Abstract
The neuronal RNA-binding protein HuD plays a critical role in the post-transcriptional regulation of short-lived mRNAs during the initial establishment and remodelling of neural connections. We have generated transgenic mice overexpressing this protein (HuD-Tg) in adult DGCs (dentate granule cells) and shown that their mossy fibres contain high levels of GAP-43 (growth-associated protein 43) and exhibit distinct morphological and electrophysiological properties. To investigate the basis for these changes and identify other molecular targets of HuD, DGCs from HuD-Tg and control mice were collected by LCM (laser capture microscopy) and RNAs analysed using DNA microarrays. Results show that 216 known mRNAs transcripts and 63 ESTs (expressed sequence tags) are significantly up-regulated in DGCs from these transgenic mice. Analyses of the 3′-UTRs (3′-untranslated regions) of these transcripts revealed an increased number of HuD-binding sites and the presence of several known instability-conferring sequences. Among these, the mRNA for TTR (transthyretin) shows the highest level of up-regulation, as confirmed by qRT–PCR (quantitative reverse transcription–PCR) and ISH (in situ hybridization). GO (gene ontology) analyses of up-regulated transcripts revealed a large over-representation of genes associated with neural development and axogenesis. In correlation with these gene expression changes, we found an increased length of the infrapyramidal mossy fibre bundle in HuD-Tg mice. These results support the notion that HuD stabilizes a number of developmentally regulated mRNAs in DGCs, resulting in increased axonal elongation.
Keywords: axonal outgrowth, dentate granule cell, gene profiling, HuD, post-transcriptional mechanisms, RNA-binding protein
Abbreviations: αCaMKII, α-Ca2+/calmodulin-dependent protein kinase II; ARE, AU-rich element; ARED, ARE database; CP, choroid plexus; Dcx, doublecortin; DGC, dentate granule cell; DIG, digoxigenin; EST, expressed sequence tag; FISH, fluorescence in situ hybridization; GAP-43, growth-associated protein 43; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GO, gene ontology; GRE, GU-rich element; IPA, ingenuity pathway analysis; IPB, infrapyramidal bundle; ISH, in situ hybridization; KO, knockout; LCM, laser capture microscopy; Mtap1b, microtubule-associated protein 1b; NIH, National Institutes of Health; PFA, paraformaldehyde; PPF, paired-pulse facilitation; qRT–PCR, quantitative reverse transcription–PCR; RNA-IP, RNA-immunoprecipitation; SMA, spinal muscular atrophy; SPB, suprapyramidal bundle; T3, 3,3′,5-tri-iodothyronine; T4, thyroxine; TTR, transthyretin; 3′-UTR, 3′-untranslated region; WT, wild-type
INTRODUCTION
Post-transcriptional mechanisms play an important role in gene regulation, particularly in neurons, where mRNAs are localized to dendrites and growing axons and regulated independently from transcription (Eberwine et al., 2001; Steward and Schuman, 2001; Martin and Zukin, 2006; Bolognani and Perrone-Bizzozero, 2008; Yoo et al., 2010). RNA-binding proteins participate in all aspects of post-transcriptional regulation, from mRNA splicing and transport to stability and translation. Among these proteins, the neuronal ELAV-like protein HuD has been shown to bind and stabilize several short-lived mRNAs, including those encoding GAP-43 (growth-associated protein 43) and other developmentally regulated proteins (for a review see Perrone-Bizzozero and Bolognani, 2002; Deschênes-Furry et al., 2006). The expression of HuD coincides with the earliest stages of neuronal differentiation and is maintained through the maturation of neurons (Okano and Darnell, 1997; Wakamatsu and Weston, 1997; Clayton et al., 1998; Bolognani et al., 2007a; Hambardzumyan et al., 2009). This pattern of expression is critical for proper neuronal differentiation, as shown by KO (knockout) and overexpression studies (Akamatsu et al., 2005; Bolognani et al., 2006). Furthermore, recent studies also demonstrate that HuD is required for mechanisms of learning and memory in mature hippocampus and peripheral nerve regeneration (Bolognani et al., 2004; Pascale et al., 2004; Anderson et al., 2003; Deschênes-Furry et al., 2007). Consistent with the role of HuD in developmental and adult plasticity, genome-wide identification of its targets revealed that many of these transcripts encode for proteins associated with axon guidance, actin dynamics and long-term potentiation (Bolognani et al., 2010).
We have previously shown that DGCs (dentate granule cells) of the hippocampus express HuD mRNA and protein during the first week of postnatal development (Bolognani et al., 2007a). However, their levels decline quickly thereafter and they are not detected in DGCs from mature mice or rats (Bolognani et al., 2006, 2007a). This developmental decline in HuD expression in DGCs correlates with that of one of its target mRNAs, GAP-43, presumably due to its instability in the absence of HuD (Namgung and Routtenberg, 2000; Bolognani et al., 2006). Supporting this idea, we have shown that both HuD and GAP-43 are re-induced in DGCs of adult rats after epileptic seizures (Bolognani et al., 2007a) and that adult transgenic mice in which HuD is overexpressed via the αCaMKII (α-Ca2+/calmodulin-dependent protein kinase II) promoter exhibit increased stabilization and accumulation of GAP-43 mRNA in DGCs (Bolognani et al., 2006). Furthermore, mossy fibres of HuD-Tg exhibit increased PPF (paired-pulse facilitation) and increased sprouting of GAP-43 containing terminals in the CA3 region (Tanner et al., 2008). To further characterize the molecular targets and mechanisms mediating the different properties of DGCs and their projections in HuD overexpressor mice, we sought to identify the mRNAs regulated by HuD in these cells using a combination of LCM (laser capture microscopy) and gene expression profiling. Our results clearly show an increased expression of development-associated genes in DGCs from adult HuD overexpressor mice relative to age-matched control littermates.
MATERIALS AND METHODS
Animals
All animal studies were approved by the University of New Mexico Animal Care and Use Committee and conformed to NIH (National Institutes of Health) guidelines for the use of animals in research. All experiments were performed using adult male HuD-Tg line 4 mice (Bolognani et al., 2006) and control littermates from 6 to 8 weeks of age. HuD-Tg mice express the HuD isoform of the human HuD protein under the control of the αCaMKII promoter [Tg(Camk2a-ELAVL4)].
LCM and microarray analysis
DGCs from both hippocampi of three pairs of HuD-Tg mice and WT (wild-type) littermates were captured in two separate experiments at the Duke DNA Microarray Facility. RNAs were then isolated and analysed by Affymetrix 430 2.0 mouse genomic arrays at Translational Genomics (T-Gen). Both facilities were part of the NIH Neuroscience DNA Microarray Consortium. Statistical analysis of transcript levels in HuD-Tg and non-transgenic control littermates was performed using GeneSpring 9.0 software, and significant differences in gene expression were determined using P<0.05.
3′-UTR (3′-untranslated regions) bioinformatics analyses
For analysis of the presence of known AREs (AU-rich elements) and 3′-UTR statistics, 3′-UTR sequences of up-regulated genes and the entire 430 2.0 Affymetrix chip were downloaded from Biomart Ensembl 61 Mus musculus gene database (NCBIM37) in FASTA format (http://www.ensembl.org/biomart). We were able to retrieve 285 3′-UTR sequences of the up-regulated genes in the dentate gyrus and 18490 3′-UTR sequences of the entire chip. Custom-written scripts in Perl 5.10.0 using BioPerl 1.5.2 modules were used to compute the presence of the motifs in the data set as described by Bolognani et al. (2010). Differences between these two datasets were analysed by χ2 test using the R statistical package version 2.7.1.
GO (gene ontology) and biological pathway analyses
Data were analysed using the Gene Set Analysis Toolkit V2 (WebGestalt) available online at the Vanderbilt University (http://bioinfo.vanderbilt.edu/webgestalt/). We compared the distribution of genes up-regulated in the dentate gyrus of HuD-Tg mice with those in the Affymetrix 430 2.0 chip gene list. The software uses the hypergeometric statistical test to analyse significance, and P<0.01 were considered as statistically significant. IPA (ingenuity pathway analysis) software (http://www.ingenuity.com/) allows for the visualization of dynamic pathways based on information manually curated from literature searches. IPA was used here to discover and visualize relevant biological networks and functional enrichments associated with HuD-DGC up-regulated and down-regulated transcripts.
FISH (fluorescence in situ hybridization)
FISH was performed as described Smith et al. (2004) with the TSA amplification modifications indicated in the GEISHA (Gene Expression In situ Hybridization Analysis project, http://geisha.arizona.edu). Briefly, after the mice were killed, brains were removed rapidly and flash-frozen in 2-methylbutane cooled to −40°C in solid CO2/methanol slurry. Sections (16 μm) were cut at −20°C on a cryostat and stored at −80°C. An antisense oligonucleotide against nucleotides 134–17 in the mouse TTR (transthyretin) mRNA (5′-GGATTCTCCAGCACCCGCGGGGCCAGCTTCAGACAC) was end-labelled using recombinant terminal transferase and DIG (digoxigenin)-dUTP (Roche, Oligonucleotide tailing kit) according to the manufacturer's protocols. Sections were hybridized overnight at 55°C with DIG-labelled TTR probes and washed with 50% formamide/1×SSC/0.1% Tween-20. Sections were then incubated in the presence of peroxidase-conjugated anti-DIG antibodies followed by tyramide-fluorescein conjugates (TSA Plus Florescence Systems Kit, PerkinElmer). After subsequent washes, fluorescent intensity in the sections was measured using NIH ImageJ.
qRT–PCR (quantitative reverse transcription–PCR)
qRT–PCR for TTR mRNA was performed as previously described (Bullock et al., 2009). Briefly, mouse TTR primers (forward 5′-AGGTCAGAAAGCAGAGTGGACCAA and reverse 5′-ACACTACTGTGCATCTACAGCCCT) were validated against mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase; forward 5′-TGTGATGGGTGTGAACCACGAGAA and reverse 5′-GAGCCCTTCCACAATGCCAAAGTT) and found to be within optimal amplification values (validation curve slopes <|0.1|). Dissociation curves of all SYBR Green primer pairs revealed no evidence of dimerization. Samples were run in triplicate in two separate plates and compared with GAPDH on the same plate as previously described (Bullock et al., 2009).
Immunohistochemistry
Four pairs of HuD-Tg and control mice were processed for immunohistochemistry as described by Tanner et al. (2008). Briefly, brains were fixed in 4% (w/v) PFA (paraformaldehyde) and cryoprotected for at least 24 h in 30% sucrose in PBS. Brains were mounted on a sliding-knife microtome and serial sections of 40 μm were placed free-floating in PBS. Sections were incubated with rabbit anti-calbindin D-28k (1:2000; Swant) followed by incubation with anti-rabbit Alexa Fluor® 568 secondary antibodies (1:100; Molecular Probes). Epifluorescent images were taken on a BioRad BX-60 microscope with an Olympus DP71 CCD-digital camera (Olympus America).
Timm's staining
Mossy fibres are characterized by the presence of high concentration of vesicular zinc (Zn2+). Visualization of this vesicular Zn2+ was achieved with slight variations to established Timm's staining protocols (Proper et al., 2000). Briefly, 300 μm vibratome sections were incubated in 4% sodium sulfide in PBS for 30 min, followed by fixation in 1.25% glutaraldehyde, 1% PFA and 10% sucrose in PBS overnight. Sections were mounted on to Vectabond (Vector Laboratories)-coated slides, desiccated and stored temporarily at −20°C. Slides were placed in Timm's developing solution (20% gum Arabic, 2 M sodium citrate, 4% dihydroquinone and 1.5% silver nitrate) for 35–45 min and rinsed with warm tap water. Sections were dehydrated with alcohols, cleared with xylenes and cover-slipped with DPX (p-xylenebispyridinium bromide).
IPB (infrapyramidal bundle) measurements
The length of the IPB was measured from images of calbindin immunofluorescence and Timm's staining using ImagePro® Plus 4. software 0 (Media Cybernetics). The length of positive mossy fibres from the hilus to the point they cross the pyramidal cell layer was divided by the total length of the most medial aspect of the hilus to the apex of the curvature of CA3 as previously described (Bagri et al., 2003).
Statistical analyses
Results are expressed as means±S.E.M. Significant differences between means were determined by Student's t tests using GraphPad Prism software package 4 (GraphPad Software). P<0.05 were considered statistically significant.
RESULTS
Laser capture and mRNA profiling of DGCs in HuD-Tg mice
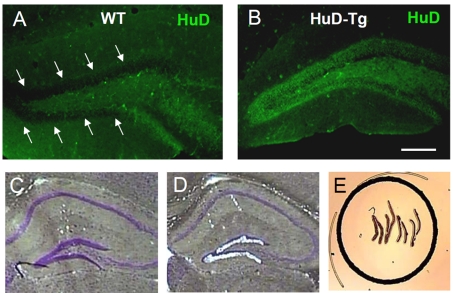
HuD is quickly down-regulated in DGCs after the first postnatal week and it is not detected in mature cells (Figure 1A). In contrast, DGCs from HuD-Tg mice express high levels of this protein even in adult mice (Figure 1B). To identify gene expression changes associated with the persistent expression of HuD, DGCs from WT and HuD-Tg mice were laser-captured (Figures 1C–1E) and RNA derived from these cells subjected to analysis using Affymetrix 430 2.0 genomic arrays. Statistical analyses revealed that 337 probe sets corresponding to 216 known transcripts and 63 ESTs (expressed sequence tags) show a significant increase in DGCs from HuD-Tg mice (termed herein HuD-DGCs) whereas 322 probe sets corresponding to 204 known transcripts and 116 ESTs were significantly decreased. Given the role of HuD in mRNA stabilization, we focused our studies on the up-regulated genes and used the list of down-regulated genes as a comparison set. The top 40 most-up-regulated mRNAs are shown in Table 1 and the complete list is available in Supplementary Tables S1 (available at http://www.asnneuro.org/an/003/an003e070add.htm). Of this complete list of up-regulated mRNAs, 73 were previously identified as HuD targets by RNA-IP (RNA-immunoprecipitation) studies of mouse forebrain transcripts (Bolognani et al., 2010, see validated target column in Supplementary Table S1) and the rest were new targets.
Figure 1. LCM of DGCs in HuD-Tg mice and control littermates.
(A, B) DGC from adult control mice (A) do not express HuD but high levels of the protein is expressed in HuD-Tg mice (B). Scale bar = 150 μm. (C–E) H/E (haematoxylin/eosin) staining demonstrating the successful laser capture of cells in the DGC layer.
Table 1. Top 40 up-regulated genes in DGCs of HuD-Tg mice.
| Affymetrix ID | Gene symbol | Gene name | Fold change | P-value |
|---|---|---|---|---|
| 1455913_x_at | TTR | Transthyretin | 57.04 | 0.0008 |
| 1454608_x_at | TTR | Transthyretin | 39.52 | 0.011 |
| 1451580_a_at | TTR | Transthyretin | 32.01 | 0.0127 |
| 1459737_s_at | TTR | Transthyretin | 23.4 | 0.002 |
| 1433474_at | Edil3 | EGF-like repeats and discoidin I-like domains 3 | 4.827 | 0.000001 |
| 1450577_at | Sstr3 | Somatostatin receptor 3 | 4.465 | 0.016 |
| 1451246_s_at | Aurkb | Aurora kinase B | 4.174 | 0.0037 |
| 1449011_at | Slc12a7 | Solute carrier family 12, member 7 | 3.778 | 0.0345 |
| 1441946_at | Itih5 | Inter-α (globulin) inhibitor H5 | 3.409 | 0.00498 |
| 1438387_x_at | Top3b | Topoisomerase (DNA) III β | 3.396 | 0.00591 |
| 1418280_at | Klf6 | Kruppel-like factor 6 | 3.285 | 0.032 |
| 1459205_at | Cpeb1 | Cytoplasmic polyadenylation element binding protein 1 (Cpeb1) | 3.075 | 0.00746 |
| 1421964_at | Notch3 | Notch gene homologue 3 (Drosophila) | 2.915 | 0.0209 |
| 1458458_at | Slfn5 | Schlafen 5 | 2.856 | 0.0273 |
| 1429011_x_at | Snrp70 | U1 small nuclear ribonucleoprotein polypeptide A | 2.826 | 0.0386 |
| 1435306_a_at | Kif11 | Kinesin family member 11 | 2.796 | 0.0287 |
| 1446819_at | Lrp1b | Low-density lipoprotein-related protein 1B (Lrp1b) | 2.773 | 0.0374 |
| 1434430_s_at | Adora2b | Adenosine A2b receptor | 2.715 | 0.0485 |
| 1456976_at | Wnt5a | Wingless-related MMTV integration site 5A | 2.71 | 0.0134 |
| 1445824_at | Zfp458 | zinc finger protein 458 | 2.708 | 0.00132 |
| 1430402_at | Sucla2 | Succinate-coenzyme A ligase, ADP-forming, β-subunit | 2.611 | 0.0439 |
| 1456936_at | Cabp4 | Calcium binding protein 4 | 2.559 | 0.0367 |
| 1456742_x_at | Tm9sf2 | Transmembrane 9 superfamily member 2 | 2.555 | 0.0134 |
| 1429462_at | Slc25a32 | Solute carrier family 25, member 32 | 2.549 | 0.0365 |
| 1419486_at | Foxc1 | Forkhead box C1 | 2.538 | 0.00644 |
| 1459116_at | Ncam2 | Neural cell adhesion molecule 2 (Ncam2), mRNA | 2.538 | 0.0327 |
| 1419193_a_at | Gmfg | Glia maturation factor, gamma | 2.535 | 0.0109 |
| 1443782_x_at | Cyp20a1 | Cytochrome P450, family 20, subfamily A, polypeptide 1 | 2.525 | 0.0255 |
| 1422637_at | Rassf5 | Ras association (RalGDS/AF-6) domain family 5 | 2.472 | 0.0171 |
| 1456312_x_at | Gsn | Gelsolin | 2.436 | 0.0307 |
| 1448768_at | Mog | Myelin oligodendrocyte glycoprotein | 2.385 | 0.0236 |
| 1433742_at | Ankrd15 | Ankyrin repeat domain 15 | 2.375 | 0.0441 |
| 1418166_at | Il12rb1 | Interleukin 12 receptor, β1 | 2.37 | 0.0379 |
| 1427829_at | Abcd4 | ATP-binding cassette, sub-family D (ALD), member 4 | 2.358 | 0.0467 |
| 1457692_at | 2010013E08Rik | THAP domain containing, apoptosis associated protein 3, mRNA (cDNA clone MGC:106590 IMAGE:5700290) | 2.338 | 0.0233 |
| 1428821_at | Agpat2 | 1-Acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, β) | 2.33 | 0.0293 |
| 1442697_at | Ipo11 | Importin 11, mRNA (cDNA clone MGC:39010 IMAGE:5364208) | 2.295 | 0.00212 |
| 1418706_at | Slc38a3 | Solute carrier family 38, member 3 | 2.292 | 0.049 |
| 1446577_at | Pde4b | C57BL/6J phosphodiesterase 4B (Pde4b) | 2.286 | 0.0448 |
| 1449009_at | Tgtp | T-cell specific GTPase | 2.277 | 0.0358 |
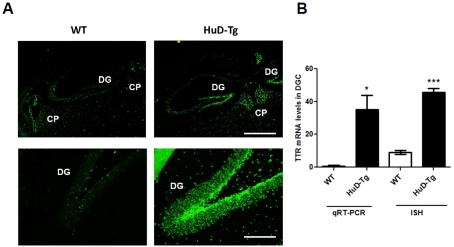
Among the new targets identified in the DGCs, the mRNA for the thyroid hormone- and retinol-binding protein TTR showed the highest levels of up-regulation and was chosen for further validation by qRT–PCR and ISH (in situ hybridization). As shown in Figure 2(A) and in agreement with previous studies (Tsai et al., 2009), high levels of TTR mRNA are observed in the CP (choroid plexus), while the hippocampus shows very low levels of expression. In comparison, DGCs of HuD-Tg mice expressed much higher levels of this mRNA (Figures 2A and 2B). The significant up-regulation of TTR mRNA in DGCs of HuD-Tg mice was confirmed by qRT–PCR using RNA from LCM-isolated DGCs from a different group of animals (Figure 2B).
Figure 2. Increased TTR mRNA levels in DGCs of HuD-Tg mice.
(A) TTR FISH images from WT and HuD-Tg mice. Low magnification taken using a ×4 objective show expression of TTR mRNA in CP in WT and HuD-Tg mice and in the dentate gyrus of HuD-Tg mice. Scale bar = 750 μm. High magnification (×20) images show increase TTR mRNA in DGC of HuD Tg mice. Scale bar = 150 μm. (B) Quantification of TTR mRNA levels in DGCs of WT and HuD-Tg by ISH and qRT–PCR was performed as described in the Materials and methods section. *P<0.05 and ***P<0.001.
Over-representation of HuD-binding motifs and other U-rich elements in the 3′-UTR of HuD-DGC mRNAs
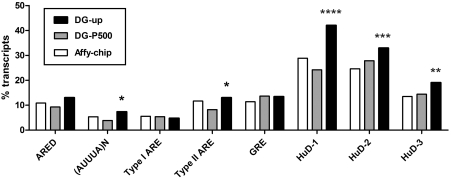
HuD and other Hu proteins are known to bind and stabilize mRNAs containing U-rich instability conferring sequences such as the classical ARE (Shaw and Kamen, 1986; Chen and Shyu, 1995) and the GRE (GU-rich element; Vlasova et al., 2008; Bolognani et al., 2010) in the 3′-UTR. Therefore subsequent analyses determined the frequencies of these elements in the 3′-UTRs of up-regulated HuD-DGC mRNAs (DG-up, Figure 3). The number of transcripts with these sequences was compared with both the total number of transcripts in the Affymetrix 430 2.0 array (Affy-chip) and the top 500 expressed (present) transcripts in DGCs whose expression levels were not significantly different between HuD-Tg and WT mice (DG-P500). As previously described (Bolognani et al., 2010), typical ARE sequences include: (i) consensus AREs motifs described in the ARED (ARE database; ARED 3.0, Bakheet et al., 2006, http://brp.kfshrc.edu.sa/ARED/), (ii) overlapping AUUUA motifs (AUUUA)n and (iii) the type I and II AREs described by Wilusz et al. (2001). Analysis of individual ARE sequences revealed that the percentage of transcripts with (AUUUA)n and type II ARE motifs in the 3′-UTR were significantly increased in HuD-DGCs (DG-up) relative to those present in the DG-P500 set, whereas there were no differences in the percentage of other ARE motifs or GRE instability-sequences (Figure 3).
Figure 3. Analyses of 3′-UTR motifs in HuD-DGCs up-regulated mRNAs.
The frequencies of the different types of ARE motifs, including the consensus motif in the ARED, GRE motif and HuD-binding motifs (see text for details) in the 3′-UTRs of HuD-DGC transcripts (DG-up, black bars) were analysed and compared with all the genes present in the Affymetrix 430 2.0 chip (Affy-chip white bars), and a list of the top 500 DG expressed transcripts that are present in control mice and whose expression levels are not significantly different in HuD-Tg mice (DG-P500, grey bars). Statistical analyses were performed using a one-tail χ2 test: ****P<0.00001, DG-up compared with both DG-P500 and Affy-chip, ***P = 0.0024, DG-up compared with Affy-chip, **P = 0.0093 DG-up compared with Affy-chip and *P = 0.040 compared with DG-P500.
Since HuD binds AU-rich and GU-rich sequences with high affinity and CU-rich sequences with less affinity (Bolognani et al., 2010), we also investigated the presence of transcripts with the three consensus-binding motifs for HuD: motif 1, CCCUCCCUCUCUC, motif 2, UUUUGUUUUGUUU and motif 3, UUUUUUUUUUAAA. As shown in Figure 3, while only 13% of all the transcripts in the chip and 14% of DG-P500 transcripts contain the AUUUA-like motif 3 in the 3′-UTR, the frequency of these sequences was significantly increased in HuD-Tg DGCs, reaching approximately 20% of all the up-regulated transcripts. Likewise, we found significant increases in the GUUUG-like (motif 2) and UCCCU-like (motif 1), which reached to approximately 33% and 42% of all DGC overexpressed transcripts, respectively (Figure 3). Overall we found that approximately 60% (161/279) of the up-regulated transcripts contained at least one of the HuD-binding motifs and 80% of DGC transcripts (223/279) contained at least one of the AU-rich or GU-rich 3′-UTR sequences described above (see Supplementary Table S1). Collectively, these findings suggest that the majority of HuD-DGCs transcripts were overexpressed because they contain binding sites for this stabilizing RNA-binding protein.
To further illustrate this point, specific examples of mRNAs containing these 3′-UTR motifs are shown in Table 2. While some transcripts like Kif11 and Gsn contain only one of these sequences, transcripts for Wnt5a, Dcx (doublecortin), Notch3, UBe3a and Jag1 contain multiple copies of these U-rich sequences and HuD-binding motifs in their 3′-UTRs. As shown in Supplementary Table S1, 51 up-regulated transcripts did not contain any of these 3′-UTR elements (e.g. Agpat2, Cabp4, Gmfg, Il12rb1, Otud3, Nol3, Dgcr2, Hesx1, Cldn5, Nfatc2, Plxnb1, Vill, Slc29a1, Neurl2, Myh14, Cd151, Adra2c, Bad, Nat6 and Sema5b) suggesting that these transcripts are indirectly regulated by HuD.
Table 2. Number of U-rich 3′-UTR motifs in select up-regulated mRNAs in DGCs of HuD-Tg mice.
| Affymetrix ID | Gene symbol | Gene name | ARE | U-rich | GRE | HuD-binding motifs* | Ensembl gene ID |
|---|---|---|---|---|---|---|---|
| 1455913_x_at | TTR | Transthyretin | 0 | 14 | 0 | 1 (3) | ENSMUSG00000061808 |
| 1433474_at | Edil3 | EGF-like repeats and discoidin I-like domains 3 | 1 | 43 | 0 | 1 (2) | ENSMUSG00000034488 |
| 1441946_at | Itih5 | Inter-α (globulin) inhibitor H5 | 1 | 4 | 1 | 7 (1) | ENSMUSG00000025780 |
| 1459205_at | Cpeb1 | Cytoplasmic polyadenylation element binding protein 1 | 0 | 0 | 9 | 0 | ENSMUSG00000025586 |
| 1421964_at | Notch3 | Notch gene homologue 3 (Drosophila) | 0 | 21 | 0 | 5 (1) | ENSMUSG00000038146 |
| 1458458_at | Slfn5 | Schlafen 5 | 0 | 0 | 0 | 14 (1) | ENSMUSG00000054404 |
| 1435306_a_at | Kif11 | Kinesin family member 11 | 0 | 5 | 0 | 1 (1) | ENSMUSG00000012443 |
| 1456976_at | Wnt5a | Wingless-related MMTV integration site 5A | 3 | 96 | 6 | 10 (1), 16 (2) | ENSMUSG00000021994 |
| 1419486_at | Foxc1 | Forkhead box C1 | 0 | 7 | 1 | 0 | ENSMUSG00000050295 |
| 1459116_at | Ncam2 | Neural cell adhesion molecule 2 | 0 | 31 | 0 | 2 (2) | ENSMUSG00000022762 |
| 1446577_at | Pde4b | Phosphodiesterase 4B | 0 | 0 | 0 | 2 (1) | ENSMUSG00000028525 |
| 1456312_x_at | Gsn | Gelsolin | 0 | 9 | 0 | 1 (2) | ENSMUSG00000026879 |
| 1418140_at | Dcx | Doublecortin | 0 | 47 | 0 | 41 (1) | ENSMUSG00000031285 |
| 1422839_at | Neurog2 | Neurogenin 2 | 1 | 12 | 0 | 4 (1), 1 (2) | ENSMUSG00000027967 |
| 1445727_at | Ube3a | Ubiquitin protein ligase E3A | 1 | 70 | 0 | 8 (2) | ENSMUSG00000025326 |
| 1421105_at | Jag1 | Jagged 1 | 9 | 74 | 0 | 1 (1), 15 (2) | ENSMUSG00000027276 |
| 1421851_at | Mtap1b | Microtubule-associated protein 1B | 3 | 18 | 0 | 1 (1), 15 (2), 1 (3) | ENSMUSG00000052727 |
| 1445634_at | Mapt | Microtubule-associated protein tau | 0 | 0 | 1 | 5 (1) | ENSMUSG00000018411 |
Number and type (in parentheses) of HuD-binding motifs in the 3′-UTR.
GO analysis of HuD-DGC of differentially expressed transcripts
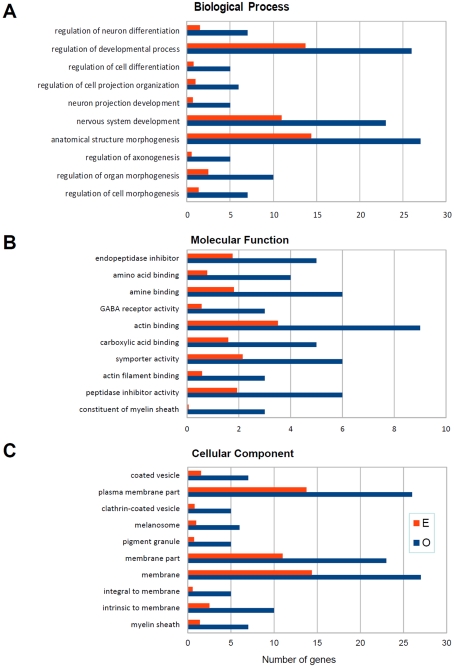
Two different bioinformatics tools were used to place HuD-induced gene expression changes into a biological context: the WebGestalt GO toolkit (http://bioinfo.vanderbilt.edu/webgestalt/) and IPA software (http://www.ingenuity.com/). The top 10 GO categories of biological processes, molecular functions, and cellular locations of the proteins encoded by HuD-DGC up-regulated mRNAs are shown in Figure 4. In each category, the number of expected (E) genes is based upon the percentage of genes on the mouse Affymetrix 430 2.0 mouse genomic array with each category and the number observed (O) genes is the actual number of up-regulated transcripts in each category. From these analyses, it is clear that this dataset is enriched in genes involved in neural development and axonogenesis (Figure 4A). Among these are positive regulators of axogenesis such as plexins B1 and B2, Mtap1b (microtubule associated protein 1b) and Sema5b, as well as regulators of neurogenesis and neuronal differentiation such as Hesx1, Dcx, Notch3 and Jagged 1, neurogenin 2 (Table 3). Consistent with these biological categories, analysis of the molecular functions (Figure 4B) and cellular components (Figure 4C) revealed that many of the genes are associated with actin binding (e.g. Gsn and Ermn), vesicle-mediated transport (e.g. Dab2 and Hip1) and myelination (e.g. MBP and PLP).
Figure 4. GO analysis of up-regulated transcripts.
GO analyses were performed as indicated in the Materials and methods section. Panels show the observed (blue bars) and expected (red bars) number of genes in the biological process, molecular function and cellular component categories. The top ten significantly enriched categories are presented.
Table 3. Genes in top enriched GO categories in DGCs of HuD-Tg mice.
| Probe set ID | Gene symbol | Gene title |
|---|---|---|
| Positive regulation of axogenesis | ||
| 1416683_at | Plxnb2 | Plexin B2 |
| 1435254_at | Plxnb1 | Plexin B1 |
| Axon ensheathment | ||
| 1456228_x_at | Mbp | Myelin basic protein |
| 1416003_at | Cldn11 | Claudin 11 |
| 1425467_a_at | Plp1 | Proteolipid protein (myelin) 1 |
| Cell development | ||
| 1422839_a_at | Neurog2 | Neurogenin 2 |
| 1437341_x_at | Cnp1 | Cyclic nucleotide phosphodiesterase 1 |
| 1420498_a_at | Dab2 | Disabled homologue 2 (Drosophila) |
| 1418140_at | Dcx | Doublecortin |
| 1416683_at | Plxnb2 | Plexin B2 |
| 1421851_at | Mtab1b | Microtubule-associated protein 1B |
| 1421964_at | Notch3 | Notch gene homologue 3 (Drosophila) |
| 1425468_at | Plp1 | Proteolipid protein (myelin) 1 |
| 1448105_at | Prm2 | Protamine 2 |
| 1448260_at | Uchl1 | Ubiquitin C-terminal hydrolase L1 |
| 1435254_at | Plxnb1 | Plexin B1 |
| Cell morphogenesis during differentiation | ||
| 1422839_at | Neurog2 | Neurogenin 2 |
| 1437341_x_at | Cnp1 | Cyclic nucleotide phosphodiesterase 1 |
| 1420498_a_at | Dab2 | Disabled homologue 2 (Drosophila) |
| 1416683_at | Plxnb2 | Plexin B2 |
| 1448260_at | Uchl1 | Ubiquitin C-terminal hydrolase L1 |
| 1435254_at | lxnb | Plexin B1 |
| Nervous system development | ||
| 1422839_at | Neurog2 | Neurogenin 2 |
| 1437341_x_at | Cnp1 | Cyclic nucleotide phosphodiesterase 1 |
| 1418140_at | Dcx | Doublecortin |
| 1416683_at | Plxnb2 | Plexin B2 |
| 1420604_at | Hesx1 | Homeo box gene expressed in ES cells |
| 1421105_at | Jag1 | Jagged 1 |
| 1456228_x_at | Mbp | Myelin basic protein |
| 1421851_at | Mtab1b | Microtubule-associated protein 1B |
| 1421964_at | Notch3 | Notch gene homologue 3 (Drosophila) |
| 1416003_at | Cldn11 | Claudin 11 |
| 1425468_at | Plp1 | Proteolipid protein (myelin) 1 |
| 1425146_at | Sema5b | Semaphorin 5B |
| 1448260_at | Uchl1 | Ubiquitin C-terminal hydrolase L1 |
| 1435254_at | Plxnb1 | Plexin B1 |
| Positive regulation of development | ||
| 1416683_at | Ptxnb2 | Plexin B2 |
| 1421105_at | Jag1 | Jagged 1 |
| 144829_at | Hps4 | Hermansky–Pudlak syndrome 4 homologue |
| 1435254_at | Plxnb1 | Plexin B1 |
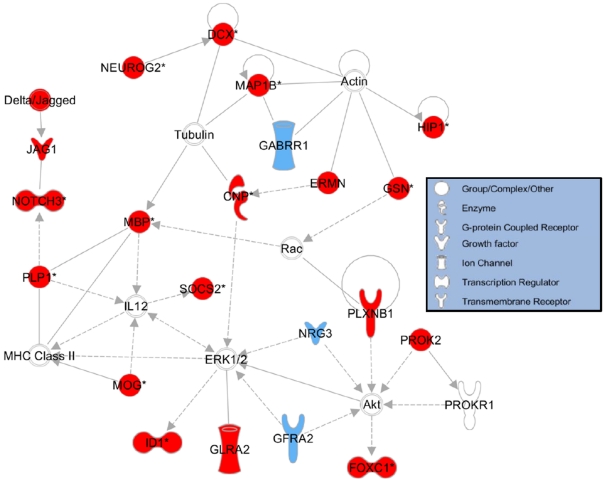
To visualize how genes in our dataset may interact with each other to influence development, we used IPA to generate a sublist of the up-regulated genes that are associated with neuronal development according to their Knowledgebase dataset. We then used this sublist of development-associated genes to perform dynamic pathway analysis. Figure 5 shows numerous interactions among the protein products of the development-associated genes present in our list (shown in red) as well as other developmentally-regulated proteins that interact with gene products from our gene list (shown in blue). In particular, several members of the Jagged/NOTCH pathway are in our list, as well as several protein products that interact with the cytoskeletal proteins actin and tubulin. The majority of the remaining proteins shown in this biological network interact with either the pro-growth kinase Akt (also known as protein kinase B) or the MAPK (mitogen-activated protein kinase) ERK (extracellular-signal-regulated kinase).
Figure 5. Biological network of genes involved in nervous system development.
IPA showing interactions of gene products from HuD-Tg overexpressed genes. Proteins that are involved in nervous system development and function and whose transcripts were up-regulated in our dataset are shown in red, whereas transcripts that were absent in our dataset but whose products are involved in nervous system development are shown in blue. Continuous lines represent direct relationships, whereas broken lines represent indirect relationships. The biological activities associated with each protein symbol are indicated in the key box.
In contrast with the up-regulated mRNAs, GO analyses of down-regulated transcripts did not show any significantly enrichment in canonical pathways at P<0.001 (Supplementary Table S2 available at http://www.asnneuro.org/an/003/an003e070add.htm). Interestingly, fold changes in this set (average −1.54) were less drastic than the fold changes in the up-regulated set, reflecting HuD's role as a stabilizer of mRNA transcripts. Furthermore, in agreement with our previous observation of compensatory decreases in other ELAV-like transcripts in the hippocampus of HuD-Tg mice (Bolognani et al., 2006) we found that HuR mRNA was also decreased in the DGCs. In contrast with HuR, mouse HuD transcripts were also increased in the DGCs (Supplementary Table S1) in the presence of the human HuD transgene that does not contain the 3′-UTR and thus cannot be detected by the mouse 3′-UTR specific probes present in the Affymetrix chip. These results suggest that HuD increases its own expression while blocking the expression of the ubiquitously expressed ELAV-like protein HuR.
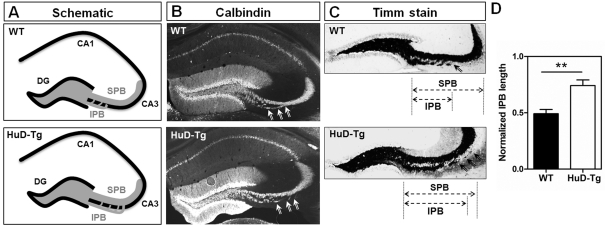
Increased length of the infrapyramidal mossy fibre bundle in HuD-Tg mice
Given the increased enrichment in genes associated with axogenesis in DGCs from HuD-Tg mice and our previous findings that these animals also show increased sprouting of GAP-43 positive mossy fibres into the CA3 region (Tanner et al., 2008), we next asked whether HuD overexpression altered mossy fibre length. As shown in Figure 6(A), mossy fibres form two separate bundles: the SPB (suprapyramidal bundle) that travels above the CA3 pyramidal layer and the IPB that originates below this layer and later crosses it to join the SPB (see arrows in Figures 6B and 6C). Mossy fibres were stained using either antibodies against the calcium-binding protein calbindin (Figure 6B) or the Timm' zinc stain (Figure 6C). The relative length of the IPB was calculated using the length of the SPB as reference as described by Bagri et al. (2003) (Figure 6C). As shown in Figure 6(D), the relative length of the IPB was significantly increased in the HuD-Tg mice, supporting the results of the gene expression studies. Overall, our data indicate that HuD target mRNAs are important for the regulation of multiple aspects of development and axonogenesis. Furthermore, our data supports the hypothesis that appropriate regulation of RNA-binding proteins is critical for normal development and adult plasticity.
Figure 6. Increased length of the infrapyramidal mossy fibre bundle in HuD-Tg mice.
(A) Shows the distribution of IPB and SPB bundles in WT and HuD transgenic mice. Mossy fibres were stained using either calbindin antibodies (B) or Timm's stain (C). The IPB length was calculated from the hilus to where the fibres cross the pyramidal cell layer (arrows), and the length of the SPB was measured from the hilus to the apex of the curvature of area CA3 (C). The length of the IPB was normalized to that of the SPB in the same section (D). Scale bar = 300 μm. **P<0.01.
DISCUSSION
The neuron-specific RNA-binding protein HuD plays a critical role in the post-transcriptional regulation of neuronal gene expression during neural development and adult synaptic plasticity. This protein is known to stabilize GAP-43 mRNA and other neuronal mRNAs containing U-rich sequences in their 3′-UTR. In this study, we examined the effect of overexpression of this RNA-binding protein in DGCs of adult mice. Normally, these cells only express HuD during the first postnatal week. Our results indicate that persistent expression of HuD in adult DGCs induced the expression of developmentally-regulated neural genes in these cells and increased the length of their axons in the IPB. Together with previous studies (reviewed by Perrone-Bizzozero and Bolognani, 2002; Deschênes-Furry et al., 2006), these results emphasize the role of HuD in controlling the expression of genes important for neuronal development and axonal outgrowth.
The most drastic gene expression change in HuD-Tg mice was a 50-fold increase in mRNA levels of the transporter of thyroxine and retinol, TTR. We confirmed this large overexpression using ISH as well as qRT–PCR. While this result was unexpected, TTR overexpression may help to account for the increased axon length observed in these mice. The most obvious explanation for the role of TTR in the observed phenotype of HuD-Tg mice is the gene product's traditional role as transporter of T4 (thyroxine) in the brain. The CP secretes TTR into the CSF (cerebrospinal fluid), where it acts to transport the prohormone T4 into the brain (Richardson et al., 2007). Once in the brain, deiodinases convert T4 into the more active hormone T3 (3,3′,5-tri-iodothyronine). Thyroid hormone levels have been shown to increase expression of NGF (nerve growth factor) as well as several neurotrophic factors (Lüesse et al., 1998). Administration of high levels of T4 to neonatal rats during the critical period of mossy fibre development (P0–P21), results in lasting changes in the distribution of IP mossy fibres in the hippocampus (Lauder and Mugnaini, 1980), which are similar to those reported here. Not surprisingly, TTR-null mice have been shown to contain only 36% of WT thyroid hormone levels in their brains (Palha et al., 1997). The critical role of T3 and TTR in hippocampal development and function is further demonstrated by the findings that prenatal and neonatal hypothyroidism cause learning and memory defects, including mental retardation in children (Zoeller and Rovet, 2004). Although thyroid hormone levels are important for growth and development, the protease activity of TTR might also help explain the increased axonal length that we observed. TTR has been shown to cause increased neurite outgrowth in PC12 cells incubated in medium from normal mice as compared with PC12 cells incubated in medium from TTR KO mice (Fleming et al., 2007; Liz et al., 2009). This increased outgrowth is reduced to TTR KO levels when the proteolytic activity of TTR is inhibited (Liz et al., 2009), although the substrate responsible for increasing neurite growth has not been identified.
As shown in Figure 4 and Table 3, many up-regulated genes encode proteins associated with axonal outgrowth (e.g. Mapt, Mtap1b, Ncam2, Cacna1a, Ermn, Plxnb1 and Plxnb2), actin dynamics (e.g. Gsn, Myh14, Tagln2 and Tnnc1) and neuronal differentiation (e.g. Aplp2, Dcx, Neurog2, Notch3, Hesx1, Jag1, Sema3e, Sema5b, Socs2 and Tgfbr2). Other HuD-up-regulated transcripts are known to be increased in the hippocampus during development (e.g. Sstr3; Stanić et al., 2009) or contextual fear conditioning (e.g. Itih5 and Edil3; Keeley et al., 2006). Besides, we noticed that the mRNAs for a couple of axogenesis-related HuD targets, GAP-43 (also known as neuromodulin) and cgp-15 (also known as neuritin; Akten et al., 2011), were not detected in the chip as being up-regulated. Although GAP-43 mRNA was detected by ISH in HuD-DGC (Bolognani, et al., 2006) and this mRNA was validated as a target of HuD by RNA-IP followed by qRT–PCR (Bolognani et al., 2010), we have previously found that the single probe available in the mouse Affymetrix 430 2.0 chip showed very low signal and failed to detect changes in this transcript in both studies. In contrast, we found that the signal of neuritin was very high in both control mice and HuD-Tg DGCs, suggesting that it may not be possible to detect increases in its levels due to ceiling effects. Besides the neuronally expressed axonal outgrowth-related proteins, some of the HuD-DGC up-regulated transcripts were found to encode myelin proteins (e.g. Plp1, Mal, Mog, Mbp, Cldn11 and Cnp). It is likely that these gene-expression changes take place in oligodendrocytes located in the DGC layer and constitute an indirect response to the increases in axonal length observed in the HuD-Tg mice. Alternatively, although HuD is not normally expressed in mature oligodendrocytes, the αCaMKII promoter used here to express transgenic HuD was shown to be active in oligodendrocyte precursor cells (Mason et al., 2003). Thus, this activity could account for some of the observed changes in myelin gene expression. Finally, it is also well established that thyroid hormone increases myelination and re-myelination and promotes the differentiation of oligodendrocyte precursor cells (Balázs et al., 1971; Fernandez et al., 2004; Barres et al., 1994) suggesting that the increased expression of TTR in HuD-Tg mice may contribute to these effects.
In conclusion our results support the notion that persistent expression of HuD in DGCs of transgenic mice has significant functional consequences on the differentiation and maturation of these cells, as shown by the increased mossy fibre length in the IPB. These results are in agreement with our previous findings of the effect of HuD overexpression on the rate of neurite outgrowth (Anderson et al., 2000, 2001) and with observations from other laboratories supporting a role of Hu proteins in neuronal development (Wakamatsu and Weston, 1997; Akamatsu et al., 1999; Kasashima et al., 1999). Furthermore, these findings are consistent with recent studies demonstrating that the interaction of HuD with the SMA (spinal muscular atrophy) protein SMN (survival of motor neurons) is critical for localizing and stabilizing mRNAs in motor neuron axons and for rescuing SMA-associated deficits (Akten et al., 2011; Fallini et al., 2011; Hubers et al., 2011). Although increased axonal outgrowth may be a desirable outcome in some conditions, such in the response to nerve injury or disease, this can also lead to problems in inappropriate innervation of target tissues and synaptic maturation. In fact, our previous studies demonstrate that, similarly to developing mossy fibres (Mori-Kawakami et al., 2003), mossy fibres in adult HuD-Tg mice have abnormally increased PPF. These maturation problems could also explain the deficits in hippocampal dependent learning observed in these animals (Bolognani et al., 2007b). Ongoing studies are characterizing the behaviour of other neuronal circuits in these mice, particularly in their response to injury.
Online data
ACKNOWLEDGEMENTS
We thank Clark Bird, Sayuri Nixon and Dr Lee Anna Cunningham for technical assistance and expertise and Dr Pate Skene and Susan Su of the Duke Microarray Facility and Kerry Ramsey, Tanya Contente-Cuomo and Dr Dietrich Stephan from T-Gen for help with the microarray studies.
Footnotes
This work was supported by the National Institutes of Health [grant number NS30255-18 (to N.P.B.)].
REFERENCES
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, Takahashi T, Noda T, Okano H. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci USA. 2005;102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B, Kye MJ, Hao le T, Wertz MH, Singh S, Nie D, Huang J, Merianda TT, Twiss JL, Beattie CE, Steen JA, Sahin M. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci USA. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KD, Morin MA, Beckel-Mitchener A, Mobarak CD, Neve RL, Furneaux HM, Burry R, Perrone-Bizzozero NI. Overexpression of HuD, but not of its truncated form HuD l+ll, promotes GAP43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J Neurochem. 2000;75:1103–1114. doi: 10.1046/j.1471-4159.2000.0751103.x. [DOI] [PubMed] [Google Scholar]
- Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero Nl. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp Neurol. 2001;168:250–258. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- Anderson KD, Merhege MA, Morin M, Bolognani F, Perrone-Bizzozero NI. Increased expression and localization of the RNA-binding protein HuD and GAP-43 mRNA to cytoplasmic granules in DRG neurons during nerve regeneration. Exp Neurol. 2003;183:100–108. doi: 10.1016/s0014-4886(03)00103-1. [DOI] [PubMed] [Google Scholar]
- Bagri A, Cheng HJ, Yaron A, Pleasure SJ, Tessier-Lavigne M. Stereotyped pruning of long hippocampal axon branches triggered by retraction inducers of the semaphorin family. Cell. 2003;113:285–299. doi: 10.1016/s0092-8674(03)00267-8. [DOI] [PubMed] [Google Scholar]
- Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázs R, Brooksbank BWL, Patel AJ, Johnson AL, Wilson DA. Incorporation of [35S]sulphate into brain constituents during development and the effects of thyroid hormone on myelination. Brain Res. 1971;30:273–293. doi: 10.1016/0006-8993(71)90079-5. [DOI] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Merhege MA, Twiss J, Perrone-Bizzozero NI. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Tanner DC, Merhege M, Deschênes-Furry J, Jasmin B, Perrone-Bizzozero NI. In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD. J Neurochem. 2006;96:790–801. doi: 10.1111/j.1471-4159.2005.03607.x. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Tanner DC, Nixon S, Okano HJ, Perrone-Bizzozero NI. Coordinated expression of HuD and GAP-43 mRNA in the hippocampus during developmental and adult plasticity. Neurochem Res. 2007a;32:2142–2151. doi: 10.1007/s11064-007-9388-8. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Qiu S, Tanner DC, Paik J, Perrone-Bizzozero NI, Weeber EJ. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol Learn Mem. 2007b;87:635–643. doi: 10.1016/j.nlm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38:117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock WM, Bolognani F, Botta P, Valenzuela CF, Perrone-Bizzozero NI. Schizophrenia-like GABAergic gene expression deficits in cerebellar Golgi cells from rats chronically exposed to low-dose phencyclidine. Neurochem Int. 2009;55:775–782. doi: 10.1016/j.neuint.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Clayton GH, Perez GM, Smith RL, Owens GC. Expression of mRNA for the ELAV-like neural-specific RNA binding protein, HuD, during nervous system development. Brain Res Dev Brain Res. 1998;109:271–280. doi: 10.1016/s0165-3806(98)00074-1. [DOI] [PubMed] [Google Scholar]
- Deschênes-Furry J, Perrone-Bizzozero NI, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. BioEssays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- Deschênes-Furry J, Mousavi K, Bolognani F, Neve RL, Parks RJ, Perrone-Bizzozero NI, Jasmin BJ. The RNA-binding protein HuD binds acetylcholinesterase mRNA in neurons and regulates its expression after axotomy. J Neurosci. 2007;27:665–675. doi: 10.1523/JNEUROSCI.4626-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Miyashiro K, Kacharmina JE, Job C. Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc Natl Acad Sci USA. 2001;98:7080–7085. doi: 10.1073/pnas.121146698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Zhang H, Su Y, Silani V, Singer RH, Rossoll W, Bassell GJ. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M, Giuliani A, Pirondi S, D'Intino G, Giardino L, Aloe L, Levi-Montalcini R, Calzà L. Thyroid hormone administration enhances remyelination in chronic demyelinating inflammatory disease. Proc Natl Acad Sci USA. 2004;101:16363–16368. doi: 10.1073/pnas.0407262101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming CE, Saraiva MJ, Sousa MM. Transthyretin enhances nerve regeneration. J Neurochem. 2007;103:831–839. doi: 10.1111/j.1471-4159.2007.04828.x. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D, Sergent-Tanguy S, Thinard R, Bonnamain V, Masip M, Fabre A, Boudin H, Neveu I, Naveilhan P. AUF1 and Hu proteins in the developing rat brain: implication in the proliferation and differentiation of neural progenitors. J Neurosci Res. 2009;87:1296–1309. doi: 10.1002/jnr.21957. [DOI] [PubMed] [Google Scholar]
- Hubers L, Valderrama-Carvajal H, Laframboise J, Timbers J, Sanchez G, Côté J. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum Mol Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- Kasashima K, Terashima K, Yamamoto K, Sakashita E, Sakamoto H. Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells. 1999;4:667–683. doi: 10.1046/j.1365-2443.1999.00292.x. [DOI] [PubMed] [Google Scholar]
- Keeley MB, Wood MA, Isiegas C, Stein J, Hellman K, Hannenhalli S, Abel T. Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn Mem. 2006;13:135–142. doi: 10.1101/lm.86906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder JM, Mugnaini E. Infrapyramidal mossy fibers in the hippocampus of the hyperthyroid rat. A light and electron microscopic study. Dev Neurosci. 1980;3:248–265. doi: 10.1159/000112397. [DOI] [PubMed] [Google Scholar]
- Liz MA, Fleming CE, Nunes AF, Almeida MR, Mar FM, Choe Y, Craik CS, Powers JC, Bogyo M, Sousa MM. Substrate specificity of transthyretin: identification of natural substrates in the nervous system. Biochem J. 2009;419:467–474. doi: 10.1042/BJ20082090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüesse HG, Roskoden T, Linke R, Otten U, Heese K, Schwegler H. Modulation of mRNA expression of the neurotrophins of the nerve growth factor family and their receptors in the septum and hippocampus of rats after transient postnatal thyroxine treatment. I. Expression of nerve growth factor, brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin 4 mRNA. Exp Brain Res. 1998;119:1–8. doi: 10.1007/s002210050313. [DOI] [PubMed] [Google Scholar]
- Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JL, Xuan S, Dragatsis I, Efstratiadis A, Goldman JE. Insulin-like growth factor (IGF) signaling through type 1 IGF receptor plays an important role in remyelination. J Neurosci. 2003;23:7710–7718. doi: 10.1523/JNEUROSCI.23-20-07710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Kawakami F, Kobayashi K, Takahashi T. Developmental decrease in synaptic facilitation at the mouse hippocampal mossy fibre synapse. J Physiol. 2003;553:37–48. doi: 10.1113/jphysiol.2003.045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgung U, Routtenberg A. Transcriptional and post-transcriptional regulation of a brain growth protein: regional differentiation and regeneration induction of GAP-43. Eur J Neurosci. 2000;12:3124–3136. doi: 10.1046/j.1460-9568.2000.00196.x. [DOI] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palha JA, Hays MT, Morreale de Escobar G, Episkopou V, Gottesman ME, Saraiva MJ. Transthyretin is not essential for thyroxine to reach the brain and other tissues in transthyretin-null mice. Am J Physiol. 1997;272:E485–E493. doi: 10.1152/ajpendo.1997.272.3.E485. [DOI] [PubMed] [Google Scholar]
- Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc Natl Acad Sci USA. 2004;101:1217–1222. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J Neurosci Res. 2002;68:121–126. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- Proper EA, Oestreicher AB, Jansen GH, Veelen CW, van Rijen PC, Gispen WH, de Graan PN. Immunohistochemical characterization of mossy fibre sprouting in the hippocampus of patients with pharmaco-resistant temporal lobe epilepsy. Brain. 2000;123:19–30. doi: 10.1093/brain/123.1.19. [DOI] [PubMed] [Google Scholar]
- Richardson SJ, Lemkine GF, Alfama G, Hassani Z, Demeneix BA. Cell division and apoptosis in the adult neural stem cell niche are differentially affected in transthyretin null mice. Neurosci Lett. 2007;421:234–238. doi: 10.1016/j.neulet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Smith CL, Afroz R, Bassell GJ, Furneaux HM, Perrone-Bizzozero NI, Burry RW. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J Neurobiol. 2004;61:222–235. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- Stanić D, Malmgren H, He H, Scott L, Aperia A, Hökfelt T. Developmental changes in frequency of the ciliary somatostatin receptor 3 protein. Brain Res. 2009;1249:101–112. doi: 10.1016/j.brainres.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Tanner DC, Qiu S, Bolognani F, Partridge LD, Weeber EJ, Perrone-Bizzozero NI. Alterations in mossy fiber physiology and GAP-43 expression and function in transgenic mice overexpressing HuD in adult dentate granule cells. Hippocampus. 2008;18:814–823. doi: 10.1002/hipo.20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KJ, Yang CH, Lee PC, Wang WT, Chiu MJ, Shen CK. Asymmetric expression patterns of brain transthyretin in normal mice and a transgenic mouse model of Alzheimer's disease. Neuroscience. 2009;159:638–646. doi: 10.1016/j.neuroscience.2008.12.045. [DOI] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu Y, Weston JA. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Yoo S, van Niekerk EA, Merianda TT, Twiss JL. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp Neurol. 2010;223:19–27. doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller RT, Rovet J. Timing of thyroid hormone action in the developing brain: clinical observations and experimental findings. J Neuroendocrinol. 2004;16:809–818. doi: 10.1111/j.1365-2826.2004.01243.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.