Abstract
Inherited dental malformations constitute a clinically and genetically heterogeneous group of disorders. Here, we report on a severe developmental dental defect that results in a dentin dysplasia phenotype with major microdontia, oligodontia, and shape abnormalities in a highly consanguineous family. Homozygosity mapping revealed a unique zone on 6q27-ter. The two affected children were found to carry a homozygous mutation in SMOC2. Knockdown of smoc2 in zebrafish showed pharyngeal teeth that had abnormalities reminiscent of the human phenotype. Moreover, smoc2 depletion in zebrafish affected the expression of three major odontogenesis genes: dlx2, bmp2, and pitx2.
Main Text
Dental development is a complex process of reiterative epithelio-mesenchymal interactions between the oral ectoderm and the mesenchymal cells of cephalic neural-crest origin. Tooth development involves numerous genes implied in various signaling pathways such as the Bone Morphogenetic Protein (BMP), Fibroblast Growth Factor (FGF), Sonic hedgehog homolog (SHH), and Wnt pathways.1, 2 Tooth developmental abnormalities can affect numbering, shape, size, hard tissue structures (such as enamel or dentin), roots, and periodontium formation, as well as global developmental processes such as dental eruption and resorption. All of these can be affected alone or together in either inherited disorders limited to the orodental sphere or more complex syndromes.
Here, we report on a unique and complex tooth malformation phenotype suggestive of autosomal-recessive inheritance in two first-degree cousins born from a highly consanguineous family of Turkish origin. Both children were referred to the Reference Center for Rare Orodental Diseases at the Strasbourg University Hospital because, compared to their healthy siblings, they exhibited extreme microdontia and were missing teeth. Both children presented with extreme microdontia, oligodontia, dental shape anomalies, double permanent-tooth formation, thin enamel, and short roots (with a thin associated alveolar bone), as seen in the spectrum of dentin dysplasia type I (Figure 1).3, 4 The eldest child (III.3) was 10 years old on last examination and presented a well-identified, moderate, X-linked, ichthyosis phenotype known to segregate in the family. The youngest child (III.4), III.3's female cousin, was 5 years old at her first visit and received followed-up examinations for the next 5 years. Both children were born after uneventful pregnancies and were normal at birth. Their developmental milestones are normal to date, and their general physical appearance is unremarkable except for obesity in III.4 (not present in III.3) and very mild bone abnormalities in III.4 (not present in III.3). The orodental findings were documented with the D[4]/phenodent Diagnosing Dental Defects Database. Oligodontia was diagnosed because III.4 was missing 13 permanent teeth and III.3 was missing 14. Anomalies of tooth size were observed, and an extreme microdontia affected both primary teeth (all present) and permanent teeth. However, some permanent teeth were macrodont. Anomalies of tooth shape concerned all existing teeth; extra cusps were visible, and crowns were tiny, globular, and malformed, especially in the primary dentition. Double tooth formation (notched and macrodont) was visible on the permanent incisors. Temporary and permanent molars exhibited taurodontism. Moreover, the molars showed tooth-structure anomalies reminiscent of the dentin dysplasia type I spectrum and had very short roots (Figure 1).3 Compared to dentin in the X-ray, the enamel was very thin and had limited contrast. The alveolar bone associated with the primary teeth was hypodeveloped. The primary teeth were mobile and exfoliated prematurely.
Figure 1.
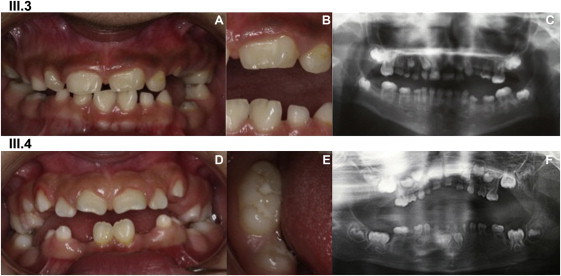
Clinical Description of the Affected Family Members
A clinical description of individuals III.3 (A, B, and C) and III.4 (D, E, and F) shows major dental developmental abnormalities in tooth number, size, shape, structure, eruption, and resorption, as seen in the intraoral photographs (A, B, D, and E) and the panoramic radiographs (C and F).
(A and B) (A) shows an intraoral view of III.3 (10 years old). Beside the microdont primary and permanent teeth, which show spaced dentition, double tooth formation (notched and macrodont) is visible on permanent central incisors 21 and 31; 21 shows a vestibular abnormal relief. These anomalies are clearly seen on (B) in an enlargement of the left incisor region.
(C) A panoramic radiograph shows III.3, who is missing the following permanent teeth: 18, 15, 24, 25, 28, 48, 45, 44, 43, 32, 33, 34, 35, and 38. The primary and permanent molars are taurodont. The roots are extremely short and are slightly more developed in the permanent dentition but are, however, conical with sharp endings. The pulp has a flame-like shape. The enamel is very thin and has limited contrast compared to the dentin in the X-Ray. Teeth 64, 65, 74, and 75 are reincluded.
(D) Intraoral view of III.4 (9.5 years old). Double tooth formation (notched and macrodont) is visible on the permanent central-upper-left incisor (21). The lower arch seems interrupted in the area of missing teeth (45, 43, 42, 32, 33, 34, and 35). Teeth 85 and 75 are reincluded, indicating ankylosis in the alveolar bone.
(E) A close-up on macrodont tooth 46 (lower-right permanent first molar) shows extra cusps and an elongated crown on its mesiodistal axis.
(F) A panoramic radiograph of III.4 at 5 years old shows oligodontia—13 permanent teeth are missing (18, 15, 25, 28, 48, 45, 43, 42, 32, 33, 34, 35, and 38)—and extreme microdontia of all the primary teeth. Note the short and sharp roots and the hypodeveloped alveolar bone.
This study—designed to identify the genetic mutations involved in the dentin dysplasia phenotype—was approved by the ethics committee of the Strasbourg University Hospital. Informed consent was obtained from all individuals who participated in the study. Homozygous mapping via GeneChip Human 250K SNP Affymetrix was performed on affected individuals III.3 and III.4 and nonaffected individuals III.1, III.2, III.5, and III.6. A unique homozygosity region was shared between the two affected individuals and was located between rs2981956 and the end of chromosome 6, defining a 3 Mb region on chromosome 6q27-ter (Figure 2A). According to Ensembl, this interval contained 69 annotated genes. Genes were selected as likely candidates either because of their known implication in inherited dental conditions or because of their potential dental expression, indicated by the following databases: Helsinki University's Gene Expression in Tooth; the UCSC Genome Browser; the 1000 Genomes Browser; the Ensembl Genome Browser; GeneHub-GEPIS; GenePaint; Eurexpress; and the Zebrafish Information Network (see Web Resources).
Figure 2.
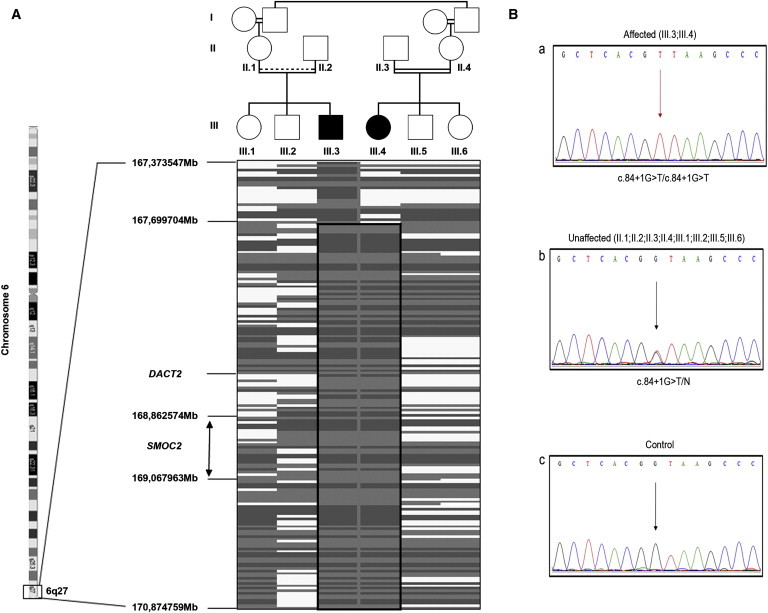
Homozygosity Mapping and Mutation Detection
(A) A simplified pedigree of the family, underlined by corresponding schematic representation of the homozygosity mapping results, shows the chromosome 6 homozygous region that is common in affected individuals: Gray shading indicates homozygous SNPs and white zones indicate heterozygous alleles.
(B) Electropherograms of a part of the SMOC2 exon1-intron1 boundary show (a) a homozygous c.84+1G>T mutation in an affected child; (b) a heterozygous c.84+1G>T mutation in a nonaffected sibling; and (c) a healthy control individual.
Two genes were selected with high priority: DACT2 (Dapper antagonist of beta-catenine 2 [OMIM 608966]) and SMOC2 (SPARC-related modular calcium-binding protein [OMIM 607223]). Dact2 modulates Wnt signaling by binding to the intracellular protein Dishevelled (Dvl) and might play an important signal-modulating role in tooth development at the level of the epithelial cells that include the enamel-knot signaling centers and the preameloblasts.5 The sequencing of DACT2 4 exons was normal in both affected individuals.
SMOC2 belongs to a family of matricellular proteins that regulate interactions between cells and the extracellular matrix. The GenePaint database indicated a high level of in situ hybridization in the craniofacial region of the mouse at embryonic day 14.5 (E14.5), especially at the level of the tooth mesenchyme. SMOC2 spans about 226 kb. The coding region of SMOC2 consists of 13 exons. Each domain of SMOC2 is encoded by one or more exons, and the domain borders coincide with splice sites. Sequencing of SMOC2 (ENST00000354536; NM_022138/hg19) revealed a homozygous mutation (c.84+1G>T) in the canonical-splice donor site of intron 1. The parents of both affected children were heterozygous for this mutation, and the children's nonaffected siblings were heterozygous for this mutation (Figure 2B). This mutation was absent in 112 ethnically matched controls. The primer sequences are detailed in Table S1, available online.
In order to confirm that SMOC2 was the only gene that carried mutations in the interval, we performed exome sequencing in collaboration with IntegraGen (Evry, France). Exons of patient III.4's DNA were captured via in-solution enrichment methodology (SureSelect Human All Exon Kit v.3, Agilent, Massy, France) with the company's biotinylated oligonucleotide probe library (Agilent Human All Exon 50 Mb Kit v.3). The genomic DNA was then sequenced on a sequencer as paired-end 75 bases (HISEQ, Illumina, San Diego, USA). Image analysis and base calling were performed with Real Time Analysis (RTA) Pipeline version 1.9, set to its default parameters (Illumina). The bioinfomatic analysis of sequencing data was based on the pipeline provided by IntegraGen (Illumina's CASAVA 1.8). CASAVA performs alignment, calls the SNPs on the basis of allele calls and read depth, and detects variants (SNPs and indels). Genetic variation annotation was performed by an in-house pipeline and provided results for the sample in tabulated text files.
Among the sequences that could be analyzed, no obvious truncating or nonsense mutation could be identified in any of the 69 genes. We identified 81 substitutions (47 intronic, or in the untranslated regions; 22 synonymous; and 12 missense, of which all were SNPs), seven deletions (all intronic and four SNPs), and one insertion (all intronic and one SNP).
However, in our 3 Mb region of interest on chromosome 6, 32 out of 279 baits could not be further analyzed because they did not provide enough coverage. A total of 6.6 kb from the 3 Mb region was not covered sufficiently and overlapped with at least one bait (average bait size was 121 bp) in each of 17 genes (from one to four baits per gene). Thus, taking into account that we had already sequenced two of these genes—SMOC2 and DACT2, which together account for six unread baits—because of their high expected impact on tooth development, 15 genes (out of the 69) that accounted for 5.5 kb were still imperfectly explored and were excluded as interesting candidates in our initial approach.
Interestingly, the region that contains our mutation is poorly covered, and we could not identify by whole-exome sequencing the SMOC2 mutation located at the end of exon 1. Moreover, this region of SMOC2 is GC rich, possibly explaining this failure. We compared exome-capture sequencing data from several independent individuals involved in other projects by applying the same setting and confirmed the deficit in the sequence coverage of this specific bait. We would like to point out that although the exome-capture approach is a true revolution in human genetics, it has to be analyzed cautiously; in our case, we would have missed the causative mutation and gene.
Although widespread expression of SMOC2 in various human tissues (skin, liver, muscle, lung, spleen, colon, pancreas, kidney) is demonstrated by quantitative reverse transcription PCR (QIAGEN Quantitect primer assay, assay name Hs_SMOC2_1_SG Cat N° QT00085687), we did not succeed in comparing the reverse transcription PCR (RT-PCR) of patients to that of controls because the SMOC2 expression seemed to be very weak in human fibroblasts.
SMOC2 was identified by way of an expressed sequence tag database search for proteins homologous to the BM-40 protein family, also known as secreted protein acidic and rich in cysteines (SPARC).6 BM-40 matricellular proteins are extracellular proteins that do not contribute structurally to the extracellular milieu but that regulate interactions between cells and the extracellular matrix.7 The SPARC/osteonectin/BM-40 family is expressed in many cell types and is highly expressed during embryogenesis, wound healing, and other instances where there is extensive tissue remodelling.8 SMOC2 shares an identical domain structure with SMOC1, another secreted modular calcium-binding protein.9 In addition to a extracellular calcium-binding (EC) domain homologous to that in BM-40, SMOC1 and SMOC2 share two thyroglobulin-like (TY) domains, an follistatin (FS) domain, and a novel domain. Mutations in SMOC1 have recently been described in patients with a rare recessive developmental disease—Waardenburg anophthalmia syndrome, which mainly involves severe eye malformations and limb defects.10, 11 SMOC2 has been reported as a risk locus for generalized vitiligo in an isolated Romanian community, but this finding has been questioned in another study.12, 13 To date, no inherited condition has been clearly related to SMOC2 mutations. The mutations identified in this family point to a major role of SMOC2 in dental development, and we aimed to gather functional data for such a role.
Mouse Smoc2 is located on chromosome 17, and its intron-exon structure is highly conserved in comparison to that of the human gene. Smoc2 is expressed in nearly all adult mouse tissues, and the highest expression is found in the heart, muscles, spleen, and ovaries.6 We next analyzed the expression of Smoc2 during mouse orodental development. We used E14.5 tooth germ cDNAs to perform a study with GeneChip Mouse Gene 1.0 ST arrays (Affymetrix). We detected greater Smoc2 expression in molar than in incisor germs; the opposite pattern was evident for Smoc1 expression. Moreover, in situ hybridization was performed on mouse embryos at E12.5, E14.5, E16.5, and E18.5, which correspond to dental lamina, cap, bell, and bell with differentiated odontoblast and preameloblast stages, respectively. Smoc2 expression was found in the oral ectoderm and the outer dental epithelium at E14.5 and in mesenchymal papilla facing the epithelial loops of molars and the only lingual loop of incisors (Figure S1).
To obtain independent functional data on the role of Smoc2 in tooth development, we turned to zebrafish by using the well-established morpholino knockdown technique.14 The development and structure of zebrafish teeth reflect the evolutionary, ancestral condition of jawed vertebrates. A distinctive feature of zebrafish dentition is the restriction of teeth to a single pair of pharyngeal bones: Teeth are absent from the oral cavity and are restricted to the fifth ceratobranchials.15 Such dentition is characteristic of the order Cypriniformes.16 Morphological signs of tooth initiation appear around the time of hatching (2 days after fertilization) in zebrafish, and the first germs become mineralized and attached to the underlying bone within 4 days after fertilization. Tooth development is similar to that of mammals.17, 18 We therefore used the zebrafish as a model to analyze the function of Smoc2 in tooth development. A search of the zebrafish genome sequence (Ensembl, zv9) revealed a 115 amino acid zebrafish Smoc2 protein (ENSDARP00000108925; named Smoc2a) that shared 68% identity with a 123 amino acid human SMOC2 splice variant (GRCh37, ENSP00000440052). We identified the full-length zebrafish smoc2, which encodes a 429 amino acid protein (Figure S2). The protein shares 67% overall identity with the longest human SMOC2 splice variant (ENSP00000346537). In particular, the C-terminal calcium-binding domains appear to be evolutionarily conserved: Human SMOC2 has numerous splice variants, all of which share the C-terminal region, which includes two calcium-binding domains. In situ hybridization of smoc2a mRNA revealed expression in the pharyngeal pouches and arches. Expression in the area from which the teeth develop was diffuse at 48 hpf (not shown) but condensed by 56 hpf to two bilateral dots, marking the position of the first pair of teeth (Figures S3C–S3D). Morpholinos are an effective way of transiently knocking down zebrafish gene function.14 Two morpholinos were created: Mo-smoc2-1 and Mo-smoc2-2. Mo-smoc2-1 was designed to target the smoc2a ATG triplet code to impair the initiation of translation and splicing within larger smoc2 transcripts. Mo-smoc2-2 was designed to target the exon2-intron2 boundary of smoc2a (Figures S2A–S2F). The efficiency of Mo-smoc2-2 was controlled by RT-PCR. The morphant transcript contained both the correctly spliced fragment and a transcript lacking part of the second exon that encodes the calcium-binding domain, leading to a premature stop codon (Figures S2F–S2H). The morpholino appears to activate a cryptic splice site, as previously noted for other morpholinos.19 We analyzed the development of the teeth in 5-day-old smoc2-2 morphants by using alizarin red to stain the calcified structures.20 In both smoc2 morphants, the first two bilateral teeth were smaller than those of the controls (Figure 3, 40 embryos were analyzed). In about 5% of the morphants, the teeth were even undetectable (Figure 3D). The size and presence of the teeth were probably dependent on the level of smoc2 depletion. In addition, although the appearance of the second tooth was already visible in the control embryos, it was undetectable in morphants (Figures 3H–3K). A close inspection of tooth shape revealed a very broad tooth base anchored within the fifth ceratobranchial bone in control larvae (Figures 3H and 3I), whereas it appeared very thin in the morphants (Figures 3J and 3K). In addition, compared to the controls, the smoc2 morphants were missing calcification of some dermal bones and the fifth ceratobranchial bone (Figures 3A–3D), indicating that the skull was affected in morphants. Injections of 0.3 mM of Mo-smoc2-2 and 0.7 mM of Mo-smoc2-1 resulted in a slightly reduced head size in 74% and 56% of the embryos, respectively (category 1, Figure S2D). This reduction was independent of the head volume, excluding the possibility that tooth development could have indirectly been affected by overall impairment of head development (Figures 3B and 3C). To further rule out developmental delay, we analyzed the head musculature in wild-type and smoc2-2 morphants at 5 dpf with a skeletal muscle reporter line (Roostalu, personal communication). All the muscles present in the wild-type were also seen in the morphant, indicating that the development of the head musculature occurred correctly in the morphant (data not shown). In addition, an immunostaining with an antibody against phosphohistone H3 marked proliferating cells. We showed that cell proliferation was not inhibited in the oropharyngeal area of 5 dpf morphants, indicating that the reduced tooth size was not a consequence of an overall reduced proliferation rate (data not shown).
Figure 3.
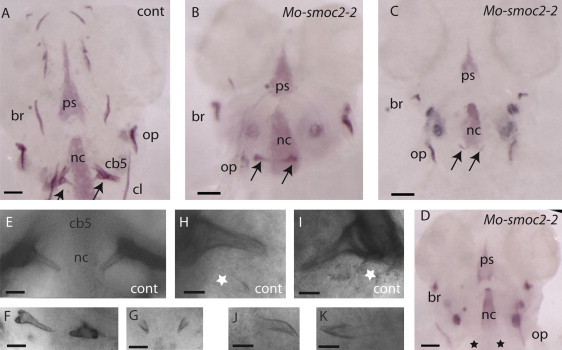
Dentition in smoc2 Morphants
The heads of the control (A) and the smoc2-2 morphants (B–D) stained with alizarin red at 5 dpf show different degrees of reduction of pharyngeal tooth size (arrows in B and C) and the complete absence of pharyngeal teeth (black stars in D) in the morphants. Control (E) and morphant (F and G) teeth are shown at higher magnification. Note the misorientation and the difference in the shape of the teeth in (F) compared to the control. Compared to the control (H and J), the morphants (J and K) show no additional emerging teeth. The white stars represent the transparent second tooth. The scale bars represent the following measurements: (A–D): 50 μm; (E–G): 25 μm; and (H–K): 12 μm. The following terms are abbreviated: branchiostegal ray (br); ceratobranchial 5 (cb5); parasphenoid (ps); ceratohyal (cl); notochord (nc); and opercle (op). All embryos are presented in ventral view, anterior up.
Next, we analyzed the developing zebrafish tooth germs by using probes of genes whose orthologs are expressed during mouse odontogenesis. dlx2 is an early marker of the dental epithelium in the mouse.21 The zebrafish possesses two semiorthologs (a duplicate gene pair equally related to a single ortholog in another species)22 of human DLX2: dlx2a and dlx2b.23, 24 Both duplicates are expressed in tooth germs from 48 hr onward.25 dlx2b is expressed initially in the thickened dental epithelium, but not in the underlying mesenchyme.26 The expression of this gene marks the location of the tooth germ undergoing morphogenesis before mineralization. dlx2b expression in 56 hpf and 72 hpf smoc2 morphants was undetectable in the pharyngeal region where teeth would normally form (Figures 4A–4D). This absence of expression was observed in 90% of smoc2-1 and smoc2-2 morphants (20 embryos were analyzed for each morpholino, Figures 4B–4D). In contrast, 100% of the control larvae (n > 30) showed normal expression (Figure 4C). Other dlx2 domains, including the forebrain, did not seem to be impaired in the morphants. Because morpholinos can cause cell death in an unspecific manner, we coinjected a morpholino targeting p53 and analyzed dlx2b expression at 72 hpf. Even when apoptosis was blocked, dlx2b expression was gone from the tooth germ area in the morphant (Figures S3I–S3M). In conclusion, apoptosis was not the cause of a lack of dlx2b expression.
Figure 4.
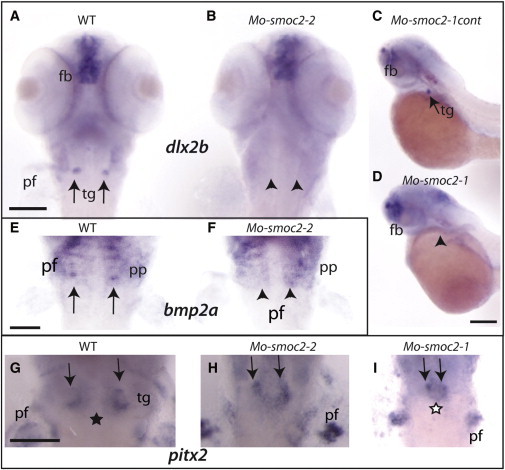
smoc2 Morphants Exhibit Tooth-Germ Defects
(A–I) In situ hybridization of both dlx2b (A–D; probe obtained from D.W. Stock, Colorado, USA), bmp2a (E and F; probe obtained from M. Hammerschmidt, Cologne, Germany) and pitx2 (G–I, pitx2 full-length cDNA was cloned into the Flc3 plasmid [Riken]) on wild-type (A, E, and G), smoc2-2 (B, F, and H), smoc2-1 (D and I), and smoc2-1cont morphants (C) shows a loss of dlx2b and bmp2 expression in smoc2 morphants (B, F, and D) and reduced and fused expression of pitx2 (H and I). The smoc2-1cont morphants do not show any defects.
The arrows represent teeth germs; the arrow heads represent missing dlx2b or bmp2a expression. The black star represents a gap within pitx2 expression, the white star represents fused pitx2 expression. Abbreviations are as follows: pf, pectoral fin; tg, teeth germs; fb, forebrain; and pp, pharyngeal pouches. Lateral (C and D) and ventral (A, B, E–I) views of embryos, 56 hpf. The scale bar represents 100 μm.
In contrast to the lack of dlx2b expression, no obvious reduction of expression of the semiortholog dlx2a in smoc2a morphants was revealed through in situ hybridization (Figures S3C and S3D), suggesting that the two orthologs were regulated differently.
Bone morphogenetic proteins (BMPs) are known to play multiple roles in tetrapod tooth development and evolution.27, 28 bmp2a was shown to be expressed in the pharyngeal tooth germ in zebrafish.28 In situ hybridization of this transcript revealed a loss of pharyngeal tooth expression of bmp2a in smoc2 morphants (100% for both morpholinos, 20 embryos were analyzed for each) compared to the control embryos (Figures 4E and 4F).
In mice, the pituitary homeobox transcription factor PITX2, a DNA- and RNA-binding protein, is expressed in the stomodeal ectoderm from which teeth are eventually derived.29 Zebrafish pitx2 is strongly expressed in bilateral patches in the pharyngeal epithelium.26 Epithelial pitx2 expression in 56 hpf morphants was not as drastically affected as that of dlx2b (Figures 4G–4I). pitx2, which is normally expressed in bilateral patches, showed a reduced expression domain (90% of the smoc2-2 and smoc2-1 morphants, n = 20). In addition, the bilateral patches were often fused (80% of the smoc2-1 morphants, 20 embryos were analyzed for each). This was never observed in the control embryos (20 embryos were analyzed). Other regions of pitx2 expression were unaffected in the morphants.
Reduction in the expression of genes considered as tooth germ markers is likely to affect the tooth development itself. Dlx2 has been shown to be involved in the patterning of murine dentition, given that the loss of function of Dlx1 and Dlx2 results in early failure of upper-molar development. Mice null for pitx2 have, among other defects, impaired determination and proliferation of tooth organogenesis.30 The effect of smoc2 knockdown on dlx2b, bmp2a, and pitx2 expression suggests that smoc2 plays a crucial role in zebrafish dental development upstream of these factors. The fact that, unlike dlx2 expression, smoc2 expression is not initially restricted to the tooth germ suggests that it defines a broader domain from which the tooth germ can develop. Similarly, it was shown that knockout of prdm1a, which is required for posterior arch development, leads to tooth depletion in zebrafish.31 Overall, the zebrafish smoc2 analysis suggests that smoc2 has an important function in oropharyngeal development. In light of the highly similar human phenotype—characterized by a very rare dental developmental abnormality—we conclude that Smoc2 plays an evolutionarily conserved role in tooth development. However, the exact role of Smoc2 during development warrants further investigation. Smoc1 and Smoc2 have been shown to be widely expressed in both embryonic and adult mice—Smoc1 mainly in basement membranes of organs and Smoc2 mainly in the extracellular matrix.9, 6, 32 Similarly, the phenotype that we observed in the heads of zebrafish morphants was not limited to teeth. Hence, in addition to tooth development, other morphogenetic events appear to require Smoc2 function.
The molecular function of Smoc2 (and Smoc1, which is often studied in conjunction) has been partially uncovered. Smoc2 has been shown to interact with ανβ1 and ανβ6 integrins and contributes to cell-cycle progression by maintaining integrin-linked kinase (ILK) activity during the G1 phase of the cell cycle.33 This suggests a role in linking the extracellular matrix with the intracellular effector ILK.
Another finding is that Smoc2 can regulate the mitogenic and angiogenic effects of vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), and FGF acting in the related pathways.34 Developmental studies in mice have shown that Smoc2 (and Smoc1) might mediate intercellular signaling and cell-type-specific differentiation during gonadal and reproductive duct development.35 The data collected here from mouse in situ hybridization shows that the ectomesenchymal Smoc2 expression is indeed localized within the proliferative compartment facing the epithelial loop at E18.5. The asymmetric mesenchymal labeling observed in the continuously growing incisor on its lingual side might be linked to the short-root anomaly.
Using a knockout mouse to further characterize Smoc2 would improve our knowledge of the exact role of this protein during dental development. Moreover, a possible interaction with other factors such as Pitx2, Dlx2, or other extracellular proteins warrants further investigation. Interestingly, in Axenfeld-Rieger syndrome,36 dental abnormalities due to PITX2 (OMIM 601542) mutations share common features with the phenotype reported herein, suggesting that PITX2 and SMOC2 may have concurrent developmental functions. The dental phenotype disclosed by the patients has been seldom reported in the literature and resembles that of dentin dysplasia type I, yet it has major differences. Teeth affected by dentin dysplasia generally appear clinically unremarkable and have normal shape and consistency. Radiographically, the roots are sharp with conical and apical constrictions. Pre-eruptive pulpal obliteration leads to a crescent-shaped pulpal remnant parallel to the cementoenamel junction in the permanent dentition and to the total pulpal obliteration in the deciduous teeth.3 When combined with certain features of dentin dysplasia type I, the phenotype described in patients III.3 and III.4 very closely matches the root phenotype but is, however, distinct because the patients' teeth were extremely microdont and presented various shape anomalies. This phenotype is very similar to the phenotype described by Ozer and already qualifies as an atypical case.4
Although the reports on the biological activities of SMOC2 suggest a widespread effect, other proteins might compensate for the absence of SMOC2. Indeed, the patients reported herein mainly presented with an orodental phenotype with very minor, if any, developmental traits.
Interestingly, a transcriptome study on human periodontal ligaments has highlighted the expression of 13 extracellular matrix genes, among which is SMOC2.37 In contrast to SMOC1, human SMOC2 appears to be particularly important for dental development but does not play a major role in eye and limb development.
In conclusion, although exome capture is a powerful approach to identifying genes, classical homozygosity mapping followed by candidate-gene selection remains an efficient process, especially for regions of the genome that are poorly covered. This is the first report showing that SMOC2 is an early dental developmental gene in human beings and highlighting this protein as potentially useful in regenerative dentistry.
Acknowledgments
We would like to thank the family and, in particular, the children, who were very participative. Our work was supported by the 2008-2009 Appel à Projet intern program at the Hôpitaux Universitaires de Strasbourg and by the 2008-2011 National Protocole Hospitalier de Recherche Clinique program from the French Ministry of Health. The Equipe d'Accueil 3949 laboratory has been part of the AVENIR INSERM program since 2007. Our work was also supported by the European Integrating Project Zebrafish Regulomics for Human Health (ZF-Health), the European network on Fish Biomedical Models (EuFishBioMed) (supported by the European Cooperation in Science and Technology [COST] Action BM0804), and the Helmholtz Association.
Published online: December 8, 2011
Footnotes
Supplemental Data include three figures and one table and can be found with this article online at http://www.cell.com/AJHG/.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Browser, http://browser.1000genomes.org
BDGP, http://www.fruitfly.org
D[4]/phenodent Diagnosing Dental Defects Database, http://www.phenodent.org
Ensembl Genome Browser, http://www.ensembl.org
Eurexpress, http://www.eurexpress.org/ee/
GenBank, http://www.ncbi.nlm.nih.gov/Genbank
Gene Expression in Tooth, http://bite-it.helsinki.fi
GeneHub-GEPIS, http://www.cgl.ucsf.edu/Research/genentech/genehub-gepis/genehubgepis-search.html
GenePaint, http://www.genepaint.org
HSF2.4.1, http://www.umd.be/HSF
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
UniGene, http://www.ncbi.nlm.nih.gov/unigene
The Zebrafish Model Organism Database (ZFIN), http://zfin.org/cgi-bin/webdriver?MIval=aa-ZDB_home.apg
Accession Numbers
The GenBank accession number for the zebrafish smoc2 sequence reported in this paper is JQ085591.
Supplemental Data
References
- 1.Fleischmannova J., Matalova E., Tucker A.S., Sharpe P.T. Mouse models of tooth abnormalities. Eur. J. Oral Sci. 2008;116:1–10. doi: 10.1111/j.1600-0722.2007.00504.x. [DOI] [PubMed] [Google Scholar]
- 2.Tummers M., Thesleff I. The importance of signal pathway modulation in all aspects of tooth development. J. Exp. Zoolog. B Mol. Dev. Evol. 2009;312B:309–319. doi: 10.1002/jez.b.21280. [DOI] [PubMed] [Google Scholar]
- 3.Barron M.J., McDonnell S.T., Mackie I., Dixon M.J. Hereditary dentine disorders: dentinogenesis imperfecta and dentine dysplasia. Orphanet J. Rare Dis. 2008;3:31. doi: 10.1186/1750-1172-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozer L., Karasu H., Aras K., Tokman B., Ersoy E. Dentin dysplasia type I: report of atypical cases in the permanent and mixed dentitions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2004;98:85–90. doi: 10.1016/j.tripleo.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Kettunen P., Kivimäe S., Keshari P., Klein O.D., Cheyette B.N., Luukko K. Dact1-3 mRNAs exhibit distinct expression domains during tooth development. Gene Expr. Patterns. 2010;10:140–143. doi: 10.1016/j.gep.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vannahme C., Gösling S., Paulsson M., Maurer P., Hartmann U. Characterization of SMOC-2, a modular extracellular calcium-binding protein. Biochem. J. 2003;373:805–814. doi: 10.1042/BJ20030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane T.F., Iruela-Arispe M.L., Johnson R.S., Sage E.H. SPARC is a source of copper-binding peptides that stimulate angiogenesis. J. Cell Biol. 1994;125:929–943. doi: 10.1083/jcb.125.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brekken R.A., Sage E.H. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–827. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 9.Vannahme C., Smyth N., Miosge N., Gösling S., Frie C., Paulsson M., Maurer P., Hartmann U. Characterization of SMOC-1, a novel modular calcium-binding protein in basement membranes. J. Biol. Chem. 2002;277:37977–37986. doi: 10.1074/jbc.M203830200. [DOI] [PubMed] [Google Scholar]
- 10.Abouzeid H., Boisset G., Favez T., Youssef M., Marzouk I., Shakankiry N., Bayoumi N., Descombes P., Agosti C., Munier F.L., Schorderet D.F. Mutations in the SPARC-related modular calcium-binding protein 1 gene, SMOC1, cause waardenburg anophthalmia syndrome. Am. J. Hum. Genet. 2011;88:92–98. doi: 10.1016/j.ajhg.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada I., Hamanoue H., Terada K., Tohma T., Megarbane A., Chouery E., Abou-Ghoch J., Jalkh N., Cogulu O., Ozkinay F., et al. SMOC1 is essential for ocular and limb development in humans and mice. Am. J. Hum. Genet. 2011;88:30–41. doi: 10.1016/j.ajhg.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkhateeb A., Al-Dain Marzouka N., Qarqaz F. SMOC2 gene variant and the risk of vitiligo in Jordanian Arabs. Eur. J. Dermatol. 2010;20:701–704. doi: 10.1684/ejd.2010.1095. [DOI] [PubMed] [Google Scholar]
- 13.Birlea S.A., Gowan K., Fain P.R., Spritz R.A. Genome-wide association study of generalized vitiligo in an isolated European founder population identifies SMOC2, in close proximity to IDDM8. J. Invest. Dermatol. 2010;130:798–803. doi: 10.1038/jid.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasevicius A., Ekker S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 15.Stock D.W., Jackman W.R., Trapani J. Developmental genetic mechanisms of evolutionary tooth loss in cypriniform fishes. Development. 2006;133:3127–3137. doi: 10.1242/dev.02459. [DOI] [PubMed] [Google Scholar]
- 16.Jackman W.R., Stock D.W. Transgenic analysis of Dlx regulation in fish tooth development reveals evolutionary retention of enhancer function despite organ loss. Proc. Natl. Acad. Sci. USA. 2006;103:19390–19395. doi: 10.1073/pnas.0609575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huysseune A., Van der heyden C., Sire J.Y. Early development of the zebrafish (Danio rerio) pharyngeal dentition (Teleostei, Cyprinidae) Anat. Embryol. (Berl.) 1998;198:289–305. doi: 10.1007/s004290050185. [DOI] [PubMed] [Google Scholar]
- 18.Van der Heyden C., Huysseune A. Dynamics of tooth formation and replacement in the zebrafish (Danio rerio) (Teleostei, Cyprinidae) Dev. Dyn. 2000;219:486–496. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1069>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 19.Draper B.W., Morcos P.A., Kimmel C.B. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 20.Debiais-Thibaud M., Borday-Birraux V., Germon I., Bourrat F., Metcalfe C.J., Casane D., Laurenti P. Development of oral and pharyngeal teeth in the medaka (Oryzias latipes): comparison of morphology and expression of eve1 gene. J. Exp. Zoolog. B Mol. Dev. Evol. 2007;308:693–708. doi: 10.1002/jez.b.21183. [DOI] [PubMed] [Google Scholar]
- 21.Thomas B.L., Tucker A.S., Qui M., Ferguson C.A., Hardcastle Z., Rubenstein J.L., Sharpe P.T. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- 22.Sharman A.C., Brand M. Evolution and homology of the nervous system: cross-phylum rescues of otd/Otx genes. Trends Genet. 1998;14:211–214. doi: 10.1016/s0168-9525(98)01488-7. [DOI] [PubMed] [Google Scholar]
- 23.Panganiban G., Rubenstein J.L. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 24.Stock D.W., Ellies D.L., Zhao Z., Ekker M., Ruddle F.H., Weiss K.M. The evolution of the vertebrate Dlx gene family. Proc. Natl. Acad. Sci. USA. 1996;93:10858–10863. doi: 10.1073/pnas.93.20.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas B.L., Liu J.K., Rubenstein J.L., Sharpe P.T. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development. 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- 26.Jackman W.R., Draper B.W., Stock D.W. Fgf signaling is required for zebrafish tooth development. Dev. Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Wise S.B., Stock D.W. Conservation and divergence of Bmp2a, Bmp2b, and Bmp4 expression patterns within and between dentitions of teleost fishes. Evol. Dev. 2006;8:511–523. doi: 10.1111/j.1525-142X.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 28.Wise S.B., Stock D.W. bmp2b and bmp4 are dispensable for zebrafish tooth development. Dev. Dyn. 2010;239:2534–2546. doi: 10.1002/dvdy.22411. [DOI] [PubMed] [Google Scholar]
- 29.Mucchielli M.L., Mitsiadis T.A., Raffo S., Brunet J.F., Proust J.P., Goridis C. Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev. Biol. 1997;189:275–284. doi: 10.1006/dbio.1997.8672. [DOI] [PubMed] [Google Scholar]
- 30.Lin C.R., Kioussi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J.C., Rosenfeld M.G. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 31.Birkholz D.A., Olesnicky Killian E.C., George K.M., Artinger K.B. Prdm1a is necessary for posterior pharyngeal arch development in zebrafish. Dev. Dyn. 2009;238:2575–2587. doi: 10.1002/dvdy.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gersdorff N., Müller M., Schall A., Miosge N. Secreted modular calcium-binding protein-1 localization during mouse embryogenesis. Histochem. Cell Biol. 2006;126:705–712. doi: 10.1007/s00418-006-0200-7. [DOI] [PubMed] [Google Scholar]
- 33.Liu P., Lu J., Cardoso W.V., Vaziri C. The SPARC-related factor SMOC-2 promotes growth factor-induced cyclin D1 expression and DNA synthesis via integrin-linked kinase. Mol. Biol. Cell. 2008;19:248–261. doi: 10.1091/mbc.E07-05-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocnik E.F., Liu P., Sato K., Walsh K., Vaziri C. The novel SPARC family member SMOC-2 potentiates angiogenic growth factor activity. J. Biol. Chem. 2006;281:22855–22864. doi: 10.1074/jbc.M513463200. [DOI] [PubMed] [Google Scholar]
- 35.Pazin D.E., Albrecht K.H. Developmental expression of Smoc1 and Smoc2 suggests potential roles in fetal gonad and reproductive tract differentiation. Dev. Dyn. 2009;238:2877–2890. doi: 10.1002/dvdy.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Idrees F., Bloch-Zupan A., Free S.L., Vaideanu D., Thompson P.J., Ashley P., Brice G., Rutland P., Bitner-Glindzicz M., Khaw P.T., et al. A novel homeobox mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome associated with brain, ocular, and dental phenotypes. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141B:184–191. doi: 10.1002/ajmg.b.30237. [DOI] [PubMed] [Google Scholar]
- 37.Nishida E., Sasaki T., Ishikawa S.K., Kosaka K., Aino M., Noguchi T., Teranaka T., Shimizu N., Saito M. Transcriptome database KK-Periome for periodontal ligament development: expression profiles of the extracellular matrix genes. Gene. 2007;404:70–79. doi: 10.1016/j.gene.2007.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


