increase between the individual disciplines to pursue the final goal of biomedical informatics; in other words, the confluence of disciplines will lead to the discovery of new relations beyond the limits of individual disciplines. Connecting one’s PHR, which records extrinsic and environmental factors affecting a person’s health, and outcome, with one’s complete DNA sequences that identify intrinsic factors is the final destination. To accomplish this, many steps and phases must be carried out, because we cannot connect genomic and clinical information so easily. We must develop specific tools to complete the steps and phases one by one. Translational research is a field with a goal to integrate biology and clinical medicine in order to bridge the gap between basic medical research and clinical care [8]. Consequently, translational research provides a vast and challenging field for biomedical informatics researchers [19].
The traditional approach in biomedical science has been knowledge-driven and aimed at generating hypotheses from domain knowledge in a top-down fashion. Instead, we are about to enter the data-intensive science era in which hypotheses are generated automatically among the enormous amount of data available by using computational science with inductive reasoning [20]. These two approaches are not conflicting, but they can be combined or integrated to discover new knowledge [21]. Thus biomedical informaticians will play a significant role in developing new methods in the field of data mining and machine learning that will then be available to domain experts.
Another role of biomedical informaticians will be as supervisors and administrators of biomedical data management. In this broad map covering the entire biomedical field, a new discipline is needed to comprehensively oversee all steps of biomedical informatics, from the micro- to the macro-level of information, and to identify which parts are unknown, which limiting factors remain to be solved, and which areas need to be linked to other areas. These administrators are different from domain experts and must fulfill their tasks to accelerate the progress of all biomedical science. As the current disciplines of biomedical informatics interconnect, new roles for biomedical and computer scientists will emerge.
Knowledge and Data Integration
As mentioned in the previous section, an increasing flood of data in electronic format nowadays characterizes a variety of human activities, including health care and biomedical research. For this reason, fifteen years after their rise, the fields of data mining and knowledge discovery in databases are still topical and represent a crucial sector of biomedical informatics [22,23]. Certainly, over the last few years, the very nature of the collected data has changed as have data mining methods and tools. Data are available in a variety of formats, including not only numeric or codified values, signals and images, but also textual reports and summaries, multivariate time series and data streams, event logs, mobility information, social networks and interaction databases [24,25]. As a consequence, a noteworthy effort has been devoted to designing and applying a number of recent technologies, such as text mining [26,27], temporal data mining [28], workflow mining [29], and networks analysis [30]. Within biomedical data mining, one of the most interesting aspects is the exploitation of domain knowledge and the integration of different data sources in the data analysis process. As a matter of fact, data analysis is strongly empowered by the knowledge available in electronic format, which can either be already formalized, say through ontology and annotation repositories, or still informal but novel, as, for example, reported in Pubmed abstracts and papers [31].
The integration of data and knowledge is being crucially stimulated by bioinformatics applications, where the joint availability of publicly available databases, annotation systems and biomedical ontologies, has given rise to the field called “integrative bioinformatics” [32].
Rather interestingly, also in medical informatics and computer science, attention has been devoted to this problem since the late nineties. Intelligent data analysis (IDA) is a research field that refers to all methods devoted to (automatically) transform data into information by exploiting the available domain knowledge. IDA and data mining have been the focus of one of the working groups of the International Medical Informatics Association since 2000 [33]. The IDA and Data Mining IMIA working group have resulted in a variety of interesting results, papers and research projects (31, 34-36).
The “natural” step forward is to build on the results obtained so far to define new methodologies able to merge data exploration, visual analytics and data mining with inductive reasoning, as also underlined in the previous section. Efforts towards the combination of reasoning approaches with data analysis have been recently published [37, 38], as have very interesting software products, including open source frameworks [39]. IDA and reasoning require different disciplines to converge. Knowledge representation, automated reasoning, statistical and mathematical methods, new algorithms, efficient and modern IT technologies, advanced interfaces based on cognitive science need to be properly integrated in this context [38]. Novel IT systems empowered by IDA tools hold the promise of leveraging biomedical research and clinical decision making.
While some of the IDA and data mining methods are now ripe and ready to be used in clinical practice, such as for example classification, regression and clustering models, other instruments still need to be more widely studied and applied [35]. Temporal reasoning and data mining represent one such interesting area, which deserves an increasing level of attention and further research. Dealing with time is a crucial and challenging problem that is widespread in biomedical applications [40,41]. Even if a variety of methods is available to deal with biomedical signals and time-varying data, none of these tools is able alone to cope with the inherent complexity of temporal information and temporal reasoning. For example, data are very often irregularly collected due to an uneven schedule of measurements and visits, which may be dependent on the organizational settings or the severity of the disease. Moreover, the interpretation of temporal data is highly context-sensitive, so that the same pattern of the same variable may assume a different meaning in different clinical problems, say in the ICU or during home monitoring. Temporal reasoning and data mining are attempting to work together to solve such a difficult task through the so-called Temporal Data Mining (TDM) [42-44] field. The main goal of TDM is to extract relevant patterns from data: a temporal pattern is thus a sequence of events that is (clinically) important in a particular problem.
Rather interestingly, TDM methods have been designed to deal with different temporal data types, including time series of physiological variables, such as arterial pressure or blood glucose levels, and sequences of clinical events, such as hospital admissions and discharges, or drug prescriptions. These methods are therefore well suited to integrate information from a variety of data sets, including clinical records, monitoring devices, and large warehouses of administrative records. The IDA IMIA working group has worked extensively in this context, proposing methods able to deal with the extraction of temporal patterns from time series data and to synthesize temporal information into temporal features [45]. Such methods strongly depend on the knowledge available about the domain, and therefore, their application requires the integration of signal processing, algorithm design, knowledge representation and formalization.
The broad coverage of biomedical informatics, which spans from the molecular to the population level, is a tremendous enabler for the cross-fertilization of its different converging disciplines. Looking again at the temporal processes domain, we can easily note that different problems can be studied with similar approaches. For example, the so-called “workflow” modeling approach can be used to model care-flow processes [46], but it can be conveniently applied also to describe and analyze the complex intertwined processes underlying molecular studies [47]. For this reason, the algorithms able to automatically analyze process data (event logs) seem to have a wide potential application in all areas covered by Biomedical Informatics [48].
Together with advances in data mining algorithms, over the last few years there has been a great growth in the number and sophistication of data warehouses and integrated data repositories. Looking at recent developments, one of the most exciting advances is represented by the implementation of complex IT infrastructures designed to support clinical and biological research. As a matter of fact, the NIH-funded i2b2 research center [49], as well as the EHR4CR project [50,51], funded by the EU-IMI initiative, have shown that it is now possible to profoundly innovate biomedical research relying on newly designed IT systems. In a nutshell, the challenge is to create IT infrastructures able to support research by providing access to data collected in a data warehouse, which is populated from different data sources, including hospital and laboratory information systems, biobanks and the variety of small databases collected for single research studies. If properly implemented, such types of infrastructure can be a great accelerator for the entire research process [52]. However, this integration poses several challenges, in terms of data and knowledge representation, standard interfaces between software systems, data access, security and privacy policies, user interfaces and data querying functionality [8]. Moreover, in order to make such systems really effective in day-to-day activities, it is necessary to implement data analysis methods to help researchers in scientific discovery and health care providers in clinical decision-making [37]. Finally, natural language processing tools should be included in order to improve data gathering from textual documents and to summarize the knowledge available in the bibliome [27]. Such an ambitious project needs the confluence of many disciplines underlying biomedical informatics, ranging from IT systems design, data base management, and software interoperability, to ontology and terminology handling, data and text mining, human-computer interfaces and finally, experts in research and clinical processes [23]. Once available, these infrastructures will clearly show that biomedical informatics may be the ultimate enabler for the applications of bioinformatics methods and algorithms within a clinical context [52, 53].
Evaluating the Performance of Biomedical Data Mining Algorithms with Statistical Tools
Novel regression and classification methods are developed in various areas of research, such as medical informatics, bioinformatics, data mining or biostatistics. The performance of several competing approaches is usually evaluated in benchmark experiments [54]. The most important question to be answered in such a classification experiment is whether two automated learning “machines” differ in a relevant magnitude and/or significantly from one another. Here, it is important to note that simple algorithms are often quite good, and they may be even superior to complicated machines [55]. And, generally, one cannot necessarily expect a pronounced superiority of a highly sophisticated approach [56].
One aspect in comparing learning machines with each other deserves specific attention. If various machines are trained on training data, their performance can only be compared in a fair manner by applying all machines to the same test data. In this case, the above described procedures lead to valid estimates.
If only a training data set is available so that machines are both trained and compared on the same data, prediction accuracy varies systematically depending on the way machines are trained [55]. For example, logistic regression utilizes all available data in the model-building step, and it is more prone to overfitting compared with ensemble methods that use only a portion of the available data and rarely overfit. Therefore, error fractions and performance estimates are more reliable for ensemble methods but may also be substantially higher. As an alternative, 5-fold or 10-fold cross validation may be used for internally validating the models that has been shown to yield satisfactory results. Here, it is important to note that all steps of model building, model-dependent data transformations, and variable selection need to be repeated for each loop of the cross validation. Overfitting could otherwise result [57]. However, even with 10-fold cross validation not the same amount of data, i.e., the same number of degrees of freedom is used as with bootstrapping. Specifically, bootstrapping–which is often used in ensemble methods–tends to use approximately 2/3 of the data set. In contrast, with 5-fold or 10 -fold cross validation, 90% or 80% of the training data is used to train the machine.
If the same set of training data is generated in the first step and if all of these data are used to train the machines, this source of bias in comparing prediction accuracies can be overcome. Specifically, all machines can be tested on the same out of bag samples, i.e., the samples not drawn in the bootstrap, and these give paired results for the machines. These can then be compared by appropriate averaging across all bootstrap samples [55], and standard statistics for comparing machines can easily be calculated.
For classification methods these are the Brier score [58,59], sensitivity, specificity, or the error fraction [55]. More specifically, the predictions from two machines for the same patient are expected to be correlated: the presence of such correlation can be formally tested with McNemar’s test. Corresponding confidence intervals for the differences in error fractions, sensitivity, or specificity can easily be calculated [60-62].
To give an example, using the notation of Table 1, Wilson’s score method – method [63] in the review of Newcombe [61]– specifically yields the following confidence interval at level 1–α for the difference of the two proportions θ = (π1 + π2) − (π1 + π3) = π2 − π3. The interval is , where δ and ε are positive values and with dl2 = (a + b)/n − l2, du2 = u2 − (a + b)/n. Here, l2 and u2 are the roots of . Similarly, dl3 = (a + c)/n − l3, du3 = u3 − (a + c)/n, where l3 and u3 are the roots of . Finally,
For a comparison of error fractions between different independent data sets, for example, to compare differences in the performance of temporal and external validation [55], appropriate tests and confidence intervals have also been developed [63].
Table 1.
Comparison of observed error frequencies (proportions in parenthesis) for two dependent machines
| Machine B |
|||||||
|---|---|---|---|---|---|---|---|
| Correct | False | Total | |||||
| Machine A | Correct | a | (π1) | b | (π2) | a+b | (πA) |
| False | c | (π3) | d | (π4) | c+d | (1−πA) | |
|
| |||||||
| Total | a+c | (πB) | b+d | (1−πB) | n | (1) | |
In regression problems, the performance of machines can easily be compared using the t-test or similar tests, and these approaches also allow the construction of appropriate confidence intervals.
In summary, it is possible to formally compare the performance of different machines using statistical tests. Even more, the relevance of the performance difference of machines can be formulated with simple to obtain parameters and confidence intervals.
Translational Bioinformatics: Leveraging the Opportunity of the Biomedical Data Deluge
Since its beginnings, biomedical informatics has striven to develop approaches to link knowledge across the entirety of biomedicine, from molecules to populations. Recent advances, especially those in bioinformatics (e.g., development of high-throughput sequencing) and health informatics (e.g., large scale deployment of electronic health records), have positioned the biomedical informatics enterprise to foster the development of a focused new area of emphasis – translational bioinformatics (TBI). TBI is a systemic approach for integrating biological and clinical knowledge with the specific goal of understanding deep questions related to human health. In contrast to the rich history of methodological advances that have been developed within the realm of bioinformatics for the full spectrum of life sciences, TBI is specifically focused on the development of approaches for a better understanding of human associated diseases [64].
In many ways, TBI is the realization of biomedical informatics principles to develop linkages between the molecular processes of disease or dysfunction with phenotypic information (e.g., symptomatology), such as described in clinical sources or catalogues in resources such as the Online Mendelian Inheritance in Man (OMIM [65]). The sheer volume of data being generated, thanks to recent and continued technological advances, is increasingly positioning the informatics community to begin the exploration of putative linkages between biological and clinical data as never before contemplated. No longer is the challenge conceptualized as “finding a disease-geneneedle in a haystack;” instead, the challenge is akin to determining the “meaningful” configurations of needles. That is to say, we are looking less for single genes or mutations that may be correlated with disease, and more for combinations of genes and mutations that collectively contribute to (or prevent) disease. Data mining techniques will be needed that can enable such “systems level” studies to be done across molecular, individual, or population levels. Previously, it would have been difficult to imagine how the seemingly separate endeavors of studying disease genes and representing clinical phenotypes in electronic health records would become intertwined towards guiding the future of medicine. Yet, we are now in the midst of discussions of how genomic features, such as those derived from Genome Wide Association Studies (GWAS), can be used to guide the next discoveries given volumes of clinical data (e.g., as exemplified in the NIH-funded eMERGE [Electronic Medical Records and Genomics] project [66]).
Heterogeneous data integration approaches have been described for incorporating an array of biological and clinical data [67,68]. To this end, hallmark initiatives like the already mentioned i2b2 project [49] have developed key infrastructure to enable inquiries that cross the “bench-to-bedside” divide [69]. We thus see less need for developing computational approaches for storing or querying biomedical data and more need for developing approaches to better understand what these data might mean in the context of human health. The concept of developing and testing in silico hypotheses is increasingly accepted by the biology community [69-71], and we are positioning the biomedical community for a new type of knowledge discovery beyond what was ever possible using in vitro or in vivo approaches (e.g., synthetic organisms may shed light on completely new perspectives of disease prophylaxis or treatment [72]). This implies that the scientific enterprise may be at the cusp of a paradigm shift from classical single gene or polymorphic mining and correlation experiments to a new cadre of studying the combination of genes or inheritable traits (e.g., as is beginning to be explored using network or graph-theoretic approaches [73]). However, the realization of this promise will require the adaptation of existing or contemplation of entirely new forms of data mining and analytic techniques.
While academic inquiry into the cause of disease is a noble and important endeavor, which can leverage a wide suite of data mining approaches for identifying and categorizing potential genes or polymorphisms of interest with respect to a given disease, an entirely different approach must be taken to be clinically meaningful. The clinical meaningfulness of a correlation is a keystone element for TBI, and the development of evaluation metrics will be essential for the clinical acceptability of putative gene(s) or polymorphism(s) of interest. Thus, while we will initially need to develop in silico approaches for developing hypotheses, there will be a need for describing the in vitro and, especially, in vivo implications. Data mining and analytic approaches thus need to not only be reliable and robust for TBI, but also be understandable and interpretable by clinicians who are faced with making actionable decisions at the point of care.
At least for the foreseeable future, advances in technology across the entire spectrum of biomedicine will increase the volume of data. There will thus continue to be a need for developing and building on the rich legacy of data mining and analytic techniques within the silos of bioinformatics and health informatics. As TBI continues to evolve from the development of infrastructure to bridge across these silos, the data mining and analytic techniques will also need to evolve and embrace a truly trans-disciplinary approach to develop and test new hypotheses with potential clinical implications. The future opportunities for data mining and analytics in the era of TBI thus promise to be rich, albeit challenging, but also have great potential for transforming the future of medicine.
Genetic Epidemiology and Bioinformatics
Genetic epidemiology may be seen as the study of the role of genetic factors in determining health and diseases at an individual, family and population level, and deals with the interplay of genetic factors with environmental factors. Alternatively, it may be seen as the science that deals with the etiology, distribution and control of diseases in individuals with some kind of relationship between them, in groups of relatives with identified causes of disease and in different populations [74]. It is closely allied to both molecular epidemiology and statistical genetics.
The study of the role of genetics in disease progress is done with the use of some analytical designs each answering slightly different questions such as: familiar aggregation studies dealing with the issue of the influence of a genetic component to the disease and the relative contribution of genes and the environment; segregation studies dealing with the pattern of inheritance of the diseases (e.g. dominant or recessive); linkage studies dealing with the location of the genes in the chromosomes; the genes that are related to specific diseases; association studies dealing with the type specific association of one-single gene with a respective disease [75].
The traditional approach has proven highly successful in identifying monogenic disorders and in locating the responsible genes [76]. Most recently, the scope of genetic epidemiology has expanded to include common diseases, for which many different genes can make a smaller contribution (polygenetic, multifactorial or multigenic disorders). This has developed rapidly in the first decade of the 21st century following completion of the human genome project, as advances in genotyping technology and bioinformatics and definitively the associated reductions in cost has made it feasible to conduct large scale genome wide association studies. These studies have revealed many thousands of single nucleotide polymorphisms in a multitude of individuals. These have led to the discovery of many genetic polymorphisms that influence the risk of developing many common diseases.
The importance of bioinformatics in genetic epidemiology is crucial and deals with collecting data and annotations and with the usage of powerful tools is capable in visualizing and searching the human DNA [77]. Bioinformatics also deals with the development of computer programs to analyze the data, because the data themselves are difficult to interpret without adequate algorithms and programs. The process of identifying the boundaries between genes and other features in a raw DNA sequence is called genome annotation and is the domain of bioinformatics. Expert biologists and genetic epidemiologists make the highest quality annotations; however, their work proceeds slowly, and computer programs are increasingly used to meet the high throughput demands of genome sequencing projects [78]. The best current technologies for annotations make use of statistical models that take advantage of parallels between DNA sequences and human language using concepts from computer science such as formal grammars. Therefore, it is imperative to note that investigations in biology and genetic epidemiology can no longer be carried out without bioinformatics methods and tools. There will in the future be many more genomic discoveries, and bioinformatics will certainly play a crucial role in interpreting and managing those discoveries.
Conclusions
Medicine and biomedical sciences are data-rich environments, and thus require sophisticated methods and approaches to be able to analyze the increasingly “big” and “complex” data sets collected so far, the availability and open distribution of such data is still uneven. As a matter of fact, while “–omics” data repositories strongly support fundamental discoveries in molecular medicine [79, 80], there is still a substantial lack of clinical data that can be used in analogous ways, with projects like eMERGE being notable exceptions [66]. Crucial issues related to data ownership, privacy protection, national and international regulations are slowing down the “phenotype” revolution, which may effectively boost biomedical research as –omics data are currently doing. Large information technology projects [68] based on the sematic web technologies [81] deal with this important topic, showing that BI, as a field, has the potential to provide the right methods and tools to foster data sharing and implementing the required data analysis steps. A “call-to-arms” is therefore needed to push forward the critical need for sharing data. As a natural corollary, there should be an increasing commitment of researchers to share the source code of their data analysis methods and algorithms, thus effectively contributing to the collective efforts of the international research community.
As a matter of fact, one of the main strengths of BI stands in its being at the intersection of multiple disciplines, and, as such, being an “inclusive” sector rather than an “exclusionary” one. Diverse scientific contributions have always been accepted and “included” if they provided advantages to the field. This has made BI not only a context for testing new methods and ideas but also the engine of innovations that has been exploited in other areas [82,83]. The main question is therefore how to preserve such openness while keeping BI, as a discipline, consistent and well founded. As far as data analysis methods and applications are concerned, it seems likely that the research scenario in biomedical data mining will progressively need a two-layer model for reaching such a complex goal. Curiosity-driven basic research will always be crucial to invent new algorithms, new methods and new software tools. Such types of research may be carried out in fields that can be quite distant from biomedicine. For this reason, when moving from basic to applied research it will be increasingly crucial to establish large and multi-disciplinary research teams, which will have the goal of selecting, tailoring, engineering and finally deploying solutions targeted to the specific needs of the intended users. Furthermore, such teams will also need to stay aware of existing methods and tools outside biomedical informatics, to propose new effective solutions.
The confluence of multiple disciplines is at the very heart of BI in general, and biomedical data mining in particular, both at a “system” level and at a “micro” level. In this respect, paraphrasing [84], biomedical informaticians will in the future be more and more like technology architects, who are able to integrate different instruments, methods and tools for the sake of health care and biomedical research.
Acknowledgements
The authors drafted individual sections based on their contributions at the Symposium: Section 1 (KT), Section 2 (RB), Section 3 (AZ), Section 4 (INS), Section 5 (MD). RB drafted the integrated paper and AM revised the final draft. The research of RB has been supported by the ITALBIONET project (Rete Italiana di Bioinformatica), funded by the Ministry of University and Research and by the ONCO-i2b2 project, funded by the Lombardia Region. Fulvia Ferrazzi is acknowledged for revising the early versions of the section on data and knowledge integration. INS is partially funded by the United States National Institutes of Health (grant R01LM009725).
Footnotes
Publisher's Disclaimer: This article is not an exact copy of the original published article in Methods of Information in Medicine. The definitive publisher-authenticated version of Bellazi R, Diomidous M, Sarkar IN, Takabayashi K, Ziegler A, McCray AT. “Data Analysis and Data Mining: Current Issues in Biomedical Informatics” will be available online at: http://www.schattauer.de/de/magazine/subject-areas/journals-a-z/methods.html.
References
- [1].Masys DR, Ellison D, Stead WW. Presentation of the 2007 Morris F. Collen award to William W. Stead, MD, including comments from recipient. J Am Med Inform Assoc. 2008 May-Jun;15(3):302–6. doi: 10.1197/jamia.M2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Haux R. Medical informatics: past, present, future. Int J Med Inform. 2010 Sep;79(9):599–610. doi: 10.1016/j.ijmedinf.2010.06.003. [DOI] [PubMed] [Google Scholar]
- [3].Haux R, Aronsky D, Leong TY, McCray AT. Methods in year 50: preserving the past and preparing for the future. Methods Inf Med. 2011;50(1):1–6. doi: 10.3414/ME10-05-0005. [DOI] [PubMed] [Google Scholar]
- [4].Hendler J. Avoiding another AI Winter, IEEE Intelligent Systems, 2-4. 2008.
- [5].Anderson JG. Social, ethical and legal barriers to e-health. Int J Med Inform. 2007 May-Jun;76(5-6):480–3. doi: 10.1016/j.ijmedinf.2006.09.016. [DOI] [PubMed] [Google Scholar]
- [6].Lau F, Kuziemsky C, Price M, Gardner J. A review on systematic reviews of health information system studies. J Am Med Inform Assoc. 2010 Nov 1;17(6):637–45. doi: 10.1136/jamia.2010.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Payne PR, Embi PJ, Niland J. Foundational biomedical informatics research in the clinical and translational science era: a call to action. J Am Med Inform Assoc. 2010 Nov 1;17(6):615–6. doi: 10.1136/jamia.2010.005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sarkar IN. Biomedical informatics and translational medicine. J Transl Med. 2010 Feb 26;8:22. doi: 10.1186/1479-5876-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Smith A, Balazinska M, Baru C, Gomelsky M, McLennan M, Rose L, Smith B, Stewart E, Kolker E. Biology and data-intensive scientific discovery in the beginning of the 21st century. OMICS. 2011 Apr;15(4):209–12. doi: 10.1089/omi.2011.0006. [DOI] [PubMed] [Google Scholar]
- [10].Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007 Jan 17;99(2):147–57. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- [11].Markie P. In: Rationalism vs. Empiricism, The Stanford Encyclopedia of Philosophy. Fall 2008 Edition. Zalta Edward N., editor. [Last accessed Aug 17th 2011]. http://plato.stanford.edu/archives/fall2008/entries/rationalism-empiricism/ [Google Scholar]
- [12].Patel VL, Shortliffe EH, Stefanelli M, Szolovits P, Berthold MR, Bellazzi R, Abu-Hanna A. The coming of age of artificial intelligence in medicine. Artif Intell Med. 2009 May;46(1):5–17. doi: 10.1016/j.artmed.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Louie B, Mork P, Martin-Sanchez F, Halevy A, Tarczy-Hornoch P. Data integration and genomic medicine. J Biomed Inform. 2007 Feb;40(1):5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- [14].Prokosch HU, Ganslandt T. Perspectives for Medical Informatics: Reusing the Electronic Medical Record for Clinical Research. Methods Inf Med. 2009;48:38–44. [PubMed] [Google Scholar]
- [15].Suzuki T, Yokoi H, Fujita S, Takabayashi K. Automatic DPC Code Selection from Electronic Medical Records : Text Mining Trial of Discharge Summary. Methods Inf Med. 2008;47:541–548. doi: 10.3414/ME9128. [DOI] [PubMed] [Google Scholar]
- [16].Haux R. Individualization, globalization and health about sustainable information technologies and the aim of medical informatics. Int J Med Inform. 2006;75:795–808. doi: 10.1016/j.ijmedinf.2006.05.045. [DOI] [PubMed] [Google Scholar]
- [17].Chung W, Oh SM, Suh T, Lee YM, Oh BH, Yoon CW. Determinants of length of stay for psychiatric inpatients: analysis of a national database covering the entire Korean elderly population. Health Policy. 2010;94:120–8. doi: 10.1016/j.healthpol.2009.09.004. [DOI] [PubMed] [Google Scholar]
- [18].Yang JY, Yang MQ, Zhu M, Arabnia HR, Deng Y. Promoting synergistic research and education in genomics and bioinformatics. BMC Genomics. 2008;9(supple I):1–5. doi: 10.1186/1471-2164-9-S1-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Burgun A, Bodenreider O. Accessing and integrating data and knowledge for biomedical research. Yearb Med Inform. 2008:91–101. [PMC free article] [PubMed] [Google Scholar]
- [20].Tran DH, Satou K, Ho TB, Pham TH. Computational discovery of miR-TF regulatory modules in human genome. Bioinform. 2010;4:371–7. doi: 10.6026/97320630004371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hey T. The fourth Pradigm: Data-intensive scientific discovery. http://research.microsoft.com/fourthparadigm/
- [22].Fayyad UM, Piatetsky-Shapiro G, Smyth P. From data mining to knowledge discovery: an overview. In: Fayyad UM, Piatetsky-Shapiro G, Smyth P, Uthurusamy R, editors. Advances in knowledge discovery and data mining. American Association for Artificial Intelligence; Menlo Park, CA, USA: 1996. pp. 1–34. [Google Scholar]
- [23].Iavindrasana J, Cohen G, Depeursinge A, Müller H, Meyer R, Geissbuhler A. Clinical data mining: a review. Yearb Med Inform. 2009:121–33. [PubMed] [Google Scholar]
- [24].Evans JA, Rzhetsky A. Advancing Science through Mining Libraries, Ontologies, and Communities. J Biol Chem. 2011 Jul 8;286(27):23659–66. doi: 10.1074/jbc.R110.176370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fernandez-Luque L, Karlsen R, Bonander J. Review of extracting information from the Social Web for health personalization. J Med Internet Res. 2011 Jan 28;13(1):e15. doi: 10.2196/jmir.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Deléger L, Grouin C, Zweigenbaum P. Extracting medical information from narrative patient records: the case of medication-related information. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):555–8. doi: 10.1136/jamia.2010.003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cohen KB, Johnson HL, Verspoor K, Roeder C, Hunter LE. The structural and content aspects of abstracts versus bodies of full text journal articles are different. BMC Bioinformatics. 2010 Sep 29;:11–492. doi: 10.1186/1471-2105-11-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bellazzi R, Sacchi L, Concaro S. Methods and tools for mining multivariate temporal data in clinical and biomedical applications. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:5629–32. doi: 10.1109/IEMBS.2009.5333788. [DOI] [PubMed] [Google Scholar]
- [29].Aalst W. van der. Process Mining: Discovery, Conformance and Enhancement of Business Processes. Springer Verlag; Berlin: 2011. [Google Scholar]
- [30].Bhavnani SK, Bellala G, Ganesan A, Krishna R, Saxman P, Scott C, Silveira M, Given C. The nested structure of cancer symptoms. Implications for analyzing co-occurrence and managing symptoms. Methods Inf Med. 2010;49(6):581–91. doi: 10.3414/ME09-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zupan B, Holmes JH, Bellazzi R. Knowledge-based data analysis and interpretation. Artif Intell Med. 2006 Jul;37(3):163–5. doi: 10.1016/j.artmed.2006.03.001. [DOI] [PubMed] [Google Scholar]
- [32].Yang JY, Niemierko A, Bajcsy R, Xu D, Athey BD, Zhang A, Ersoy OK, Li GZ, Borodovsky M, Zhang JC, Arabnia HR, Deng Y, Dunker AK, Liu Y, Ghafoor A. 2K09 and thereafter : the coming era of integrative bioinformatics, systems biology and intelligent computing for functional genomics and personalized medicine research. BMC Genomics. 2010 Dec 1;11(Suppl 3):I1. doi: 10.1186/1471-2164-11-S3-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bellazzi R, Zupan B. Intelligent data analysis--special issue. Methods Inf Med. 2001;40(5):362–4. [PubMed] [Google Scholar]
- [34].Bellazzi R, Zupan B. Towards knowledge-based gene expression data mining. J Biomed Inform. 2007 Dec;40(6):787–802. doi: 10.1016/j.jbi.2007.06.005. [DOI] [PubMed] [Google Scholar]
- [35].Bellazzi R, Zupan B. Predictive data mining in clinical medicine: current issues and guidelines. Int J Med Inform. 2008 Feb;77(2):81–97. doi: 10.1016/j.ijmedinf.2006.11.006. [DOI] [PubMed] [Google Scholar]
- [36].Holmes JH, Peek N. Intelligent data analysis in biomedicine. J Biomed Inform. 2007 Dec;40(6):605–8. doi: 10.1016/j.jbi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- [37].Nuzzo A, Riva A, Bellazzi R. Phenotypic and genotypic data integration and exploration through a web-service architecture. BMC Bioinformatics. 2009 Oct 15;10(Suppl 12):S5. doi: 10.1186/1471-2105-10-S12-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Klimov D, Shahar Y, Taieb-Maimon M. Intelligent interactive visual exploration of temporal associations among multiple time-oriented patient records. Methods Inf Med. 2009;48(3):254–62. doi: 10.3414/ME9227. [DOI] [PubMed] [Google Scholar]
- [39].Demšar J, Zupan B, Leban G, Curk T. Orange: From Experimental Machine Learning to Interactive Data Mining, Knowledge Discovery in Databases: PKDD 2004. Volume 3202/2004. 2004. pp. 537–539. (Lecture Notes in Computer Science). [Google Scholar]
- [40].Augusto JC. Temporal reasoning for decision support in medicine. Artificial Intelligence in Medicine. 2005;33(1):1–24. doi: 10.1016/j.artmed.2004.07.006. [DOI] [PubMed] [Google Scholar]
- [41].Adlassnig KP, Combi C, Das AK, Keravnou ET, Pozzi G. Temporal representation and reasoning in medicine: Research directions and challenges. Artificial Intelligence in Medicine. 2006;38(2):101–113. doi: 10.1016/j.artmed.2006.10.001. [DOI] [PubMed] [Google Scholar]
- [42].Roddick JF, Spiliopoulou M. A Survey of Temporal Knowledge Discovery Paradigms and Methods. IEEE Transactions on Knowledge and Data Engineering. 2002;14(4):750–767. [Google Scholar]
- [43].Post AR, Harrison JH. Temporal data mining. Clinics in Laboratory Medicine. 2008;28(1):83–100. doi: 10.1016/j.cll.2007.10.005. [DOI] [PubMed] [Google Scholar]
- [44].Mitsa T. Temporal Data Mining. CRC Press; 2010. [Google Scholar]
- [45].Shahar Y. A framework for knowledge-based temporal abstraction. Artificial Intelligence. 1997;90(1-2):79–133. doi: 10.1016/0933-3657(95)00036-4. [DOI] [PubMed] [Google Scholar]
- [46].Panzarasa S, Maddè S, Quaglini S, Pistarini C, Stefanelli M. Evidence-based careflow management systems: the case of post-stroke rehabilitation. J Biomed Inform. 2002 Apr;35(2):123–39. doi: 10.1016/s1532-0464(02)00505-1. [DOI] [PubMed] [Google Scholar]
- [47].Peleg M, Yeh I, Altman RB. Modelling biological processes using workflow and Petri Net models. Bioinformatics. 2002 Jun;18(6):825–37. doi: 10.1093/bioinformatics/18.6.825. [DOI] [PubMed] [Google Scholar]
- [48].Mans R, Schonenberg H, Leonardi G, Panzarasa S, Cavallini A, Quaglini S, van der Aalst W. Process mining techniques: an application to stroke care. Stud Health Technol Inform. 2008;136:573–8. [PubMed] [Google Scholar]
- [49].Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010 Mar-Apr;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. [Last accessed 14 April 2011]; http://www.ehr4cr.eu/
- [51].Zapletal E, Rodon N, Grabar N, Degoulet P. Methodology of integration of a clinical data warehouse with a clinical information system: the HEGP case. Stud Health Technol Inform. 2010;160(Pt 1):193–7. [PubMed] [Google Scholar]
- [52].Sintchenko V, Coiera E. Developing decision support systems in clinical bioinformatics. Methods Mol Med. 2008;141:331–51. doi: 10.1007/978-1-60327-148-6_18. [DOI] [PubMed] [Google Scholar]
- [53].Szalma S, Koka V, Khasanova T, Perakslis ED. Effective knowledge management in translational medicine. J Transl Med. 2010 Jul 19;8:68. doi: 10.1186/1479-5876-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hothorn T, Leisch F, Zeileis A, Hornik K. The design and analysis of benchmark experiments. J Comput Graph Statist. 2005;14:675–699. [Google Scholar]
- [55].König IR, Malley JD, Pajevic S, Weimar C, Diener H-C, Ziegler A, et al. Patient-centered yes/no prognosis using learning machines. Int J Data Min Bioinform. 2008;2:289–341. doi: 10.1504/ijdmb.2008.022149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hand D. Classifier technology and the illusion of progress. Stat Sci. 2006;21:1–14. [Google Scholar]
- [57].Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- [58].Bradley AA, Schwartz SS, Hashino T. Sampling uncertainty and confidence intervals for the Brier score and Brier skill score. Weather Forecast. 2008;23:992–1006. [Google Scholar]
- [59].Ferro CAT. Comparing probabilistic forecasting systems with the Brier score. Weather Forecast. 2007;22:1076–1088. [Google Scholar]
- [60].Tango T. Equivalence test and confidence interval for the difference in proportions for the paired-sample design. Stat Med. 1998;17:891–908. doi: 10.1002/(sici)1097-0258(19980430)17:8<891::aid-sim780>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- [61].Newcombe RG. Improved confidence intervals for the difference between binomial proportions based on paired data. Stat Med. 1998;17:2635–2650. [PubMed] [Google Scholar]
- [62].Zhou XH, Qin GS. A supplement to: “A new confidence interval for the difference between two binomial proportions of paired data”. J Stat Plan Inference. 2007;137:357–358. [Google Scholar]
- [63].Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat Med. 1998;17:873–890. doi: 10.1002/(sici)1097-0258(19980430)17:8<873::aid-sim779>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- [64].Butte AJ. Translational bioinformatics: coming of age. J Am Med Inform Assoc. 2008 Nov-Dec;15(6):709–14. doi: 10.1197/jamia.M2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009 Jan;37(Database issue):D793–6. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sarkar IN, Butte AJ, Lussier YA, Tarczy-Hornoch P, Ohno-Machado L. Translational bioinformatics: linking knowledge across biological and clinical realms. J Am Med Inform Assoc. 2011 May 10; doi: 10.1136/amiajnl-2011-000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sujansky W. Heterogeneous database integration in biomedicine. J Biomed Inform. 2001 Aug;34(4):285–98. doi: 10.1006/jbin.2001.1024. [DOI] [PubMed] [Google Scholar]
- [68].Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. Journal of the American Medical Informatics Association : JAMIA. 2009 Sep-Oct;16(5):624–30. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sarkar IN, Butte AJ, Lussier YA, Tarczy-Hornoch P, L. O-M. Translational Bioinformatics: Linking Knowledge Across Biological and Clinical Realms. J Am Med Inform Assoc. 2011 doi: 10.1136/amiajnl-2011-000245. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kollmann M, Sourjik V. In silico biology: from simulation to understanding. Curr Biol. 2007 Feb 20;17(4):R132–4. doi: 10.1016/j.cub.2006.12.034. [DOI] [PubMed] [Google Scholar]
- [71].Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. From in vivo to in silico biology and back. Nature. 2006 Oct 5;443(7111):527–33. doi: 10.1038/nature05127. [DOI] [PubMed] [Google Scholar]
- [72].Aubel D, Fussenegger M. Mammalian synthetic biology--from tools to therapies. Bioessays. 2010 Apr;32(4):332–45. doi: 10.1002/bies.200900149. [DOI] [PubMed] [Google Scholar]
- [73].Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M. Drug-target network. Nat Biotechnol. 2007 Oct;25(10):1119–26. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- [74].Morton Newton E, Chung Chin Sik., editors. Genetic Epidemiology. Academic; New York: 1978. [Google Scholar]
- [75].Morton NE. Genetic Epidemiology. Annals of Human Genetics. 1997;61(1):1–13. doi: 10.1046/j.1469-1809.1997.6110001.x. [DOI] [PubMed] [Google Scholar]
- [76].Spence MA. Encyclopedia of Biostatistics. Wiley Interscience; 2005. Genetic Epidemiology. [Google Scholar]
- [77].Hogeweg P. Searls David B., editor. The Roots of Bioinformatics in Theoretical Biology. PLoS Computational Biology. 2011;7(3) doi: 10.1371/journal.pcbi.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008 January;36:25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–69. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhang H, Morrison MA, Dewan A, Adams S, Andreoli M, Huynh N, Regan M, Brown A, Miller JW, Kim IK, Hoh J, Deangelis MM. The NEI/NCBI dbGAP database: genotypes and haplotypes that may specifically predispose to risk of neovascular age-related macular degeneration. BMC Med Genet. 2008 Jun 9;9:51. doi: 10.1186/1471-2350-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Rahmouni H Boussi, Solomonides T, Mont M Casassa, Shiu S, Rahmouni M. A Modeldriven Privacy Compliance Decision Support for Medical Data Sharing in Europe. Methods Inf Med. 2011 Aug 15;50(4):326–36. doi: 10.3414/ME10-01-0075. [DOI] [PubMed] [Google Scholar]
- [82].Bardram JE. Pervasive healthcare as a scientific discipline. Methods Inf Med. 2008;47(3):178–85. doi: 10.3414/me9107. [DOI] [PubMed] [Google Scholar]
- [83].Adams SA. Revisiting the online health information reliability debate in the wake of “web 2.0”: an inter-disciplinary literature and website review. Int J Med Inform. 2010 Jun;79(6):391–400. doi: 10.1016/j.ijmedinf.2010.01.006. [DOI] [PubMed] [Google Scholar]
- [84].Musen MA. Architectures for architects. Methods Inf Med. 1993;32(1):12–3. [PubMed] [Google Scholar]