Abstract
Tomato mosaic virus (ToMV), like other eukaryotic positive-strand RNA viruses, replicates its genomic RNA in replication complexes formed on intracellular membranes. Previous studies showed that a host seven-pass transmembrane protein TOM1 is necessary for efficient ToMV multiplication. Here, we show that a small GTP-binding protein ARL8, along with TOM1, is co-purified with a FLAG epitope-tagged ToMV 180K replication protein from solubilized membranes of ToMV-infected tobacco (Nicotiana tabacum) cells. When solubilized membranes of ToMV-infected tobacco cells that expressed FLAG-tagged ARL8 were subjected to immunopurification with anti-FLAG antibody, ToMV 130K and 180K replication proteins and TOM1 were co-purified and the purified fraction showed RNA-dependent RNA polymerase activity that transcribed ToMV RNA. From uninfected cells, TOM1 co-purified with FLAG-tagged ARL8 less efficiently, suggesting that a complex containing ToMV replication proteins, TOM1, and ARL8 are formed on membranes in infected cells. In Arabidopsis thaliana, ARL8 consists of four family members. Simultaneous mutations in two specific ARL8 genes completely inhibited tobamovirus multiplication. In an in vitro ToMV RNA translation-replication system, the lack of either TOM1 or ARL8 proteins inhibited the production of replicative-form RNA, indicating that TOM1 and ARL8 are required for efficient negative-strand RNA synthesis. When ToMV 130K protein was co-expressed with TOM1 and ARL8 in yeast, RNA 5′-capping activity was detected in the membrane fraction. This activity was undetectable or very weak when the 130K protein was expressed alone or with either TOM1 or ARL8. Taken together, these results suggest that TOM1 and ARL8 are components of ToMV RNA replication complexes and play crucial roles in a process toward activation of the replication proteins' RNA synthesizing and capping functions.
Author Summary
Many important pathogens of plants, animals, and humans are positive-strand RNA viruses. They replicate via complementary RNA in replication complexes formed on host intracellular membranes. In the replication process, not only viral replication proteins but also host factors play important roles. Although many host factors whose knockdown affects the multiplication of positive-strand RNA viruses have been identified, the function of each host factor in virus multiplication is only poorly understood in most instances. In this paper, we show that a host small GTP-binding protein ARL8 is required for the multiplication of Tomato mosaic virus (ToMV), and that it forms a complex with ToMV replication proteins and another essential host factor TOM1 that is a seven-pass transmembrane protein. We further demonstrate that the replication proteins acquire the ability to synthesize negative-strand ToMV RNA and RNA 5′ cap only in the presence of both TOM1 and ARL8. The replication proteins of ToMV are multifunctional proteins that participate in RNA replication on membranes and RNA silencing suppression in the cytosol. Our results suggest that ToMV replication proteins are programmed to express their replication-related activities only on membranes through interactions with these host membrane proteins.
Introduction
Many animal viruses of medical and veterinary importance such as Poliovirus and Hepatitis C virus, and most plant viruses, including Tobacco mosaic virus (TMV), Brome mosaic virus (BMV) and Tomato bushy stunt virus (TBSV) are positive-strand RNA viruses. These viruses have single-stranded, messenger-sense RNA genomes in virions. After infection, their genomic RNAs are released into the cytoplasm of host cells and are translated to produce viral proteins including those that are required for RNA replication (hereafter, replication proteins). The replication proteins recruit their genomic RNAs onto intracellular membranes and synthesize complementary, negative-strand RNAs. The negative-strand RNAs are sequestered with the replication proteins in membranous compartments that are isolated from the cytosol, and are used as templates to synthesize positive-strand RNA (genomic and, for certain viruses, subgenomic RNAs), which are released into the cytosol [1]. The membrane-bound complexes that synthesize viral positive-strand RNAs are called ‘replication complexes’.
The multiplication of positive-strand RNA viruses depends not only on viral replication proteins but also on host factors. To date, a large number of such host factors has been identified [2]–[6], however, their roles in the viral RNA replication are revealed only for limited instances. For example, molecular chaperones, heat shock protein 70 (HSP70), HSP40, HSP90, and cyclophilin B, are required for efficient replication of BMV, Flock house virus, TBSV, and/or Hepatitis C virus, presumably by modifying the structure of viral replication proteins [7]–[11]. Eukaryotic translation elongation factor 1A (eEF1A) and glyceraldehyde-3-phosphate dehydrogenase that can bind to positive-strand and negative-strand TBSV RNAs are recruited to TBSV replication complexes and facilitate negative-strand and positive-strand RNA synthesis, respectively [12], [13]. The reticulon homology proteins interact with BMV 1a replication protein and play important roles in the formation of both spherular and double-membrane replication compartments by regulating membrane curvature [14]. The endosomal sorting complexes required for transport (ESCRT) proteins also participate in the formation of TBSV replication compartments, but, unlike reticulon homology proteins, they are not present in the mature TBSV replication complexes [15]. Lipid composition of the membranes is also important for RNA replication of positive-strand RNA viruses [16]–[19]. The 3A proteins of Poliovirus and Coxsackievirus B3 (CVB3: a picornavirus) bind to GBF1, a guanine nucleotide exchange factor for a small GTP-binding protein ARF1, and modulates the function of GBF1-ARF1 to preferentially recruit phosphatidylinositol-4-kinase IIIβ over other effectors of ARF1 and to facilitate the formation of phosphatidylinositol-4-phosphate (PI4P) lipid-enriched organelles, which are the essential binding site for 3D polymerase [20]. Facilitation of viral RNA replication by modulation of lipid biosynthesis by viral proteins is also reported for other viruses [21]–[23].
The genus Tobamovirus includes TMV, Tomato mosaic virus (ToMV), Youcai mosaic virus (this virus is identical to TMV-Cg and, in this report, is referred to as TMV-Cg for consistency with our previous publications), and other related viruses. The genome of a tobamovirus is a non-segmented, single-stranded, 5′-capped RNA of 6.4 kilobases that encodes a replication protein of approximately 130 kDa (130K protein) and its read-through product of 180 kDa (180K protein), a cell-to-cell movement protein, and a coat protein (CP). The 130K protein has a methyltransferase-like domain that is involved in 5′ capping of progeny RNAs and a helicase-like domain, and the read-through region of the 180K protein contains a polymerase-like domain [24]. The 130K and 180K proteins are multifunctional proteins that participate in RNA replication and RNA silencing suppression [25]. A fraction of the 130K and 180K proteins co-translationally bind to the genomic RNA to form a ribonucleoprotein complex, termed pre-membrane-targeting complex (PMTC), which is targeted to membrane surfaces to form viral replication complexes. The negative-strand RNA is synthesized only after PMTC binds to membranes and kept sequestered in the replication complex [26], [27]. Another fraction of the 130K protein that stays in the cytosol (in a non-membrane-bound form) participates in RNA silencing suppression by binding to double-stranded small RNAs [28], [29]. If negative-strand RNA is synthesized in or leaks into the cytosol, it could be a source of double-stranded RNA that would trigger RNA silencing against the viral genome [15], [30]. Thus, functional specialization of the replication proteins into two alternative forms is important for the virus to multiply successfully, and should be strictly regulated by as yet undefined mechanisms.
In tobamovirus multiplication, several host factors including eEF1A, a subunit of eukaryotic translation initiation factor 3, TOM1, and TOM2A have been suggested to participate [31]–[35]. TOM1 and TOM2A encode seven-pass and four-pass transmembrane proteins, respectively. In A. thaliana, simultaneous inactivation of TOM1 and its paralog TOM3 results in complete inhibition of multiplication of both TMV-Cg and ToMV [36], [37], whereas knockout of TOM2A and its related genes results in incomplete inhibition of TMV-Cg multiplication but had little effect on ToMV multiplication [38]. TOM1 interacts with tobamoviral helicase domain polypeptides and TOM2A [33], [34], [37], and is thought to help tether tobamovirus replication proteins on membranes. To understand the roles of these host factors in tobamovirus multiplication and to further identify novel host factor genes, especially those that are difficult to identify through genetic approaches, e.g., genes that are functionally redundant and that are essential for host viability, we have developed experimental systems that enable biochemical analyses of tobamovirus RNA replication. These include a translation and replication system for tobamovirus RNA in evacuolated plant cell extract (BYL) [39] and a method to prepare and purify active tobamovirus RNA-dependent RNA polymerase (RdRp) [27]. Our previous study showed that the purified ToMV RdRp preparation contained viral replication proteins, TOM1, TOM2A, HSP70, eEF1A, and an unidentified protein of approximately 20 kDa [27]. Here, we identify the 20-kDa protein and show that the protein and TOM1 play crucial roles in the process toward activation of the replication proteins' essential functions.
Results
Association of a small GTP-binding protein ARL8 with membrane-bound tobamovirus replication proteins
In a previous study [27], we generated a transgenic tobacco (Nicotiana tabacum) BY-2 cell line in which infection of ToMV-180FS-GFP that encodes FLAG-StrepII-tagged 180K (180K-FS) (Figure 1A) could be induced by treatment with estradiol from a cDNA cassette integrated in the host genome. We prepared membrane fractions from ToMV-180FS-GFP-infected and evacuolated protoplasts, solubilized the membranes with a detergent lysophosphatidylcholine (LPC), and then purified the 180K-FS protein using an anti-FLAG antibody. The purified fraction contained viral 130K protein, host TOM1, TOM2A, HSP70, EF1A, and an unidentified protein of approximately 20 kDa ([27] and Figure 1B).
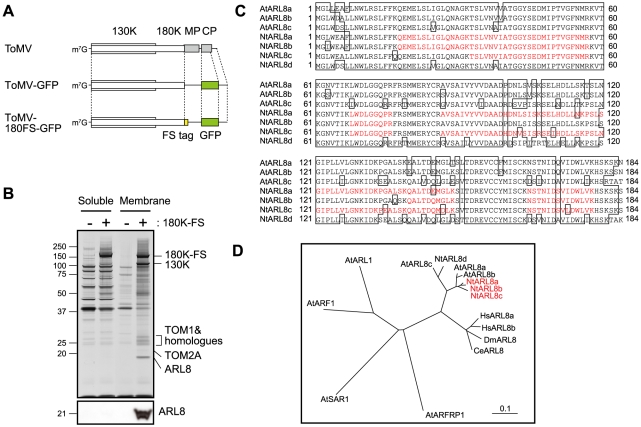
Figure 1. Analysis of proteins co-purified with the 180K-FS protein.
(A) Schematic representation of ToMV, ToMV-GFP, and ToMV-180FS-GFP. (B) Silver staining of proteins contained in the FLAG-purified fractions “Copyright © American Society for Microbiology, Journal of Virology, 80, 2006, 8459-8468, doi:10.1128/JVI.00545-06”. Soluble and membrane fractions of ToMV-GFP- or ToMV-180FS-GFP-infected and evacuolated tobacco BY-2 protoplasts were prepared, treated with LPC, clarified by centrifugation, and subjected to affinity purification with monoclonal anti-FLAG antibody. Co-purified proteins were separated by SDS-12% PAGE and silver-stained (upper panel). The positions of protein markers are shown on the left with their molecular weights (x10−3). Proteins identified by LC-MS/MS are shown on the right. The bottom panel shows an immunoblot of the fractions detected with anti-ARL8 antibodies. (C) Alignment of deduced amino acid sequences of A. thaliana (At) and N. tabacum (Nt) ARL8 proteins. The sequence data of the AtARL8 proteins were obtained from the Arabidopsis Information Resource (http://www.arabidopsis.org). Peptides identified by LC-MS/MS are indicated by red letters. (D) Phylogenetic tree of ARF family proteins. An unrooted tree was constructed using ClustalW with standard parameters and drawn using TreeView. Scale bar represents 0.1-amino acid substitutions per site. Abbreviations: At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens.
We analyzed the 20-kDa protein using liquid chromatography-tandem mass spectrometry (LC-MS/MS) and found that it was a small GTP-binding protein, NtARL8. NtARL8 co-purified with the 180K-FS protein from the solubilized membranes but not from the 15,000 x g supernatant of the extract of ToMV-180FS-GFP-infected and evacuolated protoplasts (Figure 1B, lower panel). By the degenerate PCR and RACE methods, we identified four ARL8-related genes from N. tabacum cv. Samsun and found by re-analyzing the LC-MS/MS data that the proteins from at least three of these genes, NtARL8a, NtARL8b, and NtARL8c, were co-purified with ToMV 180K-FS (Figure 1C; Genbank Accession Nos. AB644217-AB644220). ARL8 belongs to the ARF family of small GTP-binding proteins that include ARF and SAR proteins. ARF family proteins have an N-terminal amphipathic helix that participates in tight binding of the protein to membranes. ARL8 family genes are conserved in plants and animals but not in yeast S. cerevisiae (Figure 1D) [40]–[42]. Human, Drosophila melanogaster, and Caenorhabditis elegans ARL8 proteins mainly localize to lysosomes and C. elegans ARL8 mediates transport of endocytosed macromolecules to lysosomes by facilitating late endosome-lysosome fusion [43]–[45]. Arabidopsis ARL8 proteins localize on vacuolar and other organellar membranes [46]. This is intriguing because ToMV RNA replication takes place on membranes and TOM1 is also localized on vacuolar and other organellar membranes.
Interaction among tobamovirus replication proteins, TOM1, and ARL8
Tobamovirus replication proteins associated with TOM1 and ARL8 ([27] and Figure 1B); however it was unclear whether a complex containing all of the three proteins is formed or the replication proteins form distinct complexes with TOM1 and ARL8. To assess these possibilities, we generated a transgenic BY-2 cell line in which C-terminally FLAG-StrepII-tagged NtARL8a (NtARL8a-FS) was stably expressed and infection of ToMV-GFP (Figure 1A) could be induced by treatment with estradiol. Membrane fractions were then prepared from NtARL8a-FS-expressing and uninfected or ToMV-GFP-infected protoplasts, and treated with LPC. From the solubilized fractions, NtARL8a-FS was affinity-purified. As expected, ToMV 130K and 180K proteins were co-purified with NtARL8a-FS from infected cells (Figure 2A, bottom panel). NtTOM1 was also co-purified with NtARL8a-FS from ToMV-infected cells. From uninfected cells, NtTOM1 was co-purified with NtARL8a-FS, but the amount was much lower than that from ToMV-infected cells (Figure 2A, fourth panel from the top). Endogenous NtARL8 was co-purified with NtARL8a-FS from uninfected and ToMV-infected cells at similar levels (Figure 2A, second panel from the top). These results suggest that a membrane-bound complex that contains ToMV replication proteins, NtTOM1, and NtARL8 is formed in infected cells.
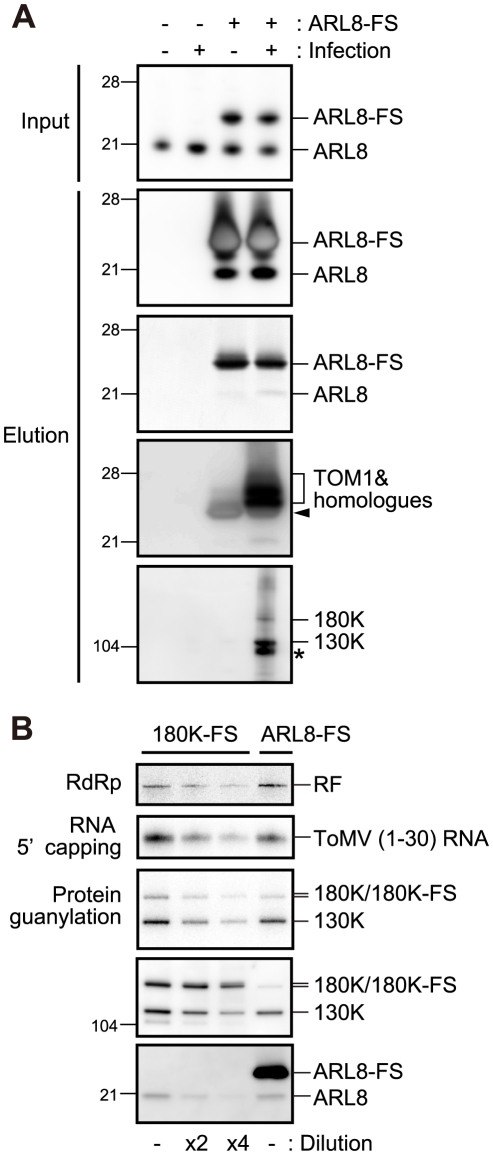
Figure 2. Affinity purification of ARL8-FS from uninfected and ToMV-infected BY-2 cells.
(A) Copurification of TOM1, 130K and 180K proteins with NtARL8a-FS. The P15 membrane fractions were prepared from the extracts of NtARL8a-FS-expressing BY-2 cells that had or had not been infected with ToMV-GFP. NtARL8a-FS was solubilized with LPC and immunopurified with the anti-FLAG antibody [27]. NtARL8a-FS and endogenous ARL8 proteins were detected by immunoblotting and Coomassie brilliant blue-staining of the blotted membrane (second and third panels from the top, respectively). TOM1 and ToMV 130K and 180K replication proteins were detected by immunoblotting. Control experiments with BY-2 cells that did not express NtARL8a-FS were performed in parallel. The arrowhead shows signals corresponding to the crossreaction of anti-TOM1 antibodies with NtARL8a-FS. The asterisk indicates a degradation product of the 130K/180K proteins. The positions of protein markers are shown on the left with their molecular weights (x10−3). (B) Copurification of ToMV RdRp and capping activities with ARL8-FS. The P15 membrane fractions were prepared from ARL8-FS-expressing and ToMV-GFP-infected BY-2 cells or from ToMV-180FS-GFP-infected BY-2 cells that were evacuolated. The fractions were solubilized with LPC and immunopurified with the anti-FLAG antibody [27]. The FLAG-purified fractions were subjected to RdRp, RNA 5′ capping, and protein guanylation assays. The fractions were also subjected to immunoblot analysis to detect ToMV replication proteins and ARL8. Where specified, the fraction was diluted 2- or 4-fold. The RdRp reaction was performed in the presence of [α-32P]CTP using exogenously added ToMV RNA as a template as described previously [27], and analyzed by PAGE. RNA 5′ capping and protein guanylation reactions were performed and products were analyzed as described in the Materials and Methods. 32P-labeled bands were detected with an image analyzer (BAS 2500, Fujifilm). The positions corresponding to double-stranded ToMV RNA (RF), ToMV (1–30) RNA and 130K, 180K (180K-FS), and ARL8 (ARL8-FS) are indicated on the right.
We previously found that only a small fraction of the membrane-bound replication proteins in plant cells participates in RNA synthesis [27]. To investigate whether ARL8-bound replication proteins are active in synthesizing RNA, we prepared FLAG-purified fractions from solubilized membranes of NtARL8a-FS-expressing and ToMV-GFP-infected BY-2 protoplasts and ToMV-180FS-GFP-infected protoplasts, and compared RdRp activity per 180K protein in these fractions (the 180K protein but not the 130K protein has the polymerase-like domain). For RdRp assay, radiolabeled ribonucleoside triphosphates and ToMV virion RNA were added because endogenous replication templates dissociate from the replication proteins by solubilization with LPC [27]. A larger amount of the 180K protein was found in the ToMV-180FS-purified fraction (Figure 2B, leftmost lane) than in the NtARL8a-FS-purified fraction (Figure 2B, rightmost lane), whereas a similar level of RdRp activity was detected in these two lanes. This suggests that ToMV 180K protein that associates with NtARL8 is actively involved in RNA synthesis.
We next performed pairwise split-ubiquitin-based yeast two-hybrid experiments, which can be used to detect interactions between a soluble protein and a membrane protein as well as between two membrane proteins [47], for tobamovirus replication proteins, TOM1, and ARL8. The results suggested that AtARL8a interacts with both AtTOM1 and a polypeptide corresponding to the helicase domain of ToMV replication proteins (amino acid residues 648-1116 of ToMV 130K protein, Figure 3A, 3B). Consistent with our previous results using the Sos recruitment system [34], [37], the ToMV helicase domain polypeptide appeared to interact with AtTOM1 (Figure 3A). The signals for ARL8 interactions with TOM1 and ToMV helicase domain were 6 and 13 times weaker than the TOM1-helicase domain interaction signal. We could not detect an interaction between TOM1 or ARL8 and the other regions of tobamovirus replication proteins by the split-ubiquitin or Sos recruitment systems (data not shown).
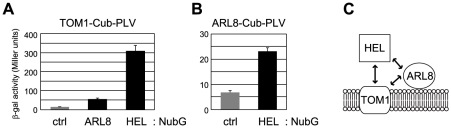
Figure 3. Interactions between TOM1, ARL8, and the helicase domain polypeptide (HEL) of ToMV replication proteins.
(A) Split-ubiquitin assay using TOM1-Cub-PLV-expressing yeast. The graph shows β-galactosidase activity in the yeast reporter strain L40 coexpressing TOM1-Cub-PLV and either NubG-ALG5 (ctrl) [47], ARL8-NubG (ARL8), or NubG-HEL (HEL). ALG5 is an unrelated yeast protein used as a negative control. (B) Split-ubiquitin assay using ARL8-Cub-PLV-expressing yeast. The graph shows β-galactosidase activity in L40 yeast coexpressing ARL8-Cub-PLV and either NubG-ALG5 (ctrl) or NubG-HEL (HEL). Averages and standard deviations of β-galactosidase activity (Miller units) for three or four independent yeast transformants are indicated in panels A and B. (C) A model of interaction between TOM1, ARL8, and ToMV HEL polypeptide.
ARL8 is necessary for tobamovirus multiplication
Because ToMV 180K protein, which shows RdRp activity, preferentially associates with NtARL8 (Figure 2B), ARL8 may positively regulate tobamovirus multiplication. To explore this further, we took advantage of A. thaliana T-DNA and transposon insertion mutants. The A. thaliana genome encodes four ARL8-related genes, designated AtARL8a-d [41], [42], [48] (Figure 1C and 1D). Quantitative RT-PCR analysis showed that mRNA levels of AtARL8a (At3g49870), AtARL8c (At5g37680), and AtARL8d (At3g49860) are about 89%, 3%, and 0.5%, respectively, of that of AtARL8b (At5g67560). AtARL8d may be a pseudogene because it lacks an N-terminal part that is conserved among the other ARL8 proteins.
We obtained A. thaliana T-DNA or transposon insertion mutants arl8a-1, arl8b-1, and arl8c-1, and constructed all possible double and triple mutants. None of these arl8 mutations had any apparent effect on plant growth or morphology (Figure 4A). For the arl8b-1 single, arl8a-1 arl8b-1 double or arl8a-1 arl8b-1 arl8c-1 triple mutants, a faint band was detected at the position of the AtARL8b protein on the immunoblot (Figure 4B, top panel). Because a transposon fragment is inserted in one of introns in the arl8b-1 mutant, a small amount of wild-type ARL8b mRNA might be produced via splicing that correctly links the exons. These mutant plants were inoculated with TMV-Cg and ToMV, and CP accumulation was examined 2 days and 7 days after inoculation, respectively. The CPs were not detected in the inoculated leaves of arl8a-1 arl8b-1 double or arl8a-1 arl8b-1 arl8c-1 triple mutants (Figure 4B), whereas in the other mutants, the CPs accumulated to a level similar to that in wild-type plants. That no single mutation in the ARL8 family members showed any detectable phenotype on tobamovirus multiplication may explain why we previously failed to identify arl8 alleles by screening randomly mutagenized A. thaliana plants. The CP of Cucumber mosaic virus (CMV) accumulated to wild-type levels in all of these arl8 mutants (Figure 4B, bottom panel). These results indicate that ARL8 plays a specific and essential role in tobamovirus multiplication in planta.
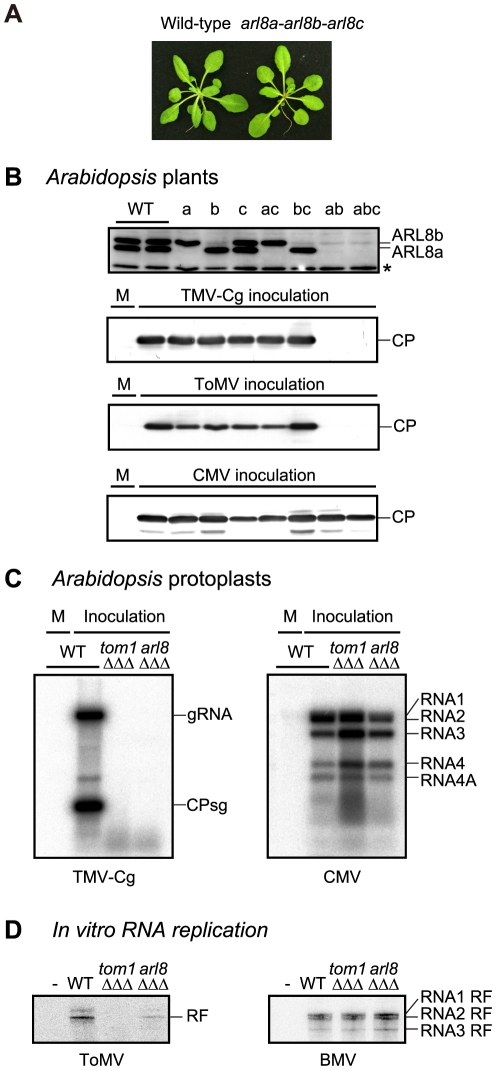
Figure 4. Effects of single and multiple mutations in A. thaliana ARL8 family members on virus multiplication.
(A) Wild-type (WT) and arl8 triple mutant plants. The photograph was taken 5 weeks after sowing. (B) Effects of arl8 mutations on the multiplication of TMV-Cg, ToMV, and CMV. Wild-type and arl8a, arl8b, and/or arl8c single, double, and triple mutant A. thaliana plants were inoculated with TMV-Cg, ToMV or CMV, as described previously [36]. Inoculated leaves were harvested 2 days after inoculation (for TMV-Cg and CMV) or 7 days after inoculation (for ToMV), and the CPs were detected by immunoblotting using a 13% acrylamide, 0.35% bisacrylamide gel. ARL8a and ARL8b proteins were also detected (top panel). The asterisk denotes a background signal derived from cross-reactivity of the anti-ARL8 antiserum to a cellular protein, which served as loading controls. M indicates mock inoculation. (C) Effects of tom1 tom3 thh1 and arl8a arl8b arl8 triple mutations on the multiplication of TMV-Cg (left panel) and CMV (right panel) in A. thaliana protoplasts. Protoplasts with each genotype were isolated from suspension-cultured cells, and inoculated with TMV-Cg and CMV RNA by electroporation [70]. Total RNA was isolated from the protoplasts 20 h after inoculation, and the accumulation of TMV-Cg and CMV RNA was analyzed by Northern blotting as described previously [70]. (D) Effect of the loss of TOM1 and ARL8 on the production of viral RF RNA in the in vitro translation-replication system. A ToMV RNA derivative TL180SF [26] and BMV virion RNAs were translated in mdBYL and ToMV PMTC were purified by 100,000 x g centrifugation and further with an anti-FLAG antibody to avoid contamination of soluble ARL8. For BMV, ribonucleoprotein complexes were purified by 100,000 x g centrifugation. The purified ribonucleoprotein fractions were mixed with membranes derived from protoplasts of suspension-cultured wild-type, tom1 or arl8 triple mutant A. thaliana cells. After the RdRp reaction, RNA samples were prepared, treated with a single-strand-specific S1 nuclease, and then analyzed by PAGE and autoradiography.
To examine whether ARL8 proteins are required for tobamovirus multiplication in single cells, we established A. thaliana suspension-cultured cells from wild-type, tom1-2 tom3-1 thh1-1 (triple) [36], and arl8a-1 arl8b-1 arl8c-1 (triple) mutants. Protoplasts were prepared from these cells and inoculated with TMV-Cg and CMV RNAs. TMV-Cg RNA accumulated in wild-type protoplasts at 20 h post-inoculation, but was not detected in tom1-2 tom3-1 thh1-1 or arl8a-1 arl8b-1 arl8c-1 mutant protoplasts (Figure 4C, left panel). In these mutant protoplasts, CMV RNA accumulated to a level similar to that in wild-type protoplasts (Figure 4C, right panel). These results indicate that TOM1 and ARL8 are necessary for intracellular tobamovirus multiplication.
Deficiency in either TOM1 or ARL8 inhibits the production of ToMV negative-strand RNA in vitro
In protoplast inoculation assays, viral replication products become detectable only after multiple rounds of RNA replication. Thus, inability of detection of progeny RNA in inoculated protoplasts does not necessarily indicate that the cells cannot support negative-stand RNA synthesis. To test the requirement of TOM1 and ARL8 for ToMV negative-strand RNA synthesis, we used an in vitro ToMV RNA translation and replication system [26], [39]. In this system, ToMV RNA is first translated in membrane-depleted evacuolated tobacco BY-2 protoplast extracts (mdBYL). By this reaction, the replication proteins are synthesized and the PMTC, a complex containing the ToMV RNA and replication proteins, is formed. The negative-strand RNA synthesis does not occur at this stage, but it does after the PMTC is incubated with membranes prepared from evacuolated plant protoplasts. In this system, negative-strand RNA synthesis, represented by the production of the double-stranded RNA called replicative form (RF) RNA, is detectable without multiple rounds of the replication cycle [26].
We translated ToMV RNA that encodes the 180K-FS protein in mdBYL and immunopurified the PMTC using the FLAG tag. The mdBYL contained small amounts of NtARL8 protein, but NtARL8 was not detectable in the purified PMTC fraction. NtTOM1 was also undetectable in the PMTC fraction. The PMTC was incubated with membranes prepared from wild-type, tom1-2 tom3-1 thh1-1, and arl8a-1 arl8b-1 arl8c-1 triple mutant A. thaliana suspension-cultured cells, and then with substrates for RNA synthesis containing [α-32P]CTP. After the reaction, total RNA was purified, treated with S1 nuclease, and separated by PAGE. This analysis showed that ToMV RF RNA, indicative of negative-RNA synthesis, was produced in the mixture to which membranes from wild-type cells were added (Figure 4D). In contrast, in samples to which membranes from the tom1-2 tom3-1 thh1-1 or arl8a-1 arl8b-1 arl8c-1 triple mutants were added, the production of ToMV RF RNA was less than 1% and 20% of that for wild-type, respectively (Figure 4D). Membranes from the mutants supported RF RNA production of BMV as did the wild-type membranes (Figure 4D), confirming their integrity. Although it is unclear whether the low-level production of ToMV RF RNA with arl8 triple mutant membranes is due to the presence of a small amount of ARL8 proteins or whether ARL8 is not absolutely required for RF RNA production, these results suggest that TOM1 and ARL8 are essential for efficient ToMV negative-strand RNA synthesis.
Co-expression of TOM1, but not ARL8, increases the relative amount of the membrane-bound tobamovirus replication proteins in yeast
To investigate the respective roles of TOM1 and ARL8 in tobamovirus multiplication, we used S. cerevisiae, which does not have genes homologous to TOM1 or ARL8 and is known to support the replication of some higher eukaryotic positive-strand RNA viruses [49]. In yeast, AtTOM1 and AtARL8a proteins and the ToMV helicase polypeptide could be expressed. However, expression of full-length 130K protein was not detected when the coding sequence for the protein was inserted downstream of a yeast promoter and introduced into yeast. We found that the original coding sequence for the ToMV 130K protein contains putative poly(A) addition-transcription termination signals [50]. Thus, we engineered the ToMV 130K-coding sequence by introducing synonymous mutations to remove the signals and to optimize codon usage for expression in yeast. When yeast carrying the modified cDNA for ToMV 130K protein under the control of the Cu2+-inducible CUP1 promoter was cultured in conditions under which the promoter was fully induced (0.2 mM CuSO4), the 130K protein accumulated to a level detectable in total cellular proteins separated by SDS-PAGE and stained with Coomassie brilliant blue dye. Because this level of 130K protein per total protein was higher than that observed in ToMV-infected plant cells, yeasts carrying the CUP1 promoter-driven modified 130K protein-coding sequences were cultured, unless otherwise specified, in the presence of a lower concentration (1 µM) of CuSO4 so that the expression level of the protein (per total protein) was comparable to that in ToMV-infected plant cells.
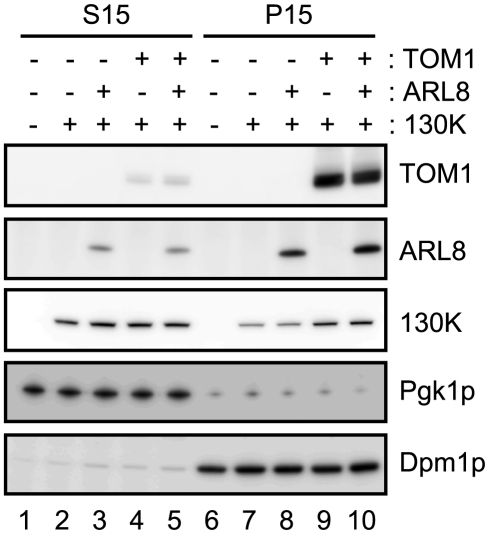
Using the expression system in yeast, we examined whether coexpression of TOM1 and/or ARL8 affects the subcellular fractionation pattern of the 130K protein. We fractionated extracts of yeast expressing the 130K protein alone or with AtTOM1 and/or AtARL8a by centrifugation (15000 x g) into supernatant (S15) and precipitate (P15) fractions. Immunoblot analysis showed that the amount of the 130K protein in the membrane-containing P15 fraction from AtTOM1-coexpressing yeast was larger than that from yeast expressing the 130K protein alone or the 130K plus AtARL8a proteins, whereas in the S15 fraction, the amount of the 130K protein was not detectably affected by coexpression of AtTOM1 or AtARL8a (Figure 5). Consistent results were obtained using the membrane-flotation assay [28]. These results suggest that TOM1 increases the relative amount of the membrane-bound 130K protein.
Figure 5. Effects of TOM1 and ARL8 coexpression on the membrane-association of ToMV 130K protein.
The soluble (S15) and membrane (P15) fractions were prepared from yeasts expressing the indicated proteins and subjected to immunoblot analysis using antibodies against TOM1, ARL8, ToMV 130K, a yeast cytoplasmic protein Pgk1p, and an ER-localized protein Dpm1p. S15 samples prepared from 0.12 OD600 units of yeast cells or P15 samples prepared from 0.6 OD600 units of yeast cells were applied in each lane.
TOM1 and ARL8 cooperatively activate tobamovirus replication proteins' capping function
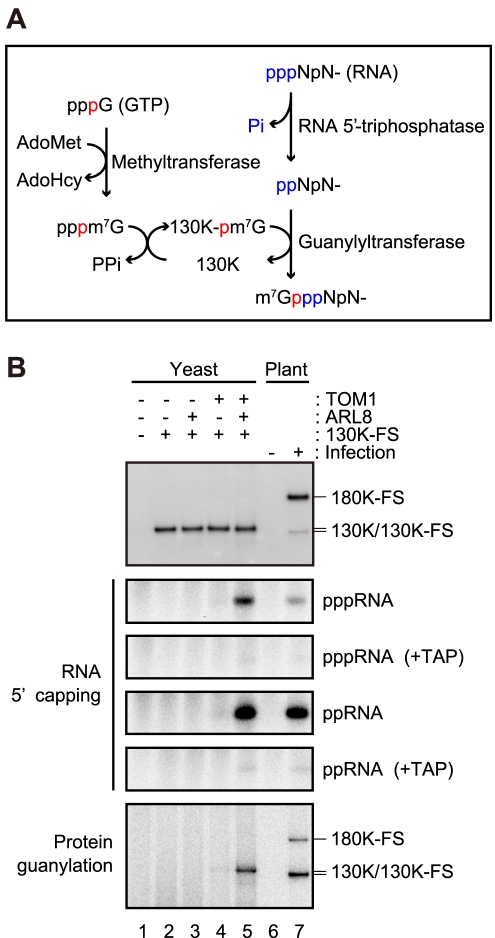
Membrane-bound tobamovirus 130K protein isolated from infected plant cells has activities to transfer a methyl group from S-adenosyl-L-methionine (AdoMet) to GTP, to form a covalent guanylate complex, and to transfer the guanylate moiety to the 5′-terminus of newly synthesized RNA to generate m7G(5′)ppp(5′)-capped RNA [51] (Figure 6A). We examined whether ToMV 130K protein, expressed alone or with TOM1 and/or ARL8, in yeast would also show this capping activity. Membrane-bound 130K protein with the FLAG-StrepII tag at the C-terminus (130K-FS) expressed in yeast with or without the plant factors were solubilized with LPC and immunopurified. As a reference, membrane-bound 180K-FS protein from ToMV-180FS-GFP-infected plant cells was also solubilized and purified in the same way (Figure 6B, top panel). The purified fractions were incubated with AdoMet, [α-32P]GTP, and uncapped (5′-triphosphate) RNA that corresponds to nucleotides 1–30 of ToMV RNA [hereafter ToMV (1–30) RNA; this RNA has only one guanylate residue at the first position from the 5′ terminus], and RNA was extracted and analyzed by PAGE. 32P-labeled RNA, indicating capped RNA, was detected in the 180K-FS-purified fraction from infected plant cells and in 130K-FS-purified fraction from yeast coexpressing AtTOM1 and AtARL8a (Figure 6B, second panel from the top, lane 5 and 7). The 32P-labeled bands disappeared after treatment with tobacco acid pyrophosphatase (TAP) (Figure 6B, third panel from the top), confirming that the signals indeed represent the 5′ cap. 32P-labeled RNA was not detected in the lanes for yeasts expressing 130K-FS alone or with either AtTOM1 or AtARL8a (Figure 6B, second panel from the top). These results suggest that TOM1 and ARL8 facilitate activation of the 5′ capping function of the 130K protein on yeast membranes. Consistent with this, the FLAG-purified fraction from the membranes of ToMV-infected, NtARL8a-FS-expressing BY-2 cells also showed RNA 5′ capping activity (Figure 2B).
Figure 6. Effects of TOM1 and ARL8 coexpression on RNA 5′ capping functions of ToMV 130K protein.
(A) A possible mechanism of RNA 5′ capping by tobamovirus replication proteins. The phosphate group at the α position of GTP is shown in red. The 5′ phosphate groups of the substrate RNA are shown in blue. See text for details. AdoHcy: S-Adenosyl-L-homocysteine. (B) Effects of the coexpression of TOM1 and ARL8 on RNA 5′ capping and guanylation of ToMV 130K protein expressed in yeast. P15 membrane fractions were prepared from the indicated yeast and BY-2 cells. The fractions were treated with LPC and subjected to immunopurification with anti-FLAG antibody [27]. The protein samples were prepared so that the concentration of the replication proteins was similar (samples for lanes 1 and 6 were prepared and analyzed in the same way as for those for lanes 2 and 7, respectively). The samples were incubated at 25°C for 60 min with [α-32P]GTP, AdoMet and uncapped ToMV (1–30) RNA with 5′-triphosphate or 5′-diphosphate. After the reaction, RNA was purified by phenol extraction and separated by 8 M urea-9% PAGE. Fractions of the purified RNA samples were treated with TAP and analyzed similarly. 32P-labeled RNA was visualized with an image analyzer (BAS2500, Fujifilm). The protein samples were also subjected to immunoblot analysis with anti-ToMV replication protein antibodies and protein guanylation assay as described in the Materials and Methods.
In bamboo mosaic virus RNA capping, newly synthesized RNA that has a 5′-triphosphate terminus accepts m7GMP after removal of the γ-phosphate group from the 5′-terminus by RNA 5′-triphosphatase activity associated with the helicase domain of the replication protein (Figure 6A) [52], [53]. Thus, we examined whether the 130K-FS protein, expressed alone or with AtTOM1 and/or AtARL8a, in yeast can add a 5′ cap to ToMV (1–30) RNA with 5′-diphosphate. Like the result with ToMV (1–30) RNA with 5′-triphosphate, 32P-labeled (i.e., capped) RNA was detected in the 130K-FS-purified fraction from yeast coexpressing AtTOM1 and AtARL8a, but not in the fractions from yeasts expressing 130K-FS alone or with either AtTOM1 or AtARL8a (Figure 6B, fourth and fifth panel from the top, lane 5 and 7).
We further examined whether 130K-FS protein co-expressed with AtTOM1 and/or AtARL8a can be guanylated. Affinity-purified 130K-FS proteins from solubilized membranes of AtTOM1- and/or AtARL8-coexpressing yeasts were incubated with AdoMet and [α-32P]GTP, and proteins were analyzed by SDS-PAGE for the production of 32P-guanylated 130K-FS protein. A 32P-labeled band at the position of the 130K-FS protein was detected in the 130K-FS-purified fraction from yeast coexpressing AtTOM1 and AtARL8a. 32P-labeled bands were also detected at the positions of the 130K and 180K-FS proteins in 180K-FS-purified fraction from infected plant cells, as expected. The 32P-labeled band was only very weakly detected for yeast expressing 130K-FS with AtTOM1 (Figure 6B, bottom panel, lane 5 and 7). When 130K-FS-purified fractions were prepared from yeast that over-expressed 130K-FS alone in the presence of a high concentration (20 µM) of CuSO4, and incubated with AdoMet and [α-32P]GTP, 32P-labeled 130K-FS was faintly detected. However, RNA 5′ capping activity was not detected in the fraction (data not shown). Taken together, these results suggest that activation of the 130K protein’s capping functions depends on coexpression of TOM1 and ARL8.
Discussion
TOM1 and ARL8 are essential components of tobamovirus replication complexes
In this study, we identified a small GTP-binding protein ARL8 that co-purifies with ToMV 180K-FS that has been solubilized by LPC from membranes of ToMV-infected plant cells (Figure 1). ToMV replication proteins and TOM1 were also efficiently co-purified with ARL8-FS from solubilized membranes of ToMV-infected cells, while TOM1 was co-purified with ARL8-FS less efficiently from solubilized membranes from uninfected cells (Figure 2). Because ToMV RNA and its complementary RNA dissociate from the replication proteins by LPC treatment [27], it seems unlikely that the observed association is mediated by RNA. Consistently, a yeast two-hybrid assay suggested that the helicase domain fragment of ToMV replication proteins, TOM1, and ARL8 interact with each other (Figure 3). Together, these results suggest that ToMV replication proteins form a complex with TOM1 and ARL8 on membranes.
Additionally, the ARL8-FS-purified fraction from solubilized membranes of ToMV-infected plant cells showed higher RdRp activity per 180K protein than the 180K-FS-purified fraction from solubilized membranes of ToMV-FS-infected plant cells did (Figure 2), suggesting that (i) most of the membrane-bound 180K protein does not have RdRp activity and that (ii) a small fraction of membrane-bound 180K protein that is associated with ARL8 and probably TOM1 does have RdRp activity. Together with the finding that mutations in the TOM1 or ARL8 genes seriously affect tobamovirus multiplication (Figure 4), these results suggest that TOM1 and ARL8 are essential components of tobamovirus replication complex.
Roles of TOM1 and ARL8 in tobamovirus RNA replication
To investigate the roles of TOM1 and ARL8 in tobamovirus RNA replication, we first used an in vitro ToMV RNA translation-replication system using a plant cell extract BYL, in combination with TOM1- or ARL8-deficient membranes from A. thaliana mutant cells. The results showed that, in the absence of TOM1 or ARL8, production of negative-strand RNA is severely inhibited, suggesting that RNA polymerase activity failed to be activated (Figure 4). With the second approach, using a heterologous expression system in yeast, we found that (i) TOM1 increases the relative amount of the membrane-bound tobamovirus replication proteins and (ii) TOM1 and ARL8 confer the guanylyltransferase activity to ToMV 130K protein (Figure 5 and Figure 6). It is unlikely that negative-strand RNA was synthesized but undetectable due to the instability of uncapped RNA in the first assay, because (i) mutants of ToMV and BMV that are defective in RNA 5′ capping functions can still produce negative-strand RNA (Figure S1 and [54]), and (ii) negative-strand RNA should be in a membranous compartment that uncapped RNA-degrading enzymes cannot enter [27]. Rather, the results suggest that both of these functions fail to be activated when either TOM1 or ARL8 is absent. If active tobamovirus replication complexes are formed through a process involving multiple successive steps, TOM1 and ARL8 would be required for one or more steps of tobamovirus replication complex formation before the replication proteins gain these enzymatic activities.
The replication proteins of tobamoviruses consist of three domains, the methyltransferase-like, helicase-like, and polymerase-like domains [24]. Because TOM1 and ARL8 interact at least with the helicase-like domain (Figure 3) and because the interaction leads to activation of both 5′ capping and polymerase functions (Figure 4 and Figure 6), changes in the helicase domain caused by binding of these host factors may induce conformational changes involving the other two domains. Alternatively, by analogy to ARF1 function in polio and other viruses [20], effectors associated with TOM1 and/or ARL8 may contribute to establishing a microenvironment on the membranes that is essential for the formation of the tobamovirus replication complexes. In support of this possibility, specific lipids are required for activation of the capping function of semliki forest virus nsP1 and polymerase function of flock house virus protein A, respectively [19], [55]. However, the fact that TOM1 and ARL8 facilitate the activation of tobamovirus replication protein's capping function in yeast, in which the effectors of ARL8 are supposed to be absent, support the possibility that the TOM1 and ARL8 proteins themselves are required for activation of the capping-related functions.
Eukaryotic cells have RNA silencing and innate immune systems that are triggered by double-strand RNA and that target positive-strand RNA viruses. Thus, for successful multiplication, positive-strand RNA viruses must avoid negative-strand RNA synthesis in the cytosol and tightly sequester negative-strand RNA replication intermediates in isolated, membranous compartments [56], [57]. Thus, it is reasonable that the replication proteins of many eukaryotic positive-strand RNA viruses have amino acid sequences or modifications that strongly facilitate membrane binding [58]–[60]. For tobamoviruses, while the replication complexes are formed in isolated, membranous compartments [27], a majority of the replication proteins (about 90%) stay in the cytosol of plant cells and play roles in suppression of RNA silencing and probably other host defense [25], [28], [61]–[64]. The host membrane protein-dependent activation of replication-related enzymatic activities may contribute to securing the functional differentiation of the replication proteins and avoiding ectopic synthesis of negative-strand RNA in the cytosol.
Materials and Methods
Plants and suspension-cultured plant cells
Transgenic Nicotiana tabacum BY-2 cell lines E182 and RT4, in which infection with GFP-expressing ToMV derivatives that encode nontagged and C-terminally FLAG-StrepII-tagged 180K proteins (ToMV-GFP and ToMV-180FS-GFP), respectively, can be induced by treatment with estradiol were prepared as described previously [27], [65]. Note that the C-terminally FLAG-StrepII-tagged ToMV 180K protein fully retains the ability to support ToMV RNA replication [26]. For expression of C-terminally FLAG-StrepII-tagged N. tabacum ARL8a (NtARL8a-FS) in BY-2 cells, a cDNA fragment including the NtARL8a-FS-coding region was amplified by the overlap extension PCR method from pNtARL8a (see below) and pTL180SF-SmaI [26] by using primers, #1 plus #2, and #3 plus #4 (Table S1). The NtARL8a-FS fragment was digested with StuI and EcoRI and then cloned between the StuI and EcoRI sites of pBICBP35 [66]. E182 cells were further transformed with an Agrobacterium tumefaciens EHA105 strain carrying pBICBP35-NtARL8aFS to establish a cell line named AF/E182. Maintenance of BY-2-derived cells and induction of ToMV infection were performed as described previously [27].
A. thaliana arl8a-1 (GABI-Kat 395G08), arl8b-1 (RIKEN 11-6303-1), and arl8c-1 (GABI-Kat 218A04) mutants were obtained from GABI-Kat and RIKEN. T-DNA and transposon insertions were confirmed by using primers, #5 and #6 (for arl8a-1), #7 and #8 (for arl8b-1), and #9 and #10 (for arl8c-1). Arabidopsis thaliana suspension-cultured cells were established from callus cultures generated from seedlings of wild-type (ecotype Col-0), tom1-2 tom3-1 thh1-1 [36], and arl8a-1 arl8b-1 arl8c-1 mutants. A. thaliana suspension-cultured cells were maintained by subculturing in fresh BY-2 growth medium at a dilution of 1∶6∼12 every 7 days. Cells were used for experiments 3 days after subculturing.
Identification of ARL8
Membrane-bound 180K-FS protein was solubilized and purified from ToMV-180FS-GFP-infected and evacuolated BY-2 protoplasts as described previously [27]. Co-purified proteins were separated by SDS-PAGE (NuPAGE 12% Bis-Tris gel: Invitrogen, Carlsbad, CA) and visualized by silver-staining (Wako, Osaka, Japan). Proteins in excised gel pieces were subjected to digestion with typsin, LC-MS/MS analysis, and MASCOT searching (this part was performed by APRO Life Science Institute, Tokushima, Japan). ARL8-related genes in N. tabacum (NtARL8s) were isolated by degenerate PCR using SMART cDNA (Clontech, Mountain View, CA) prepared from N. tabacum poly(A)+ RNA as templates. mRNA sequences of the NtARL8 genes were determined by the 5′ and 3′ random amplified cDNA ends method using the SMART cDNA kit. NtARL8a cDNA was amplified by RT-PCR from N. tabacum total RNA using primers #11 and #12, and cloned into pCR2-TOPO (Invitrogen) to obtain pNtARL8a.
Antibodies
An anti-ARL8 antiserum was prepared in rabbits using N-terminally truncated NtARL8a protein (amino acid residues 17–185) expressed in E. coli [67] as an antigen and was used for immunoblot analysis at 1∶1000 dilution. The antiserum recognized AtARL8a, AtARL8b, and AtARL8c (Figure 4B and data not shown). Rabbit antisera against TOM1 and ToMV 130K/180K proteins have been described previously [62]. Anti-Pgk1p and anti-Dpm1p were purchased from Invitrogen.
Split-ubiquitin assay
A DNA fragment containing the A. thaliana ARL8a (AtARL8a: At3g49870)-coding region was amplified by RT-PCR from A. thaliana total RNA using the primers #13 and #14, and was cloned into pCR2-TOPO (Invitrogen) to obtain pAtARL8a. DNA fragments corresponding to the ADH1 promoter and AtARL8a-coding regions were amplified from pADNS [68] and pAtARL8a by PCR using primers, #15 plus #16 and #17 plus #18, respectively, and then the two fragments were fused by the overlap extension PCR method. The fused fragment was digested with XhoI and used to replace the XhoI fragment of pRS305(Δwbp1-Cub-PLV) [47] to obtain a plasmid for ARL8-Cub-PLV expression. A DNA fragment corresponding to the AtARL8a-coding region was amplified from pAtARL8a by PCR using primers, #18 and #19. The fragment was digested with ClaI and XhoI and used to replace the ClaI-XhoI fragment of pTN-TOM1-NubG [33] to obtain a plasmid for AtARL8a-NubG expression. A DNA fragment corresponding to amino acid residues 648-1116 of ToMV 130K protein was obtained from a plasmid for 5′SOS-LHel expression [37] by NcoI and NotI digestion. The fragment with a synthetic NotI-PstI linker was inserted between the NcoI and PstI sites of pTN-NubG-TOM2A [33] to obtain a plasmid for NubG-HEL expression. NubG-ALG5 that was used as a negative control was described previously [47]. β-galactosidase assay was performed as described previously [33].
Expression of ToMV 130K, ARL8 and TOM1 proteins in yeast
A protease-deficient yeast strain BJ5465 (MATa ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1) [8] was used. The CUP1 promoter and ADH1 terminator sequences were amplified by PCR from pRS314(NubG-ALG5) [47] and pADNS using primers, #20 plus #21, and #22 plus #23, respectively. The PCR products were digested with [KasI plus SnaBI] and [SacI plus HindIII], respectively. These two fragments, a synthetic SnaBI-NotI-SacI linker (#24 and #25) and YCp22 plasmid that had been double-digested with KasI and HindIII were mixed and ligated to obtain YCp22-CUP. Synthesis of the optimized 130K protein coding fragment-containing plasmid was performed by GENEART (Regensburg, Germany)-TAKARA (Shiga, Japan; Accession number AB642602). The optimized fragment with and without the C-terminal FLAG-StrepII tag was cloned between the SnaBI and NotI sites of YCp22-CUP to obtain YCp22-CUP-130Kv2 and YCp22-CUP-130Kv2-FS, respectively. AtTOM1 and AtARL8a proteins were expressed from S. cerevisiae ADH1 and CYC1 promoter-driven gene cassettes on YEp vectors [69] (YEp195-ADH-TOM1 and YEp181-CYC-ARL8), respectively.
Isolation of tobamovirus replication proteins from yeast and plant cells
Yeasts expressing ToMV 130K protein, AtTOM1 and/or AtARL8a were treated with lyticase (125 units per mL) in 1 M sorbitol-0.1 M potassium phosphate (pH 7.6) for 30 min at 30°C, and the resulting spheroplasts were washed twice with 1 M sorbitol [60]. Yeast spheroplasts were disrupted in hypotonic buffer [10 mM Tris-HCl (pH 8), 10 mM EDTA with one tablet of Complete Mini protease inhibitors EDTA-free (Roche Diagnostics GmbH, Mannheim, Germany) per 10 ml] with 50 strokes of a tight-fitting Dounce homogenizer, and the homogenates were centrifuged at 800 x g for 10 min at 4°C to remove nuclei and cell debris. Induction of ToMV infection in tobacco BY-2-derived cell lines, preparation of protoplasts and extracts from the cells, and isolation of the P15 membrane fractions were conducted as described previously [27], except that the hypotonic buffer was used for cell disruption in Figure 5 and 6.
RNA 5′ capping and protein guanylation assays
Purification and solubilization of P15 membrane fractions from ToMV 130K-FS-expressing yeast cells and ToMV-180FS-GFP-infected BY-2 cells were performed as described previously [27], except that solubilization buffer [30 mM Hepes-KOH (pH 7.4), 150 mM NaCl, 1 mM MgOAc, 0.1 mM DTT, 0.2 mM ATP and UTP, 1% LPC (Wako), and one tablet of Complete mini protease inhibitor EDTA-free per 10 mL] was used. Uncapped RNA corresponding to the ToMV 5′ untranslated region (nucleotides 1-30) was synthesized with an AmpliScribe T7-FLASH kit (Epicentre, Madison, WI) using annealed oligonucleotides #26 and #27 as template. ToMV (1–30) RNA has only one guanylate residue at the 5′ end and 5′-triphosphate and 5′-diphosphate RNAs were generated by in vitro transcription in the presence of ATP, UTP, CTP and either GTP or GDP. The 180K protein- or 130K protein-purified fractions were mixed with AdoMet (100 µM), GTP (1 µM), [α-32P] GTP (10 µCi in 30 µL reaction mixture) and the uncapped RNA substrate (1 µg in 30 µL reaction mix), and incubated at 25°C for 1 h, followed by RNA purification by phenol extraction and alcohol precipitation. Treatment of the RNA with tobacco acid pyrophosphatase (Nippongene, Tokyo, Japan) was performed according to the manufacturer's instruction. RNA was separated with 8 M urea-9% PAGE and 32P signals were detected using a Fuji BAS-2500 bio image analyzer (Fujifilm, Tokyo, Japan). For guanylyltransferase reaction, the replication protein-purified fractions were mixed with AdoMet (100 µM), GTP (1 µM), and [α-32P] GTP (10 µCi in 30 µL reaction mixture) and incubated at 25°C for 30 min. Protein samples were precipitated with 10% trichloroacetic acid and dissolved in standard SDS-PAGE sample buffer, then separated with NuPAGE 3–8% Tris-acetate gel (Invitrogen).
Supporting Information
Effects of the H81A mutation in ToMV replication protein genes on RNA 5′ capping and RdRp functions. Wild-type and H81A ToMV RNAs (1 µg) were translated in membrane-containing BYL reaction mixtures (50 µl). Top panel shows an immunoblot of translation products detected by anti-ToMV replication protein antibodies. To eliminate excess GTP, membrane fractions of BYL reaction mixtures were collected by centrifugation at 15000 xg and subjected to protein guanylation and RdRp assays. The positions of ToMV genomic RNA (gRNA), subgenomic RNA (CPsg), and RF RNA are indicated to the right. Note that ToMV genomic RNA is a substrate of host CCA tRNA nucleotidyltransferase and a part of 32P in ToMV genomic RNA might be introduced by this enzyme.
(EPS)
List of primers used in this study.
(DOCX)
Acknowledgments
We thank GABI-Kat and RIKEN PSC for A. thaliana seeds. We also thank Drs. Y. Ohashi, I. Mitsuhara, M. Yoshikawa and members of our laboratory for helpful discussion; and Ms. Y. Gotoh; for technical assistance.
Footnotes
The authors have declared that no competing interests exist.
MI was supported by CREST-JST and PROBRAIN. MN was supported, in part, by a postdoctoral fellowship from the Japan Society for the Promotion of Science. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, et al. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 2.Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, et al. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A. 2003;100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pogany J, Stork J, Li Z, Nagy PD. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc Natl Acad Sci U S A. 2008;105:19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita Y, Mizuno T, Diez J, Naito S, Ahlquist P, et al. Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J Virol. 2003;77:2990–2997. doi: 10.1128/JVI.77.5.2990-2997.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weeks SA, Miller DJ. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J Virol. 2008;82:2004–2012. doi: 10.1128/JVI.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castorena KM, Weeks SA, Stapleford KA, Cadwallader AM, Miller DJ. A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells. J Virol. 2007;81:8412–8420. doi: 10.1128/JVI.00189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, et al. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, et al. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 2010;6:e1001175. doi: 10.1371/journal.ppat.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang RY, Nagy PD. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe. 2008;3:178–187. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Diaz A, Wang X, Ahlquist P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc Natl Acad Sci U S A. 2010;107:16291–16296. doi: 10.1073/pnas.1011105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barajas D, Jiang Y, Nagy PD. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009;5:e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee WM, Ishikawa M, Ahlquist P. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J Virol. 2001;75:2097–2106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma M, Sasvari Z, Nagy PD. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol. 2010;84:2270–2281. doi: 10.1128/JVI.02003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Ahola T, Lampio A, Auvinen P, Kaariainen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee WM, Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J Virol. 2003;77:12819–12828. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa M, Okada Y. Replication of tobamovirus RNA. Proc Jpn Acad Ser B. 2004;80:215–224. [Google Scholar]
- 25.Ishibashi K, Nishikiori M, Ishikawa M. Interactions between tobamovirus replication proteins and cellular factors: their impacts on virus multiplication. Mol Plant Microbe Interact. 2010;23:1413–1419. doi: 10.1094/MPMI-04-10-0102. [DOI] [PubMed] [Google Scholar]
- 26.Komoda K, Mawatari N, Hagiwara-Komoda Y, Naito S, Ishikawa M. Identification of a ribonucleoprotein intermediate of tomato mosaic virus RNA replication complex formation. J Virol. 2007;81:2584–2591. doi: 10.1128/JVI.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishikiori M, Dohi K, Mori M, Meshi T, Naito S, et al. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J Virol. 2006;80:8459–8468. doi: 10.1128/JVI.00545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagiwara-Komoda Y, Hirai K, Mochizuki A, Nishiguchi M, Meshi T, et al. Overexpression of a host factor TOM1 inhibits tomato mosaic virus propagation and suppression of RNA silencing. Virology. 2008;376:132–139. doi: 10.1016/j.virol.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Kurihara Y, Inaba N, Kutsuna N, Takeda A, Tagami Y, et al. Binding of tobamovirus replication protein with small RNA duplexes. J Gen Virol. 2007;88:2347–2352. doi: 10.1099/vir.0.82994-0. [DOI] [PubMed] [Google Scholar]
- 30.Aliyari R, Ding SW. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol Rev. 2009;227:176–188. doi: 10.1111/j.1600-065X.2008.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaji Y, Sakurai K, Hamada K, Komatsu K, Ozeki J, et al. Significance of eukaryotic translation elongation factor 1A in tobacco mosaic virus infection. Arch Virol. 2010;155:263–268. doi: 10.1007/s00705-009-0571-x. [DOI] [PubMed] [Google Scholar]
- 32.Yamaji Y, Kobayashi T, Hamada K, Sakurai K, Yoshii A, et al. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology. 2006;347:100–108. doi: 10.1016/j.virol.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 33.Tsujimoto Y, Numaga T, Ohshima K, Yano MA, Ohsawa R, et al. Arabidopsis TOBAMOVIRUS MULTIPLICATION (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J. 2003;22:335–343. doi: 10.1093/emboj/cdg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamanaka T, Ohta T, Takahashi M, Meshi T, Schmidt R, et al. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci U S A. 2000;97:10107–10112. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osman TA, Buck KW. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J Virol. 1997;71:6075–6082. doi: 10.1128/jvi.71.8.6075-6082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujisaki K, Ravelo GB, Naito S, Ishikawa M. Involvement of THH1, an Arabidopsis thaliana homologue of the TOM1 gene, in tobamovirus multiplication. J Gen Virol. 2006;87:2397–2401. doi: 10.1099/vir.0.81942-0. [DOI] [PubMed] [Google Scholar]
- 37.Yamanaka T, Imai T, Satoh R, Kawashima A, Takahashi M, et al. Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J Virol. 2002;76:2491–2497. doi: 10.1128/jvi.76.5.2491-2497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujisaki K, Kobayashi S, Tsujimoto Y, Naito S, Ishikawa M. Analysis of tobamovirus multiplication in Arabidopsis thaliana mutants defective in TOM2A homologues. J Gen Virol. 2008;89:1519–1524. doi: 10.1099/vir.0.2008/000539-0. [DOI] [PubMed] [Google Scholar]
- 39.Komoda K, Naito S, Ishikawa M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc Natl Acad Sci U S A. 2004;101:1863–1867. doi: 10.1073/pnas.0307131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okai T, Araki Y, Tada M, Tateno T, Kontani K, et al. Novel small GTPase subfamily capable of associating with tubulin is required for chromosome segregation. J Cell Sci. 2004;117:4705–4715. doi: 10.1242/jcs.01347. [DOI] [PubMed] [Google Scholar]
- 41.Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back' communication. EMBO Rep. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, et al. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagshaw RD, Callahan JW, Mahuran DJ. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem Biophys Res Commun. 2006;344:1186–1191. doi: 10.1016/j.bbrc.2006.03.221. [DOI] [PubMed] [Google Scholar]
- 44.Nakae I, Fujino T, Kobayashi T, Sasaki A, Kikko Y, et al. The arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2434–2442. doi: 10.1091/mbc.E09-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J Cell Sci. 2006;119:1494–1503. doi: 10.1242/jcs.02958. [DOI] [PubMed] [Google Scholar]
- 46.Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis Subcellular Database. Nucleic acids res. 2007;35:D213–218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci U S A. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen M, Albrethsen J, Larsen FH, Skriver K. The Arabidopsis ADP-ribosylation factor (ARF) and ARF-like (ARL) system and its regulation by BIG2, a large ARF-GEF. Plant Science. 2006;171:707–717. [Google Scholar]
- 49.Nagy PD. Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol. 2008;46:217–242. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- 50.Graber JH, McAllister GD, Smith TF. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merits A, Kettunen R, Makinen K, Lampio A, Auvinen P, et al. Virus-specific capping of tobacco mosaic virus RNA: methylation of GTP prior to formation of covalent complex p126-m7GMP. FEBS Lett. 1999;455:45–48. doi: 10.1016/s0014-5793(99)00856-x. [DOI] [PubMed] [Google Scholar]
- 52.Huang YL, Hsu YH, Han YT, Meng M. mRNA guanylation catalyzed by the S-adenosylmethionine-dependent guanylyltransferase of bamboo mosaic virus. J Biol Chem. 2005;280:13153–13162. doi: 10.1074/jbc.M412619200. [DOI] [PubMed] [Google Scholar]
- 53.Li YI, Shih TW, Hsu YH, Han YT, Huang YL, et al. The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5′ cap structure by exhibiting RNA 5′-triphosphatase activity. J Virol. 2001;75:12114–12120. doi: 10.1128/JVI.75.24.12114-12120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahola T, den Boon JA, Ahlquist P. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J Virol. 2000;74:8803–8811. doi: 10.1128/jvi.74.19.8803-8811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu SX, Ahlquist P, Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc Natl Acad Sci U S A. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.den Boon JA, Diaz A, Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 58.Lampio A, Kilpelainen I, Pesonen S, Karhi K, Auvinen P, et al. Membrane binding mechanism of an RNA virus-capping enzyme. J Biol Chem. 2000;275:37853–37859. doi: 10.1074/jbc.M004865200. [DOI] [PubMed] [Google Scholar]
- 59.Liu L, Westler WM, den Boon JA, Wang X, Diaz A, et al. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 2009;5:e1000351. doi: 10.1371/journal.ppat.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller DJ, Ahlquist P. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J Virol. 2002;76:9856–9867. doi: 10.1128/JVI.76.19.9856-9867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubota K, Tsuda S, Tamai A, Meshi T. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J Virol. 2003;77:11016–11026. doi: 10.1128/JVI.77.20.11016-11026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagiwara Y, Komoda K, Yamanaka T, Tamai A, Meshi T, et al. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 2003;22:344–353. doi: 10.1093/emboj/cdg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Goregaoker SP, Culver JN. Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. J Virol. 2009;83:9720–9730. doi: 10.1128/JVI.00941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J Virol. 2005;79:2549–2558. doi: 10.1128/JVI.79.4.2549-2558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dohi K, Nishikiori M, Tamai A, Ishikawa M, Meshi T, et al. Inducible virus-mediated expression of a foreign protein in suspension-cultured plant cells. Arch Virol. 2006;151:1075–1084. doi: 10.1007/s00705-005-0705-8. [DOI] [PubMed] [Google Scholar]
- 66.Mori M, Kaido M, Okuno T, Furusawa I. mRNA amplification system by viral replicase in transgenic plants. FEBS Lett. 1993;336:171–174. doi: 10.1016/0014-5793(93)81634-c. [DOI] [PubMed] [Google Scholar]
- 67.Okamura H, Nishikiori M, Xiang H, Ishikawa M, Katoh E. Interconversion of two GDP-bound conformations and their selection in an Arf-family small G protein. Structure. 2011;19:988–998. doi: 10.1016/j.str.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 68.Colicelli J, Birchmeier C, Michaeli T, O'Neill K, Riggs M, et al. Isolation and characterization of a mammalian gene encoding a high-affinity cAMP phosphodiesterase. Proc Natl Acad Sci U S A. 1989;86:3599–3603. doi: 10.1073/pnas.86.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 70.Ishikawa M, Naito S, Ohno T. Effects of the tom1 mutation of Arabidopsis thaliana on the multiplication of tobacco mosaic virus RNA in protoplasts. J Virol. 1993;67:5328–5338. doi: 10.1128/jvi.67.9.5328-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of the H81A mutation in ToMV replication protein genes on RNA 5′ capping and RdRp functions. Wild-type and H81A ToMV RNAs (1 µg) were translated in membrane-containing BYL reaction mixtures (50 µl). Top panel shows an immunoblot of translation products detected by anti-ToMV replication protein antibodies. To eliminate excess GTP, membrane fractions of BYL reaction mixtures were collected by centrifugation at 15000 xg and subjected to protein guanylation and RdRp assays. The positions of ToMV genomic RNA (gRNA), subgenomic RNA (CPsg), and RF RNA are indicated to the right. Note that ToMV genomic RNA is a substrate of host CCA tRNA nucleotidyltransferase and a part of 32P in ToMV genomic RNA might be introduced by this enzyme.
(EPS)
List of primers used in this study.
(DOCX)