Abstract
Sigma receptors are small membrane proteins implicated in a number of pathophysiological conditions including drug addiction, psychosis and cancer; thus, small molecule inhibitors of sigma receptors have been proposed as potential pharmacotherapeutics for these diseases. We previously discovered that endogenous monochain N-alkyl sphingolipids including D-erythro-sphingosine, sphinganine, and N,N-dimethyl sphingosine bind to the sigma-1 receptor at physiologically relevant concentrations [Ramachandran et al. 2009 Eur J Pharmacol. 609(1–3):19–26]. Here, we investigated several N-alkylamines of varying chain lengths as sigma receptor ligands. Although the KI values for N-alkylamines were found to be in the micromolar range, when N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl derivatives of butyl- (1a and 1b), heptyl- (2a and 2b), dodecyl- (3a and 3b), and octadecyl-amine (4a and 4b) were evaluated as sigma receptor ligands we found that these compounds exhibited nanomolar affinities with both sigma-1 and sigma-2 receptors. A screen of the high affinity ligands 2a, 2b, 3a and 3b against a variety of other receptors/transporters confirmed these four compounds to be highly selective mixed sigma-1 and sigma-2 ligands. Additionally, in HEK293 cells reconstituted with Kv1.4 potassium channel and the sigma-1 receptor, these derivatives were able to inhibit the outward current from the channel – consistent with sigma receptor modulation. Finally, cytotoxicity assays showed that 2a, 2b, 3a and 3b were highly potent against a number of cancer cell lines, demonstrating their potential utility as mixed sigma-1 and sigma-2 receptor anti-cancer agents.
Two subtypes of the sigma receptor have been described, sigma-1 and sigma-2, which are distinguishable by their pharmacological profiles, selectivity, functions, subcellular locations, and molecular weight. The sigma-1 receptor was cloned in 1996 (1) and has since been extensively characterized, while very little is known about the sigma-2 receptor that is yet to be cloned. The sigma-1 receptor is a membrane protein of 25.3 kDa and has been recognized in recent years as a small molecule operated chaperone important for the regulation of Ca2+ entry from the ER to the mitochondria (2). Additionally, sigma ligands can also modulate the function of various voltage-gated ion channels including potassium (K+) (3), sodium (Na+) (4), calcium (Ca2+) (5), and chloride (Cl−) channels (6) independent of G proteins or phosphorylation. A direct physical interaction between the sigma-1 receptor and voltage gated potassium channels (3) and between the sigma-1 receptor and the acid-sensing ion channel ASIC 1a (7) have also been demonstrated leading to the suggestion that the sigma-1 receptor is a regulatory subunit for certain ion channels.
Depending on cell types, the sigma-1 receptor has been localized to different subcellular regions including the endoplasmic reticulum (8, 9), the mitochondria associated ER membrane (MAM) (2), the ER cisternae proximal to the plasma membrane (10), in focal adhesion contacts (11), at the plasma membrane (3) and at the nuclear membranes (12). Functional modulation of the sigma-1 receptor may be achieved pharmacologically by synthetic small molecules and those present endogenously such as progesterone (13), N,N-dimethyltryptamine (14), and/or sphingolipids (8, 15). Therefore, the plethora of physiological outcomes resulting from treatments with sigma receptor ligands may be explained by the differential subcellular localization and wide tissue distribution of the sigma-1 receptor. For a review on the sigma-1 receptor as an inter-organelle signaling modulator refer to Su et al. 2010.
Both sigma receptor subtypes are expressed in high density in many human tumors and cancer cell lines (16–19). These tumors and cancer cell lines include those from the breast, lung, colon, ovaries, prostate, and brain (18, 20–22). The elevated levels of both sigma receptor subtypes in many cancer cell lines has led to a concerted effort in the development of sigma receptor targeting anti-cancer agents and imaging tools (23–25). For example, small molecule sigma-1 receptor antagonists, BD-1047 and BD-1063, have been reported to inhibit cancer cell survival while the agonist, (+)-SKF-10,047 and (+)-pentazocine, abrogated these effects (24). Additionally, a sigma-1 receptor antagonist, rimcazole, was shown to initiate tumor-selective and caspase dependent apoptosis, which could be rescued by the agonist (+)-pentazocine (26). Sigma-2 receptor ligands including siramesine, SV119 and SW43 have also been used to augment conventional chemotherapeutic agents in pre-clinical model of pancreatic cancer (25). Interestingly, the sigma-1 and sigma-2 receptors are suggested to have opposite cellular function since inhibition of the sigma-1 receptor (by treatment with antagonists) and activation of the sigma-2 receptor (with sigma-2 receptor agonists) resulted in anti-proliferative effects against cancer cells (27). Sigma-1 and sigma-2 receptor ligands have also been aggressively pursued in the area of tumor imaging in light of their elevated expressions in cancer cells. For example, Mach and co-workers compared the non-selective high affinity radioligand N-[1-(4′-[18F]fluorobenzyl)piperidin-4-yl]-3-bromophenylacetamide ([18F]FBPBPA) with 2-deoxy-2-[18F]fluoro-D-glucose ([18F]FDG) and 5-[125I]iodo-2′-deoxyuridine ([125I]IUdR) in tumor imaging study of nude mice with tumors grown from the mouse mammary adenocarcinoma cell line 66. They confirmed receptor-specific uptake of the tracer (~25%) in the tumor and in the brain (~60%) with the following order of tumor uptake [18F]FDG > [18F]FBPBPA > [125I]IUdR (16). Additionally, the tumor-to blood and tumor-to-muscle ratios were larger for [18F]FBPBPA than for both [18F]FDG and [125I]IUdR. These studies demonstrated the utility for sigma-1 receptor imaging agents as tools for anti-cancer therapy. Several recent reviews are focused on the development and use of sigma ligands for these purposes (28, 29).
Structure activity studies of sigma-1 receptor ligands have mainly focused on N,N-diarylalkylamine (30) and N,N-dialkyl-phenylpropyl amine molecules (14) but in general have overlooked long chain N-alkylamines such as tridemorph (31), D-erythro-sphingosine (15), N,N-dimethyl-sphingosines (15), and glycosphingolipids such as galactosylceramide and lactosylceramide (8) which also bind to the sigma-1 receptor. To expand our knowledge regarding the interaction of sphingosine-like molecules with the sigma-1 receptor, we designed, synthesized, and evaluated N-alkylamine derivatives with the goal of developing high affinity and selective sigma receptor ligands. In addition, we report the selectivity of these N-alkylamine derivatives compared to over 40 other membrane receptor and transporter targets. Finally, their utility as potential anti-cancer agents was analyzed through multiplex cytotoxicity assays measuring growth inhibition of various cancer cell lines.
Materials and Methods
Materials
Chemicals were purchased from Aldrich Chemical Co. (Milwaukee, WI) and utilized without further purification. [3H]-(+)-pentazocine and [3H]-ditolyl guanidine (DTG) were purchased from Perkin Elmer Life Sciences (Wellesley, MA).
Chemical syntheses
Synthesis of 1-bromo-3-(4-nitrophenyl)propane
The preparation of 1-bromo-3-(4-nitrophenyl)propane from 1-bromo-3-phenylpropane was performed according to Hajipour A.R. et al. (32). Briefly, P2O5 and silica gel was ground at room temperature for 1 min to homogeneity. P2O5/silica gel mixture (1 mmol) and 1-bromo-3-phenylpropane (1 mmol, 0.199 g) were combined and ground for 30 s before 0.5 ml of 65% HNO3 was added. Additional grinding was carried out until TLC (n-hexane/ethyl acetate 9:1) showed a complete disappearance of 1-bromo-3-phenylpropane (~ 6 min). The product was extracted from the silica gel with ethyl acetate (10 ml), washed with H2O (10 ml) and dried with anhydrous MgSO4. Solvent was evaporated under reduced pressure and the product was purified by column chromatography (n-hexane/ethyl acetate, 9:1). 1-bromo-3-(4-nitrophenyl)propane was obtained as a yellow oil (0.74 mmol, 0.18 g 74%).
Synthesis of N-alkylamine derivatives
In a round bottom flask equipped with a magnetic stirrer and a condenser, 2 mmol of 1-bromo-3-phenylpropane (0.4 g) or 1-bromo-3-(4-nitrophenyl)propane (0.49 g) and 10 mmol of appropriate amine (5 equivalent) were refluxed in ethanol (10 ml) until TLC (n-hexane/ethyl acetate 9:1) showed a complete disappearance of the 1-bromo-3-phenylpropane or 1-bromo-3-(4-nitrophenyl)propane (3 h). The reaction mixture was quenched with water and extracted 3 times with 5 ml ethyl acetate. The combined extracts were dried with anhydrous MgSO4, and evaporated under reduced pressure. The purification of the product was performed by column chromatography using silica gel and n-hexane/ethyl acetate 9:1.
Structural characterizations of N-alkylamine derivatives
Yields refer to isolated products after column chromatography. Products were characterized by 13C, 1H and IR spectroscopy and mass spectrometry. All 1H NMR spectra were recorded at 300 MHz in CDCl3 relative to TMS (0.00 ppm), and IR spectra were recorded on a Shimadzu 435 IR spectrometer and these were performed at the Research Institute of Petroleum Industry, Tehran, Iran.
N-(3-Phenylpropyl)butan-1-amine (1a)
Yellow oil, Yield (0.30 g, 78 %, 1.5 mmol). IR (KBr): 1664 cm−1. 1H NMR (CDCl3): δ 7.21–7.08 (m, 5 H), 2.4 (m, 6 H), 2.00 (m, 4 H), 1.34 (m, 4H), 0.96 (t, 3 H, J= 7.4). 13C NMR (CDCl3): δ 138.1, 128.9, 128.2, 126.1, 49.6, 49.4, 33.4, 32.8, 29.6, 20.2, 13.8. MS (CI) m/z 191 (100, M+). Anal. Calcd for C13H21N: C, 81.62; H, 11.06; N, 7.32%. Found; C, 81.50; H, 11.20; N, 7.20%.
N-(3-(4-Nitrophenyl)propyl)butan-1-amine (1b)
Red oil, Yield (0.33 g, 70 %, 1.4 mmol). IR (KBr): 1664 cm−1. 1H NMR (CDCl3): δ 8.21 (d, J=8.3, 2 H), 7.55 (d, J=8.3, 2 H), 2.4 (m, 6 H), 2.00 (m, 4 H), 1.34 (m, 4H), 0.96 (t, 3 H, J= 7.4). 13C NMR (CDCl3): δ 148.1, 145.2, 130.3, 124.0, 49.6, 49.5, 32.7, 30.8, 29.5, 20.1, 13.8. MS (CI) m/z 236 (100, M+). Anal. Calcd for C13H20N2O2: C, 66.07; H, 8.53; N, 11.85%. Found; C, 65.90; H, 8.70; N, 11. 80%.
N-(3-Phenylpropyl)heptan-1-amine (2a)
Yellow oil, yield (6.0 g, 100%, 25.7 mmol). IR (KBr): 1660 cm−1. 1H NMR (CDCl3): δ 7.40–7.28 (m, 5 H), 2.62–2.55 (m, 6 H), 1.95 (m, 3 H), 1.38–1.10 (m, 10 H), 0.88 (t, 3 H, J= 7.5). 13C NMR (CDCl3): δ 148.1, 145.2, 130.3, 122.1, 49.9, 49.5, 31.8, 30.8, 30.5, 27.1, 22.6, 14.1. MS (CI) m/z 233 (100, M+). Anal. Calcd for C16H27N: C, 82.34; H, 11.66; N, 6.00%. Found; C, 82.40; H, 11.70; N, 6.10%
N-(3-(4-Nitrophenyl)propyl)heptan-1-amine (2b)
Red oil, yield (0.57 g, 100%, 2 mmol). IR (KBr): 1660 cm−1. 1H NMR (CDCl3): δ 8.21 (d, J=8.6, 2 H), 7.55 (d, J=8.6, 2 H), 2.62–2.55 (m, 6 H), 1.95 (m, 3 H), 1.38–1.10 (m, 10 H), 0.88 (t, 3 H, J= 7.5). 13C NMR (CDCl3): δ 142.1, 128.8, 126.0, 49.9, 49.5, 31.8, 30.8, 30.5, 27.1, 22.6, 14.1. MS (CI) m/z 278 (100, M+). Anal. Calcd for C16H26N2O2: C, 69.03; H, 9.41; N, 10.06%. Found; C, 68.80; H, 9.60; N, 9.60%.
N-(3-Phenylpropyl)dodecan-1-amine (3a)
Oil, yield (0.5 g, 82 %, 1.6 mmol). IR (KBr): 1661 cm−1. 1H NMR (CDCl3): δ 7.40–7.29 (m, 5 H), 2.58 (m, 6 H), 1.98 (m, 3 H), 1.30 (m, 20 H), 0.88 (t, 3 H, J= 7.5). 13C NMR (CDCl3): δ 142.1, 128.8, 127.6, 126.4, 49.9, 49.5, 30.7, 30.5, 29.6, 29.5, 29.3, 27.0, 22.7, 14.1. MS (CI) m/z 303 (100, M+). Anal. Calcd for C21H37N: C, 83.10; H, 12.29; N, 4.61%. Found; C, 82.90; H, 12.50; N, 4.50%.
N-(3-(4-Nitrophenyl)propyl)dodecan-1-amine (3b)
Yield (0.55 g, 78 %, 1.6 mmol). IR (KBr): 1660 cm−1. 1H NMR (CDCl3): δ 8.21 (d, J=8.4, 2 H), 7.55 (d, J=8.4, 2 H), 2.56 (m, 6 H), 2.01 (m, 3 H), 1.30 (m, 20 H), 0.88 (t, 3 H, J= 7.5). 13C NMR (CDCl3): δ 148.1, 145.2, 130.3, 124.0, 49.6, 49.5, 30.7, 30.5, 29.6, 29.5, 29.3, 27.0, 22.7, 14.1. MS (CI) m/z 348 (100, M+). Anal. Calcd for C21H36N2O2: C, 72.37; H, 10.41; N, 8.04%. Found; C, 72.10; H, 10.50; N, 8.00%.
N-(3-Phenylpropyl)octadecan-1-amine (4a)
Yield (0.62 g, 80 %, 1.6 mmol). IR (KBr): 1660 cm−1. 1H NMR (CDCl3): δ 7.28 (m, 5 H), 2.58 (m, 6 H), 2.0 (m, 3 H), 1.30 (m, 32 H), 0.88 (t, 3 H, J= 7.5). 13C NMR (CDCl3): δ 142.1, 128.8, 127.6, 126.4, 49.9, 49.5, 30.7, 30.5, 29.6, 29.5, 29.3, 27.0, 22.7, 14.1. MS (CI) m/z 387 (100, M+). Anal. Calcd for C27H49N: C, 83.65; H, 12.74; N, 3.61%. Found; C, 83.70; H, 12.80; N, 3.50%.
N-(3-(4-Nitrophenyl)propyl)octadecan-1-amine (4b)
Yield (0.80 g, 92 %, 1.85 mmol). IR (KBr): 1660 cm−1. 1H NMR (CDCl3): δ 8.21 (d, J=8.4, 2 H), 7.55 (d, J=8.4, 2 H), 2.56 (m, 6 H), 2.01 (m, 3 H), 1.30 (m, 32 H), 0.88 (t, 3 H, J= 7.5). 13C NMR (CDCl3): δ 148.1, 145.2, 130.3, 124.0, 49.6, 49.5, 30.7, 30.5, 29.6, 29.5, 29.3, 27.0, 22.7, 14.1. MS (CI) m/z 432 (100, M+). Anal. Calcd for C27H48N2O2: C, 74.95; H, 11.18; N, 6.47%. Found; C, 74.80; H, 11.40; N, 6.70%.
Overexpression and purification of the sigma-1 receptor from E. coli
The guinea pig sigma-1 receptor was expressed and purified as previously described (33) using the pMal P2X plasmid (New England Biolabs, Ipswich, MA) encoding the guinea pig sigma-1 receptor fused to the maltose binding protein (MBP) at its N-terminus and carried a hexahistidine epitope tag at its C-terminus. Proteins were expressed in the E. coli strain BL21(DE3) (Novagen, Madison, WI). Cells were grown to an OD600 of 0.7 before induction with 0.6 mM IPTG for 4 h at 37°C. The collected E. coli pellet was resuspended in buffer I (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 1 mM 2-mercaptoethanol, and 1 mM EDTA) and the cell suspension was sonicated using a Branson soniWer 250 employing a 1 cm probe (output 50%, 2 s bursts, 1 s lag) for 15 min on ice. The cell lysate was centrifuged at 100,000 g for 1 h to separate total particulate and soluble proteins. The particulate fraction was extracted with a 4:1 ratio of Triton X-100 to total protein (w/w) for 3 h with stirring at 4°C. The extracted material was centrifuged at 100,000 g for 1 h and the supernatant was diluted with a volume of buffer I that reduces the Triton X-100 concentration to 0.5% – 1% before loading onto an amylose column (New England Biolabs, Ipswich, MA). After washing the resin once with 5 column volumes of buffer II (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 1 mM 2-mercaptoethanol, 1 mM EDTA, 0.5% TX-100) and once with 3 column volumes of buffer III (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 5 mM CaCl2, 0.5% TX-100), the MBP-sigma-1 receptor fusion protein was eluted with 3 column volumes of buffer IV (20 mM Tris-Cl pH 7.5, 200 mM NaCl, 5 mM CaCl2, 10 mM maltose, 0.5% TX-100).
The pure MBP-sigma-1-receptor fusion protein was cleaved with Factor Xa protease (Novagen, Madison, WI) in a volume of 5 ml at room temperature for 24 – 48 h and the cleavage monitored by SDS-polyacrylamide gel electrophoresis. The sigma-1 receptor from the Factor Xa cleavage was purified with HIS-Select HC Nickel affinity gels (Sigma, St. Louis, MO) in a batch format with overnight incubation on Ni2+ beads at 4°C, then 3 washes each lasting 15 min in buffer V (50 mM Na2HPO4 pH 8, 200 mM NaCl, 0.5% TX-100), and eluted for 1 h in buffer VI (50 mM Na2HPO4 pH 8, 200 mM NaCl, 250 mM imidazole, 0.5% TX-100) at RT. Centrifugation for each step of the Ni2+ purification was performed at 5000 rpm for 1 min at RT in a bench-top microcentrifuge.
Preparation of guinea pig liver membranes (GPLM) and rat liver membranes (RLM)
Membranes were prepared as described previously (34) from frozen tissues (Pel Freez Biologicals, Rogers, AR). The liver tissue was homogenized (10 ml buffer/g wet tissue) by 4 bursts of 10 s each using a brinkman polytron (American Laboratory Trading Inc., East Lyme, CT) on setting 6 in ice cold sodium phosphate buffer (10 mM pH 7.4) containing 0.32 M sucrose and a cocktail of protease inhibitors (20 μg/ml leupeptin, 5 μg/ml soybean trypsin inhibitor, 100 μM phenylmethylsulfonyl fluoride (PMSF), 100 μM benzamidine and 1 mM EDTA). The membrane suspension after homogenization was centrifuged for 10 min at 17,000 g and the supernatant from this first centrifugation was further centrifuged at 100,000 g to collect the membrane fraction. The pellet from the second high speed spin was resuspended in the same buffer as above, snap frozen and stored at −80°C at a protein concentration of 10 mg/ml.
Competitive displacement of [3H]-(+)-pentazocine and [3H]-DTG binding
Competition ligand binding to the pure sigma-1 receptor and the sigma-1 receptor from guinea pig liver membranes (GPLM) was performed as described previously (33). Binding was carried out in 50 mM Tris-Cl, pH 8.0 in a total volume of 100 μL containing 50 – 100 ng of the pure sigma-1 receptor, 10 nM [3H]-(+)-pentazocine and various concentrations of the inhibitors to be tested. After incubation at 32°C for 60 min, the reaction was terminated by rapid filtration through glass fiber filters (Whatman GF/B, Whatman, Maidstone, UK), using a Brandel cell harvester (Brandel, Gaithersburg, MD). The glass fiber filters were pre-soaked in 0.5% polyethyleneimine (PEI) for at least 1 h at room temperature. Filters were washed 4 times with 4 ml of ice-cold 50 mM Tris-Cl, pH 8.0. Radioactivity was quantified by liquid scintillation (Ultima Gold, Perkin Elmer, Waltham, MA) counting using a liquid scintillation counter (Packard model 1600CA, Packard Instrument Co., Downers Grove, IL). The affinity determinations for the sigma-2 receptor were performed in rat liver membranes (RLM) similarly to that described above using 20 nM [3H]-DTG as the radioligand with 100 nM (+)-pentazocine added to selectively mask the sigma-1 receptor binding sites in these assays. IC50 values were then converted to KI values using the Cheng – Prusoff correction with the following equation KI = IC50/(1 + [L]/KD) where [L] is the ligand concentration of the radioactive molecule, and KD is the previous determined dissociation constant (10 nM for [3H]-(+)-pentazocine, and 20 nM for [3H]-DTG).
Alternate receptor and transporter targets determination
KI determinations for receptor binding profiles to alternate targets were performed by the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP) directed by Bryan L. Roth MD, PhD at the University of North Carolina at Chapel Hill and Project Officer Jamie Driscol at NIMH, Bethesda MD, USA. For detailed methodology, please refer to the PDSP web site http://pdsp.med.unc.edu/. A default concentration of 10 μM of N-alkylamine derivatives was chosen for the primary screening against the binding of a radioligand to the target receptor/transporter. Selected receptor and transporter targets with affinities to the sigma-1 receptor, their specific radioligand and concentrations are listed in the order shown in Table 3: The protein source used in these assays for the sigma-1 receptor was a rat brain homogenate; for the sigma-2 receptor was PC2 cells, and for the other membrane receptors/transporters, either transient or stably transfected cells were used, e.g. COS-7, HEK293, CHO, NIH3T3. Total bound radioactivity was estimated from quadruplicate wells containing no test or reference compound and adjusted to 100%; non-specifically bound radioactivity was assessed from quadruplicate wells containing 10 μM of the suitable reference compound and adjusted to 0%. The average bound radioactivity in the presence of the test compound (10 μM) quadruplicate determinations was then expressed on a percentage scale. The percentage inhibition of radioligand binding was calculated as follows: % inhibition = 100% − % radioactivity bound.
Table 3. KI values of selected N-alkylamine derivatives for alternate receptor/transporter targets.
KI values of 2a, 2b, 3a and 3b for alternate receptor/transporter targets. Sig-1R, sigma-1 receptor; Sig-2R, sigma-2 receptor; 5HT (5-hydroxytryptamine/serotonin receptor) – subtype 1A, 1B, 2B and 7; α (α – adrenergic receptor) subtype 1A and 1D; β (β-adrenergic receptor) subtype 2 and 3; D (dopamine receptor) type 1 and 4; H (histamine receptor) type 1, 2 and 3; M (muscarinic acetylcholine receptor) type 1, 2, 3, 4 and 5; NET (norepinephrine transporter); SERT (serotonin transporter).
| Receptors/Transporters | Radioligand/Concentration (nM) | KI (μM) | |||
|---|---|---|---|---|---|
| 2a | 2b | 3a | 3b | ||
| Sig-1R | [3H]-(+) Pentazocine/3 nM | 0.09 | 0.02 | 1.5 | 0.20 |
| Sig-2R | [3H]-DTG/3 nM | 0.25 | 0.014 | 3.6 | 0.27 |
| 5HT1A | [3H]8-OH-DPAT/0.5 nM | 0.13 | 0.05 | >10.0 | 0.04 |
| 5HT1B | [3H]GR127543/0.3 nM | >10.0 | >10.0 | >10.0 | >10.0 |
| 5HT2B | [3H]LSD/1 nM | >10.0 | 1.1 | >10.0 | 7.9 |
| 5HT7 | [3H]LSD/1 nM | >10.0 | 0.9 | >10.0 | >10.0 |
| α-1A | [3H]Prazosin/0.7 nM | >10.0 | >10.0 | >10.0 | >10.0 |
| α-1D | [125I]HEAT/0.1 nM | >10.0 | 6.9 | >10.0 | 9.0 |
| β-2 | [125I]Iodopindolol/0.1 nM | >10.0 | >10.0 | >10.0 | >10.0 |
| β-3 | [125I]Iodopindolol/0.1 nM | >10.0 | >10.0 | >10.0 | >10.0 |
| D1 | [3H]SCH233930/0.2 nM | >10.0 | >10.0 | >10.0 | >10.0 |
| D4 | [3H]N-methylspiperone/0.3 nM | 0.39 | 0.24 | >10.0 | 1.8 |
| H1 | [3H]Pyrilamine/0.9 nM | >10.0 | 9.2 | >10.0 | >10.0 |
| H2 | [3H]Tiotidine/3 nM | >10.0 | 0.6 | >10.0 | 3.9 |
| H3 | [3H]a-methylhistamine/0.4 nM | >10.0 | >10.0 | >10.0 | >10.0 |
| M1 | [3H]QNB/0.5 nM | >10.0 | 1.9 | >10.0 | >10.0 |
| M2 | [3H]QNB/0.5 nM | >10.0 | 3.2 | >10.0 | 5.6 |
| M3 | [3H]QNB/0.5 nM | >10.0 | 1.1 | >10.0 | 3.3 |
| M4 | [3H]QNB/0.5 nM | >10.0 | 1.4 | >10.0 | 4.6 |
| M5 | [3H]QNB/0.5 nM | >10.0 | 0.3 | >10.0 | 3.6 |
| NET | [3H]Nisoxetine/0.5 nM | >10.0 | 4.0 | >10.0 | >10.0 |
| SERT | [3H]Citalopram/0.5 nM | >10.0 | 1.1 | >10.0 | 0.7 |
The secondary assays to determine the KI values were performed only for targets with ≥50% inhibition in the primary screen. The radioligands used in the secondary screen were the same as in the primary screen.
Patch Clamp Recordings
A pIRES bicistronic mammalian expression plasmid (BD Biosciences Clonetech, Mountain View, CA) was constructed which carried the rat Kv1.4 in the first multiple cloning site and the rat sigma-1 receptor in the second multiple cloning site. Human Embryonic Kidney (HEK-293) cells were cultured on 60 mm dishes and transfected with 3 μL of Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and 2 μg of pIRES KvS1R and 0.3 μg of pEGFP (BD Biosciences Clonetech, Mountain View, CA) following manufacturers instructions. Thirty six to forty eight hours after transfection, whole cell K+ currents were recorded from GFP positive HEK cells under voltage clamp using standard patch clamp techniques (35), using patch pipettes filled with (in mM) 120 KCl, 10 NaCl, 10 EGTA, 2 MgCl2, 2–4 MgATP, 0.3 NaGTP, 10 HEPES, pH 7.3. The external solution was composed of (in mM) 130 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, pH 7.4 at room temperature (22–24°C). Patch pipettes with resistances from 1–3 MΩ were fabricated from borosilicate glass (Garner Glass Co., Claremont, CA). K+ channels are activated by voltage step from −120 mV to 20 mV for 250 ms. Axopatch 200B, patch clamp amplifier interfaced to a PC running PCLAMP 7 (Axon Instruments, Foster City, CA) was used for data acquisition. Drugs were applied by a gravity feed system at a flow rate of 1–3 ml/min. Series resistance compensation was used for cells with series resistance values higher than 10 MΩ. We were able to achieve 80–90% series resistance compensation with such cells.
Multiplex Cytotoxicity Assays against a panel of cancer cell lines
The multiplex cytotoxicity assays on an array of cancer cell lines were performed at the Small Molecule and Medicinal Chemistry Screening Facility at the University of Wisconsin School of Medicine and Public Health with funding from the Wisconsin Alumni Research Foundation (WARF) Lead Discovery Initiative from WARF.
Compound handling
Working stocks were prepared at a final concentration of 100X in anhydrous DMSO. Serial dilutions were made in DMSO in 96 well polypropylene plates using the Precision XS liquid handler (BioTek, Inc. Winooski, VT). Compounds were divided equally into the corresponding wells of a 384 well plate in all 4 quadrants using a Biomek FX liquid handler with 96 channel pipetting head (Beckman-Coulter Inc., Miami, FL). Compounds were stored at −20°C in 100% DMSO until the day of the assay(s). Freeze-thaw cycles were minimized to a maximum of 10 per plate.
Cytotoxicity assays
All cell lines were maintained as previously reported (36). Cells were harvested by trypsinization using 0.25% trypsin and 0.1% EDTA and then counted in a Cellometer Auto T4 cell counter (Nexcelom LLC, Lawrence, MA), before dilution for assay plating. Cell plating, compound handling and assay set up were performed as previously reported (36) except that cells were plated in 50 μL volumes in 384 well clear bottom, tissue culture plates (Corning-Costar, Cambridge, MA). Compounds were added from the 384-well compound stock plates at a 1:100 dilution using a Biomek FX liquid handler equipped with a 384 channel head (Beckman Coulter Inc., Miami, FL). Acetoxymethyl ester (Calcein AM, Molecular Probes, Eugene, OR) reagent and ethidium homodimer -1 (EthD-1, Molecular Probes, Eugene, OR) at 30 μL, 10 μM CAM and 100 μM EthD-1 were added and the cells were incubated for 30 min at 37°C. Plates were read for emission by using the appropriate wavelengths for Calcein AM and EthD-1 following manufacturers’ instructions. 15 μL of CellTiter-Glo reagent (Promega Corporation, Madison, WI) was added and incubated for 10 min at room temperature with gentle agitation to lyse the cells. Each plate was re-read for luminescence to confirm the inhibition observed in the fluorescent Calcein AM/EthD-1 assay.
Data analysis
IC50 for cytotoxicity assays were determined for Calcein AM, the ratio of LIVE/DEAD cells (Calcein AM/EthD-1) and CellTiter-Glo using XLfit 4.0 as previously reported in Langenhan, et al. (36). The IC50 determined using the best-fit curve for dosage response from any of the 3 assays is reported as the final IC50. The best-fit curve is defined as the curve that has the lowest standard error. If the standard error exceeds 10% of the IC50, the assay was deemed a failure and repeated.
Results
N-alkylamines bind to the sigma-1 receptor
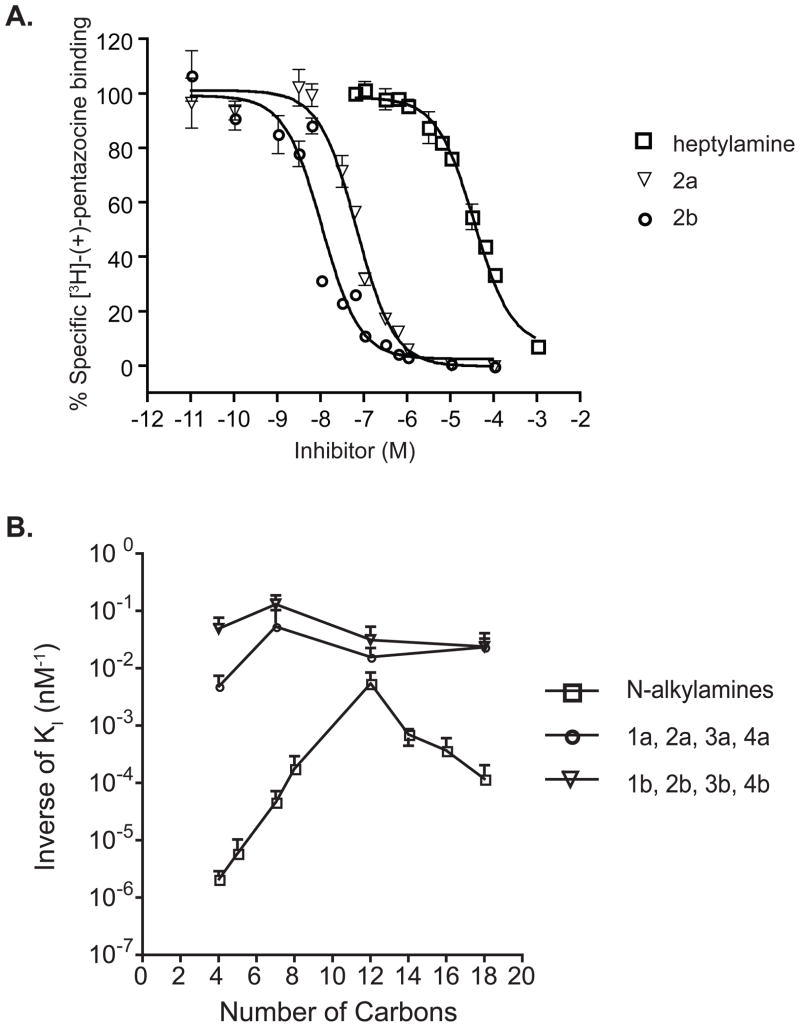
The discovery that D-erythro-sphingosine binds to the sigma-1 receptor with relatively high affinity (15) prompted us to study monochain sphingosine-like N-alkylamine molecules in order to determine the relationship between sigma-1 receptor affinity and the number of alkyl carbons on the molecule. We performed competitive displacement of [3H]-(+)-pentazocine binding to the pure guinea pig sigma-1 receptor (33) and found that affinities of N-alkylamines for the sigma-1 receptor increased in the series (Table 1). The data show a positive correlation between the number of carbons and affinity for the sigma-1 receptor from butylamine to dodecylamine. However, further increase in carbon chain length showed a decrease in affinity above dodecylamine (Table 1).
Table 1.
KI values of N-alkylamine for the pure sigma-1 receptor.
| Compounds | KI (μM) pure sigma-1 receptor |
|---|---|
| Butylamine | 477 ± 67 |
| Amylamine | 167 ± 35 |
| Heptylamine | 21 ± 9 |
| Octylamine | 6 ± 3 |
| Dodecylamine | 0.2 ± 0.02* |
| Tretradecylamine | 1.0 ± 0.03 |
| Hexadecylamine | 3.0 ± 0.36 |
| Octadecylamine | 8.5 ± 5.5* |
Values obtained from Ramachandran et al. (2010) Eur J Pharmacol 609, 19–26.
Addition of N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl to N-alkylamines resulted in an increase in affinity to both sigma-1 and sigma-2 receptors
The presence of a nitrogen has been reported to be an important pharmacophoric element for sigma-1 receptor binding (37). Furthermore, N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl substitution on the obligate nitrogen of sigma-1 receptor ligands was previously shown to enhance the affinity of sigma-1 receptor ligands (38). Using the alkylation of alkylhalide methodology depicted in the Materials and Methods section (synthetic scheme), butylamine, heptylamine, dodecylamine, and octadecylamine were selected to highlight the effect of adding the N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl moiety on the affinity of these compounds for the sigma-1 receptor. Binding curves to the sigma-1 receptor for heptylamine and heptylamine derivatives are summarized in Figure 1A and KI values are shown in Table 2. While heptylamine has a 21 ± 9 μM affinity, the addition of the N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl shifted the competitive displacement curves 3 to 4 orders of magnitude to the left (2a, 18 ± 14 nM; 2b, 7.5 ± 1 nM) indicating a dramatic increase in affinity for each compound (Figure 1A). Similarly, affinities for the other six N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl derivatives at the pure sigma-1 receptor were all higher than their N-alkylamine counterparts (Table 2). Figure 1B illustrates the relationship between carbon chain length (plotted on the abscissa) and the inverse of KI of N-alkylamines and their derivatives (plotted on the ordinate). Interestingly, the addition of the pharmacophore moiety was most dramatic with butylamine and less so with dodecylamine, suggesting a combined effect of the aromatic feature and the aliphatic hydrocarbon moieties.
Synthetic Scheme.
Figure 1.
Interaction of N-alkylamine and N-alkylamine derivatives with the pure sigma-1 receptor. A. Representative competitive displacement of [3H]-(+)-pentazocine binding curves for heptylamine and its analogs (2a and 2b) on the pure sigma-1 receptor. Data points were obtained in triplicate and 10 μM haloperidol were used to determine non-specific binding. Error bars represent Mean ± SEM (n = 3). B. The reciprocal of KI values of N-alkylamines and their derivatives at the pure sigma-1 receptor (KI values are listed in Table 2) plotted against the number of carbon atoms. The vertical axis is shown in log scale illustrating the orders of magnitude change in affinity when the N-3-phenylpropyl or N-3-(4-nitrophenyl)propyl group was substituted on the nitrogen of the N-alkylamines. Error bars represent Mean ± SEM (n = 3).
Table 2.
KI values of N-alkylamine derivatives for the pure sigma-1 receptor and the sigma-1 and the sigma-2 receptors in membranes.
| Compounds | KI (μM)
|
sigma-2/sigma-1c | ||
|---|---|---|---|---|
| pure sigma-1 receptor | sigma-1 sitesa | sigma-2 sitesb | ||
| 1a | 0.20 ± 0.02 | 0.073 ± 0.002 | 0.015 ± 0.01 | 2 |
| 1b | 0.020 ± 0.005 | 0.003 ± 0.0005 | 0.022 ± 0.01 | 7 |
| 2a | 0.018 ± 0.014 | 0.015 ± 0.009 | 0.033 ± 0.01 | 2 |
| 2b | 0.0075 ± 0.001 | 0.010 ± 0.002 | 0.011 ± 0.005 | 1 |
| 3a | 0.063 ± 0.011 | 5.3 ± 0.1 | 2.1 ± 1.0 | 0.4 |
| 3b | 0.032 ± 0.022 | 5.1 ± 0.2 | 2.4 ± 1.0 | 0.5 |
| 4a | 0.042 ± 0.002 | 34.4 ± 0.6 | 20.0 ± 2.0 | 0.6 |
| 4b | 0.042 ± 0.007 | 26.9 ± 0.7 | 9.0 ± 0.4 | 0.3 |
Sigma-1 sites KI values were determined using guinea pig liver membranes.
Sigma-2 sites KI values were determined using rat liver membranes in the presence of 100 nM (+)-pentazocine.
Ratio values were obtained from the KI value of sigma-1 and sigma-2 sites.
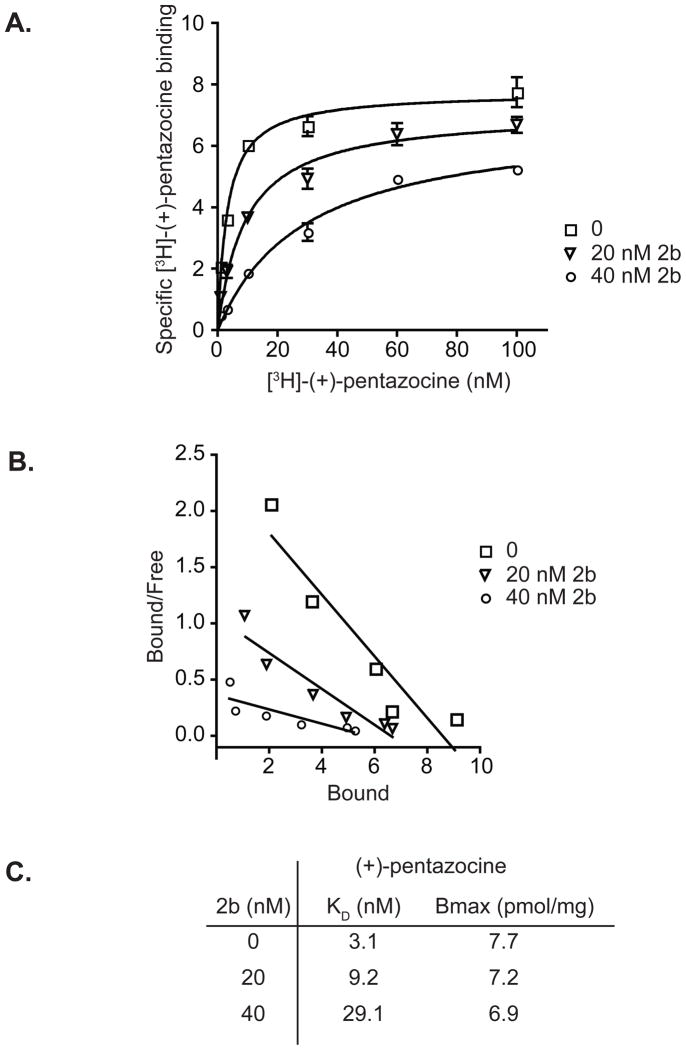
In guinea pig liver membranes (GPLM), KI values for shorter chain N-alkylamine derivatives were comparable to the KI values obtained with the pure sigma-1 receptor in that all showed nanomolar affinities (Table 2). For longer chain N-alkylamine derivatives, however, values obtained from GPLM were in the micromolar range (Table 2). Since the longer chain N-alkylamines (3a, 3b, 4a, and 4b) are structurally similar to lipids, the apparent low affinities of these molecules may be attributable to their partitioning in membranes consequently reducing the free ligand concentrations available for binding. As was demonstrated previously for D-erythro-sphingosine (15), N-alkylamine derivatives also showed a competitive binding mechanism against [3H]-(+)-pentazocine binding to the sigma-1 receptor in GPLM (Figures 2). As depicted, the presence of increasing concentrations of 2b caused the KD of [3H]-(+)-pentazocine to change dramatically, from 3 nM for the control to 9 nM and 29 nM with the addition of 20 nM and 40 nM 2b respectively. On the other hand the Bmax remained unchanged (Figure 2D), characteristic of a competitive binding mechanism.
Figure 2.
Binding mechanism of N-alkylamine derivatives. A. Saturation binding of [3H]-(+)-pentazocine in the presence of increasing concentration of 2b. Data points were obtained in duplicates and the non-specific binding (in the presence of 10 μM haloperidol) was subtracted from the total binding (without haloperidol). B. Scatchard transformation of the binding curves shown in B. C. Bmax and KD values of [3H]-(+)-pentazocine in the presence of 20 nM and 40 nM of 2b.
We also determined the affinities of 1a, 1b, 2a, 2b, 3a, 3b, 4a, and 4b for the sigma-2 receptor in rat liver membranes (RLM) using the competitive displacement of [3H]-DTG binding. In these assays, 100 nM cold (+)-pentazocine was used to block the sigma-1 receptor sites. Again, KI values of shorter chain N-alkylamine derivatives (1a, 1b, 2a, and 2b) determined for the sigma-2 receptor were in nanomolar affinities while longer chain (3a, 3b, 4a, and 4b) were all in the micromolar affinities. N-alkylamine derivatives were fairly non-selective between the sigma-1 and sigma-2 receptor indicating that the two receptor subtypes may share some similarities in their binding characteristics for monochain lipid-like molecules.
2a and 2b are selective sigma-1 and sigma-2 receptors ligands
To determine whether N-alkylamine derivatives are selective for sigma-1 and sigma-2 receptors, we selected four of the eight synthetic N-alkylamine derivatives – 2a, 2b, 3a and 3b – and submitted them to the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP) to screen for alternate receptor and transporter targets. Figure S-2 shows a heat map of the data from a primary screen of 44 membrane receptors and transporters (shown on the vertical axis) tested with 2a, 2b, 3a, or 3b (shown on the horizontal axis). The values in each box correspond to the percent inhibition at 10 μM (a default concentration) of 2a, 2b, 3a, or 3b against a radioligand that binds to the membrane receptor/transporter under test. In the illustration of these data, the more intense red color represents a greater percent inhibition and the beige color indicates lower inhibition. From the primary screen results, there were number of protein targets that N-alkylamine derivatives bind to.
Receptor/transporter targets identified to have ≥50% inhibition at 10 μM in the primary screen were subjected to a secondary assay to obtain the KI values for these targets. While the primary screen showed numerous targets that were inhibited by 50% or greater at 10 μM of 2a, 2b, 3a, or 3b, the KI values obtained in the secondary screen showed that these four molecules have much lower affinities for alternate receptors/transporters than to the sigma-1 and sigma-2 receptors. This is especially true for 2b since there are only two other targets, 5HT1A and D4 receptors, with KI values below 50 nM. In addition, for 3b, 5HT1A was the only other target demonstrating a KI value comparable to both the sigma-1 and sigma-2 receptors. Together, the data from the primary and secondary screens showed that of the four N-alkylamine derivatives tested, 2b and 3b are high affinity and relatively high selectivity mixed sigma-1/sigma-2 receptor ligands.
It is important to note that the KI values of the four N-alkylamine derivatives for the sigma-1 and sigma-2 receptors measured by NIMH PDSP were slightly different from our determinations (compare Table 2 and Table 3). The difference in the affinities of N-alkylamine derivatives obtained from NIMH PDSP and our results is likely due to the difference in the source of the receptors – NIMH PDSP used RLM as a source of sigma-1 receptor and PC12 cells for sigma-2 receptor, while our standard source of sigma-1 and sigma-2 receptors were GPLM and RLM, respectively. Nonetheless, KI values for 2a, 2b, 3a and 3b obtained from NIHM PDSP confirmed our general conclusion that an electron-withdrawing moiety (4-nitro) is an important structural element of high affinity sigma-1 and sigma-2 receptors ligands.
1a and 2a functionally inhibit Kv1.4 potassium channel current
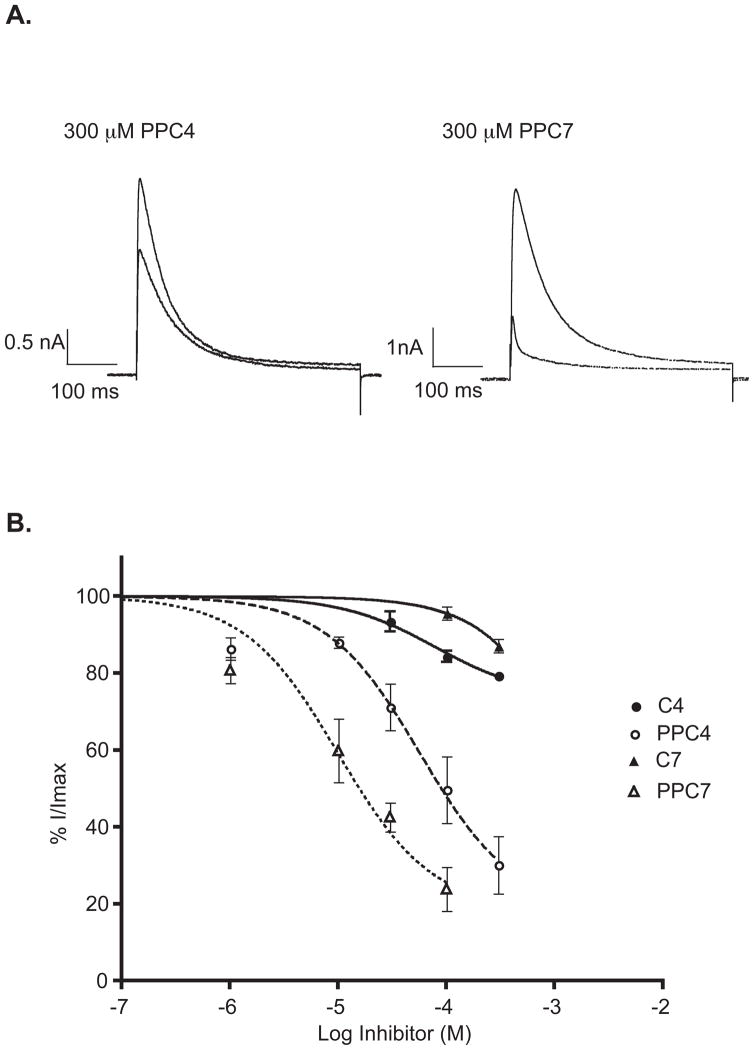
The sigma-1 receptor has been shown to modulate the activity of ion channels such as K+, Na+, Ca2+ and Cl− channels (3–6). To test the functional property of the N-3-phenylpropyl derivatives on a known effector of the sigma-1 receptor, we co-expressed Kv1.4 potassium channel and the sigma-1 receptor in human embryonic kidney (HEK-293) cells. Coexpression of both the Kv1.4 potassium channel and the sigma-1 receptor was achieved by the use of a pIRES vector that carried both the Kv1.4 and sigma-1 receptor sequences. Potassium currents were recorded using whole cell patch clamp technique. Examples of channel current traces from the control samples with no drug treatment and current traces resulted from the application of 1a and 2a are shown in Figure 3A. 1a and 2a at concentrations up to 300 μM did not significantly inhibit the Kv1.4 potassium channel with average inhibition being 21% for butylamine and 15% heptylamine (Figure 3B). However, the N-3-phenylpropyl derivatives of the butylamine and heptylamine showed profound inhibition of the Kv1.4 potassium channel at the same concentrations. The IC50 for inhibition of the Kv1.4 potassium channel was 50 μM for 1a and 10 μM for 2a. This dramatic increase in the potency of both 1a and 2a to inhibit the Kv1.4 potassium channel is consistent with their increase in apparent affinity for the sigma-1 receptor.
Figure 3.
Inhibition of Kv1.4 potassium channel by 1a and 2a. A. Representative K+ channel current traces from three experiments showing inhibition of Kv1.4 potassium channel by N-3-phenylpropyl derivatives of N-alkylamines. HEK-293 cells were transfected with both the sigma-1 receptor and Kv1.4 potassium channel. K+ channels were activated by 400 ms pulses to +20 mV from a resting potential of −80 mV. B. Concentration response curves of butylamine, heptylamine and their N-3-phenylpropyl derivatives. The leftward shift of the response curve for the N-3-phenylpropyl derivatives corresponds with the relative increase in affinity at the sigma-1 receptor.
N-alkylamine derivatives are cytotoxic against a number of cancer cell lines
Since both the sigma-1 and the sigma-2 receptors are overexpressed in a number of human tumors and cancer cell lines (18), 2a, 2b, 3a and 3b were tested for their cytotoxicity against multiple types of cancer cells. Three different cytotoxicity assays were used, Calcein AM, CellTiter-Glo and EthD-1, with concentrations of N-alkylamine derivatives ranging from 1 μM to 100 μM (36, 39). All three assays produced similar results; however, only IC50 values from CellTiter-Glo are reported in Table 4. A three dimensional illustration of the reciprocal of IC50 in μM are plotted for clarity. Since we (and others) have shown that the sigma-1 receptor is predominantly in the ER and ER cisternae (2, 10, 40) and the binding site is highly likely to be intra luminal in the ER, the micromolar concentrations of N-alkylamine derivatives required for cytotoxicity responses in these assays may be due to interference from membrane partitioning of the drugs as they pass through multiple layers of biologic membranes before accessing the functional sigma-1 receptor binding site. Nevertheless, 3b was found to be highly cytotoxic against the majority of cancer cell lines whereas 3a treatments were significantly less effective. The cytotoxicity patterns observed for 3b indicates a general mechanism of toxicity that is dependent on the presence of the 4-nitro moiety and agrees with the enhanced targeting of sigma receptors.
Table 4.
Summary of the cancer cells cytotoxicity measurements with 2a, 2b, 3a and 3c. IC50 values were obtained from CellTiter-Glo assays measuring ATP, an indicator of metabolically active cells.
| Cell lines | IC50 (μM) | |||
|---|---|---|---|---|
| 2a | 2b | 3a | 3b | |
| MDA-MB-231 | 33±2 | 30±2 | 54±3 | 15±1 |
| MCF-7 | 20±1 | 21±1 | 50±18 | 12±1 |
| MCF-10A | 70±5 | 53±6 | 53±18 | 15±1 |
| NCI-H460 | 45±5 | 38±4 | 55±3 | 18±2 |
| A549 | 83±11 | 55±5 | 87±70 | 29±1 |
| H1299 | 39±6 | 28±1 | 39±0.1 | 13±1 |
| HCT-15 | 18±12 | >100 | >100 | 25±290 |
| HT-29 | 40±1 | 30±1 | 42±21 | 14±0.2 |
| SK-OV-3 | >100 | 73±8 | 60±7 | 16±1 |
| Du145 | 25±1 | 29±1 | 51±4 | 17±0.4 |
| SF-268 | 63±2 | 40±3 | 60±14 | 16±1 |
IC50 values ± standard errors (in μM) are presented in tabular form. MDA-MB-231 and MCF-7, human breast adenocarcinoma; MCF-10A, human immortalized breast cell line; NCI-H460, human lung carcinoma; A549, human lung adenocarcinoma; H1299, human non-small cell lung carcinoma; HCT-15, human colorectal adenocarcinoma; HT-29, human colorectal adenocarcinoma; SK-OV-3, human ovary adenocarcinoma; Du145, human prostate carcinoma; SF-268, human CNS glioblastoma.
In contrast, 2a and 2b showed some selectivity in cytotoxicity against a number of cancer cell lines. For example, 2a and 2b were relatively more potent against MCF-7 and MDA-MB-231 – both of which are human mammary adenocarcinoma – as compared to the MCF-10A – a non-tumorigenic mammary epithelial cell line. These results are consistent with a previous report indicating that the sigma-1 receptor levels in breast cancer cells are elevated as compared to normal cells (41). However, simple correlation between toxicity and expression levels of sigma receptor may not provide a complete explanation of the data since a lung cancer cell line H1299 that was previously shown to have increased levels of sigma-1 receptor (17, 42) was insensitive to 2a and 2b. Nevertheless, a similar toxicity pattern of 2a and 2b across the panel of cancer cell lines suggests that these two compounds are likely to function through the same mechanism that is likely to involve sigma receptors.
Since sigma-1 and sigma-2 receptor ligands have been shown to induce apoptosis through the activation of caspases (25, 26), 2a, 2b, 3a and 3b were determined for their caspase activation potential (this work was performed at UW-Madison SMMCF). These measurements showed that all four compounds did not activate caspases 3/7, 8, and 9 (data not shown). Rimcazole has been previously shown to activate caspases 3/7 in MCF-7 and MDA MB 468 mammary carcinoma (Spruce et al. 2004); but contrary to earlier reports, in our experiments, rimcazole was not found to activate caspases 3/7 in any cancer cell lines tested, including MCF-7. It is therefore unclear whether the cytotoxic effect of 2a, 2b, 3a and 3b is through apoptosis or another currently unknown mechanism of cell death.
Discussion
The binding of tridemorph (15, 31), (2R, 5R)-2-butyl-5-heptylpyrrolidine (43) and monochain sphingolipids (15) to the sigma-1 receptor warranted the current systematic study to examine features of sphingosine-like molecules that strengthen their binding affinities for the sigma-1 receptor. Upon testing a series of N-alkylamines for their affinity at the sigma-1 receptor, we found a dependence on the hydrocarbon chain length, whereby dodecylamine exhibited maximum binding. Earlier studies based on structure-activity relationships of known sigma-1 receptor ligands indicated that there is a requirement for at least one arylalkyl substitution on a nitrogen atom for high affinity to the sigma-1 receptor (14, 30, 44–46). Furthermore, electron-withdrawing groups such as a para-nitro substitution to the phenyl ring have also been shown to enhance the affinity of a ligand (47). In agreement with these earlier reports, we found that both N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl derivatives of N-alkylamines have 2 to 6 orders of magnitude higher in affinity than their corresponding amines for the pure sigma-1 receptor. The enhancement in affinity with the addition of N-3-phenylpropyl or N-3-(4-nitrophenyl)propyl groups is more profound with shorter chain length butylamine than with longer chain length dodecylamine for the pure sigma-1 receptor (Table 2). Studies by Glennon et al. 2005 proposed a pharmacophore for high affinity sigma-1 receptor ligands to be composed of two hydrophobic regions flanking a central nitrogen atom (30). The primary hydrophobic region is situated optimally at five carbons (6 – 10 Å) from the nitrogen. The secondary hydrophobic site is located closer to the obligate nitrogen than the primary site and is separated from the nitrogen by 2.5 – 3.9 Å, approximately the length of 3 carbons. It is likely that the N-3-phenylpropyl and N-3-(4-nitrophenyl)propyl moieties of N-alkylamine derivatives occupy the secondary hydrophobic site (closer to the nitrogen). The results with butyl- and heptylamine derivatives (1a – 2b) correlate well with these earlier studies by Glennon et al. where it was shown that compounds containing aliphatic hydrocarbon chain from 4 to 8 carbons can bind to the sigma-1 receptor in the primary hydrophobic region of the Glennon model (30). The data presented here provide further support for the conclusion that the sigma-1 receptor binding site(s) can accommodate molecules with aliphatic hydrocarbon chain as long as 18 carbons (octadecane) with a preference for 12 carbons (dodecane). Because the longer carbon chain of N-alkylamines bears resemblance to lipids such as D-erythro-sphingosine, we propose that the alkyl chain intercalates against the transmembrane (TM) regions of the sigma-1 receptor possibly the TM2 region.
Our earlier results showed that sphingosine, N,N-dimethyl-sphingosine, and sphinganine weakly interact with the sigma-2 receptor (15) therefore it was somewhat unexpected that N-alkylamine derivatives also bind to the sigma-2 receptor with KI values comparable to those obtained for the sigma-1 receptor in GPLM (Table 2). These data reflects a continue challenge in the field to design selective sigma-2 receptor ligands.
Together these systematic binding studies of N-alkylamines to the pure sigma-1 receptor and N-alkylamine derivatives to both the sigma-1 and the sigma-2 receptors have expanded our understanding of features of both sigma binding sites that may have been neglected since the discovery that tridemorph (Figure S-1) binds with high affinity to the sigma-1 receptor. Further studies are needed to identify the binding region of the alkyl chain moiety of these molecules in order to better understand the essential structural features of sigma-2 receptors.
Both sigma-1 and sigma-2 receptors are overexpressed in numerous cancer cell lines and sigma ligands inhibit cancer cell growth presumably by activating caspase dependent apoptosis (17–21, 25, 26, 41). Multiplex cytotoxicity assays revealed that 2a, 2b, 3a and 3b inhibited growth of a number of cancer cell lines including breast (MCF-7 and MDA-MB-231), lung (NCI-H460, A549, and H1299), prostate (Du145), ovarian (SK-OV-3), colorectal (HCT-15 and HT-29), and CNS (SF-268). Whether N-alkylamine derivatives in these studies inhibit cancer cell growth via the activation of caspases remains unclear since rimcazole – shown previously to enhance the activity of caspases 3 and 7 (26) – did not give a positive caspase signal in our studies. Functional characterization of 1a and 2a showed that these molecules actively inhibit outward current from the Kv1.4 potassium channel with relative IC50 values that closely correlate with the relative binding affinities at the sigma-1 receptor. There has been a focus in recent years directed at understanding the mechanism by which cellular membrane electrical potential affects cancer cell growth and proliferation; for example, numerous reports showed that progression through the cell cycle is dependent on ion translocation across the plasma membrane (for a review see (48)). Thus, pharmacological blockade of ion channel activity may lead to inhibition of cell proliferation. The coupling of the sigma-1 receptor to multiple ion channels may provide a mechanism by which sigma ligands are cytotoxic to cancer cells. Of further importance is the finding that N-alkylamine derivatives in this study including 2a, 2b, 3a and 3b are highly selective for the sigma-1 and sigma-2 receptors. Therefore, N-alkylamine derivatives may be valuable tools for the functional and biochemical studies of both sigma receptor subtypes and may prove useful as potential diagnostic and/or anti-cancer therapeutic agents.
Supplementary Material
Acknowledgments
Research funding: This research was supported by National Institute of Health Grants MH065503 and GM-33138 (to A.E.R.), the Retina Research Foundation Edwin and Dorothy Gamewell Professorship (to A.E.R.), the UW-Madison Advanced Opportunity Fellowships (AOF) and the Gates Millennium Scholarship (to U.B.C). The cytotoxicity assays were performed at the University of Wisconsin – Madison Small Molecule and Medicinal Chemistry Facility and were funded by the Wisconsin Alumni Research Foundation (WARF). The alternate target assays were carried out by the National Institute of Mental Health’s Psychoactive Drug Screening Program, University of North Carolina Chapel Hill, Department of Pharmacology School of Medicine, CB 7365, 8032 Burnett-Womack Building, Chapel Hill, NC 27599, phone: 919-966-7535, bryan_roth@med.unc.edu.
We thank the Retina Research Foundation Edwin and Dorothy Gamewell Professorship (to A.E.R.). We acknowledge invaluable discussion during these experiments with Dr. Lianwang Guo (Department of Pharmacology, University of Wisconsin – Madison), we thank Dr. Nicholas Cozzi (Department of Pharmacology, University of Wisconsin – Madison) for arranging the alternate target screenings with NIMH PDSP, Noel Peters and Dr. F Michael Hoffmann from the UW-Madison Small Molecule and Medicinal Chemistry Screening Facility, Dr. Meyer B. Jackson (Department of Neuroscience, University of Wisconsin – Madison) for his expert advice on the electrophysiology experiments, and Dr. Jay Yang (Department of Anesthesiology, University of Wisconsin-Madison) for his assistance with the 3-dimensional graphical depictions of N-alkylamine and N-alkylamine derivatives.
ABBREVIATIONS
- DMT
N,N-dimethyltryptamine
- DTG
ditolylguanidine
- ER
endoplasmic reticulum
- GPLM
guinea pig liver membranes
- IP3R-3
inositol trisphophate receptor type 3
- KO
knock-out
- MAM
mitochondria associated ER membrane
- MBP
maltose-binding protein
- NIMH PDSP
National Institute of Mental Health’s Psychoactive Drug Screening Program
- SBDL
steroid-binding domain liked
- SKF-10
047, N-allyl-normetazocine
- TM
transmembrane
- RLM
rat liver membranes
- TLC
thin layer chromatography
- SMMCSF
Small Molecule and Medicinal Chemistry Screening Facility
Footnotes
Structure of tridemorph (Figure S-1) and the results of the primary alternate target screening (Figure S-2) are provided in the Supporting Information section. These data may be accessed free of charge via the internet at http://pubs.acs.org.
References
- 1.Hanner M, Moebius FF, Flandorfer A, Knaus HG, Striessnig J, Kempner E, Glossmann H. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. PNAS. 1996;93:8072–8077. doi: 10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 3.Aydar E, Palmer CP, Klyachko VA, Jackson MB. The sigma receptor as a ligand-regulated auxiliary potassium channel subunit. Neuron. 2002;34:399–410. doi: 10.1016/s0896-6273(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 4.Johannessen M, Ramachandran S, Riemer L, Ramos-Serrano A, Ruoho AE, Jackson MB. Voltage-gated sodium channel modulation by sigma-receptors in cardiac myocytes and heterologous systems. Am J Physiol Cell Physiol. 2009;296:C1049–1057. doi: 10.1152/ajpcell.00431.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Cuevas J. Sigma receptors inhibit high-voltage-activated calcium channels in rat sympathetic and parasympathetic neurons. J Neurophysiol. 2002;87:2867–2879. doi: 10.1152/jn.2002.87.6.2867. [DOI] [PubMed] [Google Scholar]
- 6.Renaudo A, L’Hoste S, Guizouarn H, Borgese F, Soriani O. Cancer cell cycle modulated by a functional coupling between sigma-1 receptors and Cl- channels. J Biol Chem. 2007;282:2259–2267. doi: 10.1074/jbc.M607915200. [DOI] [PubMed] [Google Scholar]
- 7.Carnally SM, Johannessen M, Henderson RM, Jackson MB, Edwardson JM. Demonstration of a direct interaction between sigma-1 receptors and acid-sensing ion channels. Biophys J. 2010;98:1182–1191. doi: 10.1016/j.bpj.2009.12.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi T, Fujimoto M. Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol Pharmacol. 2010;77:517–528. doi: 10.1124/mol.109.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha Y, Dun Y, Thangaraju M, Duplantier J, Dong Z, Liu K, Ganapathy V, Smith SB. Sigma receptor 1 modulates endoplasmic reticulum stress in retinal neurons. Invest Ophthalmol Vis Sci. 2011;52:527–540. doi: 10.1167/iovs.10-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavlyutov TA, Epstein ML, Andersen KA, Ziskind-Conhaim L, Ruoho AE. The sigma-1 receptor is enriched in postsynaptic sites of C-terminals in mouse motoneurons. An anatomical and behavioral study. Neuroscience. 2010;167:247–255. doi: 10.1016/j.neuroscience.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mavlyutov TA, Ruoho AE. Ligand-dependent localization and intracellular stability of sigma-1 receptors in CHO-K1 cells. J Mol Signal. 2007;2:8. doi: 10.1186/1750-2187-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang G, Mysona B, Dun Y, Gnana-Prakasam JP, Pabla N, Li W, Dong Z, Ganapathy V, Smith SB. Expression, subcellular localization, and regulation of sigma receptor in retinal muller cells. Invest Ophthalmol Vis Sci. 2006;47:5576–5582. doi: 10.1167/iovs.06-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su TP, London ED, Jaffe JH. Steroid binding at sigma receptors suggests a link between endocrine, nervous, and immune systems. Science. 1988;240:219–221. doi: 10.1126/science.2832949. [DOI] [PubMed] [Google Scholar]
- 14.Fontanilla D, Johannessen M, Hajipour AR, Cozzi NV, Jackson MB, Ruoho AE. The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science. 2009;323:934–937. doi: 10.1126/science.1166127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran S, Chu UB, Mavlyutov TA, Pal A, Pyne S, Ruoho AE. The sigma1 receptor interacts with N-alkyl amines and endogenous sphingolipids. Eur J Pharmacol. 2009;609:19–26. doi: 10.1016/j.ejphar.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mach RH, Huang Y, Buchheimer N, Kuhner R, Wu L, Morton TE, Wang L, Ehrenkaufer RL, Wallen CA, Wheeler KT. [[(18)F]N-(4′-fluorobenzyl)-4-(3-bromophenyl) acetamide for imaging the sigma receptor status of tumors: comparison with [(18)F]FDG, and [(125)I]IUDR. Nucl Med Biol. 2001;28:451–458. doi: 10.1016/s0969-8051(01)00201-3. [DOI] [PubMed] [Google Scholar]
- 17.John CS, Bowen WD, Varma VM, McAfee JG, Moody TW. Sigma receptors are expressed in human non-small cell lung carcinoma. Life Sci. 1995;56:2385–2392. doi: 10.1016/0024-3205(95)00232-u. [DOI] [PubMed] [Google Scholar]
- 18.Vilner BJ, John CS, Bowen WD. Sigma-1 and sigma-2 receptors are expressed in a wide variety of human and rodent tumor cell lines. Cancer Res. 1995;55:408–413. [PubMed] [Google Scholar]
- 19.Wheeler KT, Wang LM, Wallen CA, Childers SR, Cline JM, Keng PC, Mach RH. Sigma-2 receptors as a biomarker of proliferation in solid tumours. Br J Cancer. 2000;82:1223–1232. doi: 10.1054/bjoc.1999.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bem WT, Thomas GE, Mamone JY, Homan SM, Levy BK, Johnson FE, Coscia CJ. Overexpression of sigma receptors in nonneural human tumors. Cancer Res. 1991;51:6558–6562. [PubMed] [Google Scholar]
- 21.Barbieri F, Sparatore A, Alama A, Novelli F, Bruzzo C, Sparatore F. Novel sigma binding site ligands as inhibitors of cell proliferation in breast cancer. Oncol Res. 2003;13:455–461. doi: 10.3727/000000003108747974. [DOI] [PubMed] [Google Scholar]
- 22.Maneckjee R, Minna JD. Biologically active MK-801 and SKF-10,047 binding sites distinct from those in rat brain are expressed on human lung cancer cells. Mol Biol Cell. 1992;3:613–619. doi: 10.1091/mbc.3.6.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abate C, Niso M, Lacivita E, Mosier PD, Toscano A, Perrone R. Analogues of sigma receptor ligand 1-cyclohexyl-4-[3-(5-methoxy-1,2,3,4-tetrahydronaphthalen-1-yl)propyl]pipe razine (PB28) with added polar functionality and reduced lipophilicity for potential use as positron emission tomography radiotracers. J Med Chem. 2011;54:1022–1032. doi: 10.1021/jm1013133. [DOI] [PubMed] [Google Scholar]
- 24.Vilner BJ, Bowen WD. Modulation of cellular calcium by sigma-2 receptors: release from intracellular stores in human SK-N-SH neuroblastoma cells. J Pharmacol Exp Ther. 2000;292:900–911. [PubMed] [Google Scholar]
- 25.Hornick JR, Xu J, Vangveravong S, Tu Z, Mitchem JB, Spitzer D, Goedegebuure P, Mach RH, Hawkins WG. The novel sigma-2 receptor ligand SW43 stabilizes pancreas cancer progression in combination with gemcitabine. Mol Cancer. 2010;9:298. doi: 10.1186/1476-4598-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spruce BA, Campbell LA, McTavish N, Cooper MA, Appleyard MV, O’Neill M, Howie J, Samson J, Watt S, Murray K, McLean D, Leslie NR, Safrany ST, Ferguson MJ, Peters JA, Prescott AR, Box G, Hayes A, Nutley B, Raynaud F, Downes CP, Lambert JJ, Thompson AM, Eccles S. Small molecule antagonists of the sigma-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 2004;64:4875–4886. doi: 10.1158/0008-5472.CAN-03-3180. [DOI] [PubMed] [Google Scholar]
- 27.Colabufo NA, Berardi F, Contino M, Niso M, Abate C, Perrone R, Tortorella V. Antiproliferative and cytotoxic effects of some sigma2 agonists and sigma1 antagonists in tumour cell lines. Naunyn Schmiedebergs Arch Pharmacol. 2004;370:106–113. doi: 10.1007/s00210-004-0961-2. [DOI] [PubMed] [Google Scholar]
- 28.van Waarde A, Rybczynska AA, Ramakrishnan N, Ishiwata K, Elsinga PH, Dierckx RA. Sigma receptors in oncology: therapeutic and diagnostic applications of sigma ligands. Curr Pharm Des. 2010;16:3519–3537. doi: 10.2174/138161210793563365. [DOI] [PubMed] [Google Scholar]
- 29.Megalizzi V, Le Mercier M, Decaestecker C. Sigma receptors and their ligands in cancer biology: overview and new perspectives for cancer therapy. Med Res Rev. 2010 doi: 10.1002/med.20218. [DOI] [PubMed] [Google Scholar]
- 30.Glennon RA. Pharmacophore identification for sigma-1 (sigma1) receptor binding: application of the “deconstruction-reconstruction-elaboration” approach. Mini Rev Med Chem. 2005;5:927–940. doi: 10.2174/138955705774329519. [DOI] [PubMed] [Google Scholar]
- 31.Moebius FF, Reiter RJ, Hanner M, Glossmann H. High affinity of sigma 1-binding sites for sterol isomerization inhibitors: evidence for a pharmacological relationship with the yeast sterol C8-C7 isomerase. Br J Pharmacol. 1997;121:1–6. doi: 10.1038/sj.bjp.0701079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajipour AR, Ruoho AE. Nitric acid in the presence of P2O5 supported on silica gel - a useful reagent for nitration of aromatic compounds under solvent-free conditions. Tetrahedron Letters. 2005;46:8301–8310. [Google Scholar]
- 33.Ramachandran S, Lu H, Prabhu U, Ruoho AE. Purification and characterization of the guinea pig sigma-1 receptor functionally expressed in Escherichia coli. Protein Expr Purif. 2007;51:283–292. doi: 10.1016/j.pep.2006.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahoun JR, Ruoho AE. (125I)iodoazidococaine, a photoaffinity label for the haloperidol-sensitive sigma receptor. PNAS. 1992;89:1393–1397. doi: 10.1073/pnas.89.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson MB. Whole-cell voltage clamp recording. Curr Protoc Neurosci. 2001;Chapter 6(Unit 6):6. doi: 10.1002/0471142301.ns0606s00. [DOI] [PubMed] [Google Scholar]
- 36.Langenhan JM, Peters NR, Guzei IA, Hoffmann FM, Thorson JS. Enhancing the anticancer properties of cardiac glycosides by neoglycorandomization. PNAS. 2005;102:12305–12310. doi: 10.1073/pnas.0503270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ablordeppey SY, Fischer JB, Glennon RA. Is a nitrogen atom an important pharmacophoric element in sigma ligand binding? Bioorg Med Chem. 2000;8:2105–2111. doi: 10.1016/s0968-0896(00)00148-6. [DOI] [PubMed] [Google Scholar]
- 38.Fontanilla D, Hajipour AR, Pal A, Chu UB, Arbabian M, Ruoho AE. Probing the steroid binding domain-like I (SBDLI) of the sigma-1 receptor binding site using N-substituted photoaffinity labels. Biochemistry. 2008;47:7205–7217. doi: 10.1021/bi800564j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langa F, Codony X, Tovar V, Lavado A, Gimenez E, Cozar P, Cantero M, Dordal A, Hernandez E, Perez R, Monroy X, Zamanillo D, Guitart X, Montoliu L. Generation and phenotypic analysis of sigma receptor type I (sigma 1) knockout mice. Eur J Neurosci. 2003;18:2188–2196. doi: 10.1046/j.1460-9568.2003.02950.x. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi T, Su TP. Intracellular dynamics of sigma-1 receptors (sigma(1) binding sites) in NG108-15 cells. J Pharmacol Exp Ther. 2003;306:726–733. doi: 10.1124/jpet.103.051292. [DOI] [PubMed] [Google Scholar]
- 41.Aydar E, Onganer P, Perrett R, Djamgoz MB, Palmer CP. The expression and functional characterization of sigma (sigma) 1 receptors in breast cancer cell lines. Cancer Lett. 2006;242:245–257. doi: 10.1016/j.canlet.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Megalizzi V, Mathieu V, Mijatovic T, Gailly P, Debeir O, De Neve N, Van Damme M, Bontempi G, Haibe-Kains B, Decaestecker C, Kondo Y, Kiss R, Lefranc F. 4-IBP, a sigma1 receptor agonist, decreases the migration of human cancer cells, including glioblastoma cells, in vitro and sensitizes them in vitro and in vivo to cytotoxic insults of proapoptotic and proautophagic drugs. Neoplasia. 2007;9:358–369. doi: 10.1593/neo.07130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumagai K, Shono K, Nakayama H, Ohno Y, Saji I. (2R-trans)-2-butyl-5-heptylpyrrolidine as a potent sigma receptor ligand produced by Streptomyces longispororuber. J Antibiot (Tokyo) 2000;53:467–473. doi: 10.7164/antibiotics.53.467. [DOI] [PubMed] [Google Scholar]
- 44.Zampieri D, Mamolo MG, Laurini E, Florio C, Zanette C, Fermeglia M, Posocco P, Paneni MS, Pricl S, Vio L. Synthesis, biological evaluation, and three-dimensional in silico pharmacophore model for sigma(1) receptor ligands based on a series of substituted benzo[d]oxazol-2(3H)-one derivatives. J Med Chem. 2009;52:5380–5393. doi: 10.1021/jm900366z. [DOI] [PubMed] [Google Scholar]
- 45.Laggner C, Schieferer C, Fiechtner B, Poles G, Hoffmann RD, Glossmann H, Langer T, Moebius FF. Discovery of high-affinity ligands of sigma1 receptor, ERG2, and emopamil binding protein by pharmacophore modeling and virtual screening. J Med Chem. 2005;48:4754–4764. doi: 10.1021/jm049073+. [DOI] [PubMed] [Google Scholar]
- 46.Gilligan PJ, Kergaye AA, Lewis BM, McElroy JF. Piperidinyltetralin sigma ligands. J Med Chem. 1994;37:364–370. doi: 10.1021/jm00029a008. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Hajipour AR, Sievert MK, Arbabian M, Ruoho AE. Characterization of the cocaine binding site on the sigma-1 receptor. Biochemistry. 2007;46:3532–3542. doi: 10.1021/bi061727o. [DOI] [PubMed] [Google Scholar]
- 48.Le Guennec JY, Ouadid-Ahidouch H, Soriani O, Besson P, Ahidouch A, Vandier C. Voltage-gated ion channels, new targets in anti-cancer research. Recent Pat Anticancer Drug Discov. 2007;2:189–202. doi: 10.2174/157489207782497244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.