Abstract
Synthesis of the translation apparatus is a central activity in growing and/or proliferating cells. Because of its fundamental importance and direct connection to cell proliferation, ribosome synthesis has been a focus of ongoing research for several decades. As a consequence, much is known about the essential factors involved in this process. Many studies have shown that transcription of the ribosomal DNA by RNA polymerase I is a major target for cellular regulation of ribosome synthesis rates. The initiation of transcription by RNA polymerase I has been implicated as a regulatory target, however, recent studies suggest that the elongation step in transcription is also influenced and regulated by trans-acting factors. This review describes the factors required for rRNA synthesis and focuses on recent works that have begun to identify and characterize factors that influence transcription elongation by RNA polymerase I and its regulation.
1. Introduction
Synthesis of eukaryotic ribosomes is a complex process that is intimately connected to many aspects of cell metabolism. Previous calculations have shown that ribosome synthesis is the most energetically costly activity in actively growing cells (Warner, 1999). Indeed, more than 60% of total cellular transcription is derived from RNA polymerase I (Pol I), the enzyme specialized for transcription of the ribosomal DNA (rDNA). Given the energetic commitment that cells make to this process, proper control of transcription by Pol I is critical.
In addition to its fundamental relevance to cell biology, ribosome synthesis has gained attention in recent years as a target for the control of cancer cell growth. For more than 100 years, the relationship between ribosome synthesis and cell growth and proliferation rates has been appreciated [initially by observation of nucleolar size in tumor-derived cells; (Pianese, 1896)]. Many oncogenes and tumor suppressors have been shown to influence Pol I-dependent transcription [e.g. P53 (Zhai and Comai, 2000; Rubbi and Milner, 2003; Trere et al., 2004), Myc (Poortinga et al., 2004; Arabi et al., 2005; Barna et al., 2008), Rb (Voit et al., 1997; Hannan et al., 2000; Trere et al., 2004), and PTEN (Zhang et al., 2005)]. Indeed, several cancer chemotherapeutics (in development and in use) target transcription of the ribosomal DNA due to its intimate connection to cell proliferation (Drygin et al., 2010; Drygin et al., 2011). Taken together, these observations demonstrate that a deeper understanding of the mechanisms that influence ribosome synthesis is fundamentally and therapeutically important.
Biosynthesis of ribosomes involves all three nuclear transcription apparatuses. Pol I synthesizes the bulk of the ribosome: three of the four ribosomal RNAs [25S (28S in mammals), 18S and 5.8S]. Pol III synthesizes the 5S rRNA, and Pol II transcribes the genes that encode the 78 ribosomal proteins (Warner, 1999). The molecular mechanisms that control transcription by Pols II and III have been reviewed recently in detail (White, 2008; Dumay-Odelot et al., 2010; Bosio et al., 2011). This review focuses exclusively on Pol I-dependent transcription of rDNA, with a particular emphasis on factors that influence the elongation step in transcription. Furthermore, studies on rRNA synthesis have been conducted using many species; however, most recent, detailed studies have been performed in Saccharomyces cerevisiae or mammalian cell culture models; thus these systems are described more thoroughly herein.
2. Ribosomal DNA and factors required for its transcription
Transcription of rDNA by Pol I has been a focus of study for many years. As a consequence, the essential factors involved have been identified in multiple eukaryotic systems. In general, the factors that influence rRNA synthesis are functionally conserved among eukaryotic species, but several important differences exist.
2.3. Ribosomal DNA
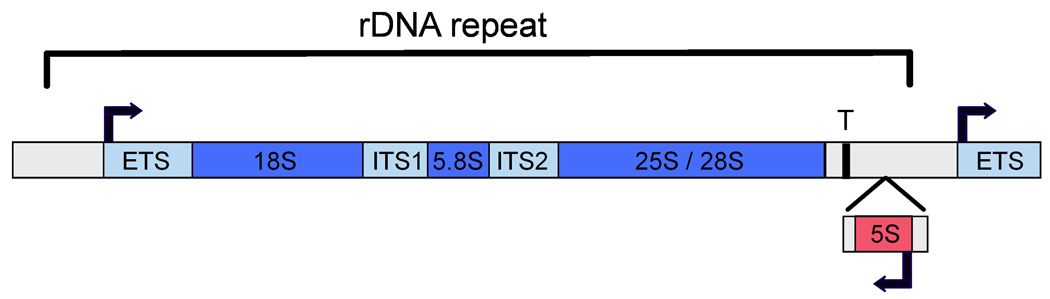
In all eukaryotes, the ribosomal DNA exists as a tandemly repeated array. Although the number of rDNA repeats and the size of a single rDNA gene varies between species, the general layout of each repeat is conserved (Figure 1). The rRNA transcript encodes the three largest rRNA species (25S, 18S, and 5.8S). A series of co-transcriptional and post-transcriptional processing events produces the mature rRNAs. Together with the 5S rRNA (synthesized by Pol III) and 78 r-proteins, ribosomes are assembled and exported from the nucleus. In eukaryotes, hundreds of accessory factors are required to efficiently assemble ribosomes, and this process has been the focus of several recent reviews (Staley and Woolford, 2009; Kressler et al., 2010).
Figure 1.
Diagram of the ribosomal DNA repeat. The promoter for Pol I is indicated by a bent arrow. Mature rRNAs (indicated in dark blue) are processed out of the precursor RNA (35S pre-rRNA in yeast; 45S, in human). RNAs that are not present in the ribosome (external transcribed sequence, “ETS” and internally transcribed sequences “ITS1” and “ITS2” are in light blue). In yeast, the gene that encodes 5S rRNA is interspersed between 35S rRNA repeats, whereas in other eukaryotes the 5S gene is located in a separate locus.
In yeast, there are approximately 200 repeats of the rDNA in a single locus per haploid genome (Nomura et al., 2004), whereas in humans, there are >200 repeats located in 5 different chromosomes (Prieto and McStay, 2008). It is known that only a fraction of these repeats is actively transcribed; many (or most) are maintained in a transcriptionally inactive state (Birch and Zomerdijk, 2008). Although studies in yeast and mammalian cell culture have shown that alteration of the ratio of active:inactive repeats is not essential for control of rRNA synthesis, alteration of accessibility of the rDNA to transcription factors could influence rRNA synthesis rates under some developmental or growth conditions (Conconi et al., 1989; French et al., 2003; Claypool et al., 2004; Oakes et al., 2006; Stefanovsky and Moss, 2006; Sanij et al., 2008). Recent studies in yeast suggest that a primary role for the inactive rDNA repeats may be in maintenance of genome stability, rather than in transcriptional control (Ide et al., 2010). Thus, cells may maintain excess inactive repeats for genome integrity rather than for regulation of rRNA synthesis.
2.2. Pol I
RNA polymerase I is a 14 subunit enzyme. In all eukaryotes, the three nuclear RNA polymerases share some peripheral subunits with one another, but the core catalytic subunits are unique to each polymerase (Table 1). There are no high resolution structures for RNA polymerases I or III, but based on the structural homology between prokaryotic RNA polymerase and yeast Pol II (Zhang et al., 1999; Cramer et al., 2000), it is certain that the three polymerases will share a high degree of structural similarity (Kuhn et al., 2007; Werner et al., 2009). Nevertheless, the differences that exist between the polymerases appear to be functionally important for the unique roles of the enzymes. These intrinsic activities of Pol I are discussed further below (Section 3.1).
Table 1.
S. cerevisiae Pol I subunit composition with homologous or shared genes for subunits of Pols II and III indicated. Shared subunits are shaded.
| Subunit name | Mass (kD) | Pol I | Pol II | Pol III |
|---|---|---|---|---|
| A190 | 186.4 | RPA190 | RPB1 | RPC160 |
| A135 | 135.7 | RPA135 | RPB2 | RPC128 |
| A43 | 36.2 | RPA43 | RPB7 | RPC25 |
| A14 | 14.6 | RPA14 | RPB4 | RPC17 |
| A12 | 13.7 | RPA12 | RPB9 | RPC11 |
| AC40 | 37.7 | RPC40 | RPB3 | RPC40 |
| AC19 | 16.1 | RPC19 | RPB11 | RPC19 |
| ABC27 | 25.1 | RPB5 | RPB5 | RPB5 |
| ABC23 | 17.9 | RPB6 | RPB6 | RPB6 |
| ABC14.5 | 16.5 | RPB8 | RPB8 | RPB8 |
| ABC10α | 7.7 | RPB10 | RPB10 | RPB10 |
| ABC10β | 8.3 | RPB12 | RPB12 | RPB12 |
| A49 | 46.6 | RPA49 | RPC34 | |
| A34.5 | 26.9 | RPA34 | RPC31 | |
| RPC37 | ||||
| RPC53 | ||||
| RPC82 |
2.3. Initiation factors
Initiation is the best characterized step in Pol I transcription. Genetic and biochemical approaches have identified the factors that are required for Pol I initiation in yeast and mammals (Nomura et al., 2004). Compared to Pol II, initiation of Pol I transcription is remarkably simple. There are clear functional similarities between the yeast and human proteins, however, in most cases sequence homology between functional counterparts is very low, if detectable.
In yeast there are four essential transcription initiation factors: Rrn3, core factor, TATA-binding protein (TBP), and upstream activating factor (UAF; Figure 2). Rrn3 functions as a monomer and its association with the A43 subunit of Pol I is required to render the polymerase competent for transcription initiation (Peyroche et al., 2000). This association of Rrn3 with the polymerase is a target for regulation of ribosome synthesis. Previous studies have suggested that covalent modifications of A43 or Rrn3 may account for this regulation (Milkereit and Tschochner, 1998; Fath et al., 2001; Claypool et al., 2004). To date, no essential sites of modification or candidate kinases have been identified for regulation of yeast Rrn3 activity (Gerber et al., 2008).
Figure 2.
Cartoon diagram of factors required for transcription initiation by Pol I. (A) Yeast factors are illustrated with subunit names indicated. Subunits of individual factors are colored the same. Separable elements in the promoter DNA sequence have been characterized and are indicated. HmoI is indicated downstream of the core promoter, but may also influence transcription initiation. (B) Mammalian factors are colored to reflect their functional analogy to the yeast factors. UBF is shown at the promoter, but it also associates with the coding region of the rDNA. Notably, TBP is required in yeast and mammals, but original characterization of SL1 included TBP as a subunit, thus it is colored as such. TAF41 and TAF12 were recently shown to bind SL1 (Denissov et al., 2007; Gorski et al., 2007); however their direct requirement for activity of SL1 in vitro has not yet been demonstrated. The transcription termination factor, TTF-I, also affects transcription initiation by Pol I and is shown bound to one of several sites upstream of the Pol I promoter.
Rrn3 is conserved in mammals. The Reeder lab showed that expression of human Rrn3 could compensate for deletion of RRN3 in S. cerevisiae (Moorefield et al., 2000). However, unlike in the yeast model, several specific sites of phosphorylation have been characterized as targets for activation or inactivation of mammalian Rrn3 function [TIF-1A in mouse; (Zhao et al., 2003; Mayer et al., 2004; Mayer et al., 2005)].
Core factor is a three subunit complex (Figure 2) that, like Rrn3, is essential even for basal levels of transcription from the rDNA promoter (Lalo et al., 1996; Keener et al., 1998). Although core factor lacks detectable affinity for DNA using standard assays (e.g. gel shift analysis; unpublished results), it can clearly direct promoter-specific transcription in the absence of UAF or TBP (Keener et al., 1998). Thus, either through direct DNA-association or through allosteric effects on the Rrn3/Pol I complex, core factor enhances polymerase binding to the promoter and properly positions the transcription start site. TBP binds to the Rrn6 subunit of core factor and can further stimulate binding to the promoter DNA (Steffan et al., 1996). TBP cannot activate Pol I transcription without core factor, however, in the presence of core factor, addition of TBP increases the rate of Pol I transcription (Aprikian et al., 2000).
In mammalian cells, the homologue of core factor is selectivity factor 1 (SL1). SL1 consists of at least 4 subunits, including TBP, and is required for promoter-dependent transcription by human Pol I (Figure 2). As with core factor, SL1 is required for transcriptional start site selection (Russell and Zomerdijk, 2006). Unlike core factor, SL1 has been identified as a target for covalent modification (e. g. phosphorylation) and control of rRNA synthesis (Zhai and Comai, 2000; Zhang et al., 2005). Whether similar regulatory strategies are employed in lower eukaryotes remains to be determined.
The last essential factor for transcription initiation in yeast is the six subunit UAF complex. Histones H3 and H4 are subunits of UAF and likely provide much of the complex’s affinity for DNA whereas the Uaf30 subunit is thought to provide sequence specificity (Keys et al., 1996; Keener et al., 1997; Hontz et al., 2008). The Rrn9 subunit of UAF associates with TBP directly, leading to core factor recruitment (Steffan et al., 1996). Additionally, biochemical studies revealed an association between the Rrn9 subunit of UAF and the Rrn7 subunit of core factor (Steffan et al., 1996). Thus, a series of protein:protein interactions results in robust recruitment of Pol I to the rDNA promoter. Under optimal growth conditions, electron microscope analysis of Pol I transcription (visualized by Miller chromatin spreading) demonstrated that transcription initiation by Pol I is exceptionally strong, resulting in ~half maximal occupancy of Pol I over the length of the rDNA (French et al., 2003). Thus, transcription initiation by Pol I is an excellent, if not the best, model for understanding mechanisms that permit strong eukaryotic transcription.
To date, no UAF analogue has been identified in mammalian cells. However, mammalian Pol I requires another critical factor called upstream binding factor (UBF). UBF plays many potential roles in rRNA synthesis affecting Pol I recruitment to the promoter, promoter escape and transcription elongation (Jantzen et al., 1990; Panov et al., 2006; Stefanovsky et al., 2006a). UBF is an HMG box-containing protein and has been shown to have sequence specific and non-specific affinity for the rDNA. Although the names are similar, UBF and UAF are not functional analogues. In fact another HMG-containing protein from yeast, Hmo1, may be the UBF counterpart (Berger et al., 2007). UBF, like SL1, is a known target for regulation of rRNA synthesis in mammalian cells (Hannan et al., 2000; Hannan et al., 2003).
Transcription initiation by Pol I is a robust, but comparatively simple step in the transcription cycle. Until recently, very little attention was paid to the elongation step in Pol I transcription. However, emerging evidence suggests that this step is important for the overall control of rRNA synthesis rate as well as for ensuring efficient processing of rRNA (Stefanovsky et al., 2006a; Schneider et al., 2007; Zhang et al., 2010). Thus, the remainder of this review will highlight recent discoveries regarding the elongation phase of Pol I transcription.
3. Elongation properties of the polymerase
As RNA polymerase I elongates through the rDNA it likely encounters a variety of intrinsic (and potentially extrinsic) kinetic barriers in the chromatin template. To negotiate these barriers and maintain the high synthesis rate that is required, Pol I must possess unique properties relative to the other nuclear polymerases and rely on trans-acting factors. Ongoing studies continue to reveal the unique properties of Pol I that render it capable of accomplishing this task.
3.1 Subunits of Pol I function as intrinsic elongation factors
The intrinsic activities of Pols I and II are substantially different in vitro, and potentially in vivo. The biggest difference between these enzymes, however, may be in their requirements for trans-acting factors. Pol I has intrinsic RNA hydrolysis activity and elongation rate-enhancing subunits (Kuhn et al., 2007; Geiger et al., 2010). These qualities potentially render Pol I less sensitive than Pol II to external kinetic barriers to transcription elongation.
The Cramer lab demonstrated that the A12.2 subunit of Pol I is capable of promoting transcript cleavage in an arrested complex (Kuhn et al., 2007). Thus, Pol I does not require association of a trans-acting factor to clear potential arrests that occur during transcription elongation. RNA polymerase II requires TFIIS [or possibly CCR4/NOT1;(Kruk et al., 2011)] to clear transcription arrest sites in vitro or in vivo [for review see (Cheung and Cramer, 2011)]. When an arrest is encountered, and Pol II back-tracks, the nascent transcript is misaligned relative to the active site and may enter the funnel of the polymerase. TFIIS must associate with Pol II to activate the hydrolytic activity of the enzyme, leading to cleavage of the RNA, correct alignment of the 3’ end of the transcript, and resumption of transcription elongation. Although TFIIS influences Pol II and potentially Pol III transcription (Ghavi-Helm et al., 2008), it is not required for Pol I.
Pol I also contains subunits that are functional analogues of TFIIF. TFIIF is a transcription factor for Pol II that serves several roles, but perhaps the best characterized role for TFIIF is in promoting transcription elongation of the enzyme (Conaway et al., 2000; Dvir et al., 2001). TFIIF’s effects on Pol II transcription elongation have been characterized in vitro and in vivo. It was known that the A49 and A34 subunits of Pol I were not essential for growth (Liljelund et al., 1992; Gadal et al., 1997), but that mutations in the genes that encode these subunits, particularly RPA49, impaired the growth of yeast cells. Structural studies in the Cramer lab identified regions of A49 that resembled the tandem-winged helix domain present in TFIIF (Geiger et al., 2010). Indeed, transcription elongation assays on synthetic templates in vitro demonstrated that Pol I complexes lacking A49 and A34 were impaired for transcription elongation. Furthermore, recombinant A49 could rescue the defect (Kuhn et al., 2007). Genetic studies from the Thuriaux lab are consistent with this conclusion (Beckouet et al., 2008).
Taken together, these data may suggest that Pol I is relatively resistant to challenges during transcription elongation. However, ongoing studies in yeast and mammalian cells suggest that additional factors influence Pol I transcription elongation in diverse, important ways.
3.2 Mutations that impair elongation
Random mutagenesis of the gene encoding the A135 subunit of Pol I led to the isolation of mutations that rendered cells sensitive to the transcription elongation inhibitor, 6-azauracil (Schneider et al., 2007). One of these mutations caused an amino acid substitution of a glycine for an aspartate at position 784 in the subunit [rpa135(D784G)]. An acidic residue at that position in the polymerase is conserved in all multi-subunit RNA polymerases and is involved in loading the incoming NTP substrate. This mutation reduced both the transcription elongation and initiation rates of Pol I in vivo and in vitro. Like deletion of the RPA49 gene (Kuhn et al., 2007; Beckouet et al., 2008), this point mutation in RPA135 resulted in clear defects in the elongation rate of the enzyme (Schneider et al., 2007). Together, these mutations have enabled us to better characterize the influence of Pol I transcription elongation on ribosome synthesis and to identify new factors that influence this step in transcription.
3.3 Coupling of elongation to rRNA processing
Visual and standard molecular analyses have demonstrated that processing of rRNA begins on nascent rRNA transcripts (Osheim et al., 2004; Kos and Tollervey, 2010). Thus, defects in transcription elongation could directly influence early processing steps. The rpa135(D784G) mutation enabled our lab to directly test this model (Schneider et al., 2007). We found that impairment of Pol I transcription elongation by mutation of RPA135 resulted in defects in multiple rRNA processing steps compared to WT. Interestingly, the observed defects in processing were more severe for large subunit rRNAs than for the 18S rRNA. It is not clear whether there is tighter coupling between processing events for the 5.8S or 25S rRNAs compared to the 18S, or whether assembly of the large subunit is simply more complex and therefore more prone to disruption. Although details of this model remain to be tested, the relationship between transcription elongation rate and the fidelity of rRNA processing emphasizes the need to better characterize this step in transcription by Pol I.
3.4 Topology effects
It has long been known that transcription leads to local disruption of DNA topology. Topoisomerase activity is required to relieve topological strain induced by transcription and other DNA metabolism. More than 20 years ago, it was shown that mutations in the genes that encode topoisomerases I and II lead to defects in synthesis of ribosomal RNA (Brill et al., 1987). Recently, the Beyer and Tollervey labs characterized the mechanism by which DNA topology can affect Pol I. They showed that partial impairment of topoisomerase II activity in cells carrying a deletion of the gene that encodes topoisomerase I resulted in a total loss of rDNA transcription (El Hage et al., 2010; French et al., 2011). In the double mutant cells, inactivation of topoisomerase II resulted in a complete block to transcription elongation by Pol I approximately 1/3 of the way through the gene. These data demonstrate that Pol I transcription results in accumulation of positive supercoiling ahead of the polymerase and this topological stress requires the action of topoisomerases for relief.
Topoisomerase inhibitors like Camptothecin have been used in cancer chemotherapy for some time. The mechanism of action for this class of inhibitors was generally considered to be dependent on cell cycle or DNA replication (Ulukan and Swaan, 2002). Perhaps this dependence of Pol I transcription on topoisomerase action and the direct relationship between rRNA synthesis and cell growth/proliferation rate might indicate a functional link between these chemotherapeutics and ribosome synthesis.
4. Trans acting factors that influence transcription elongation
Over the past 15 years, several studies have implicated the transcription initiation step as a major target for the regulation of rRNA synthesis. Although intense effort was appropriated to determining how transcription initiation can be regulated, very little work was focused on the characterization of factors that influence transcription elongation by Pol I.
During the same time, dozens of factors were identified for their roles in influencing Pol II transcription elongation, and emerging models suggest that this step in the transcription cycle may be the dominant target for control of gene expression (Selth et al., 2010; Nechaev and Adelman, 2011). In the last five years, four factors have been characterized for their roles in Pol I transcription elongation. Even this relatively small sample of studies demonstrates that the elongation step in Pol I transcription is influenced by trans-acting factors across eukaryotic species.
4.1 Spt4 and Spt5
Spt4 and Spt5 form a heterodimer (Spt4/5) that influences Pol II transcription elongation (Swanson and Winston, 1992; Wada et al., 1998). Homologues of SPT5 are present in all domains of life (Hirtreiter et al., 2010). It was known that Spt4/5 can associate with Pol II, but mass spectrometry data from the Hartzog lab demonstrated that Spt5 associates with proteins apart from Pol II, including chromatin modifiers, mRNA capping machinery and Pol I (Lindstrom et al., 2003). We confirmed that Spt4 and Spt5 co-purify with Pol I, and we further demonstrated that deletion of SPT4 led to defects in Pol I transcription elongation (Schneider et al., 2006). Specifically, we observed a small increase in the amount of rRNA synthesized per transcribing RNA polymerase as a function of time in the mutant cells, compared to WT. Thus, we concluded that Spt4/5 is a negative regulator of Pol I transcription elongation. Together with the UBF results from the Moss lab discussed below (Stefanovsky et al., 2006a), this work reignited the interest in factors that influence transcription elongation, after years of silence on the topic.
SPT5 is an essential gene in yeast. Recently, we have characterized point mutations in SPT5 that partially inactivate the protein. Genetic data using these mutations supported the model that Spt4/5 is a negative regulator of Pol I transcription elongation; however, several lines of evidence also showed that Spt5 can positively influence Pol I transcription elongation, directly or indirectly (Anderson et al., 2011). These data, and reference to established roles for Spt5 in Pol II transcription, led to the model that Spt4/5 inhibits transcription but can also activate elongation rate, perhaps after one or more covalent modifications of the complex.
In a separate study, the binding of Spt5 to Pol I was characterized in vitro. Using recombinantly expressed domains of Spt5 and purified Pol I and Pol II, it was shown that Spt5 binds directly to both polymerases and that the same domains of the protein are involved in both interactions (Viktorovskaya et al., 2011). It was further shown that the largest subunits of Pol I and Pol II interact with Spt5. Additional contacts may occur between Spt5 and the subunits A135, A49 and A34 of Pol I. These findings were consistent with previous studies that examined human Spt5 and human Pol II. These data demonstrate that association of Spt5 with polymerases is conserved between man and yeast and that binding of Spt5 to both polymerases is direct.
4.2 Paf1 complex
Like Spt4/5, the Paf1 complex (Paf1C) also influences Pol I transcription elongation in yeast (Mueller and Jaehning, 2002; Squazzo et al., 2002). However, unlike Spt4/5, Paf1C acts exclusively to activate Pol I transcription elongation (Zhang et al., 2009; Zhang et al., 2010).
Paf1C consists of 5 subunits and is conserved among eukaryotes (Mueller and Jaehning, 2002). In yeast, none of the genes for Paf1C subunits is essential; however, deletion of CTR9 or PAF1 results in severe growth defects (Zhang et al., 2009). Many previous studies have revealed the physical and genetic interactions between subunits of Paf1C and Pol II (Krogan et al., 2002; Squazzo et al., 2002). Based on these and many other data, it is widely accepted that Paf1C directly associates with Pol II and influences Pol II transcription and mRNA processing (Mueller et al., 2004).
We showed that subunits of Paf1C also associate with the rDNA in yeast. Deletion of CTR9 or PAF1 resulted in poor growth, reduced rRNA synthesis and defects in rRNA processing (Zhang et al., 2009). However, despite reduced rRNA synthesis rate, there was no reduction in the number of Pol I complexes engaged in transcription. After controlling for potential changes in the rDNA copy number or the degradation rate of rRNA, it was concluded that mutation of Paf1C resulted in reduced Pol I transcription elongation rate in vivo.
It is known that mutations in genes for Paf1C lead to changes in the mRNA expression pattern in cells. Thus, to determine if Paf1C could directly influence Pol I transcription, the complex was purified and included in transcription elongation rate assays for Pol I (Zhang et al., 2010). Since Paf1C increased the transcription elongation rate of Pol I in a purified transcription assay, it was clear that the complex could directly influence Pol I transcription elongation.
In mammalian cells, Paf1C has been closely tied to cell proliferation and growth. The Batra lab showed that the human PAF1 gene (PD2) is over-produced in some tumor cells (Moniaux et al., 2006). They went on to show that overexpression of PD2 is sufficient to induce tumor growth in mice. Later, it was also shown that Paf1C subunits are expressed highly in stem cells, however, during differentiation, the expression level is reduced (Ding et al., 2009; Ponnusamy et al., 2009). Given the connection between cell proliferation and rRNA synthesis rates, all of these data are consistent with Paf1C affecting Pol I in human cells. However, this model has not yet been tested.
4.3 UBF
UBF is a well-characterized HMG-box containing protein that was originally identified by the Tjian lab as an activator of Pol I transcription in human cells (Jantzen et al., 1990). Many subsequent studies have shown that UBF is critical for expression of rRNA in mammals and that it serves as a regulatory target for control of rRNA synthesis. For many years, it was thought that UBF functions solely in initiation of Pol I transcription; however, two recent papers from the Moss and Zomerdijk labs challenge this model.
Using a combination of in vitro and in vivo analyses, the Moss lab concluded that UBF can inhibit Pol I transcription, but that this inhibition is not due to reduced Pol I occupancy of the rDNA (Stefanovsky et al., 2006a). The simplest explanation for this observation is that UBF influences Pol I transcription elongation. This model is consistent with previous data that identified UBF association with the coding region of the rDNA in addition to the promoter (O'Sullivan et al., 2002). Later studies suggested that UBF association with the rDNA may result in a pseudo-nucleosome structure that is altered by ERK-dependent phosphorylation and directly influences Pol I progress through the gene (Stefanovsky et al., 2006b).
Around the same time, the Zomerdijk lab presented data suggesting that UBF is neither a transcription initiation factor nor an effector of transcription elongation. Their in vitro data suggest that UBF increases the rate of promoter escape for Pol I (Panov et al., 2006). Like transcription elongation, the promoter escape step in transcription has been largely ignored, however, given the high density of polymerases on the rDNA during active growth, the escape step must be efficient. These data suggest that UBF plays a direct role in this step of Pol I transcription.
The importance of UBF to Pol I transcription has long been appreciated. These data suggest that UBF directly affects post-initiation steps in Pol I transcription. Beyond influencing Pol I transcription directly, the Hannan lab has shown that UBF expression levels determine the ratio of active versus inactive rDNA repeats in human cells (Sanij et al., 2008). Thus, UBF can control both the accessibility of the rDNA for transcription factors and the activity of the polymerase once bound. More work is required to determine the mechanism(s) by which UBF affects transcription of rDNA and how these various properties of UBF interrelate.
4.4 Elongator 3b
In most eukaryotes, the sole role of Pol I is in production of rRNA. However, in trypanosomes, Pol I also transcribes protein coding genes (Lee and Van der Ploeg, 1997). Even in this unique cellular setting, the elongation step in Pol I transcription appears to be controlled by trans-acting factors.
Elongator is a six subunit complex with known roles in Pol II transcription elongation, and is conserved between yeast and mammals. In African trypanosomes, Elongator is also localized to the nucleolus. Furthermore, mutation or down-regulation of the Elp3b subunit of the complex results in increased synthesis of rRNA by Pol I (Alsford and Horn, 2011). Based on these observations and the effects of ELP3b mutations on trypanosome sensitivity to drugs that affect transcription elongation, the Horn lab concluded that Pol I transcription is negatively influenced by Elongator in trypanosomes. This finding expands the potential mechanisms by which Elongator may influence transcription and adds to the growing list of factors across eukaryotic species that influence Pol I transcription elongation.
5. rDNA chromatin
The number of trans-acting factors with known roles in Pol I transcription elongation is growing. However, most of these factors appear to directly affect Pol I activity. Another potential mechanism for regulation of transcription elongation is by influencing the chromatin state of the rDNA. Many examples of this type of influence on Pol II transcription have been described [for review see (Selth et al., 2010)]. Furthermore, the Grummt and Langst labs have shown that chromatin modifications influence transcription initiation by Pol I in mammals (Langst et al., 1997; Strohner et al., 2001; Nemeth et al., 2004). However, few factors have been identified for a role in modification of rDNA chromatin. Part of this lag in discovery may be due to an ongoing controversy regarding the chromatin state in actively transcribed rDNA repeats (Jones et al., 2007; Merz et al., 2008).
5.1 Yeast rDNA chromatin
Eukaryotic cells carry hundreds of copies of the rDNA in tandem repeats. Only a fraction of these repeats is actively transcribed whereas the rest are transcriptionally silent. In yeast, the ratio of active to inactive repeats is ~1:1 during exponential growth. It is generally accepted that the inactive repeats are densely packed with nucleosomes and likely resemble heterochromatin found elsewhere in the nucleus. However, the chromatin structure of the active repeats remains unresolved in the literature.
Because of the tandem repeating structure on the rDNA, recombination between repeats can lead to rapid alteration of the rDNA copy number in the cell. The Nomura lab showed that deletion of the FOB1 gene in yeast ultimately resulted in stabilizing the rDNA copy number (Kobayashi et al., 1998). This observation enabled the construction of strains with reduced rDNA repeat numbers. In cells with very few rDNA repeats [e.g. 20–40 copies; (Cioci et al., 2003)], all of the repeats are actively transcribed (French et al., 2003). Even under these conditions, chromatin immunoprecipitation (ChIP) detects histone occupancy of the rDNA (Jones et al., 2007). These data led to the conclusion that actively transcribed rDNA genes normally carry nucleosomes, although the arrangement of those nucleosomes on the DNA does not mirror that of the rest of the genome.
Of course, ChIP is not the only method for probing protein occupancy of DNA. The Griesenbeck lab employed an assay that uses micrococcal nuclease fusions to proteins of interest (chromatin endogenous cleavage; ChEC). Cells are lysed and the fused nuclease is activated by incubation with calcium. Using primer extension, the cleavage of specific sites within the genome is quantified. More cleavage is indicative of more occupancy of that region by the fusion protein. ChEC analysis detected no occupancy of histones in the active rDNA repeats. Instead of histones, Hmo1 (the yeast analogue of UBF) bound the actively transcribed genes (Merz et al., 2008). These data led to the conclusion that actively transcribed genes lack histones.
To date, there has been no resolution of this controversy in the literature. The Proudfoot lab concluded that the nucleosome structure of the rDNA is dynamic and unphased (Jones et al., 2007). Perhaps this dynamism is partially responsible for the differences in signal detected using these two different methods. More investigation of this phenomenon is required to determine the structure of the actively transcribed as well as the silent rDNA repeats in yeast.
5.2 Mammalian chromatin
It is known that RNA polymerase III can transcribe through nucleosomal DNA in vitro (Studitsky et al., 1997), whereas Pol II needs assistance from multiple trans-acting factors to accomplish this task (Belotserkovskaya et al., 2003). Until recently, the question of whether Pol I could independently negotiate chromatin remained open. The Zomerdijk lab employed a series of biochemical and genetic tests to show that Pol I does not efficiently transcribe nucleosomal templates in vitro. However, the FACT complex (facilitates chromatin transcription) enhances the ability of Pol I to transcribe through nucleosomes (Birch et al., 2009). Furthermore, FACT associates with Pol I in cell extracts and localizes to the rDNA in growing cells. Thus, FACT can directly influence the ability of Pol I to elongate in vivo.
Nucleolin is an abundant nucleolar protein that affects rDNA transcription, pre-rRNA processiong and ribosome assembly (Mongelard and Bouvet, 2007). Interestingly, nucleolin has also been shown to influence chromatin remodeling [indirectly, by association with the remodelers SWI/SNF and ARC; (Angelov et al., 2006)] as well as Pol I transcription in vitro and in vivo (Rickards et al., 2007). Perhaps nucleolin and FACT fill similar roles in transcription of Pol I through rDNA chromatin. The molecular mechanisms by which nucleolin influences Pol I and Pol II remain unclear.
The role of chromatin and its covalent modifications in transcription elongation by Pol I will be a topic of substantial future study. Several previous studies have characterized differences between the chromatin state at actively transcribed rDNA repeats relative to transcriptionally silent repeats (McKnight and Miller, 1976; Dammann et al., 1993; Stefanovsky and Moss, 2006; Sanij et al., 2008); however, consequences of covalent modifications of histones within the rDNA coding region remain unclear. The simplest proposal is that these modifications will mirror those of Pol II transcribed genes; however, continued effort is required to test this hypothesis. Additionally, DNA methylation was recently shown to influence Pol I transcription in mammalian cells (Gagnon-Kugler et al., 2009). The functional relationship between the rDNA template and its transcription requires substantial further characterization.
6. Is rRNA synthesis regulated at the elongation step?
It is well known that ribosome synthesis rates and cell growth/proliferation rates are proportional. Because of this relationship, there is much interest in identifying the molecular mechanisms that regulate Pol I transcription in order to understand and potentially control cell proliferation (Drygin et al., 2010). Transcription initiation by Pol I has been clearly described as a regulatory target for the control of ribosome synthesis, but is the elongation rate also regulated in response to growth stimuli? Multiple studies have shown that under conditions when rRNA synthesis is inhibited almost completely, Pol I loading on the rDNA is reduced but not eliminated (Claypool et al., 2004; Tongaonkar et al., 2005; Philippi et al., 2010). These data suggest that either rRNA is made by the polymerase and degraded, or the elongation rate of those engaged polymerases is reduced. Indeed, the cellular response may include both inhibition of rDNA transcription and increased rRNA degradation (Reiter et al., 2011). Two studies, discussed below, have identified factors that potentially influence regulation of Pol I elongation in response to growth signaling (Stefanovsky et al., 2006a; Zhang et al., 2010). Ongoing studies may also identify the ribonucleases required for rapid degradation of rRNA upon nutrient limitation.
6.1 Mammalian UBF affects Pol I transcription elongation
The best characterized nutrient-responsive regulator of Pol I transcription elongation is UBF, a factor first identified as an initiation factor for Pol I. As described above, the Moss lab used a series of biochemical and genetic experiments to show that UBF can also inhibit Pol I transcription elongation (Stefanovsky et al., 2006a). Importantly, they also showed that after extended serum starvation, addition of growth factors to the culture medium results in de-repression of Pol I transcription. This activation of rRNA synthesis was independent of increased transcription initiation. They showed that this de-repression results from ERK-dependent phosphorylation of the HMG boxes of UBF. Mutation of phosphorylated residues within a truncated UBF construct eliminated the ability of ERK to activate Pol I elongation in vitro using templates coated with UBF. Thus, these data suggest UBF is a direct, nutrient responsive regulator of Pol I transcription elongation in human cells.
6.2 The Paf1 complex in yeast
The Paf1 complex influences Pol I and Pol II transcription elongation, but its function is not essential for survival. In WT yeast cells, inhibition of TOR signaling (with rapamycin) results in fast and robust inhibition of Pol I transcription. After 40 minutes exposure to rapamycin, rRNA transcription is almost undetectable with standard metabolic labeling approaches. In Paf1 mutants, however, inhibition of Pol I transcription in response to rapamycin is not complete (Zhang et al., 2010). This dependence on Paf1C function for efficient regulation is not specific to TOR signaling, since amino acid limitation showed the same trend. Thus, directly or indirectly, Paf1C function is required for efficient regulation of rRNA synthesis. Since we know that Paf1C can directly affect Pol I transcription, the simplest model to explain these observations is that Paf1C itself is directly modified in response to nutrients.
7. Summary
This review has highlighted issues concerning Pol I transcription, specifically focusing on the elongation step. Emerging interest in Pol I transcription elongation has led to identification of a small number of factors that can potentially affect Pol I transcription elongation directly. Two of these factors influence Pol I transcription in a nutrient or growth-responsive manner. Certainly, many more factors will be identified with roles in Pol I transcription elongation. Recent studies suggest that Pol II transcription may be primarily regulated at the elongation step rather than through recruitment to the promoter. Given the robust data that support regulation of Pol I transcription initiation, complete regulation of rRNA synthesis at the elongation step is highly unlikely. However, considering the strength of Pol I transcription and the magnitude by which it must be modulated in growing versus quiescent cells, it is obvious that cells must employ multiple overlapping regulatory mechanisms to control this robust “player” in cell metabolism. Thus, the field can look forward to the identification of several modulators of elongation and ever increasing complexity in the quest to define and control rRNA synthesis and cell proliferation.
Acknowledgements
First, I wish to apologize to colleagues whose work was not cited in this review. Every attempt was made to fairly refer to examples of each topic, but the depth of the field precludes complete inclusion. I would like to thank Dr. Ann L. Beyer and all members of the Schneider lab for critical evaluation of the manuscript. The work of the Schneider lab is supported by the National Institutes of Health grant #GM84946.
Abbreviations
- Pol
RNA polymerase
- rRNA
ribosomal RNA
- rDNA
ribosomal DNA
- Spt4/5
Spt4 and Spt5
- Paf1C
Paf1 complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Alsford S, Horn D. Elongator protein 3b negatively regulates ribosomal DNA transcription in african trypanosomes. Mol. Cell Biol. 2011;31:1822–1832. doi: 10.1128/MCB.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Sikes ML, Zhang Y, French SL, Salgia S, Beyer AL, Nomura M, Schneider DA. The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J. Biol. Chem. 2011 doi: 10.1074/jbc.M110.202101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongelard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, Bouvet P. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. Embo J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprikian P, Moorefield B, Reeder RH. TATA binding protein can stimulate coredirected transcription by yeast RNA polymerase I. Mol. Cell Biol. 2000;20:5269–5275. doi: 10.1128/mcb.20.14.5269-5275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckouet F, Labarre-Mariotte S, Albert B, Imazawa Y, Werner M, Gadal O, Nogi Y, Thuriaux P. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol. Cell Biol. 2008;28:1596–1605. doi: 10.1128/MCB.01464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Berger AB, Decourty L, Badis G, Nehrbass U, Jacquier A, Gadal O. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell Biol. 2007;27:8015–8026. doi: 10.1128/MCB.01102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, Russell J, Lee SC, Zomerdijk JC. FACT facilitates chromatin transcription by RNA polymerases I and III. Embo J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem. Soc. Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio MC, Negri R, Dieci G. Promoter architectures in the yeast ribosomal expression program. Transcription. 2011;2:71–77. doi: 10.4161/trns.2.2.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Cioci F, Vu L, Eliason K, Oakes M, Siddiqi IN, Nomura M. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell. 2003;12:135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. Embo J. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I, Theis M, Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle. 2010;9:3687–3699. doi: 10.4161/cc.9.18.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A, Conaway JW, Conaway RC. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev. 2001;11:209–214. doi: 10.1016/s0959-437x(00)00181-7. [DOI] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S, Milkereit P, Peyroche G, Riva M, Carles C, Tschochner H. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14334–14339. doi: 10.1073/pnas.231181398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, El Hage A, Smith MM, Tollervey D, Smith JS, Beyer AL. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol. Cell Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Mariotte-Labarre S, Chedin S, Quemeneur E, Carles C, Sentenac A, Thuriaux P. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell Biol. 1997;17:1787–1795. doi: 10.1128/mcb.17.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon-Kugler T, Langlois F, Stefanovsky V, Lessard F, Moss T. Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell. 2009;35:414–425. doi: 10.1016/j.molcel.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- Gerber J, Reiter A, Steinbauer R, Jakob S, Kuhn CD, Cramer P, Griesenbeck J, Milkereit P, Tschochner H. Site specific phosphorylation of yeast RNA polymerase I. Nucleic Acids Res. 2008;36:793–802. doi: 10.1093/nar/gkm1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JC. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. Embo J. 2007;26:1560–1568. doi: 10.1038/sj.emboj.7601601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Hannan RD, Smith SD, Jefferson LS, Lun M, Rothblum LI. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene. 2000;19:4988–4999. doi: 10.1038/sj.onc.1203875. [DOI] [PubMed] [Google Scholar]
- Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontz RD, French SL, Oakes ML, Tongaonkar P, Nomura M, Beyer AL, Smith JS. Transcription of multiple yeast ribosomal DNA genes requires targeting of UAF to the promoter by Uaf30. Mol. Cell Biol. 2008;28:6709–6719. doi: 10.1128/MCB.00703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- Jantzen HM, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Jones HS, Kawauchi J, Braglia P, Alen CM, Kent NA, Proudfoot NJ. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Dodd JA, Lalo D, Nomura M. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Josaitis CA, Dodd JA, Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- Keys DA, Lee BS, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 1803;2010:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- Lalo D, Steffan JS, Dodd JA, Nomura M. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J. Biol. Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- Langst G, Blank TA, Becker PB, Grummt I. RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. Embo J. 1997;16:760–768. doi: 10.1093/emboj/16.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Van der Ploeg LH. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu. Rev. Microbiol. 1997;51:463–489. doi: 10.1146/annurev.micro.51.1.463. [DOI] [PubMed] [Google Scholar]
- Liljelund P, Mariotte S, Buhler JM, Sentenac A. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9302–9305. doi: 10.1073/pnas.89.19.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, 3rd, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19:933–941. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL, Miller OL., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008;22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Tschochner H. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. Embo J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Moniaux N, Nemos C, Schmied BM, Chauhan SC, Deb S, Morikane K, Choudhury A, Vanlith M, Sutherlin M, Sikela JM, Hollingsworth MA, Batra SK. The human homologue of the RNA polymerase II-associated factor 1 (hPaf1), localized on the 19q13 amplicon, is associated with tumorigenesis. Oncogene. 2006;25:3247–3257. doi: 10.1038/sj.onc.1209353. [DOI] [PubMed] [Google Scholar]
- Moorefield B, Greene EA, Reeder RH. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4724–4729. doi: 10.1073/pnas.080063997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 1809;2011:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Strohner R, Grummt I, Langst G. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 2004;32:4091–4099. doi: 10.1093/nar/gkh732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Nogi Y, Oakes M. Transcription of rDNA in the Yeast Saccharomyces cerevisiae. In: Oakes MOJ, editor. The Nucleolus. London: Kluwer Academic / Plenum Publishers; 2004. pp. 128–153. [Google Scholar]
- O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell Biol. 2002;22:657–668. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes ML, Siddiqi I, French SL, Vu L, Sato M, Aris JP, Beyer AL, Nomura M. Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast. Mol. Cell Biol. 2006;26:3889–3901. doi: 10.1128/MCB.26.10.3889-3901.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Russell J, Zomerdijk JC. UBF activates RNA polymerase I transcription by stimulating promoter escape. Embo J. 2006;25:3310–3322. doi: 10.1038/sj.emboj.7601221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. Embo J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippi A, Steinbauer R, Reiter A, Fath S, Leger-Silvestre I, Milkereit P, Griesenbeck J, Tschochner H. TOR-dependent reduction in the expression level of Rrn3p lowers the activity of the yeast RNA Pol I machinery, but does not account for the strong inhibition of rRNA production. Nucleic Acids Res. 2010;38:5315–5326. doi: 10.1093/nar/gkq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianese G. Beitr. Pathol. Anat. Allgem. Pathol. 1896;142:1–193. [Google Scholar]
- Ponnusamy MP, Deb S, Dey P, Chakraborty S, Rachagani S, Senapati S, Batra SK. RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells. 2009;27:3001–3011. doi: 10.1002/stem.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K, Wall M, Brandenburger Y, Palatsides M, Pearson RB, McArthur GA, Hannan RD. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. Embo J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. Pseudo-NORs: a novel model for studying nucleoli. Biochim. Biophys. Acta. 2008;1783:2116–2123. doi: 10.1016/j.bbamcr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Reiter A, Steinbauer R, Philippi A, Gerber J, Tschochner H, Milkereit P, Griesenbeck J. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Mol. Cell Biol. 2011;31:803–817. doi: 10.1128/MCB.01227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006:203–216. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, Moss T, Rothblum L, Hannan KM, McArthur GA, Pearson RB, Hannan RD. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12707–12712. doi: 10.1073/pnas.0605686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu. Rev. Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. Embo J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell. 2006a;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Stefanovsky V, Moss T. Regulation of rRNA synthesis in human and mouse cells is not determined by changes in active gene count. Cell Cycle. 2006;5:735–739. doi: 10.4161/cc.5.7.2633. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Langlois F, Bazett-Jones D, Pelletier G, Moss T. ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF. Biochemistry. 2006b;45:3626–3634. doi: 10.1021/bi051782h. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Keys DA, Dodd JA, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. Embo J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongaonkar P, French SL, Oakes ML, Vu L, Schneider DA, Beyer AL, Nomura M. Histones are required for transcription of yeast rRNA genes by RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10129–10134. doi: 10.1073/pnas.0504563102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trere D, Ceccarelli C, Montanaro L, Tosti E, Derenzini M. Nucleolar size and activity are related to pRb and p53 status in human breast cancer. J. Histochem. Cytochem. 2004;52:1601–1607. doi: 10.1369/jhc.4A6454.2004. [DOI] [PubMed] [Google Scholar]
- Ulukan H, Swaan PW. Camptothecins: a review of their chemotherapeutic potential. Drugs. 2002;62:2039–2057. doi: 10.2165/00003495-200262140-00004. [DOI] [PubMed] [Google Scholar]
- Viktorovskaya OV, Appling FD, Schneider DA. Yeast transcription elongation factor SPT5 associates with RNA polymerase I and RNA polymerase II directly. J. Biol. Chem. 2011 doi: 10.1074/jbc.M110.202119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol. Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Werner M, Thuriaux P, Soutourina J. Structure-function analysis of RNA polymerases I and III. Curr. Opin. Struct. Biol. 2009;19:740–745. doi: 10.1016/j.sbi.2009.10.005. [DOI] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Comai L, Johnson DL. PTEN represses RNA Polymerase I transcription by disrupting the SL1 complex. Mol. Cell Biol. 2005;25:6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sikes ML, Beyer AL, Schneider DA. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 2009 doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Smith ADt, Renfrow MB, Schneider DA. The RNA polymerase-associated factor 1 complex (Paf1C) directly increases the elongation rate of RNA polymerase I and is required for efficient regulation of rRNA synthesis. J. Biol. Chem. 2010;285:14152–14159. doi: 10.1074/jbc.M110.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan X, Frodin M, Grummt I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]