Abstract
South Asia harbors one of the highest levels genetic diversity in Eurasia, which could be interpreted as a result of its long-term large effective population size and of admixture during its complex demographic history. In contrast to Pakistani populations, populations of Indian origin have been underrepresented in previous genomic scans of positive selection and population structure. Here we report data for more than 600,000 SNP markers genotyped in 142 samples from 30 ethnic groups in India. Combining our results with other available genome-wide data, we show that Indian populations are characterized by two major ancestry components, one of which is spread at comparable frequency and haplotype diversity in populations of South and West Asia and the Caucasus. The second component is more restricted to South Asia and accounts for more than 50% of the ancestry in Indian populations. Haplotype diversity associated with these South Asian ancestry components is significantly higher than that of the components dominating the West Eurasian ancestry palette. Modeling of the observed haplotype diversities suggests that both Indian ancestry components are older than the purported Indo-Aryan invasion 3,500 YBP. Consistent with the results of pairwise genetic distances among world regions, Indians share more ancestry signals with West than with East Eurasians. However, compared to Pakistani populations, a higher proportion of their genes show regionally specific signals of high haplotype homozygosity. Among such candidates of positive selection in India are MSTN and DOK5, both of which have potential implications in lipid metabolism and the etiology of type 2 diabetes.
Introduction
Understanding the genetic structure of mankind globally and the role of natural selection in shaping it are complex tasks that require data from multiple populations to represent the geographic range and environmental diversity of the inhabited world. Previous studies on South Asia have highlighted this region as having one of the highest levels of genetic diversity, second only to Africa.1, 2 Studies of haploid loci (mtDNA and the nonrecombining region of Y Chromosome [NRY]) have revealed that the South Asian genetic makeup is dominated by largely autochthonous lineages testifying for low levels of admixture with other parts of Eurasia because the peopling of the subcontinent some 50,000 to 70,000 years ago.3, 4, 5, 6 Notably, these genetic dates are earlier than the oldest confirmed human fossil in the subcontinent, found in Sri Lanka and dated to 31,000 years before present (YBP),7 but postdate the archaeological evidence below and above the layers of ash from the Mount Toba volcanic supereruption associated with the Middle Palaeolithic tools that could have been produced by anatomically modern humans.8, 9 Recent archaeological evidence from the Jebel Faya site in the Arabian Peninsula permitted the authors to consider that the manufacturers of these tools could have dispersed into India as early as 125,000 YBP.10, 11 Whether the genes of the crafters of these Middle Palaeolithic tools still persist among modern populations is a lingering question.
Although the HapMap,12 the Human Genome Diversity Project,13 the 1000 Genomes Project14 and the Human Genome Organisation (HUGO) Pan-Asian SNP Consortium15 have all significantly improved our understanding of the global genetic diversity of humans, there are still significant gaps in their coverage. India remains one such region, where large genetic diversity and vast population sizes have so far gone underrepresented in genome-wide studies of human genetic diversity despite some important recent advances.1, 12, 16, 17, 18, 19, 20, 21 Most studies highlight the elevated genetic diversity of the South Asian populations and their general clustering by language group and/or geography. Relying on extensive resequencing rather than on genotyping panel data1 showed that 30% of SNPs found in Indian populations were not seen in HapMap populations and that compared to these populations (including Africans) some Indian populations displayed higher levels of genetic variation, whereas some others showed unexpectedly low diversity. Operating with a thin set of genome-wide polymorphisms,19 identified lower than expected levels of variation across geographically and linguistically distinct populations when sampling Indian immigrants living in the USA. Others have, contrary to this finding, shown high levels of intergroup genetic differentiation of Indian populations sampled in India.17, 18 Furthermore, Reich et al.18 reported higher than expected levels of homozygosity within Indian groups when examining a high density genome-wide SNP data set and attributed this pattern to population stratification born out of the endogamy associated with the caste system. Reich et al.18 have also made an argument for a sizeable contribution from West Eurasia to a putative ancestral north Indian (ANI) gene pool. Through admixture between an ancestral south Indian (ASI) gene pool, this ANI variation was found to have contributed significantly to the extant makeup of not only north (50%–70%) but also south Indian populations (>40%). This is in contrast with the results from mtDNA studies, where the percentage of West Eurasian maternal lineages is substantial (up to 50%) in Indus Valley populations but marginal (<10%) in the south of the subcontinent.5, 22 Because any potential genetic impact into South Asia from the west would involve at least one of the immediately adjacent regions—Central Asia, the Caucasus, or West Asia (including Iran)—assessment of the extent of admixture in South Asia and its sources is crippled without genetic data from those regions. Genome-wide scans on the Human genome diversity panel (HGDP) data involving 51 global populations have revealed that South Asia, represented by Pakistani populations, shares most signals of recent positive selection with populations from Europe, the Near East, and North Africa.23 Given the environmental differences between Europe and Pakistan and the possible depth of human habitation in South Asia, this result is surprising, but considering the lack of Indian data it remains to be determined whether South Asian-specific signals of positive selection do exist.
To shed more light on the nature of genetic continuity and discontinuity between South and West Asia, the Near East, the Caucasus, and Central Asia, we applied FST, principal component analysis (PCA) and model-based structure-like approaches to a genome-wide sample of ca 530,000 SNPs in a sample set combining published data on India,12, 21, 24 relevant global reference populations,24, 25, 26 and 142 newly genotyped Indian individuals of various linguistic, geographic and social affiliations (Table S1).
Material and Methods
Samples, Genotyping, and Quality Control
We introduce here 142 Indian samples from 30 populations that we have genotyped with Illumina 650K SNP array according to manufacturers' specifications. All subjects filled and signed personal informed consents and the study was approved by scientific council of the Estonian Biocentre. The data can be accessed through The National Center for Biotechnology Information -Gene Expression Omnibus (NCBI GEO) (GSE33489) and by request to the authors. We analyzed these data together with published data from12, 21, 24, 25, 26 on Indian and other populations used as background. The overlap of SNPs between the different Illumina (610K, 650K, and 660K) arrays in published and new data was ca. 530,000 SNPs; overlap between this data set and the HapMap 3 was 480,000 SNPs. Depending on the analyses (e.g., computational optimization), we included different number of reference populations from these sources (Table S1). We excluded published data on South Asian populations genotyped on different platforms (Affimetrix)16, 18 from most of the analyses because cross-platform overlap in SNPs is limited (ca. 95,000 SNP) for haplotype based analyses. However, we used data from Reich et al.18 to validate our FST and PCA results. Sampling locations for populations analyzed here are shown on Figure S10 (available online) together with a comparison to sampling from the previous study.18
We filtered the combined data sets by using PLINK software 1.0527 to include only SNPs on the 22 autosomal chromosomes with a minor allele frequency > 1% and genotyping success > 97%. Only individuals with a genotyping success rate > 97% were used. Because background linkage disequilibrium (LD) can affect both principal component and structure-like analysis,28 we thinned the marker set further by excluding SNPs in strong LD (pairwise genotypic correlation r2 > 0.4) in a window of 200 SNPs (sliding the window by 25 SNPs at a time). Depending on the number of reference populations, this yielded data sets of ca. 200,000 SNPs that were used for the respective analyses.
Phylodemographic Analyses
We calculated mean pairwise FST values between populations (and regional population groups) for all autosomal SNPs by using the approach of Weir et al.29 assembled into an in-house R script. For FST calculation, the combined data set was filtered to include only populations with n > 4. In some cases geographically close populations with a smaller sample size were grouped. Given the high levels of population structure within India, resulting from restricted gene flow between populations, genetic drift in small endogamous units, and our small sample sizes, the interpretation of FST distances between pools of samples from different, although genetically closely related, populations might not, necessarily, be straightforward. To validate our results, we recalculated FST values excluding population pools and setting a threshold of minimum of seven samples per population. To increase population coverage, we included here data from Reich et al.18 and the resulting cross-platform SNP panel consisted of 95,001 post quality control SNPs (Table S1).
PCA was carried out in the smartpca program28 on the Eurasian populations. Here too, we repeated the analysis on the data set that included more Indian samples18 but fewer SNPs (Table S1). Geographic spread of principal components ([PCs] averaged to population level) was visualized with kriging procedure in Surfer package of Golden Software. Spatial autocorrelation and modified t test that estimate correlation of spatially located variables and correct for spatial autocorrelation were carried out in Passage 2.30 Geographic distances between populations were calculated as Eucleidian distances between x and y coordinates on a Conformal Conic Asia Lambert projection.
To monitor convergence between individual runs, we ran ADMIXTURE31 100 times at K = 2 to K = 18 (Figure S4A). The lowest cross validation indexes that point to the best K were observed at K = 15, but there was no significant difference above K = 10 (Figure S4C). However, judging by low level of variation in Loglikelihood scores (LLs < 1) within a fraction (10%) of runs with the highest LLs,24, 32 we assume that the global maximum was reached at K = 2 to K = 8, K = 12, and K = 13 (Figure S4B) rendering these practical representations of genetic structure at different levels of resolution. We also verified that all runs within these 10% of runs at these values of K did indeed produce a very similar (indistinguishable) ancestry proportions pattern.
Haplotype Diversity Associated with Ancestry Informative Markers
We used the individual ancestry proportion inferred by using ADMIXTURE as a quantitative trait and tested for association. Allele dosage for an SNP associated with a given ancestry is expected to increase with an increasing proportion of ancestry. Assuming such a relationship between the genotype and trait value, we used regression analysis to estimate how strongly each SNP is associated with a given ancestry. We expected a large number of SNPs to be associated with a given ancestry component, therefore occasional false positive SNPs are negligible, and we chose not to apply any multiple testing correction procedures. Instead, we chose to filter out statistically significant regression coefficients (beta values) by using arbitrarily chosen significance threshold. In order to select only strongly associated SNPs, we further filtered SNPs to retain only those exceeding 90 or 95 percentile points of positive beta-value distribution. The haplotype diversity flanking each associated SNP was then summarized with the number of distinct haplotypes. A summary statistic derived from the number of distinct haplotypes across genomic windows has been shown to be informative about past population demography.33 In this study, Lohmueller et al.33 considered the joint distribution of two haplotype based statistics—the number of distinct haplotypes and the count of the most common haplotype. Here, we use only the number of distinct haplotypes to measure haplotype diversity. Genomic windows of different size—0.45, 0.33, 0.26, 0.1, and 0.05 centiMorgans—were defined around each associated SNP, and the number of distinct haplotypes within each window was counted. We followed33 and randomly selected a subset of nSNP SNPs from each window to ensure that all windows have the same number of SNPs and that the resulting statistics are not affected by the unequal distribution of markers across the genome. Within each window we randomly sampled nSNP SNPs multiple times, counted the number of distinct haplotypes each time, and took the average as a summary. For each population, we randomly chose ten individuals and counted the total number of windows having 0, 1, 2,.., nmax number of haplotypes and plotted this summary statistic by using heatmap.
Nucleotide substitutions arising in one population and then introduced to other populations are expected to show different levels of haplotype diversity in the source and recipient populations. However, this difference gets diluted because hybrid haplotypes arise through recombination in the recipient population. Their number will increase each generation, and it is therefore important to explore how the number of generations since the migration into new population will affect our ability to detect source and recipient populations for a given mutation on the basis of haplotype diversity differences.
We generated population samples by simulating admixture events between European and Asian populations as described in the next section. We explored haplotype diversity flanking SNPs associated with Asian ancestry in these samples from admixed populations. Our simulated data set shows that when European population is the recipient and Asian population the is source, then
-
(1)
Haplotype diversity flanking Asian alleles in admixed recipient populations is lower than in source Asian populations for all the simulated admixture events except for the oldest one that occurred 750 generations ago. The latter case confirms our expectation that immigrant alleles will be flanked with a higher number of hybrid haplotypes (those having both Asian and European ancestry blocks) with an increasing number of generations since admixture.
-
(2)
Haplotype diversity flanking European alleles in admixed populations can be comparable (for those populations having 70% of European ancestry) or even higher (for those having 90% European ancestry) than in the original European population despite the fact that admixed populations always have lower European ancestry (90% or 70%) than the original European population. This might be because of novel hybrid haplotypes produced by the recombination process. Our simulations show that haplotype diversity flanking autosomal SNPs can be used to infer source population even when populations dispersed these alleles 288, 400, or 500 generations ago. Assuming an average human generation interval of 25 years, this is 7,200, 10,000 or 12,500 years, which roughly overlaps with the Neolithic period.
Demographic Model for Simulations
We used MaCS coalescent simulator34 to generate simulated data for three nonadmixed and 18 admixed populations by modifying the demographic model originally published in.35 In this study a series of population genetic statistics were used to fit demographic history of simulated populations to those observed for African, Asian, and European populations. Here, we used these demographic parameters to simulate samples of sequences drawn from African, Asian, and European populations. An additional 18 admixed populations were generated by simulating admixture events between European and Asian populations at different times in the past (measured in generations) and using different proportions: 50/50, 70/30, and 90/10 of sequences from European and Asian populations, respectively:
-
(1)
Admixture 750 generations ago; assuming one generation to be 25 years, this is roughly 18,750 years ago
-
(2)
Admixture 500 generations, ∼12,500 years, ago
-
(3)
Admixture 400 generation,∼10,000 years, ago
-
(4)
Neolithic admixture 288 generations ago; that is 62 generations after Neolithic expansion in a European population as defined in the best fit model of Schaffner et al.35
-
(5)
Late Bronze Age/Iron Age admixture 138 generations, ∼3,450 years, ago
-
(6)
Historical time admixture 70 generations, ∼1,750 years, ago
We used the recombination rate ratio (cM/Mb) mappings for the first chromosome from HapMap project36 to model variation in recombination rate in simulated sequences. The total physical length of simulated sequences was 250 megabases. From each simulated population a sample of 30 sequences were drawn to construct 15 genotypes that were then subjected to quality control and LD pruning steps as for the Illumina genotyped populations analyzed in this study. Admixture proportions for each simulated individual were then inferred with structure-like analysis assuming three populations. SNPs associated with Asian and European ancestry and haplotype diversity flanking them were identified as described above.
Testing for Selection
The combined data set was filtered to include Indian populations and a comprehensive set of reference populations that yielded a data set of 990 individuals and 531,315 autosomal SNPs (Table S1). This data was phased with Beagle 3.1.37 Although integrated haplotype score (iHS) and cross population extended haplotype homozygosity (XP-EHH) have already been calculated for the HGDP-Centre d'Etude du Polymorphisme Humaine (CEPH) panel,23 we recalculated all statistics by using our 531,315 SNPs to allow for unbiased comparisons between India and other geographic regions. XP-EHH and iHS were calculated as previously described with tools provided by J. Pickrell. Genetic distances between markers were calculated with the HapMap genetic map.36 For iHS, ancestral and derived states for each SNP were established by comparison to the UCSC snp128OrthoPanTro2RheMac2 table. Where the chimpanzee allele was known, it was assumed to be the ancestral allele; where the allele was unknown (17,868 SNPs, 3.36% of the data), the SNP was excluded from all subsequent calculations. XP-EHH and FST require two populations. Because the Mandenka, Yoruba, and Bantu farmers have clustered together in previous analyses of population structure,25, 38 they were grouped together in our analyses and were used as the outgroup population for all comparisons; HGDP Europeans were used as the outgroup for analyses where the focal population was African farmers. Both XP-EHH and iHS scores were normalized and windowed as in Pickrell;23 however, we chose not merge any adjacent outlier windows because this procedure can be very conservative and significantly affect the ranking of windows (data not shown).
Enrichment Testing
We retrieved the list of RefSeq genes from the UCSC table browser and mapped the starting and ending coordinates of all genomic transcripts to our windows. The longest transcript length was used for genes with multiple transcripts. On the basis of this list, we performed searches for gene enrichment for all Gene Ontology (GO) terms by using DAVID 6.739, 40 on all genes in the top 1% and 5% windows of the iHS and XP-EHH test statistic distributions.
Results
We have based our analyses of human genetic variation on a sample of 1310 individuals that belong to 112 populations. The sample set includes 142 previously unpublished samples from India and published compatible data from South Asia and beyond (Table S1), chosen to represent the global and regional contexts of human genetic variation. For some analyses we also included published data on Indian populations18 genotyped on a different platform; adding these sources yielded a combined data set of 1,442 individuals but only ca. 95,000 SNPs (Table S1).
Mean pairwise FST values29 within and among continental regions (Figure 1) reveal that the South Asian autosomal gene pool falls into a distinct geographic cluster, characterized internally, like other continental regions, by short interpopulation genetic distances (<0.01). At the interregional scale, the South Asian cluster shows somewhat shorter genetic distances with West Eurasian (average FST = 0.042) than with East Asian (average FST = 0.051) populations. Importantly, the Pakistani (Indus Valley) populations differ substantially from most of the Indian populations and show comparably low genetic differentiation (within the FST range of 0.008–0.020) from European, Near Eastern, Caucasian, and Indian populations (Figure 1 and Figures S1 and S11). In agreement with previous Y-chromosome studies,41, 42 the Brahmin and Kshatriya from Uttar Pradesh stand out by being closer to Pakistani (FST = 0.006 on average) and West Eurasian populations (FST = 0.030) than to other Indian populations (average FSTs 0.017 and 0.046, respectively) from the same geographic area (Figures S1 and S11).
Figure 1.
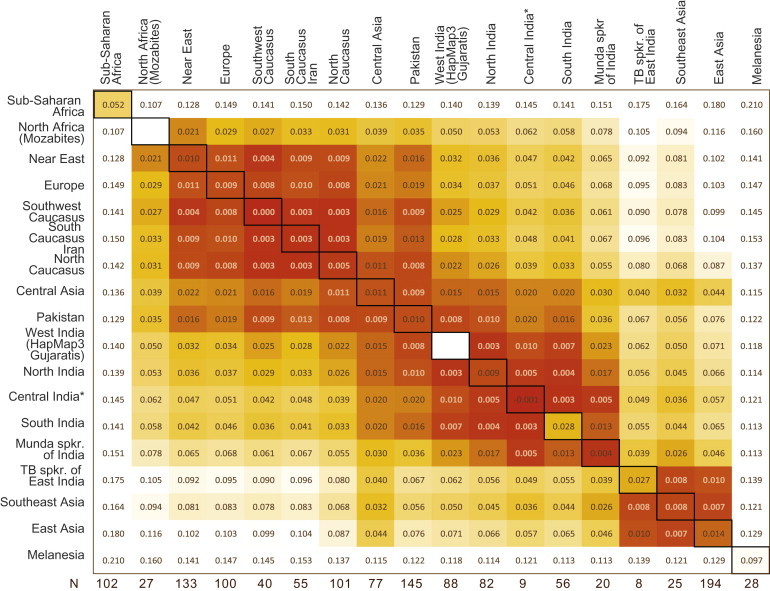
Matrix of Pairwise Mean FST Values of Regional Groupings of the Studied Populations
Average of intergroup FST values (where the regional group is composed of multiple populations) is given in the diagonal. Central India is itself a composite of two regional groupings of samples from different populations that makes the negative intergroup FST uninformative.
Similar to the patterns revealed by the pairwise FST results, PCA of the Eurasian populations clusters them by geographic proximity with the first component separating West from East Eurasia and the second component differentiating South Asian populations from the rest (Figure 2A and Figures S12 and S13). Consistent with their geographic location, Pakistanis are positioned between Indian and West Eurasian populations on this plot. However, whereas Reich et al.18 identified a cline of Indian populations toward Europe with no corresponding cline within the Europeans, we observe a more complex picture. The inclusion of more populations from Europe and the Caucasus24, 26 reveals a cline within the West Eurasian cluster on the PCA (Figure 2A), where both PC1 (r = 0.59) and notably PC2 (r = 0.87) display significant correlation with distance from Spain and Iran, respectively (Figure S2). On this PC1 × PC2 composite cline, most of the Indian populations form a disperse cluster, an edge of which is formed by a subset of the Hapmap Gujaratis and Uttar Pradesh Brahmins and Kshatriyas. Compared to Gujaratis, the Uttar Pradesh samples are more widely dispersed, overlapping substantially with most of the samples from the southern, Dravidic speaking states of Tamil Nadu and Andhra Pradesh. Furthest on the PC2 axis lay samples from the southern Indian states of Karnataka, Kerala, and the Pulliyar population from Tamil Nadu.
Figure 2.
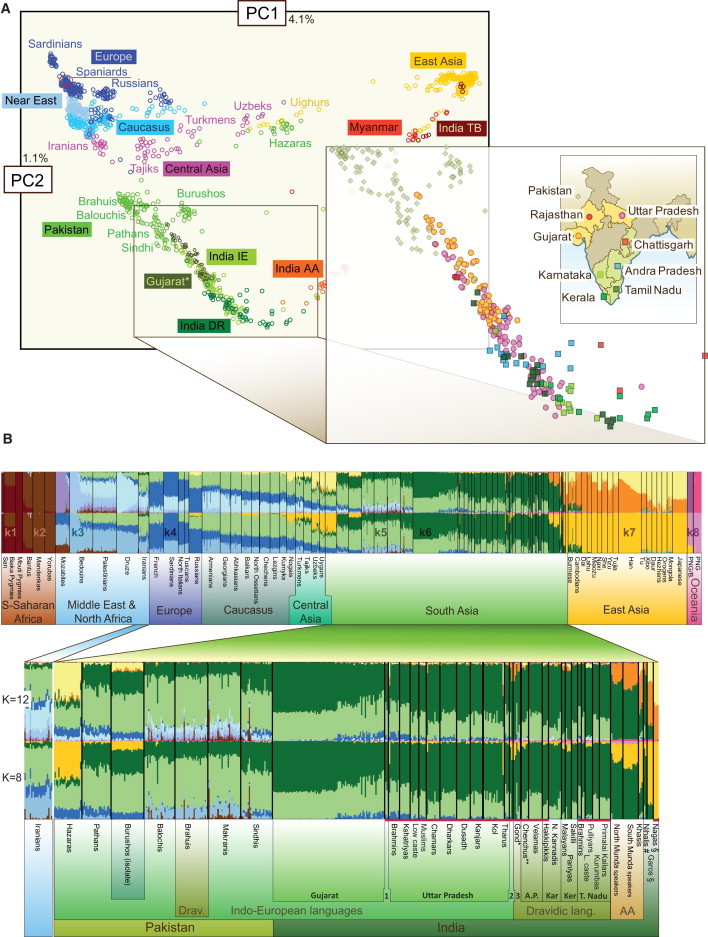
Genome-Wide Structure of the Studied Populations Revealed by 530,000 SNPs
(A) principal component analysis of the Eurasian populations. The following abbreviations are used: IE, Indo European speakers; DR, Dravidic speakers; AA, Austroasiatic speakers; TB, Tibeto Burman speakers; ∗, data from Hapmap.
(B) ADMIXTURE analysis at K = 8 and 12. The following symbols are used: ∗, contains one Dhurwa; ∗∗, contains one Lambadi; 1, Rajasthan; 2, Chattisgarh and Jharkhand; 3, Chattisgarh, Orissa, and Madhya Pradesh. A.P., Andhra Pradesh; Kar, Karnataka; Ker, Kerala; T. Nadu, Tamil Nadu; #, Nihali language isolate speakers from Maharasthra; §, Tibeto Burman speakers from east Indian states Meghalaya and Nagaland; AA, Austroasiatic languages.
Notably, within South Asia (India and Pakistan), PC1 is strongly correlated (r = 0.69) with longitude and PC2 with latitude (r = 0.60). Both remain significant after correcting for spatial autocorrelation. These relations are identifiable also from spatial representations of the principal components (Figure S2). The third PC differentiates West Eurasia by latitude, and we find Bedouins and Lithuanians on either end of the PC3 axis (Figure S3). The fourth PC is of particular interest because it connects Baluchistan, the Caucasus, and Central Asia (Figures S2 and S3). The spread of PC4 in West Eurasia is not concentric and thus difficult to explain by correlation with geographic distance from any one point. The strongest correlation is with distance from Iran (r = 0.69), but this is to a large extent explained by spatial autocorrelation because correcting for that renders a p value slightly over 0.05. Notable, however, is that PC4 has nonmarginal values also in northeast China, which is difficult to absorb into current models of human demographic history. Overall, PCA reveals that the genetic landscape of South Asia is characterized by two principal components of which PC2 is specific to India and PC4 to a wider area encompassing Pakistan, the Caucasus, and Central Asia.
In order to study this duality in more detail, we used the model-based structure-like algorithm ADMIXTURE31 that computes quantitative estimates for individual ancestry in constructed hypothetical ancestral populations. Most South Asians bear membership in only two of the constructed ancestral populations at K = 8. These two main ancestry components—k5 and k6, colored light and dark green in Figure 2B—are observed at all K values between K = 6 and K = 17 (Figure S4). These correlate (r > 0.9; p < 0.00001) perfectly with PC4 and PC2 in West Eurasia, respectively. Looking at the Pakistani populations (0.51) and Baluchistan (Balochi, Brahui, and Makrani) in particular (0.59), the proportion of the light green component (k5) is significantly higher than in the Indian populations, (on average 0.26) (Figure S5). Importantly, the share of this ancestry component in the Caucasus populations (0.50) is comparable to the Pakistani populations. There are a few populations in India who lack this ancestry signal altogether. These are the thus-far sampled Austroasiatic tribes from east India, who originated in Southeast Asia and represent an admixture of Indian and East Asian ancestry components,21 and two small Dravidian-speaking tribes from Tamil Nadu and Kerala. However, considering the geographic spread of this component within India, there is only a very weak correlation (r = 0.4) between probability of membership in this cluster and distance from its closest core area in Baluchistan (Figure S6). Instead, a more steady cline (correlation r = 0.7 with distance from Baluchistan) of decrease of probability for ancestry in the k5 light green ancestral population can be observed as one moves from Baluchistan toward north (north Pakistan and Central Asia) and west (Iran, the Caucasus, and, finally, the Near East and Europe).
If the k5 light green ancestry component (Figure 2B) originated from a recent gene flow event (for example by a demic diffusion model) with a single center of dispersal where the underlying alleles emerged, then one would expect different levels of associated haplotypic diversity to suggest the point of origin of the migration. To assess diversity within the ancestry components revealed by the ADMIXTURE analyses at K = 8, we counted the number of unique haplotypes in genomic windows surrounding SNPs in strong positive association with this ancestry component. Because recombination on autosomal chromosomes will over time erase the signal and thus limit the utility of this approach, we used simulations to explore how deep in time one can go to trace directionality of migration (Figure S7). Our simulations show that differences in haplotype diversity between source and recipient populations can be detected even for migration events that occurred 500 generations ago (∼12,500 years ago assuming one generation to be 25 years). For alleles associated with k5, haplotype diversity is comparable among all studied populations across West Eurasia and the Indus basin (Figure S8). However, we found that haplotypic diversity of this ancestry component is much greater than that of those dominating in Europe (k4, depicted in dark blue) and the Near East (k3, depicted in light blue), thus pointing to an older age of the component and/or long-term higher effective population size (Figure S8). Haplotype diversity flanking Asian alleles (k7) is twice greater than that of European alleles—this is probably because the k7 ancestry component is a composite of two Asian components (see Figure S4, K > 10).
In contrast to widespread light green ancestry, the dark green ancestry component, k6 is primarily restricted to the Indian subcontinent with modest presence in Central Asia and Iran. Haplotype diversity associated with dark green ancestry is greatest in the south of the Indian subcontinent, indicating that the alleles underlying it most likely arose there and spread northwards. It is notable that this ancestry component also exhibits greater haplotype diversity than European or Near Eastern components despite the fact that the Illumina genotyped markers were principally ascertained in a sample of European individuals. This observation shows again that haplotype based measures of diversity can be relatively robust to ascertainment bias.43
Long-standing human habitation of the Indian subcontinent should have provided ample opportunity for the action of positive selection and the emergence of adaptations to the local environment. To examine this possibility in greater detail, we calculated iHS44 and XP-EHH,45 two haplotype-based tests that detect positive natural selection, for all Dravidian and Indo-European speaking Indian individuals in our combined data set (n = 154). After dividing the autosomal genome into 13,274 nonoverlapping 200 kb windows covered by our SNP data set (see Material and Methods), we calculated the fraction of windows in the top 1% of the Indian test statistic distribution shared with the top 5% windows in other populations (Figure 3). Our results largely agree with the recent description of three main patterns underlying selective sweeps in continental Eurasian populations following the out-of-Africa event46 and suggest that Indian sweep signals have more in common with those detected in West rather than East Eurasia. However, when we compare the fraction of outlying Indian signals also found in European or East Asian populations to the fraction of outlying Pakistani signals shared with the same regions, we find Pakistan consistently appearing markedly more similar to West Eurasian than to Indian populations (Figure 3). This result remains when we examine signals of recent positive selection in north and south India separately. Combined with our ADMIXTURE and PCA results, this is powerful evidence that Pakistan is a poor proxy for South Asian genetic diversity, despite having often fulfilled this role in previous publications.
Figure 3.
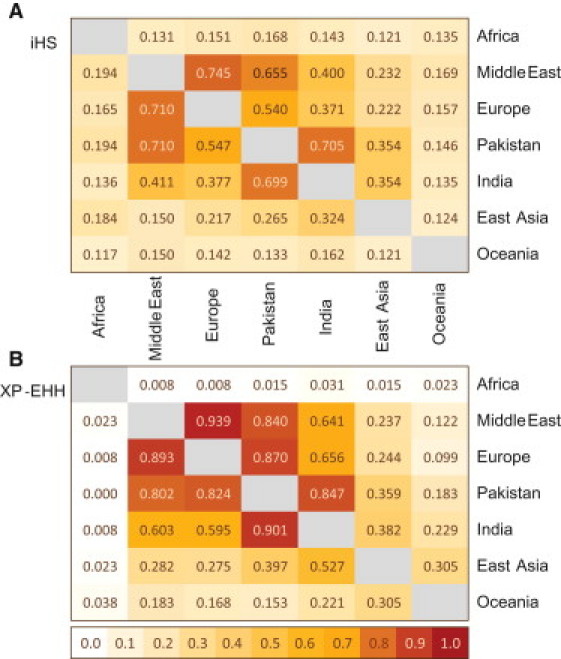
Sharing Signals for Selection between Continental Populations
(A) iHS signal sharing between continental populations. The fraction of signals found in the top 1% of iHS scores in population i and the top 5% of population j is given in cell (i,j). Africa refers to Yoruba, Mandenka, and Bantu individuals from the HGDP-CEPH panel.
(B) XP-EHH signal sharing between continental populations. The fraction of signals found in the top 1% of XP-EHH scores in population i and the top 5% of population j is given in cell (i,j). Africa refers to Yoruba, Mandenka, and Bantu individuals from the HGDP-CEPH panel.
To gain insight into the type of biological processes likely to have come under positive selection in India, we tested for overrepresentation of GO47 terms in the countrywide results. These analyses revealed that 20 GO terms were overrepresented in our windowed top 1% iHS results and 27 were overrepresented in the top 1% XP-EHH results when an individual 0.05 significance level was used (Table S2. Significantly Enriched GO Terms, Before FDR Correction, in the Top 1% iHS All India Results, Reported by DAVID Analysis, Table S3. Significantly Enriched GO Terms, Before FDR Correction, in the Top 1% XP-EHH All India Results, Reported by DAVID Analysis). These results include terms such as lipid metabolism and catabolism, which are associated with genes implicated in the etiology of type 2 diabetes (MIM 125853), the incidence of which is rapidly growing in India and could represent maladaptations to recent changes in the environment, diet, and lifestyle following industrialization.48, 49, 50 However, after false-discovery-rate (FDR) correction for multiple testing, no terms associated with genes found in the top 1% of either test remained significant. Nevertheless, and because positive selection does not necessarily entail pathway enrichment, we note that one of the strongest XP-EHH signals (Table S4. Top 20 Most Significant iHS Windows in the All India Results, Table S5. Top 20 Most Significant iHS Windows in the All India Results) is a region in chromosome 20 containing the DOK5 (MIM 608334), a member of the insulin signaling pathway.51 A three SNP haplotype in this gene has been associated with increased risk of obesity and type 2 diabetes in a large homogeneous north Indian sample,52 although this association has yet to be replicated in another cohort. The gene is the seventh strongest signal in the countrywide results (empirical p = 0.0007), and the seventh and 16th most significant signal in south and north Indian, respectively. Notably, the window is also present in the top 5% results in Europe and East Asia, but nowhere else is evidence for positive selection for this gene nearly as powerful as it is in the Indian subcontinent. Also strongly outlying (XP-EHH empirical p = 0.0015) is CLOCK (MIM 601851), a key regulator of circadian rhythms in humans, which shows strong evidence of selection in all populations, although principally in West Eurasia—it is also within the top 20 European windows but only at the tail end of the top 5% in East Asia. Its disruption has been shown to associate with the development of type 2 diabetes53 and the etiology of metabolic syndrome (MIM 605552)54 as well as with general energy intake in overweight subjects.55, 56 Other genes in the window are TMEM165, a transmembrane protein of no known function and SRD5A3 (MIM 611715), a steroid reductase implicated in androgen signaling in some types of prostate cancer.57 Finally, an interesting candidate for selection according to both XP-EHH and iHS results is MSTN (MIM 601788), a negative regulator of skeletal muscle tissue development expressed in utero and also associated with body fat accumulation and expressed throughout gestation in the human placenta, where it plays a role in glucose uptake.58, 59, 60, 61 The gene shares a window with an uncharacterized reading frame, C2orf88, and HIBCH (MIM 610690), a component in the propionate catabolism pathway;62 the window is associated with extremely significant empirical p values in both iHS and XP-EHH scans (Table S4). MSTN has been identified as a target of strong positive selection twice already on the basis of an excess of derived alleles that indicate the action of positive diversifying selection, especially in African individuals,63, 64 although neither of the implicated SNPs are included in our data, rendering successful reconstruction of the haplotypes presented by Saunders64 in our data impossible without additional genotyping. Nonetheless, FST at the genomic window associated with MSTN is high when compared to genomic averages between Indians and Europeans, and between Indians and African farmers, although low between Indians and East Asians.
Discussion
Relative to East and West Eurasia, the populations of the Indian subcontinent have been underrepresented in genome-wide data sets that have been compiled in attempts to address global patterns of variation at common SNPs. In this study we have asked how representative of South Asian genetic variation are the available and widely used data sets including populations of Pakistan from the HGDP25 and Gujaratis from HapMap Phase 3 data.12 While combining the new data we generated for north and south Indian populations with these public resources, we confirmed the existence of a general principal component cline stretching from Europe to south India.18 Pakistani populations are in the middle of this cline (Figure 1) and show similar FST distances both to populations of Europe and to those of south India, suggesting that they might represent only a fraction of genetic variation in South Asia just as they represent only a fraction of genetic variation in Europe. Additionally, the relatively low genetic diversity among Pakistani populations (average pairwise FST 0.0056, although this measure excludes the Hazara, who show substantial admixture with Central Asian populations; see Figure 2) is less than one third of the diversity observed among all South Asian populations (0.0184), even when excluding the most divergent Austroasiatic and Tibeto-Burman speaking groups of east India. Although the Pakistani and Indian populations have largely nonoverlapping distributions on our PC plot (Figure 2), the HapMap Gujaratis show genetic distances to other global populations, similar to those estimated for other populations of India and appear on the Indian cline between Pakistanis and south Indians, thus being better representatives of the genetic diversity of South Asia than Pakistanis. However, although the geographic representation of Indian populations on our PC plot is neither comprehensive nor balanced, we note that on average the Gujarati samples position 0.78 standard deviations from the location of the Indian mean (excluding the outlying Austroasiatic and Tibeto-Burman speakers). This is about five times more than the mean value from samples from Uttar Pradesh, for example, which appear very close to our all-Indian mean. For comparison, on average the Pakistani and Tamil Nadu samples are located 3.06 and 0.95 standard deviations away from the Indian mean, respectively.
Notably, all South Asian populations, except for Indian Tibeto-Burman speakers, show lower FST distances to Europe than to East Asia (Figure 1). This could be either because of Indian populations sharing a common ancestry with West Eurasian populations because of recent gene flow or because East Asian populations have relatively high pairwise FST with other non-African populations, probably because of their history of genetic bottlenecks.46, 65, 66, 67 Similarly, the clines we detect between India and Europe (e.g., PC1 and PC2 in Figure 2 and Figure S2) might not necessarily reflect one major episode of gene flow but be rather a reflection of complex demographic processes involving drift and isolation by distance. Nevertheless, the correlation of PC1 with longitude within India might be interpreted as a signal of moderate introgression of West Eurasian genes into western India, which is consistent with previous studies on uniparental5, 6 and autosomal markers.18 Overall, the contrasting spread patterns of PC2 and PC4, and of k5 and k6 in the ADMIXTURE analysis (Figure 2 and Figures S2 and S6), could be seen as consistent with the recently advocated model where admixture between two inferred ancestral gene pools (ancestral northern Indians [ANI] and ancestral southern Indians [ASI]) gave rise to the extant South Asian populace.18 The geographic spread of the Indian-specific PC2 (or k6) could at least partly correspond to the genetic signal from the ASI and PC4 (or k5), distributed across the Indus Valley, Central Asia, and the Caucasus, might represent the genetic vestige of the ANI (Figure S2). However, within India the geographic cline (the distance from Baluchistan) of the Indus/Caucasus signal (PC4 or k5) is very weak, which is unexpected under the ASI-ANI model, according to which the ANI contribution should decrease as one moves to the south of the subcontinent. This can be interpreted as prehistorical migratory complexity within India that has perturbed the geographic signal of admixture.
Overall, the locations of the Indian populations on the PC1/PC2 plot (Figure 2A and Figure S12) reflect the correlated interplay of geography and language. In concordance with the geographic spread of the respective language groups, the Indian Indo-European- and Dravidic-speaking populations are placed on a north to south cline. The Indian Austroasiatic-speaking populations are, in turn, in agreement with their suggested origin in Southeast Asia21 drawn away from their Indo-European speaking neighbors toward East Asian populations. In this respect, it is interesting to note that, although represented by only one sample each, the positions of Indo-European-speaking Bhunjia and Dhurwa amidst the Austroasiatic speakers probably corroborates the proposed language change for these populations.68
In structure-like analyses, membership in multiple ancestry components can be interpreted as admixture, shared ancestry, or even unresolved ancestry.25, 69 However, some heuristic interpretations of the ancestry proportions palette in terms of past migrations seem too obvious to be ignored. For example, it was first suggested by the German orientalist Max Müller that ca. 3,500 years ago a dramatic migration of Indo-European speakers from Central Asia (the putative Indo Aryan migration) played a key role in shaping contemporary South Asian populations and was responsible for the introduction of the Indo-European language family and the caste system in India. A few studies on mtDNA and Y-chromosome variation have interpreted their results in favor of the hypothesis,70, 71, 72 whereas others have found no genetic evidence to support it.3, 6, 73, 74 However, any nonmarginal migration from Central Asia to South Asia should have also introduced readily apparent signals of East Asian ancestry into India (see Figure 2B). Because this ancestry component is absent from the region, we have to conclude that if such a dispersal event nevertheless took place, it occurred before the East Asian ancestry component reached Central Asia. The demographic history of Central Asia is, however, complex, and although it has been shown that demic diffusion coupled with influx of Turkic speakers during historical times has shaped the genetic makeup of Uzbeks75 (see also the double share of k7 yellow component in Uzbeks as compared to Turkmens and Tajiks in Figure 2B), it is not clear what was the extent of East Asian ancestry in Central Asian populations prior to these events. Another example of an heuristic interpretation appears when we look at the two blue ancestry components (Figure 2B) that explain most of the genetic diversity observed in West Eurasian populations (at K = 8), we see that only the k4 dark blue component is present in India and northern Pakistani populations, whereas, in contrast, the k3 light blue component dominates in southern Pakistan and Iran. This patterning suggests additional complexity of gene flow between geographically adjacent populations because it would be difficult to explain the western ancestry component in Indian populations by simple and recent admixture from the Middle East.
Several aspects of the nature of continuity and discontinuity of the genetic landscape of South Asia and West Eurasia still elude our understanding. Whereas the maternal gene pool of South Asia is dominated by autochthonous lineages, Y chromosome variants of the R1a clade are spread from India (ca 50%) to eastern Europe and their precise origin in space or time is still not well understood.76 In our analysis we find genetic ancestry signals in the autosomal genes with somewhat similar spread patterns. Both PC2 and k5 light green at K = 8 extend from South Asia to Central Asia and the Caucasus (but not into eastern Europe). In an attempt to explore diversity gradients within this signal, we investigated the haplotypic diversity associated with the ancestry components revealed by ADMIXTURE. Our simulations show that one can detect differences in haplotype diversity for a migration event that occurred 500 generations ago, but chances to distinguish signals for older events will apparently decrease with increasing age because of recombination. In terms of human population history, our oldest simulated migration event occurred roughly 12,500 years ago and predates or coincides with the initial Neolithic expansion in the Near East. Knowing whether signals associated with the initial peopling of Eurasia fall within our detection limits requires additional extensive simulations, but our current results indicate that the often debated episode of South Asian prehistory, the putative Indo-Aryan migration 3,500 years ago (see e.g., Abdulla15) falls well within the limits of our haplotype-based approach. We found no regional diversity differences associated with k5 at K = 8. Thus, regardless of where this component was from (the Caucasus, Near East, Indus Valley, or Central Asia), its spread to other regions must have occurred well before our detection limits at 12,500 years. Accordingly, the introduction of k5 to South Asia cannot be explained by recent gene flow, such as the hypothetical Indo-Aryan migration. The admixture of the k5 and k6 components within India, however, could have happened more recently—our haplotype diversity estimates are not informative about the timing of local admixture.
Both k5 and k6 ancestry components that dominate genetic variation in South Asia at K = 8 demonstrate much greater haplotype diversity than those that predominate in West Eurasia. This pattern is indicative of a more ancient demographic history and/or a higher long-term effective population size underlying South Asian genome variation compared to that of West Eurasia. Given the close genetic relationships between South Asian and West Eurasian populations, as evidenced by both shared ancestry and shared selection signals, this raises the question of whether such a relationship can be explained by a deep common evolutionary history or secondary contacts between two distinct populations. Namely, did genetic variation in West Eurasia and South Asia accumulate separately after the out-of-Africa migration; do the observed instances of shared ancestry component and selection signals reflect secondary gene flow between two regions, or do the populations living in these two regions have a common population history, in which case it is likely that West Eurasian diversity is derived from the more diverse South Asian gene pool.
Similar to observed patterns of neutral genetic diversity, one could ask whether Indian populations contain a reservoir of selective signals hitherto unidentified in other Old World groups, akin to what has been found in uniparentally inherited markers, or whether the region fits into the Eurasian landscape of positive selection signals.23 At the global level, our haplotype-based scans of positive selection showed similar patterns of signal sharing to those revealed by FST comparisons, and Indian as well as Pakistani populations share more signals with West Eurasia than with the rest of the world. In fact, barring the actual numbers on them, Figure 1, Figure 3 bear a striking similarity to each other. Despite this, the results leave ample room for the existence of local adaptation to the Indian environment, both recent and old. XP-EHH, by its nature, detects older or stronger sweeps acting on alleles that have reached high frequency in a given population. Previous studies23, 77 have shown that the vast majority of XP-EHH signals are shared across extended geographic distances. Compared to Pakistani populations (87%), both north (66%) and south Indian (52%) populations share substantially less signals of complete selective sweep with European populations (Figure S9). Sharing of the complete sweep signals between India and East Asia is even lower (53%). In the case of iHS, Indian signals sharing with Europe and East Asia was less pronounced (37% and 32%, respectively), probably stemming from the nature of iHS, as it detects younger, on-going sweeps and is therefore more likely to highlight recent, private signals of local adaptation that have not yet become widespread by gene flow.
Our analysis of the genes contained within the top 1% of selective signals in the countrywide data suggested that 25 GO terms were overrepresented among our strongest selection candidates, although none were significant after Benjamini correction. We also tested the top 5% of results in the Indian data and found that five GO terms related to cell-cell binding and metal ion binding remained highly significant after multiple testing corrections (data not shown). However, examination of the genes associated with these terms revealed that all significant results could be ascribed to positional gene clustering, whereby multiple genes associated with the same GO term, generally members of a single gene family, fell within the same 200 kb window but were treated as independent findings by the gene set enrichment analysis tool we used. It is worth recalling that gene-enrichment tools were originally devised for the assessment of gene expression changes in microarray RNA work, where individual genes could be unequivocally identified. Given the degree of resolution provided by the data sets that we have used here, any attempts to use automated annotation tools to understand signals of positive selection extending over multiple genes is fraught with interpretative perils. Alternatives include the precise CMS test that often is applicable on dense HapMap2 data78 or a windowing approach, whereby ontological associations are mapped not to individual genes, but rather to the windows they occupy. The latter approach could successfully correct for the clustering effect we identify and more generally for the effect of gene size on enrichment results, whereby long genes are more likely to be statistical outliers simply because they contain more SNPs than short genes, and GO categories associated with long genes are therefore more likely to appear enriched. We believe that collapsing annotations to the window level could reduce the false-positive rate in enrichment scans, although at the same time it would be far more conservative and risk obscuring genuine signals. In our data, for example, none of the five significant GO terms at the genic level are significant when examined at the windowed level (data not shown).
In the wake of these results, we chose to examine the contents of the 20 strongest iHS and XP-EHH signals, which can be expected to contain candidates for adaptation via classical sweeps. Within these regions we find four genes—DOK5, MSTN, CLOCK, and PPARA—implicated in lipid metabolism and etiology of type 2 diabetes, although one of them, PPARA, is in a window that contains seven other genes. Variation in DOK5 and CLOCK has been previously associated with type 2 diabetes and metabolic disorders, whereas MSTN is not an obvious candidate for involvement in disease etiology because its main function is negative regulation of muscle development in utero; it also plays a significant role in glucose uptake. Interestingly, Indian newborns weigh on average 700 g less than their European counterparts yet have a similar absolute fat mass.79 At birth, these children are already adipose and exhibit some degree of insulin resistance when compared to European babies;80 this difference persists into adulthood, such that the average age of diagnosis of diabetes in India is 10 years lower than in Europe.81
It bears recalling that India has one of the world's fastest growing, and soon greatest in absolute terms, incidence of type 2 diabetes,82, 83 as well as a sizeable number of cases of the metabolic syndrome,84 both of which have been linked to recent rapid urbanization.81, 85, 86 Phenotypically, even nonobese Asian Indians have been shown to exhibit increased levels of insulin resistance compared to European controls.87 They also have increased levels of both subcutaneous and visceral adipose tissue at the expense of lean tissue when compared to matched-age and -weight European controls88, 89 and show differences in adipocyte morphology.90 In this context, it is tempting to hypothesize that past natural selection might have influenced genetic variation at these loci to increase infant survival, a change that became disadvantageous after changes in diet and lifestyle. Therefore, the loci we identify could be theoretically considered responsible for some of the present type 2 diabetes epidemic in India, making them worthy candidates for further functional examination. However, because relevant life-history traits, lipid metabolism and type 2 diabetes are all complex traits and the effect of natural selection would be expected to be fragmented across multiple genes,23, 45, 77 it would be naive to expect that a relationship between past selective processes and present-day disease would be mechanistically simple and explainable by variation at a handful of genetic loci.
Summing up, our results confirm both ancestry and temporal complexity shaping the still on-going process of genetic structuring of South Asian populations. This intricacy cannot be readily explained by the putative recent influx of Indo-Aryans alone but suggests multiple gene flows to the South Asian gene pool, both from the west and east, over a much longer time span. We highlight a few genes as candidates of positive selection in South Asia that could have implications in lipid metabolism and etiology of type 2 diabetes. Further studies on data sets without ascertainment and allele frequency biases such as sequence data will be needed to validate the signals for selection.
Acknowledgments
We thank A. Migliano, S. Raj, and P. Underhill for discussion; J. Pickrell and J. Barna for help calculating iHS and XP-EHH scores; A. Aasa, I. Hilpus, T. Reisberg, V. Soo, and L. Anton for technical assistance. R.V., M.M., G.C. and C.B.M. thank the European Union European Regional Development Fund through the Centre of Excellence in Genomics to Estonian Biocentre, and University of Tartu. This research was supported by Estonian Basic Research grant SF0270177As08 to R.V. and SF0180026s09 to M.R. and R.M.; Tartu University grant (PBGMR06901) to T.K.; Estonian Science Foundation grants (7858) to E.M. and (8973) to M.M.; Estonian Ministry of Education and Research (0180142s08) and European Commission grant 245536 (OPENGENE) to M.N.; European Commission grant (ECOGENE 205419) to M.M., I.G.R., B.Y., G.H, R.M., and R.V.; and Council of Scientific and Industrial Research, Government of India to L.S. and K.T. Calculations were carried out in the High Performance Computing Center, University of Tartu and with University of Cambridge Bioinformatics and Computational Biology services.
Published online: December 8, 2011
Footnotes
Supplemental Data include 13 figures and six tables and can be found with this article online at http://www.cell.com/AJHG/.
Web Resources
The URLs for data presented herein are as follows:
The 1000 Genomes Project, http://www.1000genomes.org/home
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Supplemental Data
Includes information on both, new data and used public data used in the study.
References
- 1.Xing J., Watkins W.S., Hu Y., Huff C.D., Sabo A., Muzny D.M., Bamshad M.J., Gibbs R.A., Jorde L.B., Yu F. Genetic diversity in India and the inference of Eurasian population expansion. Genome Biol. 2010;11:R113. doi: 10.1186/gb-2010-11-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson Q.D., Gray R.D., Drummond A.J. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol. Biol. Evol. 2008;25:468–474. doi: 10.1093/molbev/msm277. [DOI] [PubMed] [Google Scholar]
- 3.Kivisild T., Bamshad M.J., Kaldma K., Metspalu M., Metspalu E., Reidla M., Laos S., Parik J., Watkins W.S., Dixon M.E., et al. Deep common ancestry of indian and western-Eurasian mitochondrial DNA lineages. Curr. Biol. 1999;9:1331–1334. doi: 10.1016/s0960-9822(00)80057-3. [DOI] [PubMed] [Google Scholar]
- 4.Kivisild T., Kaldma K., Metspalu M., Parik J., Papiha S.S., Villems R. In: Genomic diversity. Papiha S.S., Deka R., Chakraborty R., editors. Kluwer Academic/Plenum Publishers; 1999. The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the Old World; pp. 135–152. [Google Scholar]
- 5.Metspalu M., Kivisild T., Metspalu E., Parik J., Hudjashov G., Kaldma K., Serk P., Karmin M., Behar D.M., Gilbert M.T., et al. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 2004;5:26. doi: 10.1186/1471-2156-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahoo S., Singh A., Himabindu G., Banerjee J., Sitalaximi T., Gaikwad S., Trivedi R., Endicott P., Kivisild T., Metspalu M., et al. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc. Natl. Acad. Sci. USA. 2006;103:843–848. doi: 10.1073/pnas.0507714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deraniyagala S.U. Department of Archaeological Survey; Sri Lanka: 1992. The prehistory of Sri Lanka; an ecological perspective. [Google Scholar]
- 8.Petraglia M., Korisettar R., Boivin N., Clarkson C., Ditchfield P., Jones S., Koshy J., Lahr M.M., Oppenheimer C., Pyle D., et al. Middle Paleolithic assemblages from the Indian subcontinent before and after the Toba super-eruption. Science. 2007;317:114–116. doi: 10.1126/science.1141564. [DOI] [PubMed] [Google Scholar]
- 9.Petraglia M.D., Haslam M., Fuller D.Q., Boivin N., Clarkson C. Out of Africa: new hypotheses and evidence for the dispersal of Homo sapiens along the Indian Ocean rim. Ann. Hum. Biol. 2010;37:288–311. doi: 10.3109/03014461003639249. [DOI] [PubMed] [Google Scholar]
- 10.Armitage S.J., Jasim S.A., Marks A.E., Parker A.G., Usik V.I., Uerpmann H.P. The southern route “out of Africa”: evidence for an early expansion of modern humans into Arabia. Science. 2011;331:453–456. doi: 10.1126/science.1199113. [DOI] [PubMed] [Google Scholar]
- 11.Petraglia M.D. Archaeology: Trailblazers across Arabia. Nature. 2011;470:50–51. doi: 10.1038/470050a. [DOI] [PubMed] [Google Scholar]
- 12.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 14.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdulla M.A., Ahmed I., Assawamakin A., Bhak J., Brahmachari S.K., Calacal G.C., Chaurasia A., Chen C.H., Chen J.M., Chen Y.T., et al. HUGO Pan-Asian SNP Consortium. Indian Genome Variation Consortium Mapping human genetic diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 16.Auton A., Bryc K., Boyko A.R., Lohmueller K.E., Novembre J., Reynolds A., Indap A., Wright M.H., Degenhardt J.D., Gutenkunst R.N., et al. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19:795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Indian Genome Variation Consortium Genetic landscape of the people of India: a canvas for disease gene exploration. J. Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- 18.Reich D., Thangaraj K., Patterson N., Price A.L., Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg N.A., Mahajan S., Gonzalez-Quevedo C., Blum M.G., Nino-Rosales L., Ninis V., Das P., Hegde M., Molinari L., Zapata G., et al. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2006;2:e215. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watkins W.S., Rogers A.R., Ostler C.T., Wooding S., Bamshad M.J., Brassington A.-M.E., Carroll M.L., Nguyen S.V., Walker J.A., Prasad B.V.R., et al. Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–1618. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaubey G., Metspalu M., Choi Y., Mägi R., Romero I.G., Soares P., van Oven M., Behar D.M., Rootsi S., Hudjashov G., et al. Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol. Biol. Evol. 2011;28:1013–1024. doi: 10.1093/molbev/msq288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quintana-Murci L., Chaix R., Wells R.S., Behar D.M., Sayar H., Scozzari R., Rengo C., Al-Zahery N., Semino O., Santachiara-Benerecetti A.S., et al. Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am. J. Hum. Genet. 2004;74:827–845. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickrell J.K., Coop G., Novembre J., Kudaravalli S., Li J.Z., Absher D.M., Srinivasan B.S., Barsh G.S., Myers R.M., Feldman M.W., Pritchard J.K. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G., et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 25.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 26.Yunusbayev B., Metspalu M., Järve M., Kutuev I., Rootsi S., Metspalu E., Behar D.M., Varendi K., Sahakyan H., Khusainova R., et al. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol. Biol. Evol. 2011 doi: 10.1093/molbev/msr221. Published online September 13, 2011. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weir B.S., Cockerham C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg M.S., Andersen C.D. PASSaGE: Pattern Analysis, Spatial Statistics and Geographic Exegesis. Version 2. Methods in Ecology and Evolution. 2011;2:229–232. [Google Scholar]
- 31.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen M., Li Y.R., Lindgreen S., Pedersen J.S., Albrechtsen A., Moltke I., Metspalu M., Metspalu E., Kivisild T., Gupta R., et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohmueller K.E., Bustamante C.D., Clark A.G. Methods for human demographic inference using haplotype patterns from genomewide single-nucleotide polymorphism data. Genetics. 2009;182:217–231. doi: 10.1534/genetics.108.099275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G.K., Marjoram P., Wall J.D. Fast and flexible simulation of DNA sequence data. Genome Res. 2009;19:136–142. doi: 10.1101/gr.083634.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaffner S.F., Foo C., Gabriel S., Reich D., Daly M.J., Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 39.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Huang W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bamshad M., Kivisild T., Watkins W.S., Dixon M.E., Ricker C.E., Rao B.B., Naidu J.M., Prasad B.V., Reddy P.G., Rasanayagam A., et al. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma S., Rai E., Sharma P., Jena M., Singh S., Darvishi K., Bhat A.K., Bhanwer A.J.S., Tiwari P.K., Bamezai R.N.K. The Indian origin of paternal haplogroup R1a1∗ substantiates the autochthonous origin of Brahmins and the caste system. J. Hum. Genet. 2009;54:47–55. doi: 10.1038/jhg.2008.2. [DOI] [PubMed] [Google Scholar]
- 43.Conrad D.F., Jakobsson M., Coop G., Wen X.Q., Wall J.D., Rosenberg N.A., Pritchard J.K. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 44.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., et al. International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coop G., Pickrell J.K., Novembre J., Kudaravalli S., Li J., Absher D., Myers R.M., Cavalli-Sforza L.L., Feldman M.W., Pritchard J.K. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diamond J. The double puzzle of diabetes. Nature. 2003;423:599–602. doi: 10.1038/423599a. [DOI] [PubMed] [Google Scholar]
- 49.Prentice A.M., Rayco-Solon P., Moore S.E. Insights from the developing world: thrifty genotypes and thrifty phenotypes. Proc. Nutr. Soc. 2005;64:153–161. doi: 10.1079/pns2005421. [DOI] [PubMed] [Google Scholar]
- 50.Ramachandran A., Ma R.C., Snehalatha C. Diabetes in Asia. Lancet. 2010;375:408–418. doi: 10.1016/S0140-6736(09)60937-5. [DOI] [PubMed] [Google Scholar]
- 51.Cai D., Dhe-Paganon S., Melendez P.A., Lee J., Shoelson S.E. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J. Biol. Chem. 2003;278:25323–25330. doi: 10.1074/jbc.M212430200. [DOI] [PubMed] [Google Scholar]
- 52.Tabassum R., Mahajan A., Chauhan G., Dwivedi O.P., Ghosh S., Tandon N., Bharadwaj D. Evaluation of DOK5 as a susceptibility gene for type 2 diabetes and obesity in North Indian population. BMC Med. Genet. 2010;11:35. doi: 10.1186/1471-2350-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcheva B., Ramsey K.M., Buhr E.D., Kobayashi Y., Su H., Ko C.H., Ivanova G., Omura C., Mo S., Vitaterna M.H., et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garaulet M., Lee Y.C., Shen J., Parnell L.D., Arnett D.K., Tsai M.Y., Lai C.Q., Ordovas J.M. CLOCK genetic variation and metabolic syndrome risk: modulation by monounsaturated fatty acids. Am. J. Clin. Nutr. 2009;90:1466–1475. doi: 10.3945/ajcn.2009.27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garaulet M., Corbalán M.D., Madrid J.A., Morales E., Baraza J.C., Lee Y.C., Ordovas J.M. CLOCK gene is implicated in weight reduction in obese patients participating in a dietary programme based on the Mediterranean diet. Int J Obes (Lond) 2010;34:516–523. doi: 10.1038/ijo.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garaulet M., Lee Y.C., Shen J., Parnell L.D., Arnett D.K., Tsai M.Y., Lai C.Q., Ordovas J.M. Genetic variants in human CLOCK associate with total energy intake and cytokine sleep factors in overweight subjects (GOLDN population) Eur. J. Hum. Genet. 2010;18:364–369. doi: 10.1038/ejhg.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uemura M., Tamura K., Chung S., Honma S., Okuyama A., Nakamura Y., Nakagawa H. Novel 5 alpha-steroid reductase (SRD5A3, type-3) is overexpressed in hormone-refractory prostate cancer. Cancer Sci. 2008;99:81–86. doi: 10.1111/j.1349-7006.2007.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McPherron A.C., Lawler A.M., Lee S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 59.McPherron A.C., Lee S.-J. Suppression of body fat accumulation in myostatin-deficient mice. J. Clin. Invest. 2002;109:595–601. doi: 10.1172/JCI13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bass J., Oldham J., Sharma M., Kambadur R. Growth factors controlling muscle development. Domest. Anim. Endocrinol. 1999;17:191–197. doi: 10.1016/s0739-7240(99)00036-3. [DOI] [PubMed] [Google Scholar]
- 61.Mitchell M.D., Osepchook C.C., Leung K.-C., McMahon C.D., Bass J.J. Myostatin is a human placental product that regulates glucose uptake. J. Clin. Endocrinol. Metab. 2006;91:1434–1437. doi: 10.1210/jc.2005-2361. [DOI] [PubMed] [Google Scholar]
- 62.Hawes J.W., Jaskiewicz J., Shimomura Y., Huang B., Bunting J., Harper E.T., Harris R.A. Primary structure and tissue-specific expression of human beta-hydroxyisobutyryl-coenzyme A hydrolase. J. Biol. Chem. 1996;271:26430–26434. doi: 10.1074/jbc.271.42.26430. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen R., Hubisz M.J., Hellmann I., Torgerson D., Andrés A.M., Albrechtsen A., Gutenkunst R., Adams M.D., Cargill M., Boyko A., et al. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 2009;19:838–849. doi: 10.1101/gr.088336.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saunders M.A., Good J.M., Lawrence E.C., Ferrell R.E., Li W.-H., Nachman M.W. Human adaptive evolution at Myostatin (GDF8), a regulator of muscle growth. Am. J. Hum. Genet. 2006;79:1089–1097. doi: 10.1086/509707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramachandran S., Deshpande O., Roseman C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keinan A., Mullikin J.C., Patterson N., Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manica A., Prugnolle F., Balloux F. Geography is a better determinant of human genetic differentiation than ethnicity. Hum. Genet. 2005;118:366–371. doi: 10.1007/s00439-005-0039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mukherjee N., Mitra M., Chakraborty M., Majumder P.P. Congruence of genomic and ethnolinguistic affinities among five tribal populations of Madhya Pradesh (India) J. Genet. 2000;79:41–46. [Google Scholar]
- 69.Weiss K.M., Long J.C. Non-Darwinian estimation: my ancestors, my genes' ancestors. Genome Res. 2009;19:703–710. doi: 10.1101/gr.076539.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wells R.S., Yuldasheva N., Ruzibakiev R., Underhill P.A., Evseeva I., Blue-Smith J., Jin L., Su B., Pitchappan R., Shanmugalakshmi S., et al. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc. Natl. Acad. Sci. USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Basu A., Mukherjee N., Roy S., Sengupta S., Banerjee S., Chakraborty M., Dey B., Roy M., Roy B., Bhattacharyya N.P., et al. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cordaux R., Aunger R., Bentley G., Nasidze I., Sirajuddin S.M., Stoneking M. Independent origins of Indian caste and tribal paternal lineages. Curr. Biol. 2004;14:231–235. doi: 10.1016/j.cub.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 73.Chaubey G., Metspalu M., Kivisild T., Villems R. Peopling of South Asia: investigating the caste-tribe continuum in India. Bioessays. 2007;29:91–100. doi: 10.1002/bies.20525. [DOI] [PubMed] [Google Scholar]
- 74.Kivisild T., Rootsi S., Metspalu M., Mastana S., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V., et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am. J. Hum. Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martínez-Cruz B., Vitalis R., Ségurel L., Austerlitz F., Georges M., Théry S., Quintana-Murci L., Hegay T., Aldashev A., Nasyrova F., Heyer E. In the heartland of Eurasia: the multilocus genetic landscape of Central Asian populations. Eur. J. Hum. Genet. 2011;19:216–223. doi: 10.1038/ejhg.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Underhill P.A., Myres N.M., Rootsi S., Metspalu M., Zhivotovsky L.A., King R.J., Lin A.A., Chow C.E., Semino O., Battaglia V., et al. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur. J. Hum. Genet. 2010;18:479–484. doi: 10.1038/ejhg.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pritchard J.K., Pickrell J.K., Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grossman S.R., Shylakhter I., Karlsson E.K., Byrne E.H., Morales S., Frieden G., Hostetter E., Angelino E., Garber M., Zuk O., et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 79.Yajnik C.S., Fall C.H.D., Coyaji K.J., Hirve S.S., Rao S., Barker D.J.P., Joglekar C., Kellingray S. Neonatal anthropometry: the thin-fat Indian baby. The Pune Maternal Nutrition Study. Int. J. Obes. Relat. Metab. Disord. 2003;27:173–180. doi: 10.1038/sj.ijo.802219. [DOI] [PubMed] [Google Scholar]
- 80.Yajnik C.S., Lubree H.G., Rege S.S., Naik S.S., Deshpande J.A., Deshpande S.S., Joglekar C.V., Yudkin J.S. Adiposity and hyperinsulinemia in Indians are present at birth. J. Clin. Endocrinol. Metab. 2002;87:5575–5580. doi: 10.1210/jc.2002-020434. [DOI] [PubMed] [Google Scholar]
- 81.Ramachandran A., Mary S., Yamuna A., Murugesan N., Snehalatha C. High prevalence of diabetes and cardiovascular risk factors associated with urbanization in India. Diabetes Care. 2008;31:893–898. doi: 10.2337/dc07-1207. [DOI] [PubMed] [Google Scholar]
- 82.King H., Aubert R.E., Herman W.H. Global burden of diabetes, 1995-2025: prevalence, numerical estimates, and projections. Diabetes Care. 1998;21:1414–1431. doi: 10.2337/diacare.21.9.1414. [DOI] [PubMed] [Google Scholar]
- 83.Wild S., Roglic G., Green A., Sicree R., King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 84.Misra A., Khurana L. The metabolic syndrome in South Asians: epidemiology, determinants, and prevention. Metab. Syndr. Relat. Disord. 2009;7:497–514. doi: 10.1089/met.2009.0024. [DOI] [PubMed] [Google Scholar]
- 85.Fall C.H.D. Non-industrialised countries and affluence. Br. Med. Bull. 2001;60:33–50. doi: 10.1093/bmb/60.1.33. [DOI] [PubMed] [Google Scholar]
- 86.Snehalatha C., Ramachandran A. Cardiovascular risk factors in the normoglycaemic Asian-Indian population—influence of urbanisation. Diabetologia. 2009;52:596–599. doi: 10.1007/s00125-009-1279-x. [DOI] [PubMed] [Google Scholar]
- 87.Banerji M.A., Faridi N., Atluri R., Chaiken R.L., Lebovitz H.E. Body composition, visceral fat, leptin, and insulin resistance in Asian Indian men. J. Clin. Endocrinol. Metab. 1999;84:137–144. doi: 10.1210/jcem.84.1.5371. [DOI] [PubMed] [Google Scholar]
- 88.Lear S.A., Humphries K.H., Kohli S., Chockalingam A., Frohlich J.J., Birmingham C.L. Visceral adipose tissue accumulation differs according to ethnic background: results of the Multicultural Community Health Assessment Trial (M-CHAT) Am. J. Clin. Nutr. 2007;86:353–359. doi: 10.1093/ajcn/86.2.353. [DOI] [PubMed] [Google Scholar]
- 89.Lear S.A., Kohli S., Bondy G.P., Tchernof A., Sniderman A.D. Ethnic variation in fat and lean body mass and the association with insulin resistance. J. Clin. Endocrinol. Metab. 2009;94:4696–4702. doi: 10.1210/jc.2009-1030. [DOI] [PubMed] [Google Scholar]
- 90.Chandalia M., Lin P., Seenivasan T., Livingston E.H., Snell P.G., Grundy S.M., Abate N. Insulin resistance and body fat distribution in South Asian men compared to Caucasian men. PLoS ONE. 2007;2:e812. doi: 10.1371/journal.pone.0000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Includes information on both, new data and used public data used in the study.


