Abstract
HIV-1 can establish a state of latent infection at the level of individual T cells. Latently infected cells are rare in vivo and appear to arise when activated CD4+ T cells, the major targets cells for HIV-1, become infected and survive long enough to revert back to a resting memory state, which is nonpermissive for viral gene expression. Because latent virus resides in memory T cells, it persists indefinitely even in patients on potent antiretroviral therapy. This latent reservoir is recognized as a major barrier to curing HIV-1 infection. The molecular mechanisms of latency are complex and include the absence in resting CD4+ T cells of nuclear forms of key host transcription factors (e.g., NFκB and NFAT), the absence of Tat and associated host factors that promote efficient transcriptional elongation, epigenetic changes inhibiting HIV-1 gene expression, and transcriptional interference. The presence of a latent reservoir for HIV-1 helps explain the presence of very low levels of viremia in patients on antiretroviral therapy. These viruses are released from latently infected cells that have become activated and perhaps from other stable reservoirs but are blocked from additional rounds of replication by the drugs. Several approaches are under exploration for reactivating latent virus with the hope that this will allow elimination of the latent reservoir.
A subset of resting memory CD4+ T cells harboring integrated but transcriptionally silent HIV-1 proviruses currently poses a barrier to viral eradication in infected subjects.
HISTORY AND DEFINITIONS
Viral latency is a state of reversibly nonproductive infection of individual cells. For some viruses, notably select members of the Herpes virus family, latency provides an important mechanism for viral persistence and escape from immune recognition (Perng and Jones 2010). Several Herpes viruses have elaborate genetic programs that allow persistence of viral genomes with minimal viral gene expression. For retroviruses, stable integration of reverse transcribed viral cDNA into host cell chromosomes is an essential step in the life cycle that allows persistence of viral genomes for the lifespan of infected cells. Some retroviruses also establish a state of latent infection. In early studies of sheep infected with the visna lentivirus, a restricted pattern of gene expression from proviral DNA was observed that suggested latency (Brahic et al. 1981). For HIV-1, the term latency was initially used in the clinical sense to describe the long asymptomatic period between initial infection and the development of AIDS. However, with the advent of sensitive RT-PCR assays for viremia (Piatak et al. 1993), it became clear that HIV-1 replicates actively throughout the course of the infection, even during the asymptomatic period. The major mechanism by which HIV-1 evades immune responses is not latency but rather through rapid evolution of escape mutations that abrogate recognition by neutralizing antibodies and cytolytic T lymphocytes (Bailey et al. 2004). Nevertheless, it has become clear that HIV-1 can establish a state of latent infection at the level of individual T cells.
Initial evidence suggesting HIV-1 latency came from in vitro infections of transformed cells lines. Surviving infected cells showed low or absent HIV-1 gene expression that could be up-regulated by various stimuli, including those causing T-cell activation (Folks et al. 1986). Molecular studies had established that transcription from the HIV-1 LTR was stimulated by inducible host transcription factors, such as NFκB. This factor rapidly translocates into the nucleus in response to T-cell-activating stimuli because of stimulus-coupled degradation of its cytoplasmic inhibitor, IκBα (Nabel and Baltimore 1987; Siekevitz et al. 1987; Bohnlein et al. 1988; Duh et al. 1989). Thus, a connection between latent infection and resting but not activated CD4+ T cells seemed quite likely.
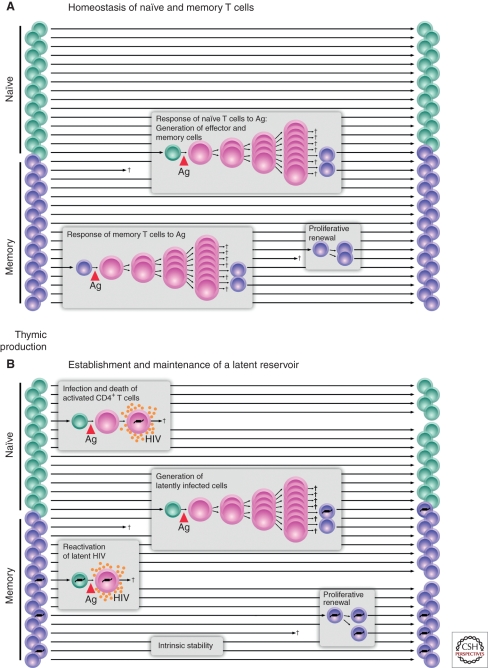
Interestingly, HIV-1 does not efficiently establish productive infection in resting CD4+ T cells (Zack et al. 1990; Bukrinsky et al. 1991; Zhou et al. 2005). How then can a stable state of latent infection develop in these resting cells? One plausible hypothesis is based on the normal physiology of CD4+ T cells (Fig. 1A). In response to antigen, resting CD4+ T cells undergo a burst of cellular proliferation and differentiation, giving rise to effector cells. Most effector cells die quickly, but a subset survives and reverts to a resting G0 state. They persist as memory cells, with an altered pattern of gene expression enabling long-term survival and rapid responses after reexposure to antigen. Activated CD4+ T cells are highly susceptible to HIV-1 infection and typically die quickly as a result of the cytopathic effects of the virus or host immune responses (Ho et al. 1995; Wei et al. 1995). However, some activated CD4+ T cells may become infected and then survive long enough to revert back to a resting state (Fig. 1B). Because HIV-1 gene expression is dependent on inducible host transcription factors that are only transiently activated after exposure to antigen, HIV-1 gene expression may be extinguished as the cells revert to a resting memory state. The result is a stably integrated but transcriptionally silent form of the virus in a cell whose function it is to survive for long periods of time. With time, additional epigenetic mechanisms discussed below may enforce latency. In the latent state, the virus persists simply as information (in the form of 10 kb of integrated HIV-1 DNA), and it is thus unaffected by antiretroviral drugs or immune responses. However, if the host cell becomes activated by an encounter with antigen or other activating stimuli, latency may be reversed, and the cell may begin to produce a virus. Therefore, by this hypothesis, HIV-1 latency inadvertently exploits the most fundamental characteristic of the immune system, the immunological memory that resides in long-lived resting lymphocytes.
Figure 1.
Establishment of a latent reservoir for HIV-1. HIV-1 latency may be viewed as a consequence of the tropism of the virus for activated CD4+ T cells. (A) Generation of memory CD4+ T cells. A fraction of the CD4+ T cells that respond to a given antigen survive and revert back to a resting state as long-lived memory T cells. (B) Generation of latently infected cells. Latency is established when activated CD4+ T cells become infected and survive long enough to revert back to a resting memory state that is nonpermissive for viral gene expression. The resulting latent reservoir is intrinsically stable because memory cells have a long lifespan and can undergo a process of proliferative renewal through homeostatic proliferation.
A prediction of this model is that a stable latent form of HIV-1 should reside in the memory subset of resting CD4+ T cells (Fig. 1). Proving that latently infected cells were present in vivo required the development of methods for isolating extremely pure populations of resting CD4+ T cells and demonstrating that within these populations were cells carrying the HIV-1 genome stably integrated into host chromosomes. In addition, it was necessary to show that replication-competent virus could be rescued from resting CD4+ T cells by cellular activation. This was particularly important because the mere presence of viral nucleic acids in resting CD4+ T cells could also be compatible with the presence of defective viruses. In 1995, the presence of integrated HIV-1 DNA in highly purified populations of resting CD4+ T cells was definitively shown by inverse PCR (Chun et al. 1995). Replication-competent virus could be rescued by cellular activation (Chun et al. 1995). This study established that latently infected cells were present in infected individuals. Interestingly, the frequency of latently infected cells was low. A critical further advance was the development of a culture assay that allowed quantitation of cells harboring replication-competent virus (Chun et al. 1997a). In this assay, serial dilutions of purified resting CD4+ T cells are stimulated with a mitogen that induces 100% of the cells to undergo blast transformation. This cellular activation reverses latency, allowing virus production. Released viruses are expanded in CD4+ T lymphoblasts from normal donors that are added to the culture. Virus growth is detected by ELISA of HIV-1 p24 antigen in the supernatant, and frequencies are calculated using limiting dilution statistics. With this approach, resting CD4+ T cells harboring replication-competent virus were detected at low frequency (1/106) in the blood and lymph nodes of all infected individuals studied. Although several mechanisms have been proposed for the origin of latently infected cells, the finding that latent HIV-1 resides predominantly in the memory subset of resting CD4+ T cells (Chun et al. 1997a; Chomont et al. 2009) is consistent with the hypothesis presented above. This model is further supported by recent studies describing the in vitro production of latently infected resting CD4+ T cells from activated primary CD4+ T cells that are infected and then allowed to return to a quiescent state (reviewed in Yang 2011).
Shortly after the in vivo detection of latently infected cells was first reported, combinations of antiretroviral drugs known as highly active antiretroviral therapy, or HAART, were shown to reduce plasma virus levels to below the limit of detection, raising hopes for viral eradication (Gulick et al. 1997; Hammer et al. 1997; Perelson et al. 1997). The question immediately became whether the pool of latently infected cells would persist in patients who had suppression of viremia to undetectable levels on HAART. Using the culture assay approach described above, three groups simultaneously showed in 1997 that latently infected cells persisted in patients who were responding well to HAART (Chun et al. 1997b; Finzi et al. 1997; Wong et al. 1997). Subsequent longitudinal studies showed that the decay rate of the pool of latently infected cells was extremely slow, with a half-life of 44 months (Finzi et al. 1999; Siliciano et al. 2003). At this rate, over 70 years of treatment would be required to eradicate the latent reservoir. These studies led to the proposal that the latent reservoir in resting memory CD4+ T cells guarantees lifetime persistence of the virus, even in patients on suppressive HAART regimens (Finzi et al. 1999; Siliciano et al. 2003; Strain et al. 2003). Because of the existence of stable viral reservoirs, a rebound in viremia inevitably occurs within weeks after interruption of therapy (Davey et al. 1999). Although multiple reservoirs may exist, replication-competent HIV-1 can be isolated from resting CD4+ T cells of all patients on HAART, regardless of the duration of treatment, making this reservoir the best-established barrier to HIV-1 eradication.
MOLECULAR MECHANISMS OF HIV-1 LATENCY
As discussed, a leading theory for how HIV-1 latency is initially established involves infection of activated CD4+ T cells as they are returning to a resting state to form long-lived memory T cells (Fig. 1B). Although this transition may give rise to proviral latency, it remains unclear how the latent state is maintained within these cells. Many studies have been performed exploring the mechanistic basis for latency. All of these studies share the ultimate aim of improving our understanding of this process sufficiently to allow a successful rational attack on the latent reservoir.
Chromatin and HIV-1 Latency
One possible mechanism for maintaining the latent state involves epigenetic changes in chromatin that suppress HIV-1 gene expression. Expression of host genes is dynamically regulated through changes in chromatin structure, chiefly reflecting altered states of chromosomal DNA compaction (Felsenfeld and Groudine 2003). DNA compaction can be more than four orders of magnitude greater in a condensed metaphase chromosome than in an actively transcribed gene. Transcribed genes are usually found within “relaxed” chromosomal DNA, termed euchromatin; nonexpressed genes commonly reside within more condensed chromosomal DNA, termed heterochromatin (Tamaru 2010). Heterochromatin formation impairs gene expression by impeding transcription factor access to the underlying DNA (Fig. 2). In an informative cell line model of HIV-1 latency called J-Lat (Jordan et al. 2001), integrated latent HIV-1 proviruses were identified within heterochromatic regions, including centromeric alphoid repeats (Jordan et al. 2003). Nevertheless, these latent viruses could be successfully activated when cells were exposed to various stimuli, including phorbol esters or tumor necrosis factor α. Further studies with this system revealed that inducible HIV-1 gene expression with integration into gene deserts, centromeric heterochromatin or highly expressed cellular genes (Lewinski et al. 2005). Although the notion of integration within heterochromatic sites was interesting from a mechanistic perspective, extensive studies of HIV-1 integration sites in cell lines infected in vitro with HIV-1 had established that HIV-1 generally integrates within cellular genes (Schroder et al. 2002). In vivo studies of HIV-1 integrations sites in resting CD4+ T cells from patients on HAART showed that most of the integrated HIV-1 DNA was within cellular genes that are actively transcribed in resting CD4+ T cells (Han et al. 2004). These findings prompted a general reconsideration of the underlying mechanisms of HIV-1 latency and raised the notion that the mechanisms might in fact be multifactoral (Lassen et al. 2004).
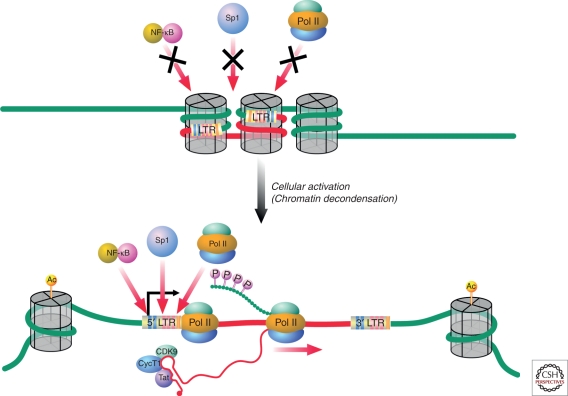
Figure 2.
Compaction of chromatin around integrated HIV-1 proviruses may restrict access of key transcription factors to the 5′ long terminal repeat. However, this condensed chromatin state is reversed when cells are activated. Activation leads to transcription factor access and effective RNA Pol II elongation, giving rise to high-level virus production when HIV-1 Tat is produced.
Although integration within heterochromatin does not appear to be a major mechanism of HIV-1 latency, other aspects of chromatin biology are clearly involved. Nucleosomes are the fundamental structural unit of chromatin (Bai and Morozov 2010). A single nucleosome comprises 147 base pairs of DNA wrapped in 1.67 left-handed superhelical turns around an octet of histone proteins that includes H2A, H2B, H3, and H4. Nucleosomes are separated by ∼80 nucleotides of linker DNA, where histone H1 binds. Nucleosomes are regulated by both SWI/SNF remodeling complexes (Liu et al. 2011) and posttranslational modifications of the histone tails, including acetylation, methylation, ubiquitylation, phosphorylation, sumoylation, and poly ADP-ribosylation (Goldberg et al. 2007). Unique combinations of these modifications have been proposed to generate a “histone code” that may provide a more universal set of operating instructions governing chromatin assembly and gene expression (Fischle et al. 2003).
Strikingly, two nucleosomes, nuc-0 and nuc-1, consistently form within the 5′ long terminal repeat (LTR) of HIV even when the provirus is integrated within euchromatin (Verdin et al. 1993). These nucleosomes regulate the basal transcriptional activity of the 5′ LTR because they overlap with the binding sites for many key transcription factors that drive HIV-1 gene expression. Reactivation of latent HIV-1 proviruses by external stimuli is consistently associated with remodeling of nuc-1. Maintenance of nuc-1 is highly dependent on the state of histone acetylation based on the finding that histone deacetylase (HDAC) inhibitors promote effective remodeling of nuc-1 and transcriptional activation of HIV-1 (Laughlin et al. 1993; Van Lint et al. 1996). By analogy, it seems likely that one or more HDACs are active on the LTR of HIV-1 proviruses where nuc-1 is maintained. HDACs do not directly bind to DNA; rather these enzymes assemble with other proteins that bind DNA. The resulting protein complexes exert repressive effects on transcription. Subsequent studies revealed that HDAC-1 is effectively recruited to the HIV-1 LTR through its interactions with multiple host factors, including p50 homodimers of NF-κB, LSF1, YY1, thyroid hormone receptor, and other DNA binding partners (Coull et al. 2000; Hsia and Shi 2002; Williams et al. 2006). Importantly, addition of HDAC inhibitors not only promotes effective remodeling of nuc-1 but also increases the effective recruitment of RNA Pol II, leading to the commencement of RNA synthesis (Williams et al. 2006). Administration of valproic acid, a weakly active HDAC inhibitor, to HIV-1-infected patients receiving antiretroviral therapy was initially reported to result in modest decreases in the latent reservoir (Lehrman et al. 2005). Disappointingly, in subsequent studies, this agent did not produce durable effects on the latent reservoir (Siliciano et al. 2007; Archin et al. 2010).
DNA Methylation and HIV Latency
DNA methylation is another mechanism that reinforces HIV-1 latency. The HIV-1 transcription start site is flanked by two CpG islands that are methylated in J-Lat T cells and a primary CD4+ T cells model of HIV-1 latency (Blazkova et al. 2009; Kauder et al. 2009). Methyl CpG binding domain protein-2 (MBD2) and HDAC2 are detectable at one of these CpG islands during latency, and inhibition of methylation on deoxycytosines by addition of 5-aza-2′- deoxycytidine (aza-CdR) prevents recruitment of MBD2 and HDAC2 (Kauder et al. 2009). Of note, synergistic reactivation of latent HIV-1 gene expression occurs when cells are cocultured with aza-CdR and inducers of NFκB (e.g., prostratin or tumor necrosis factor α). CpG methylation may, in fact, correspond to a relatively late silencing event in a sequential program of modifications that serve to reinforce latency. Consistent with this model, proviruses in memory CD4+ T cells of some patients on HAART contain a high proportion of nonmethylated cytosines within the 5′ LTR (Blazkova et al. 2009). The renewed appreciation of the potential regulatory role of DNA methylation will likely prompt new studies of DNA methylation inhibitors in combination with other latency antagonists.
Transcriptional Interference as a Potential Driver of HIV Latency
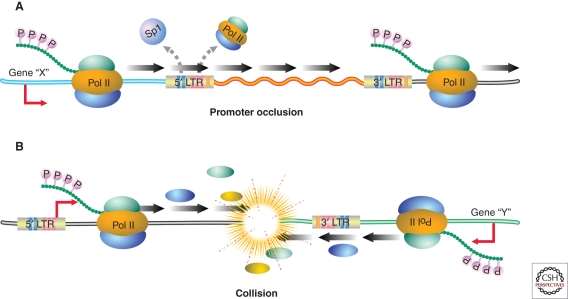
In general, HIV-1 favors integration into actively transcribed genes (Schroder et al. 2002), likely because the host factor LEDGF/p75 (Meehan et al. 2009) binds to integrase and directs integration to intronic regions within these highly expressed genes. Nevertheless, the finding that latent proviruses are frequently integrated within highly expressed genes in both J-Lat cells (Lewinski et al 2005) and primary CD4+ T cells (Han et al. 2004) was quite surprising. These findings suggest that some forms of latency mechanistically involve transcriptional interference (Fig. 3). Different forms of transcriptional interference occur, depending on the orientation of the HIV-1 provirus relative to the host gene. If both share the same polarity, promoter occlusion can occur in which read-through by an upstream elongating RNA Pol II displaces key transcription factors on HIV-1 LTR, thereby reinforcing viral latency (Greger et al. 1998; Lenasi et al. 2008). In a primary cell model of HIV-1 latency, a modest preference for integration with the same polarity was observed (Shan et al. 2011). Conversely, if the cellular gene and provirus are arranged in opposite polarity, RNA Pol II collisions may occur, leading to premature termination of transcription at one or both promoters (Han et al. 2008). This form of transcriptional interference could also generate double-stranded viral RNAs that might participate in RNA interference, lead to RNA-mediated methylation, or result in biologically active antisense RNAs. Despite the mechanism of transcriptional interference, activation of the HIV-1 LTR converts this promoter into a very strong transcription unit possibly because of the tight binding affinity of NF-κB that overcomes interference and results in effective proviral reactivation (De Marco et al. 2008).
Figure 3.
Two scenarios in which transcriptional interference may promote HIV-1 latency. In the first case, the HIV-1 provirus is integrated in the same polarity as an upstream gene within an intron of this gene. Read through by RNA Pol II initiating at the upstream promoter occludes the 5′LTR and displaces key transcription factors thereby promoting viral latency. Alternatively, the HIV-1 provirus and the cellular gene may be arranged in opposite polarity. In this situation, initiating polymerases collide, leading to decreased expression of one or both transcription units. The fact that latent HIV-1 proviruses are commonly found in actively transcribed genes suggests that transcriptional interference may be important in the maintenance of proviral latency.
RNA Pol II Initiation
Initiation of HIV-1 gene expression is critically dependent on a range of host transcription factors whose activities are induced by extracellular stimuli, including T-cell receptor ligation or the action of various cytokines (Colin and Van Lint 2009). Key inducible transcription factors include NF-κB, NFAT, and AP-1, which interact with their cognate enhancers within the HIV-1 LTR. In resting cells, both NF-κB and NFAT are sequestered in the cytoplasm and thus are not available in the nucleus to promote the activation of latent HIV-1 proviruses. Sp1 also plays a key role by binding at three sites in the core promoter. Elimination of one or more of these sites greatly impairs both basal and Tat-dependent activation of the HIV-1 LTR (Ross et al. 1991; Sune and Garcia-Blanco 1995). Sp1 may act, at least in part, through its associations with TATA-box-binding protein (TBP), TBP-associated factor (TAF) 250, and TAF55 (Emili et al. 1994; Chiang and Roeder 1995; Shao et al. 1995). The juxtaposition of the Sp1 and κB sites also appears to be critically important for full LTR activation (Perkins et al. 1993). Of note, both NF-κB and NFAT bind to the κB sites (Kinoshita et al. 1998). Which of these factors is more important for activation of latent HIV-1 proviruses continues to be hotly debated in the field, but it seems likely that cellular context is critical. In contrast to the key roles of the κB enhancers and Sp1 sites in the reactivation of latent provirus, binding sites for other described transcription factors (e.g., LEF-1, COUP-TF, YY1, Ets-1, and USF) likely play more supporting roles (Rohr et al. 2003).
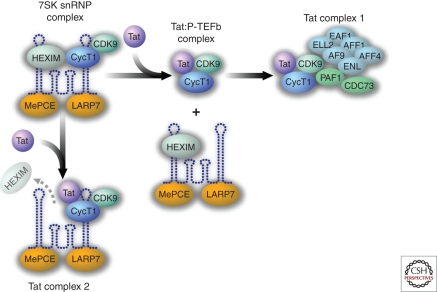
Members of the NF-κB/Rel family of transcription factors exert both activating and inhibitory effects on the HIV-1 LTR. For example, the binding of homodimers of p50 recruits HDAC1 and leads to transcriptional inhibition because of histone deacetylation and chromatin condensation (Williams et al. 2006). Conversely, p50/RelA heterodimers (the prototypical NF-κB complex) readily displace p50 homodimers and promote strong transcriptional activation. These effects of NF-κB, in part, involve the recruitment of p300/CBP (Gerritsen et al. 1997), which mediates acetylation of histone tails and leads to increased RNA Pol II and TFIIH/CDK7 binding. The latter complex helps mediate promoter clearance and phosphorylates the carboxy-terminal domain (CTD) of RNA Pol II on serine-5 residues. Finally, RelA has also been implicated in the recruitment of P-TEFb to the HIV LTR (Barboric et al. 2001), although BRD4 also may play a role (Yang et al. 2005). The PTEFb complex, comprising cyclin T1 and CDK9, phosphorylates serine 2 in the CTD of RNA Pol II, thereby promoting effective Pol II elongation. This P-TEFb complex is hijacked by HIV Tat to drive high-level Pol II elongation (Fig. 4) (Wei et al. 1998). Like the negative regulation of NF-κB and NFAT, P-TEFb is also partially sequestered in an inactive ribonucleoprotein complex composed of 7SK RNA and HEXIM-1 (7SK snRNP complex) (Yang et al. 2001).
Figure 4.
HIV-1 Tat effectively antagonizes HIV-1 latency by liberating P-TEFb from an inactive 7SK snRNP complex. In addition to the Tat-Cyclin T1-CDK9 complex that binds to TAR and promotes serine-2 phosphorylation of the carboxy-terminal domain of RNA Pol II, thereby eliminating promoter proximal pausing, two additional Tat complexes have recently been discovered. Tat complex 1 corresponds to the assembly of Tat-Cyclin T1-CDK9 with AFF1, AFF4, AFR, ENL, EAF1, and ELL2. This complex has been termed a “super-elongation complex.” ELL2 blocks backtracking by RNA Pol II. AFR, ENL, and AF9 correspond to transcription factors/coactivators; AF9 potentiates CDK9 kinase activity. Tat complex 2 corresponds to the 7SK snRNP complex lacking HEXIM. This RNP complex may correspond to an additional reservoir of Tat-CyclinT1-CDk9 within cells. However, its precise function remains to be delineated.
Just as posttranslational modification of histone tails plays a key role in regulating HIV-1 gene expression, reversible acetylation of nonhistone substrates, notably the NF-κB RelA subunit, also regulates HIV-1 LTR activation (Chen and Greene 2004). The RelA subunit is targeted by p300/CBP for acetylation at multiple sites, including lysines 218, 221, and 310. These modifications regulate distinct NF-κB functions (Chen et al. 2001, 2002), including DNA binding, assembly with IκBα, and the overall transcriptional activity of NF-κB. Acetylation of RelA is reversed by the actions of two deacetylases, HDAC3 (Chen et al. 2001), and SIRT1 (Yeung et al. 2004). Of note, SIRT1-mediated deacetylation of RelA at lysine 310 is blocked by HIV Tat. Because acetylation of lysine 310 in RelA greatly enhances transcriptional activity, this inhibitory effect of Tat could markedly boost NF-κB action and increase viral gene expression as well as expression of genes involved in inflammation and immune activation (Kwon et al. 2008). Importantly, HDAC3-mediated deacetylation of lysine 221 restores the ability of newly synthesized IκBα to bind to RelA (Sun et al. 1993), leading in turn to nuclear export of NF-κB, thereby terminating its transcriptional activity.
These studies highlight how dynamic changes in the acetylation state of RelA serve as an intranuclear molecular switch regulating both the strength and duration of the NF-κB transcriptional response and consequently the activation of its target genes, including integrated latent HIV-1 proviruses. Many agents being considered for purging of the latent reservoir correspond to inducers of the NF-κB pathway. Unfortunately, because NF-κB is a “master regulator” of so many key responses in mammals, side effects will almost certainly occur.
RNA Pol II Elongation
HIV-1 transcription can be viewed as occurring in two phases: an early Tat-independent phase that promotes at best low-level expression of viral genes but importantly allows production of Tat, and a later Tat-dependent phase that mediates high-level viral transcription (Barboric and Peterlin 2005). The relative absence of Tat may serve as a central driver of HIV-1 latency (Karn 2011). Tat acts by binding to the transactivation-responsive element (TAR), a rather simple RNA stem-loop structure located at the 5′ end of all viral transcripts (Fig. 2). The Tat-TAR complex in turn binds P-TEFb, which mediates serine-2 phosphorylation of the CTD of Pol II (Parada and Roeder 1996; Kim et al. 2002) and phosphorylation of Spt5, a factor that prevents premature Pol II release at terminator sequences (Bourgeois et al. 2002). These events result in highly effective RNA Pol II elongation. In the absence of Tat, the elongating Pol II complex transcripts are naturally obstructed because of binding of two factors, termed negative elongation factor (NELF) and DRB sensitivity-inducing factor (DSIF). This obstruction results in the production of short viral transcripts of ∼60 nucleotides in length. Phosphorylation of NELF by P-TEFb (Fujinaga et al. 2004) and the promoter-clearing action of TFIIH/CDK7 remove these inhibitors and, when coupled with the positive effects of Pol II and Spt5 phosphorylation, set the stage for unimpeded high-level Pol II elongation. In resting cells, cyclin T1 levels are often low, resulting in decreased P-TEFb activity (Liou et al. 2002). This could also contribute to latency in resting cells.
Recently, the first crystal structure for Tat bound to P-TEFb was reported (Tahirov et al. 2010). This structure, resolved at 2.1 angstroms, reveals intimate contacts between Tat and the T loop of CDK9 and marked interactions of Tat with cyclin T1. Tat-induced changes in the conformation of P-TEFb likely explain how this viral protein liberates this complex from HEXIM-1 and 7SK RNA (Fig. 4). This structure could lead to the rational design of a new class of HIV-1 inhibitors targeting Tat. Because Tat plays such a pivotal role in controlling viral gene expression, it is understandable how small stochastic changes in its expression can determine whether viral latency or productive infection ensues (Weinberger et al. 2005).
As noted, Tat cannot produce its effects in the absence of P-TEFb, a kinase complex that is normally bound in a ribonucleoprotein complex of 7SK RNA and HEXIM-1 or HEXIM-2 (Fig. 4). Tat effectively liberates P-TEFb from this complex. However, the Tat-P-TEFb complex might not be as simple as originally thought. Rather than just three proteins, a much larger complex of the coactivators AFF4, ENL, and AF9 and the elongation factor ELL2 has recently been detected by two groups (He et al. 2010; Sobhian et al. 2010) (Fig. 4, Tat complex 1). ELL2 promotes elongation by curbing RNA Pol II “backtracking,” and as such, its presence in the Tat complex is quite intriguing. A second complex of Tat, P-TEFb, and 7SK RNA but lacking Hexim-1 has also been isolated (Sobhian et al. 2010) (Fig. 4, Tat complex 2). The function of this complex is not yet clear, but it could correspond to a partially active pool of P-TEFb (Karn 2011).
RNA Splicing and Export
Considerable attention has focused on the role of chromatin and transcription in the control of HIV-1 latency. However, another rather unexpected mechanism has emerged. Specifically, resting CD4+ T cells from HIV-1-infected patients on antiretroviral therapy retain both Tat and Rev transcripts in their nuclei (Lassen et al. 2006). This finding stands in sharp contrast to the effective export of these multiply spliced viral transcripts in activated CD4+ T cells. This defect in viral RNA export can be rescued by ectopic expression of polypyrimidine tract binding protein (PTB), an RNA export protein. Of note, a short form of PTB is present in resting CD4+ T cells, whereas full-length PTB is virtually undetectable. Whether this export difference is rooted in changes in the splicing of the export protein or a selective degradation of PTB in resting CD4+ T cells remains to be determined.
CLINICAL SIGNIFICANCE OF HIV-1 LATENCY
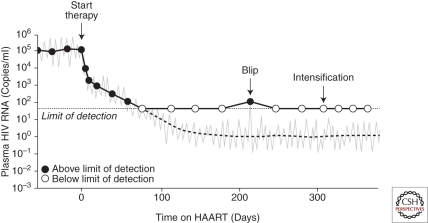
The existence of a latent reservoir for HIV-1 has implications for eradication efforts and also for understanding the clinical response to antiretroviral therapy, in particular the decay in viremia after initiation of treatment. In 1995, George Shaw and David Ho showed that treatment with a potent antiretroviral drug caused plasma viral levels to fall exponentially (Fig. 5) (Ho et al. 1995; Wei et al. 1995). They reasoned that the newer antiretroviral drugs were powerful enough to stop all new infection of susceptible cells. Importantly, all antiretroviral drugs in clinical use block new infection of susceptible cells but not the release of virions by cells that contain integrated proviruses. If the drugs completely block de novo infection, virus appearing in the plasma must come from cells infected before therapy. Because the decay rate of free virus is very fast, the decrease in plasma virus levels after initiation of effective therapy is determined mainly by the decay rate of previously infected cells. The rapid initial drop in plasma virus level directly reflects the short half-life (∼1 day) of the CD4+ T lymphoblasts that produce most of the plasma virus (Fig. 5). If the initial therapy is monotherapy, viremia rebounds because of the outgrowth of preexistent resistant variants (Wei et al. 1995). Conversely, when appropriate combinations of three antiretroviral drugs are started, plasma virus levels almost always fall to below the limit of detection (Gulick et al. 1997; Hammer et al. 1997; Perelson et al. 1997). After the rapid initial decay during the first 1–2 weeks of treatment, plasma virus levels decline at a slower rate, reflecting the release of virus from another population of infected cells with a slower decay rate (Fig. 5). Interestingly, the nature of the cells responsible for this second-phase decay is still unclear (Spivak et al. 2011). Initially, it was hoped that the second-phase decay would continue, eventually leading to eradication (Perelson et al. 1997). However, studies with especially sensitive assays for plasma HIV-1 revealed at least one more phase in the decay curve (Fig. 5). In many patients, viremia appears to level off at around 1 copy/ml and remain at that level indefinitely (Dornadula et al. 1999; Maldarelli et al. 2007; Palmer et al. 2008). This persistent viremia at levels below the limit of detection of the standard clinical assay (50 copies/ml) is termed residual viremia.
Figure 5.
Dynamics of viral decay in patients on HAART medications. After initiation of HAART, levels of plasma virus fall rapidly, reflecting the exponential decay of the activated CD4+ T cells that produce most of the plasma virus and the slower decay of a second population of infected cells that have a half-life of about 2 weeks. The second phase decay brings the level of plasma virus down to a new steady state that is below the limit of detection of clinical assays (50 copies/ml). The average level of residual viremia is around 1 copy/ml. Residual viremia appears to reflect release of virus from stable reservoirs and is not reduced further by treatment intensification. Biological and statistical fluctuations in the level of this residual viremia are occasionally captured in clinical measurements as “blips,” but these transient elevations do not reflect the evolution of resistant virus.
Initially, residual viremia was thought to result from ongoing rounds of replication despite drug treatment (Dornadula et al. 1999). Such a scenario raises the possibility of the eventual emergence of drug-resistant virus and viral rebound. However, viral rebound is not seen in adherent patients, and as discussed below, direct sequence analysis of residual viremia suggests another explanation, namely, the release of virus from stable reservoirs, such as that found in resting CD4+ T cells. Every day, a small fraction of the cells in this latent reservoir may become activated by encounter with antigen or cytokines. Although the virus produced by these activated cells will not infect other cells if antiretroviral treatment completely blocks new infection events, the released virus can still be detected in the plasma by sensitive RT-PCR assays.
Evidence that this residual viremia is derived at least in part from the reactivation of latent virus in resting CD4+ T cells comes from studies in which the rare viruses in the plasma of patients on suppressive HAART regimens have been directly sequenced. These viruses lack new resistance mutations (Hermankova et al. 2001; Kieffer et al. 2004; Persaud et al. 2004; Nettles et al. 2005; Bailey et al. 2006), even during the blips or transient elevations in viremia that many patients experience (Fig. 5). When phylogenetic analyses are performed, sequences from the plasma virus and those from viruses present in the latent reservoir in resting CD4+ T cells are intermingled and, in some cases, identical. These findings indicate that at least some of the residual viremia may be derived from latently infected cells that have become activated (Tobin et al. 2005; Bailey et al. 2006). However, several recent studies suggest that the relationship between the free virus particles detected in the plasma of patients on HAART and the virus archived in the latent reservoir in resting CD4+ T cells is more complex (see below). In any event, direct analysis of residual viremia has provided little evidence for the notion that these viruses are derived from ongoing productive rounds of viral replication. Rather, these viruses are largely derived from stable reservoirs.
This conclusion is strongly supported by the results of intensification studies. Dinoso et al. (2009a,b) showed that intensification of optimal three-drug HAART regimens by addition of a fourth drug, either the potent reverse transcriptase inhibitor efavirenz or boosted forms of the protease inhibitors atazanavir or lopinavir, had no effect on the level of residual viremia (Dinoso et al. 2009a). Subsequent studies showed that intensification with the integrase inhibitor raltegravir similarly did not reduce residual viremia (Gandhi et al. 2010a; McMahon et al. 2010). Intensification also fails to reduce the size of the latent reservoir (Gandhi et al. 2010b). These findings are consistent with the idea that residual viremia reflects release of virus from stable reservoirs rather than ongoing viral replication. The highly cooperative dose-response curves of some classes of antiretroviral drugs allow extraordinarily high levels (10 logs) of inhibition of viral replication (Shen et al. 2008), providing a pharmacodynamic explanation for the ability of combinations of antiretroviral drugs to completely block viral replication.
From a clinical standpoint, the latent reservoir for HIV-1 in resting CD4+ T cells is also important as a repository for drug-resistant viruses (Persaud et al. 2000; Noe et al. 2005). During periods of active viral replication, new sequences are continuously deposited in the latent reservoir. In patients who are failing therapy with drug-resistant virus, these resistant viruses are deposited in the latent reservoir and can potentially reemerge if treatment with the failed regimen is reinstituted. Interestingly, the original wild-type viruses also persist in the latent reservoir and can reemerge if all treatment is stopped in patients who are failing therapy (Deeks et al. 2001; Ruff et al. 2002).
OTHER VIRAL RESERVOIRS
Since the original description of the latent reservoir in resting CD4+ T cells, several cell types and anatomical sites have been proposed as additional reservoirs. Unfortunately, the terms latency and reservoir have been used rather loosely. An accepted definition of latency is given above. A practical definition for an HIV-1 reservoir is a cell type or anatomical site that allows persistence of replication-competent HIV-1 for long periods of time in patients on optimal HAART regimens. In a practical sense, reservoirs are barriers to eradication. Eradication strategies will likely be attempted only in patients who have had suppression of active viral replication with HAART for prolonged periods (greater than 1 year, for example). Thus, the only reservoirs that are clinically significant are those that allow persistence on a time scale of years. To date, only the latent reservoir in resting CD4+ T cells has met this criterion (Finzi et al. 1999; Siliciano et al. 2003; Strain et al. 2003).
Mullins and colleagues proposed an elegant phylogenetic approach for defining reservoirs. A reservoir allows long-term persistence of viral genomes, and thus, sequences obtained from a reservoir show a distinct lack of temporal structure in that sequences isolated at later time points do not show greater divergence from the most recent common ancestor. The latent reservoir in resting CD4+ T cells meets this criteria (Ruff et al. 2002). For example, wild-type sequences are preserved in the latent reservoir even in patients failing therapy with drug-resistant viruses (Ruff et al. 2002).
As discussed above, another approach to identifying reservoirs is to examine the sequences from the residual viremia in patients on HAART. Because the half-life of virions in the plasma is on the order of minutes, the continued presence of free virus in the plasma indicates ongoing virus production. As discussed above, this does not appear to reflect new cycles of replication because residual viremia is not reduced by intensification of HAART. Rather it reflects release of virus from stable reservoirs, and therefore, analysis of residual viremia might provide important clues to the nature of these reservoirs. In general, the residual viremia resembles viral sequences in resting CD4+ T cells (Bailey et al. 2006). However, in some patients, the residual viremia is dominated by a small number of viral clones that are not well represented in circulating CD4+ T cells (Bailey et al. 2006). In addition, analysis of viral sequences in resting CD4+ T cell and in the residual viremia with phylogenetic methods designed to detect population structure revealed that some of the viruses in the plasma represent a distinct population not found in resting CD4+ T cells in the blood (Ruff et al. 2002; Nickle et al. 2003). These studies suggest that, in addition to the latent reservoir in resting CD4+ T cells, an additional source of residual viremia may exist in patients on HAART.
Cells of the monocyte-macrophage lineage are clearly infected by HIV-1 (Gartner et al. 1986; Igarashi et al. 2001). The role of macrophages in HIV-1 pathogenesis is described in Brennan et al. (2009), Sahu et al. (2009), and Swanstrom and Coffin (2011). Infected macrophages may constitute a particularly important source of virus late in the course of disease when CD4+ T cells have been largely depleted (Igarashi et al. 2001). It is less clear whether a true state of latency is established in macrophages in vivo; the infection is typically productive in these cells. There is some evidence for infection of circulating monocytes (Zhu et al. 2002; Arfi et al. 2008), but these cells cannot be considered a reservoir because they circulate only for about 1 day before entering the tissues and differentiating into macrophages. Importantly, monocytes can cross the blood–brain barrier and differentiate into macrophages and microglial cells. Thus, infection of monocytes may provide a mechanism for entry of virus into the central nervous system (CNS) (Gras and Kaul 2010). Infected macrophages and microglial cells clearly contribute to viral persistence and CNS pathology in HIV-1 infection (Schnell et al. 2009), and there is evidence in the SIV model for latent infection (Clements et al. 2002). In determining the importance of these cells as long-term reservoirs for HIV-1 in patients on HAART, it is important to understand their turnover rates. These cells are continually replaced by monocytes, but their half-lives are poorly understood. There is considerable controversy over the question of whether progenitor cells in the monocyte-macrophage lineage or hematopoietic stem cells (HSC) are infected in vivo. Interest in this issue has been heightened by recent evidence that HIV-1 can infect hematopoietic progenitor cells and establish a state of latent infection (Carter et al. 2010). The extent to which these cells survive in patients on HAART remains unclear.
ERADICATION STRATEGIES
There is now great interest in finding a way to eliminate the latent reservoir in resting CD4+ T cells (Richman et al. 2009). Some investigators have suggested that the stability of the latent reservoir is a reflection of continued reseeding of the reservoir by new rounds of infection despite the presence of potent antiviral drugs (Chun et al. 2005). If this were true, then intensification of HAART might reduce residual viremia and accelerate the decay of the latent reservoir. However, as discussed above, intensification does not affect residual viremia. The remarkable stability of the reservoir is more likely because of the fact that it resides in long-lived resting memory CD4+ T cells (Chun et al. 1997a). Recent results suggest that the low level of homeostatic proliferation that maintains the pool of memory T cells may contribute to the stability of the reservoir (Chomont et al. 2009).
Because intensification of HAART is unlikely to lead to eradication, current efforts have focused on finding ways to reactivate latent HIV-1 in patients on HAART. The presumption is that infected cells in which latency is reversed will die from viral cytopathic effects or be killed by HIV-1-specific cytolytic T lymphocytes. It is also presumed that HAART will prevent new rounds of infections by virus released in the process. Initial efforts to reverse latency involved the cytokine IL-2. Although treated patients appeared to have reduced numbers of latently infected cells, rapid rebound in viremia was observed when treatment was interrupted (Davey et al. 1999). Attempts to induce global T-cell activation with anti-CD3 antibodies were also unsuccessful. There is now interest in the cytokine IL-7 that is more effective in reversing latency than IL-2 (Brooks et al. 2003). Unfortunately, IL-7 also drives homeostatic proliferation of memory T cells and could lead to expansion of the latent reservoir (Chomont et al. 2009). More recent efforts have taken advantage of advances in our understanding of the molecular mechanisms regulating HIV-1 latency. Agonists of protein kinase C (Kulkosky et al. 2001; Korin et al. 2002; Brooks et al. 2003; Williams et al. 2004) and inhibitors of histone deacetylases (Ylisastigui et al. 2004; Lehrman et al. 2005; Williams et al. 2006; Archin et al. 2009) have shown promise in in vitro models. These approaches are discussed more fully in Hoxie and June (2011). A major problem with all of these approaches is their lack of specificity for infected cells.
Appropriate experimental models are likely to be critical to the development of approaches for eliminating the latent reservoir. Recently, several laboratories developed primary cell models for HIV-1 latency (reviewed in Gulick et al. 1997). These models appear to recapitulate the biology of the quiescent G0 cells that harbor latent HIV-1 in vivo better than models based on continuously proliferating cell lines. However, it is not yet clear what models most accurately reflect the behavior of latently infected cells in vivo. Another major advance has been the development of SIV/macaque models for HIV-1 latency. SIV establishes a state of latent infection in resting CD4+ T cells in the same way that HIV-1 does (Shen et al. 2003). Several groups recently developed SIV or SHIV models that are sensitive to HAART (Shen et al. 2003; Dinoso et al. 2009b; North et al. 2010). In these models, agents that might effectively purge virus from the latent reservoir can be evaluated.
CONCLUDING PERSPECTIVE
A subset of resting memory CD4+ T cells harboring integrated but transcriptionally silent HIV-1 proviruses currently poses an insurmountable barrier to viral eradication in infected subjects. Achieving the lofty goal of viral eradication will certainly not be easy because of both the intrinsic biological properties of latent proviruses and the nature of their cellular hosts. However, a better understanding of the mechanisms that underlie viral latency and a fuller appreciation for the range of cells participating in meaningful cellular reservoirs could result in a rational attack on latent HIV-1 reservoirs. Achieving the goal of complete viral eradication or alternatively a functional cure, in which subjects continue to harbor virus but do not require antiretroviral therapy, could be critical for the millions of infected individuals in the developing world where the availability of lifelong antiretroviral therapy is uncertain at best.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retrov 25: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Cheema M, Parker D, Wiegand A, Bosch RJ, Coffin JM, Eron J, Cohen M, Margolis DM 2010. Antiretroviral intensification and valproic acid lack sustained effect on residual HIV-1 viremia or resting CD4+ cell infection. PLoS One 5: e9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfi V, Riviere L, Jarrosson-Wuilleme L, Goujon C, Rigal D, Darlix JL, Cimarelli A 2008. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J Virol 82: 6557–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Morozov AV 2010. Gene regulation by nucleosome positioning. Trends Genet 26: 476–483 [DOI] [PubMed] [Google Scholar]
- Bailey J, Blankson JN, Wind-Rotolo M, Siliciano RF 2004. Mechanisms of HIV-1 escape from immune responses and antiretroviral drugs. Curr Opin Immunol 16: 470–476 [DOI] [PubMed] [Google Scholar]
- Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, et al. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 80: 6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Peterlin BM 2005. A new paradigm in eukaryotic biology: HIV Tat and the control of transcriptional elongation. PLoS Biol 3: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 8: 327–337 [DOI] [PubMed] [Google Scholar]
- Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, et al. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog 5: e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnlein E, Lowenthal JW, Siekevitz M, Ballard DW, Franza BR, Greene WC 1988. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-α gene and type 1 HIV. Cell 53: 827–836 [DOI] [PubMed] [Google Scholar]
- Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol Cell Biol 22: 1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahic M, Stowring L, Ventura P, Haase AT 1981. Gene expression in visna virus infection in sheep. Nature 292: 240–242 [DOI] [PubMed] [Google Scholar]
- Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, Wilke CO 2009. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J Virol 83: 8470–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Hamer DH, Arlen PA, Gao L, Bristol G, Kitchen CM, Berger EA, Zack JA 2003. Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19: 413–423 [DOI] [PubMed] [Google Scholar]
- Bukrinsky MI, Stanwick TL, Dempsey MP, Stevenson M 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CC, Onafuwa-Nuga A, McNamara LA, Riddell J IV, Bixby D, Savona MR, Collins KL 2010. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med 16: 446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Greene WC 2004. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol 5: 392–401 [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J 21: 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC 2001. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293: 1653–1657 [DOI] [PubMed] [Google Scholar]
- Chiang CM, Roeder RG 1995. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267: 531–536 [DOI] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, et al. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF 1995. In vivo fate of HIV-1-infected T cells: Quantitative analysis of the transition to stable latency. Nat Med 1: 1284–1290 [DOI] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, Digiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. 1997a. Quantitation of latent tissue reservoirs and total body load in HIV-1 infection. Nature 387: 183–188 [DOI] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS 1997b. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci 94: 13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Nickle DC, Justement JS, Large D, Semerjian A, Curlin ME, O’Shea MA, Hallahan CW, Daucher M, Ward DJ, et al. 2005. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest 115: 3250–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JE, Babas T, Mankowski JL, Suryanarayana K, Piatak M Jr, Tarwater PM, Lifson JD, Zink MC 2002. The central nervous system as a reservoir for simian immunodeficiency virus (SIV): Steady-state levels of SIV DNA in brain from acute through asymptomatic infection. J Infect Dis 186: 905–913 [DOI] [PubMed] [Google Scholar]
- Colin L, Van Lint C 2009. Molecular control of HIV-1 postintegration latency: Implications for the development of new therapeutic strategies. Retrovirology 6: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol 74: 6790–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey RT Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, et al. 1999. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci 96: 15109–15114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Wrin T, Liegler T, Hoh R, Hayden M, Barbour JD, Hellmann NS, Petropoulos CJ, McCune JM, Hellerstein MK, et al. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N Engl J Med 344: 472–480 [DOI] [PubMed] [Google Scholar]
- De Marco A, Biancotto C, Knezevich A, Maiuri P, Vardabasso C, Marcello A 2008. Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 5: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, O’Shea A, Callender M, Spivak A, Brennan T, et al. 2009a. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci 106: 9403–9408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, Siliciano RF, Zink MC, Clements JE 2009b. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J Virol 83: 9247–9257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L Jr, Ingerman MJ, Witek J, Kedanis RJ, Natkin J, DeSimone J, et al. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282: 1627–1632 [DOI] [PubMed] [Google Scholar]
- Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB 1989. Tumor necrosis factor α activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-κB sites in the long terminal repeat. Proc Natl Acad Sci 86: 5974–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Greenblatt J, Ingles CJ 1994. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol Cell Biol 14: 1582–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M 2003. Controlling the double helix. Nature 421: 448–453 [DOI] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278: 1295–1300 [DOI] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5: 512–517 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475–479 [DOI] [PubMed] [Google Scholar]
- Folks T, Powell DM, Lightfoote MM, Benn S, Martin MA, Fauci AS 1986. Induction of HTLV-III/LAV from a nonvirus-producing T-cell line: Implications for latency. Science 231: 600–602 [DOI] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi RT, Zheng L, Bosch RJ, Chan ES, Margolis DM, Read S, Kallungal B, Palmer S, Medvik K, Lederman MM, et al. 2010a. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: A randomized controlled trial. PLoS Med 7: 1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi RT, Bosch RJ, Aga E, Albrecht M, Demeter LM, Dykes C, Bastow B, Para M, Lai J, Siliciano RF, et al. 2010b. No evidence for decay of the latent reservoir in HIV-1-infected patients receiving intensive enfuvirtide-containing antiretroviral therapy. J Infect Dis 201: 293–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233: 215–219 [DOI] [PubMed] [Google Scholar]
- Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T 1997. CREB-binding protein/p300 are transcriptional coactivators of 65. Proc Natl Acad Sci 94: 2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E 2007. Epigenetics: A landscape takes shape. Cell 128: 635–638 [DOI] [PubMed] [Google Scholar]
- Gras G, Kaul M 2010. Molecular mechanisms of neuroinvasion by monocytes-macrophages in HIV-1 infection. Retrovirology 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Demarchi F, Giacca M, Proudfoot NJ 1998. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucl Acids Res 26: 1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, et al. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med 337: 734–739 [DOI] [PubMed] [Google Scholar]
- Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, Eron JJ Jr, Feinberg JE, Balfour HH Jr, Deyton LR, et al. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med 337: 725–733 [DOI] [PubMed] [Google Scholar]
- Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 78: 6122–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Lin YB, An W, Xu J, Yang HC, O’Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF 2008. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermankova M, Ray SC, Ruff C, Powell-Davis M, Ingersoll R, D’Aquila RT, Quinn TC, Siliciano JD, Siliciano RF, Persaud D 2001. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 286: 196–207 [DOI] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373: 123–126 [DOI] [PubMed] [Google Scholar]
- *.Hoxie JA, June CH. 2011. Novel cell and gene therapies for HIV. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia SC, Shi YB 2002. Chromatin disruption and histone acetylation in regulation of the human immunodeficiency virus type 1 long terminal repeat by thyroid hormone receptor. Mol Cell Biol 22: 4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci 98: 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20: 1726–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J 2011. The molecular biology of HIV latency: Breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS 6: 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 5: e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer TL, Finucane MM, Nettles RE, Quinn TC, Broman KW, Ray SC, Persaud D, Siliciano RF 2004. Genotypic analysis of HIV-1 drug resistance at the limit of detection: Virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis 189: 1452–1465 [DOI] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J 2002. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol 22: 4622–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S, Chen BK, Kaneshima H, Nolan GP 1998. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell 95: 595–604 [DOI] [PubMed] [Google Scholar]
- Korin YD, Brooks DG, Brown S, Korotzer A, Zack JA 2002. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J Virol 76: 8118–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Culnan DM, Roman J, Dornadula G, Schnell M, Boyd MR, Pomerantz RJ 2001. Prostratin: Activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98: 3006–3015 [DOI] [PubMed] [Google Scholar]
- Kwon HS, Brent MM, Getachew R, Jayakumar P, Chen LF, Schnolzer M, McBurney MW, Marmorstein R, Greene WC, Ott M 2008. Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell Host Microbe 3: 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF 2004. The multifactorial nature of HIV-1 latency. Trends Mol Med 10: 525–531 [DOI] [PubMed] [Google Scholar]
- Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF 2006. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog 2: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin MA, Zeichner S, Kolson D, Alwine JC, Seshamma T, Pomerantz RJ, Gonzalez-Scarano F 1993. Sodium butyrate treatment of cells latently infected with HIV-1 results in the expression of unspliced viral RNA. Virology 196: 496–505 [DOI] [PubMed] [Google Scholar]
- Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, et al. 2005. Depletion of latent HIV-1 infection in vivo: A proof-of-concept study. Lancet 366: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T, Contreras X, Peterlin BM 2008. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol 79: 6610–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou LY, Herrmann CH, Rice AP 2002. Transient induction of cyclin T1 during human macrophage differentiation regulates human immunodeficiency virus type 1 Tat transactivation function. J Virol 76: 10579–10587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Balliano A, Hayes JJ 2011. Mechanism(s) of SWI/SNF-induced nucleosome mobilization. Chembiochem 12: 196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, et al. 2007. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog 3: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon D, Jones J, Wiegand A, Gange SJ, Kearney M, Palmer S, McNulty S, Metcalf JA, Acosta E, Rehm C, et al. 2010. Short-course raltegravir intensification does not reduce persistent low-level viremia in patients with HIV-1 suppression during receipt of combination antiretroviral therapy. Clin Infect Dis 50: 912–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan AM, Saenz DT, Morrison JH, Garcia-Rivera JA, Peretz M, Llano M, Poeschla EM 2009. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog 5: e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Baltimore D 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326: 711–713 [DOI] [PubMed] [Google Scholar]
- Nettles RE, Kieffer TL, Kwon P, Monie D, Han Y, Parsons T, Cofrancesco J Jr, Gallant JE, Quinn TC, Jackson B, et al. 2005. Intermittent HIV-1 viremia (Blips) and drug resistance in patients receiving HAART. JAMA 293: 817–829 [DOI] [PubMed] [Google Scholar]
- Nickle DC, Jensen MA, Shriner D, Brodie SJ, Frenkel LM, Mittler JE, Mullins JI 2003. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol 77: 5540–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe A, Plum J, Verhofstede C 2005. The latent HIV-1 reservoir in patients undergoing HAART: An archive of pre-HAART drug resistance. J Antimicrob Chemother 55: 410–412 [DOI] [PubMed] [Google Scholar]
- North TW, Higgins J, Deere JD, Hayes TL, Villalobos A, Adamson L, Shacklett BL, Schinazi RF, Luciw PA 2010. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J Virol 84: 2913–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci 105: 3879–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada CA, Roeder RG 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384: 375–378 [DOI] [PubMed] [Google Scholar]
- Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387: 188–191 [DOI] [PubMed] [Google Scholar]
- Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM, Nabel GJ 1993. A cooperative interaction between NF-κ B and Sp1 is required for HIV-1 enhancer activation. EMBO J 12: 3551–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Jones C 2010. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip Perspect Infect Dis 2010: 262415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud D, Pierson T, Ruff C, Finzi D, Chadwick KR, Margolick JB, Ruff A, Hutton N, Ray S, Siliciano RF 2000. A stable latent reservoir for HIV-1 in resting CD4+ T lymphocytes in infected children. J Clin Invest 105: 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud D, Siberry GK, Ahonkhai A, Kajdas J, Monie D, Hutton N, Watson DC, Quinn TC, Ray SC, Siliciano RF 2004. Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads. J Virol 78: 968–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatak M Jr, Saag MS, Yang LC, Clark SJ, Kappes JC, Luk KC, Hahn BH, Shaw GM, Lifson JD 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259: 1749–1754 [DOI] [PubMed] [Google Scholar]
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ 2009. The challenge of finding a cure for HIV infection. Science 323: 1304–1307 [DOI] [PubMed] [Google Scholar]
- Rohr O, Marban C, Aunis D, Schaeffer E 2003. Regulation of HIV-1 gene transcription: From lymphocytes to microglial cells. J Leukocyte Biol 74: 736–749 [DOI] [PubMed] [Google Scholar]
- Ross EK, Buckler-White AJ, Rabson AB, Englund G, Martin MA 1991. Contribution of NF-κB and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: Distinct patterns of viral growth are determined by T-cell types. J Virol 65: 4350–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff CT, Ray SC, Kwon P, Zinn R, Pendleton A, Hutton N, Ashworth R, Gange S, Quinn TC, Siliciano RF, et al. 2002. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol 76: 9481–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu GK, Paar D, Frost SD, Smith MM, Weaver S, Cloyd MW 2009. Low-level plasma HIVs in patients on prolonged suppressive highly active antiretroviral therapy are produced mostly by cells other than CD4 T-cells. J Med Virol 81: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R 2009. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog 5: e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110: 521–529 [DOI] [PubMed] [Google Scholar]
- Shan L, Yang HC, Rabi SA, Bravo HC, Irizarry RA, Zhang H, Margolick JB, Siliciano JD, Siliciano RF 2011. Influence of host gene transcription level and orientation on HIV-1 latency in a primary cell model. J Virol 10.1128/JVI.02536-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Ruppert S, Robbins PD 1995. The retinoblastoma-susceptibility gene product binds directly to the human TATA-binding protein-associated factor TAFII250. Proc Natl Acad Sci 92: 3115–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, Li M, Clements JE, Siliciano RF 2003. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol 77: 4938–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, Zhang H, Zhou Y, Pitt E, Anderson KS, Acosta EP, et al. 2008. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med 14: 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekevitz M, Josephs SF, Dukovich M, Peffer N, Wong-Staal F, Greene WC 1987. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science 238: 1575–1578 [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9: 727–728 [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Lai J, Callender M, Pitt E, Zhang H, Margolick JB, Gallant JE, Cofrancesco J Jr, Moore RD, Gange SJ, et al. 2007. Stability of the latent reservoir for HIV-1 in patients receiving valproic acid. J Infect Dis 195: 833–836 [DOI] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak AM, Rabi SA, McMahon MA, Shan L, Sedaghat AR, Wilke CO, Siliciano RF 2011. Dynamic constraints on the second phase compartment of HIV-infected cells. AIDS Res Hum Retrov 10.1089/aid.2010.0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain MC, Gunthard HF, Havlir DV, Ignacio CC, Smith DM, Leigh-Brown AJ, Macaranas TR, Lam RY, Daly OA, Fischer M, et al. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: Intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci 100: 4819–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC 1993. NF-κB controls expression of inhibitor I κ B α: Evidence for an inducible autoregulatory pathway. Science 259: 1912–1915 [DOI] [PubMed] [Google Scholar]
- Sune C, Garcia-Blanco MA 1995. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol 69: 6572–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Swanstorm R, Coffin J 2011. HIV-1 pathogenesis: The virus. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH 2010. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 465: 747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H 2010. Confining euchromatin/heterochromatin territory: Jumonji crosses the line. Genes Dev 24: 1465–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, et al. 2005. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: Expression of archival virus and replication of virus. J Virol 79: 9625–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Ott M, Verdin E 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J 15: 1112–1120 [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Paras P Jr, Van Lint C 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 12: 3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373: 117–122 [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92: 451–462 [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122: 169–182 [DOI] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Fenard D, Bisgrove D, Verdin E, Greene WC 2004. Prostratin antagonizes HIV latency by activating NF-κB. J Biol Chem 279: 42008–42017 [DOI] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC 2006. NF-κB pp50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 25: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278: 1291–1295 [DOI] [PubMed] [Google Scholar]
- Yang HC 2011. Primary cell models of HIV latency. Curr Opin HIV AIDS 6: 62–67 [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414: 317–322 [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545 [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW 2004. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 23: 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM 2004. Coaxing HIV-1 from resting CD4 T cells: Histone deacetylase inhibition allows latent viral expression. AIDS 18: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS 1990. HIV-1 entry into quiescent primary lymphocytes: Molecular analysis reveals a labile, latent viral structure. Cell 61: 213–222 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang H, Siliciano JD, Siliciano RF 2005. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol 79: 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Muthui D, Holte S, Nickle D, Feng F, Brodie S, Hwangbo Y, Mullins JI, Corey L 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J Virol 76: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]