Abstract
Acquired immunodeficiency syndrome (AIDS) of humans is caused by two lentiviruses, human immunodeficiency viruses types 1 and 2 (HIV-1 and HIV-2). Here, we describe the origins and evolution of these viruses, and the circumstances that led to the AIDS pandemic. Both HIVs are the result of multiple cross-species transmissions of simian immunodeficiency viruses (SIVs) naturally infecting African primates. Most of these transfers resulted in viruses that spread in humans to only a limited extent. However, one transmission event, involving SIVcpz from chimpanzees in southeastern Cameroon, gave rise to HIV-1 group M—the principal cause of the AIDS pandemic. We discuss how host restriction factors have shaped the emergence of new SIV zoonoses by imposing adaptive hurdles to cross-species transmission and/or secondary spread. We also show that AIDS has likely afflicted chimpanzees long before the emergence of HIV. Tracing the genetic changes that occurred as SIVs crossed from monkeys to apes and from apes to humans provides a new framework to examine the requirements of successful host switches and to gauge future zoonotic risk.
Approximately one century ago in southeastern Cameroon, cross-species transmission of SIVcpz from chimpanzees gave rise to HIV-1 group M, the principal cause of the AIDS pandemic in humans.
Acquired Immune Deficiency Syndrome (AIDS) was first recognized as a new disease in 1981 when increasing numbers of young homosexual men succumbed to unusual opportunistic infections and rare malignancies (CDC 1981; Greene 2007). A retrovirus, now termed human immunodeficiency virus type 1 (HIV-1), was subsequently identified as the causative agent of what has since become one of the most devastating infectious diseases to have emerged in recent history (Barre-Sinoussi et al. 1983; Gallo et al. 1984; Popovic et al. 1984). HIV-1 spreads by sexual, percutaneous, and perinatal routes (Hladik and McElrath 2008; Cohen et al. 2011); however, 80% of adults acquire HIV-1 following exposure at mucosal surfaces, and AIDS is thus primarily a sexually transmitted disease (Hladik and McElrath 2008; Cohen et al. 2011). Since its first identification almost three decades ago, the pandemic form of HIV-1, also called the main (M) group, has infected at least 60 million people and caused more than 25 million deaths (Merson et al. 2008). Developing countries have experienced the greatest HIV/AIDS morbidity and mortality, with the highest prevalence rates recorded in young adults in sub-Saharan Africa (http://www.unaids.org/). Although antiretroviral treatment has reduced the toll of AIDS- related deaths, access to therapy is not universal, and the prospects of curative treatments and an effective vaccine are uncertain (Barouch 2008; Richman et al. 2009). Thus, AIDS will continue to pose a significant public health threat for decades to come.
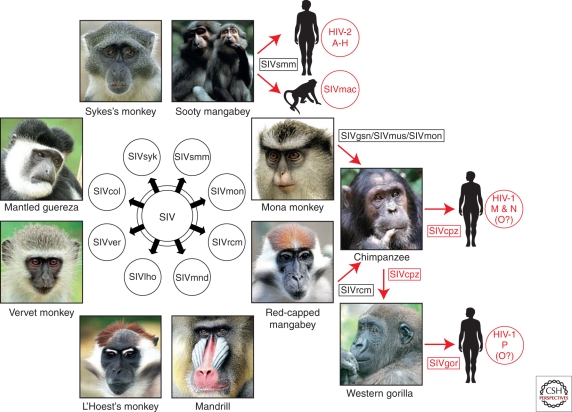
Ever since HIV-1 was first discovered, the reasons for its sudden emergence, epidemic spread, and unique pathogenicity have been a subject of intense study. A first clue came in 1986 when a morphologically similar but antigenically distinct virus was found to cause AIDS in patients in western Africa (Clavel et al. 1986). Curiously, this new virus, termed human immunodeficiency virus type 2 (HIV-2), was only distantly related to HIV-1, but was closely related to a simian virus that caused immunodeficiency in captive macaques (Chakrabarti et al. 1987; Guyader et al. 1987). Soon thereafter, additional viruses, collectively termed simian immunodeficiency viruses (SIVs) with a suffix to denote their species of origin, were found in various different primates from sub-Saharan Africa, including African green monkeys, sooty mangabeys, mandrills, chimpanzees, and others (Fig. 1). Surprisingly, these viruses appeared to be largely nonpathogenic in their natural hosts, despite clustering together with the human and simian AIDS viruses in a single phylogenetic lineage within the radiation of lentiviruses (Fig. 2). Interestingly, close simian relatives of HIV-1 and HIV-2 were found in chimpanzees (Huet et al. 1990) and sooty mangabeys (Hirsch et al. 1989), respectively. These relationships provided the first evidence that AIDS had emerged in both humans and macaques as a consequence of cross-species infections with lentiviruses from different primate species (Sharp et al. 1994). Indeed, subsequent studies confirmed that SIVmac was not a natural pathogen of macaques (which are Asian primates), but had been generated inadvertently in US primate centers by inoculating various species of macaques with blood and/or tissues from naturally infected sooty mangabeys (Apetrei et al. 2005, 2006). Similarly, it became clear that HIV-1 and HIV-2 were the result of zoonotic transfers of viruses infecting primates in Africa (Hahn et al. 2000). In this article, we summarize what is known about the simian precursors of HIV-1 and HIV-2, and retrace the steps that led to the AIDS pandemic.
Figure 1.
Origins of human AIDS viruses. Old World monkeys are naturally infected with more than 40 different lentiviruses, termed simian immunodeficiency viruses (SIVs) with a suffix to denote their primate species of origin (e.g., SIVsmm from sooty mangabeys). Several of these SIVs have crossed the species barrier to great apes and humans, generating new pathogens (see text for details). Known examples of cross-species transmissions, as well as the resulting viruses, are highlighted in red.
Figure 2.
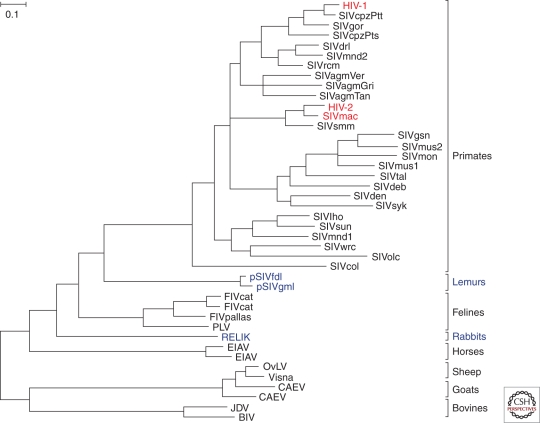
Phylogeny of lentiviruses. The evolutionary relationships among Pol sequences (∼ 770 amino acids) derived from various mammalian lentiviruses; host species are indicated at the right. Exogenous viruses are depicted in black, with HIV-1, HIV-2, and SIVmac highlighted in red; endogenous viruses are shown in purple. The phylogenetic tree was estimated using maximum likelihood methods (Guindon and Gascuel 2003). The scale bar represents 0.10 amino acid replacements per site.
PRIMATE LENTIVIRUSES
Lentiviruses cause chronic persistent infections in various mammalian species, including bovines, horses, sheep, felines, and primates. The great majority of lentiviruses are exogenous, meaning that they are transmitted horizontally between individuals. However, it has recently become clear that, on several occasions in the past, lentiviruses have infiltrated their hosts’ germlines and become endogenous, vertically transmissible, genomic loci (Fig. 2). Examples include the rabbit endogenous lentivirus type K (RELIK), which became germ-line embedded approximately 12 million years ago (Katzourakis et al. 2007; van der Loo et al. 2009), and two prosimian endogenous lentiviruses, which independently invaded the germ-lines of both the grey mouse lemur (pSIVgml) and the fat-tailed dwarf lemur (pSIVfdl) about 4 million years ago (Gifford et al. 2008; Gilbert et al. 2009). These “viral fossils” are of particular interest because they provide direct evidence of the timescale of lentivirus evolution. Molecular clocks derived from extant SIV sequences suggested that ancestral SIVs existed only a few hundreds of years ago (Wertheim and Worobey 2009), but it has long been suspected that such analyses may grossly underestimate deeper evolutionary timescales (Sharp et al. 2000; Holmes 2003). Recent studies of SIV-infected monkeys on Bioko Island, Equatorial Guinea, partly substantiated this conclusion, showing that geographically isolated subspecies have been infected with the same type of SIV for at least 30,000 years and probably much longer (Worobey et al. 2010). The endogenous viruses in lemurs reveal that the span of evolutionary history of primate lentiviruses as a whole is at least two orders of magnitude greater still. Thus, it is possible that at least some SIVs, such as those infecting four closely related species of African green monkeys (Chlorocebus species), have coevolved with their respective hosts for an extended period of time, perhaps even before these hosts diverged from their common ancestor (Jin et al. 1994a). So far, SIV infections have only been found in African monkeys and apes, and so it seems likely that primate lentiviruses emerged in Africa sometime after the splits between lineages of African and Asian Old World monkeys, which are believed to have occurred around 6–10 million years ago (Fabre et al. 2009). However, because neither Asian nor New World primates have been sampled exhaustively, the conclusion that SIVs are restricted to African primates must remain tentative, especially because none of these primate species has been examined for endogenous forms of SIV (Ylinen et al. 2010). Thus, our understanding of the evolutionary history of primate lentiviruses is still incomplete.
To date, serological evidence of SIV infection has been reported for over 40 primate species, and molecular data have been obtained for most of these (also see Klatt et al. 2011). The latter studies have shown that the great majority of primate species harbor a single “type” or “strain” of SIV. That is, viral sequences from members of the same species form a monophyletic clade in evolutionary trees. This host-specific clustering indicates that the great majority of transmissions occur among members of the same species; however, there are also numerous documented instances when SIVs have crossed between species. Examples range from incidental “dead-end” infections (e.g., SIVver infections of baboons) (Jin et al. 1994b; van Rensburg et al. 1998) to the generation of new SIV lineages with substantial secondary spread (e.g., SIVgor infection of gorillas) (Van Heuverswyn et al. 2006). In addition, cross-species transmissions have generated mosaic SIV lineages through superinfection and recombination in species that already harbored an SIV (e.g., SIVsab infection of sabaeus monkeys) (Jin et al. 1994a). In both mandrills (Mandrillus sphinx) and moustached monkeys (Cercopithecus cephus), such recombination events have led to the emergence of a second SIV strain that cocirculates with the original virus (Souquiere et al. 2001; Aghokeng et al. 2007). Thus, it is clear that in addition to more long-standing virus/host relationships, a number of naturally occurring SIVs have emerged more recently as a result of cross-species transmission and recombination. What remains unknown is when and how often these cross-species transfers have occurred, what impact they had on virus and host biology, and whether AIDS is a frequent consequence of SIV host switching. The prevalence of naturally occurring SIV infections varies widely, ranging from 1% in some species to over 50% in others (Aghokeng et al. 2010), and it is tempting to speculate that less ubiquitous SIVs were acquired more recently and/or may be more pathogenic.
ORIGIN AND DISTRIBUTION OF SIVcpz
Of the many primate lentiviruses that have been identified, SIVcpz has been of particular interest because of its close genetic relationship to HIV-1 (Fig. 2). However, studies of this virus have proven to be challenging because of the endangered status of chimpanzees. The first isolates of SIVcpz were all derived from animals housed in primate centers or sanctuaries, although infection was rare in these populations. Collective analyses of nearly 2,000 wild-caught or captive-born apes identified fewer than a dozen SIVcpz positive individuals (Sharp et al. 2005). Because other primate species, such as sooty mangabeys and African green monkeys, are much more commonly infected, both in captivity and in the wild (Fultz et al. 1990; Phillips-Conroy et al. 1994; Santiago et al. 2005), this finding raised doubts about whether chimpanzees represented a true SIV reservoir. To resolve this conundrum, our laboratory developed noninvasive diagnostic methods that detect SIVcpz specific antibodies and nucleic acids in chimpanzee fecal and urine samples with high sensitivity and specificity (Santiago et al. 2003; Keele et al. 2006). These technical innovations, combined with genotyping methods for species and subspecies confirmation as well as individual identification, permitted a comprehensive analysis of wild-living chimpanzee populations throughout central Africa.
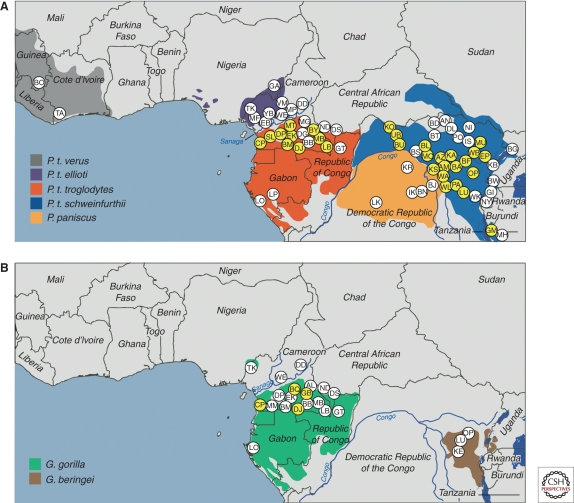
Chimpanzees are classified into two species, the common chimpanzee (Pan troglodytes) and the bonobo (Pan paniscus). Common chimpanzees have traditionally been further subdivided into a number of geographically differentiated subspecies (Groves 2001). Four subspecies were defined on the basis of mitochondrial DNA sequences (Gagneux et al. 1999), namely western (P. t. verus), Nigeria-Cameroonian (P. t. ellioti, formerly termed P. t. vellerosus), central (P. t. troglodytes), and eastern (P. t. schweinfurthii) chimpanzees. To determine the distribution of SIVcpz among these populations, fecal (and in some cases urine) samples were collected at different field sites and tested for the presence of virus specific antibodies. Antibody positive fecal specimens were then subjected to RNA extraction and reverse transcriptase polymerase chain reaction (RT-PCR) amplification to molecularly characterize the infecting virus strain. At select field sites, mitochondrial and microsatellite analyses of host DNA were also used to confirm sample integrity and to determine the number of tested individuals. Figure 3A summarizes current molecular epidemiological data derived from the analysis of over 7,000 chimpanzee fecal samples collected at nearly 90 field sites (Santiago et al. 2002, 2003; Worobey et al. 2004; Keele et al. 2006; Van Heuverswyn et al. 2007; Li et al. 2010; Rudicell et al. 2010). These studies have identified common chimpanzees as a natural SIVcpz reservoir, but also revealed important differences between the epidemiology of SIVcpz and that of other primate lentiviruses. First, only two of the four chimpanzee subspecies were found to harbor these viruses. SIVcpz was detected at multiple sites throughout the ranges of both central and eastern chimpanzees in an area ranging from Cameroon to Tanzania, but there was no evidence of infection in western and Nigeria-Cameroonian chimpanzees, nor in bonobos, despite testing of multiple communities. In addition, SIVcpz prevalence rates among central and eastern chimpanzees varied widely, ranging from 30% to 50% in some communities to rare or absent infection in others. In contrast, other SIVs, such as those of sooty mangabeys and African green monkeys, are much more widely and evenly distributed and infect their hosts at generally higher prevalence rates (Phillips-Conroy et al. 1994; Santiago et al. 2005). Nonetheless, the puzzle of why SIVcpz was so scarce among captive chimpanzees was finally resolved: As it turned out, most of these apes were imported from West Africa and thus were members of the P. t. verus subspecies, which does not harbor SIVcpz (Prince et al. 2002; Switzer et al. 2005).
Figure 3.
Geographic distribution of SIVcpz and SIVgor infections in sub-Saharan Africa. Field sites where wild-living (A) chimpanzees and bonobos, and (B) gorillas have been sampled are shown (each site is identified by a two-letter code; because of space limitations, only a subset is depicted). Sites where SIV infections were detected are highlighted in yellow. The upper panel depicts the ranges of the four subspecies of the common chimpanzee (Pan troglodytes verus, gray; P. t. ellioti, magenta; P. t. troglodytes, red; and P. t. schweinfurthii, blue) and of the bonobo (P. paniscus, orange). The lower panel depicts the ranges of western (Gorilla gorilla, green) and eastern (G. beringei, brown) gorillas (map courtesy of Lilian Pintea, The Jane Goodall Institute). Data were compiled from several studies (Santiago et al. 2002, 2003; Worobey et al. 2004; Keele et al. 2006; Van Heuverswyn et al. 2007; Li et al. 2010; Rudicell et al. 2010).
The absence of SIVcpz from two of the four subspecies suggested that chimpanzees had acquired this virus more recently, after their divergence into different subspecies. Indeed, phylogenetic analyses of full-length proviral sequences revealed that SIVcpz represents a complex mosaic, generated by recombination of two lineages of SIVs that infect monkeys (Bailes et al. 2003). In the 5′ half of the genome, as well as the nef gene and 3′ LTR, SIVcpz is most closely related to SIVrcm from red-capped mangabeys (Cercocebus torquatus); however, in the vpu, tat, rev, and env genes, SIVcpz is most closely related to a clade of SIVs infecting several Cercopithecus species, including greater spot-nosed (C. nictitans), mustached (C. cephus), and mona (C. mona) monkeys (Bailes et al. 2003). Chimpanzees are known to hunt and kill other mammals, including monkeys (Goodall 1986), suggesting that they acquired SIV in the context of predation. The current range of the central chimpanzee overlaps those of red-capped mangabeys and the various Cercopithecus species, and so it is likely that the cross-species transmission events that led to the emergence of SIVcpz occurred in that area, and that SIVcpz later spread to eastern chimpanzees, although it is unclear whether this occurred during or subsequent to their divergence from the central subspecies. Importantly, all of more than 30 sequenced SIVcpz strains show an identical mosaic genome structure. Moreover, there is no evidence that chimpanzees harbor any other SIV, although they, as well as bonobos, are routinely exposed to SIVs through their hunting behavior (Mitani and Watts 1999; Surbeck and Hohmann 2008; Leendertz et al. 2011).
NATURAL HISTORY OF SIVcpz INFECTION
Initially, SIVcpz was thought to be harmless for its natural host. This was because none of the few captive apes that were naturally SIVcpz infected suffered from overt immunodeficiency, although in retrospect this conclusion was based on the immunological and virological analyses of only a single naturally infected chimpanzee (Heeney et al. 2006). In addition, SIV-infected sooty mangabeys and African green monkeys showed no sign of disease despite high viral loads in blood and lymphatic tissues (Paiardini et al. 2009), leading to the belief that all naturally occurring SIV infections are nonpathogenic. However, the sporadic prevalence of SIVcpz, along with its more recent monkey origin, suggested that its natural history might differ from that of other primate lentiviruses. To address this, a prospective study was initiated in Gombe National Park, Tanzania, the only field site where SIVcpz infected chimpanzees are habituated and so can be observed in their natural habitat.
Gombe is located in northwestern Tanzania on the shores of Lake Tanganyika. The park is home to three communities, termed Kasekela, Mitumba, and Kalande, which have been studied by Goodall and colleagues since the 1960s, 1980s, and 1990s, respectively (Pusey et al. 2007). Prospective studies of SIVcpz in Gombe began in 2000 (Santiago et al. 2002). By 2009, infections were documented in all three communities, with mean biannual prevalence rates of 13%, 12%, and 46% in Mitumba, Kasekela, and Kalande, respectively (Rudicell et al. 2010). Analysis of epidemiologically linked infections revealed that SIVcpz spreads primarily through sexual routes, with an estimated transmission probability per coital act (0.0008–0.0015) that is similar to that of HIV-1 among heterosexual humans (0.0011) (Gray et al. 2001; Rudicell et al. 2010). SIVcpz also appears to be transmitted from infected mothers to their infants, and in rare cases, possibly by aggression (Keele et al. 2009). Migration of infected females constitutes a major route of virus transmission between communities (Rudicell et al. 2010).
Behavioral and virological studies also provided insight into the pathogenicity of SIVcpz. Age-corrected mortality analyses revealed that infected chimpanzees had a 10- to 16-fold increased risk of death compared to uninfected chimpanzees (Keele et al. 2009). SIVcpz-infected females were less likely to give birth and had a much higher infant mortality rate than uninfected females. Postmortem analyses revealed significant CD4+ T-cell depletion in three infected individuals, but not in either of two uninfected individuals. One infected female, who died within 3 years of acquiring the virus, had histopathological findings consistent with end-stage AIDS. Taken together, these findings provided compelling evidence that SIVcpz was pathogenic in its natural host. Subsequent studies of both wild and captive chimpanzees confirmed these findings. By the end of 2010, the Kasekela and Mitumba communities had experienced three additional deaths, all SIVcpz related. One case concerned an infant born to an infected mother, whereas the other two were adult females, one of whom died with severe CD4+ T cell depletion within 5 years of acquiring SIVcpz (KA Terio et al., submitted). Moreover, demographic studies revealed that the Kalande community, which showed the highest SIVcpz prevalence rates (40%–50%), had suffered a catastrophic population decline, whereas the sizes of the Mitumba and Kasekela communities, which were infected at a much lower level (12%–13%), remained stable (Rudicell et al. 2010). It has been suggested that only members of the P. t. schweinfurthii subspecies, or more particularly the chimpanzees of Gombe, are susceptible to SIVcpz-associated pathogenicity (Weiss and Heeney 2009; Soto et al. 2010). However, a prospective study of orphaned chimpanzees in Cameroon identified an SIVcpz infected P. t. troglodytes ape that suffered from progressive CD4+ T cell loss, severe thrombocytopenia, and clinical AIDS (Etienne et al. 2011). Thus, it seems likely that SIVcpz has a substantial negative impact on the health, reproduction, and lifespan of all chimpanzees that harbor SIVcpz in the wild.
ORIGIN AND DISTRIBUTION OF SIVgor
Noninvasive testing also led to the unexpected finding of a new SIV lineage in wild-living gorillas (Fig. 4). Analysis of ∼ 200 fecal samples from southern Cameroon identified several HIV/SIV antibody positive gorillas, and amplification of viral sequences revealed the existence of a new SIV lineage, termed SIVgor (Van Heuverswyn et al. 2006). This lineage fell within the radiation of SIVcpz, clustering with strains from P. t. troglodytes apes, suggesting that gorillas had acquired SIVgor by cross-species infection from sympatric chimpanzees (Fig. 4). Phylogenetic analyses of full-length SIVgor sequences confirmed this conclusion, indicating that SIVgor resulted from a single chimpanzee-to-gorilla transmission event estimated to have occurred at least 100 to 200 years ago (Takehisa et al. 2009). Subsequent screening of over 2500 fecal samples from 30 field sites across central Africa uncovered additional SIVgor infections, but only in western lowland gorillas (Gorilla gorilla gorilla) and not in eastern gorillas (Gorilla beringei). Although virus-positive apes were present at field sites more than 400 km apart, only four such sites were identified, and prevalence rates in these communities did not exceed 5% (Fig. 3B). Thus, SIVgor appears to be much less common in gorillas than SIVcpz is in chimpanzees (Neel et al. 2010). Whether SIVgor is more prevalent in communities in parts of west central Africa that have not yet been tested is not known. It is also unclear whether SIVgor is pathogenic for its host, because there has been no opportunity to study the natural history of this infection in either captive or wild-living gorillas. Finally, it remains a mystery how gorillas acquired SIVgor, because they are herbivores and do not hunt or kill other mammals. Nonetheless, gorillas and chimpanzees feed in the same forest areas, which must have led to at least one encounter that allowed transmission.
Figure 4.
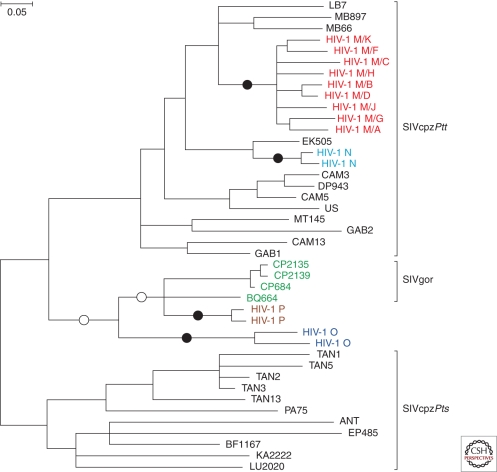
HIV-1 origins. The phylogenetic relationships of representative SIVcpz, HIV-1, and SIVgor strains are shown for a region of the viral pol gene (HIV-1/HXB2 coordinates 3887–4778). SIVcpz and SIVgor sequences are shown in black and green, respectively. The four groups of HIV-1, each of which represents an independent cross-species transmission, are shown in different colors. Black circles indicate the four branches where cross-species transmission-to-humans has occurred. White circles indicate two possible alternative branches on which chimpanzee-to-gorilla transmission occurred. Brackets at the right denote SIVcpz from P. t. troglodytes (SIVcpzPtt) and P. t. schweinfurthii (SIVcpzPts), respectively. The phylogenetic tree was estimated using maximum likelihood methods (Guindon and Gascuel 2003). The scale bar represents 0.05 nucleotide substitutions per site.
ORIGINS OF HIV-1
HIV-1 has long been suspected to be of chimpanzee origin (Gao et al. 1999); however, until recently, the perceived lack of a chimpanzee reservoir left the source of HIV-1 open to question. These uncertainties have since been resolved by noninvasive testing of wild-living ape populations. It is now well established that all naturally occurring SIVcpz strains fall into two subspecies-specific lineages, termed SIVcpzPtt and SIVcpzPts, respectively, that are restricted to the home ranges of their respective hosts (Figs. 3 and 4). Viruses from these two lineages are quite divergent, differing at about 30%–50% of sites in their Gag, Pol, and Env protein sequences (Vanden Haesevelde et al. 1996). Interestingly, population genetic studies have shown that central and eastern chimpanzees are barely differentiated, calling into question their status as separate subspecies (Fischer et al. 2006; Gonder et al. 2011). However, the fact that they harbor distinct SIVcpz lineages suggests that central and eastern chimpanzees have been effectively isolated for some time. In addition, molecular epidemiological studies in southern Cameroon have shown that SIVcpzPtt strains show phylogeographic clustering, with viruses from particular areas forming monophyletic lineages, and the discovery of SIVgor has identified a second ape species as a potential reservoir for human infection (Van Heuverswyn et al. 2006). Collectively, these findings have allowed the origins of HIV-1 to be unraveled (Keele et al. 2006; Van Heuverswyn et al. 2007).
HIV-1 is not just one virus, but comprises four distinct lineages, termed groups M, N, O, and P, each of which resulted from an independent cross-species transmission event. Group M was the first to be discovered and represents the pandemic form of HIV-1; it has infected millions of people worldwide and has been found in virtually every country on the globe. Group O was discovered in 1990 and is much less prevalent than group M (De Leys et al. 1990; Gurtler et al. 1994). It represents less than 1% of global HIV-1 infections, and is largely restricted to Cameroon, Gabon, and neighboring countries (Mauclere et al. 1997; Peeters et al. 1997). Group N was identified in 1998 (Simon et al. 1998), and is even less prevalent than group O; so far, only 13 cases of group N infection have been documented, all in individuals from Cameroon (Vallari et al. 2010). Finally, group P was discovered in 2009 in a Cameroonian woman living in France (Plantier et al. 2009). Despite extensive screening, group P has thus far only been identified in one other person, also from Cameroon (Vallari et al. 2011). Although members of all of these groups are capable of causing CD4+ T-cell depletion and AIDS, they obviously differ vastly in their distribution within the human population.
Figure 4 depicts a phylogenetic tree of representative HIV-1, SIVcpz, and SIVgor strains. It shows that all four HIV-1 groups, as well as SIVgor, cluster with SIVcpzPtt from central chimpanzees, identifying this subspecies as the original reservoir of both human and gorilla infections. HIV-1 groups N and M are very closely related to SIVcpzPtt strains from southern Cameroon, indicating that they are of chimpanzee origin. It has even been possible to trace their ape precursors to particular P. t. troglodytes communities. HIV-1 group N appears to have emerged in the vicinity of the Dja Forest in south-central Cameroon, whereas the pandemic form, group M, likely originated in an area flanked by the Boumba, Ngoko, and Sangha rivers in the southeastern corner of Cameroon (Keele et al. 2006; Van Heuverswyn et al. 2007). Existing phylogenetic data support a gorilla origin of HIV-1 group P, but too few SIVgor strains have been characterized to identify the region where this transmission might have occurred. In contrast, the immediate source of HIV-1 group O remains unknown, because there are no ape viruses that are particularly closely related to this group (Fig. 4). Thus, HIV-1 group O could either be of chimpanzee or gorilla origin. Nonetheless, the fact that group O and P viruses are more closely related to SIVcpzPtt than to SIVcpzPts suggests that both groups originated in west central Africa, which is consistent with their current distributions.
How humans acquired the ape precursors of HIV-1 groups M, N, O, and P is not known; however, based on the biology of these viruses, transmission must have occurred through cutaneous or mucous membrane exposure to infected ape blood and/or body fluids. Such exposures occur most commonly in the context of bushmeat hunting (Peeters et al. 2002). Whatever the circumstances, it seems clear that human–ape encounters in west central Africa have resulted in four independent cross-species transmission events. Molecular clock analyses have dated the onset of the group M and O epidemics to the beginning of the twentieth century (Korber et al. 2000; Lemey et al. 2004; Worobey et al. 2008). In contrast, groups N and P appear to have emerged more recently, although the sequence data for these rare groups are still too limited to draw definitive conclusions.
Eastern chimpanzees are endemically infected with SIVcpzPts throughout central Africa (Fig. 3A). Although prevalence rates have not been determined for all field sites, the P. t. schweinfurthii communities that have been studied show infection rates that are very similar to those found in P. t. troglodytes (Keele et al. 2006, 2009; Rudicell et al. 2010). Given that SIVcpzPtt strains have been transmitted to gorillas and humans on at least five occasions, it is striking that evidence of similar transmissions from eastern chimpanzees is lacking. There are a number of possible explanations. First, the risk of human exposure to SIVcpzPts may be lower, perhaps because of differences in the frequencies or types of human–ape interactions in central and east Africa. Second, SIVcpzPts infections of humans may have occurred, but gone unrecognized, because of limited human sampling and a lack of lineage-specific serological tests. Finally, as discussed below, SIVcpzPtt has evolved to overcome human restriction factors, such as tetherin, which may pose a barrier to cross-species transmission; because SIVcpzPts is highly divergent from SIVcpzPtt, viruses from this lineage may not have been able to adapt in the same way. Although SIVcpzPtt and SIVcpzPts strains replicate with similar kinetics in human CD4+ T cells in vitro (Takehisa et al. 2007), such cultures are unlikely to accurately recapitulate the conditions of viral replication and transmission in vivo.
ORIGINS OF HIV-2
Since its first discovery, HIV-2 has remained largely restricted to West Africa, with its highest prevalence rates recorded in Guinea-Bissau and Senegal (de Silva et al. 2008). However, overall prevalence rates are declining, and in most West African countries HIV-2 is increasingly being replaced by HIV-1 (van der Loeff et al. 2006; Hamel et al. 2007). Viral loads tend to be lower in HIV-2 than HIV-1 infected individuals, which may explain the lower transmission rates of HIV-2 and the near complete absence of mother-to-infant transmissions (Popper et al. 2000; Berry et al. 2002). In fact, most individuals infected with HIV-2 do not progress to AIDS, although those who do, show clinical symptoms indistinguishable from HIV-1 (Rowland-Jones and Whittle 2007). Thus, it is clear that the natural history of HIV-2 infection differs considerably from that of HIV-1, which is not surprising given that HIV-2 is derived from a very different primate lentivirus.
A sooty mangabey origin of HIV-2 was first proposed in 1989 (Hirsch et al. 1989) and subsequently confirmed by demonstrating that humans in West Africa harbored HIV-2 strains that resembled locally circulating SIVsmm infections (Gao et al. 1992; Chen et al. 1996). SIVsmm was found to be highly prevalent, both in captivity and in the wild, and to be nonpathogenic in its natural host (Silvestri 2005). In a wild-living sooty mangabey community, SIVsmm was primarily found in higher-ranking females, suggesting that virus infection had no appreciable negative effect on reproductive behavior or success (Santiago et al. 2005). Finally, the fact that sooty mangabeys are frequently hunted as agricultural pests in many areas of West Africa provided plausible routes of transmission.
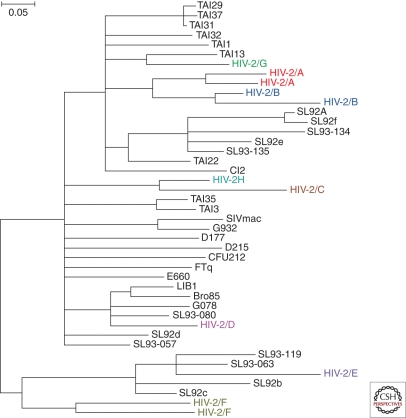
Since its first isolation, at least eight distinct lineages of HIV-2 have been identified, each of which appears to represent an independent host transfer (Fig. 5). By analogy with HIV-1, these lineages have been termed groups A–H, although only groups A and B have spread within humans to an appreciable degree. Group A has been found throughout western Africa (Damond et al. 2001; Peeters et al. 2003), whereas group B predominates in Cote d’Ivoire (Pieniazek et al. 1999; Ishikawa et al. 2001). All other HIV-2 “groups” were initially identified only in single individuals, suggesting that they represent incidental infection with very limited or no secondary spread. Of these, groups C, G, and H have been linked to SIVsmm strains from Cote d’Ivoire, group D is most closely related to an SIVsmm strain from Liberia, and groups E and F resemble SIVsmm strains from Sierra Leone (Gao et al. 1992; Chen et al. 1996, 1997; Santiago et al. 2005). Because of their sporadic nature, groups C–H have been assumed to represent “dead-end” transmissions. However, a second divergent HIV-2 strain has recently been placed in group F (Fig. 5). This virus was identified in an immigrant in New Jersey, who came from the same geographic area in Sierra Leone where this lineage was first discovered (Smith et al. 2008). Unlike the original index case, the newly identified group F infection was associated with reduced CD4 T cell counts and high viral loads (Smith et al. 2008). It is presently unknown whether group F has been spreading cryptically in humans, or whether the two group F viruses represents independent transmissions from sooty mangabeys.
Figure 5.
HIV-2 origins. The phylogenetic relationships of representative SIVsmm and HIV-2 strains are shown for a region of the viral gag gene (SIVmac239 coordinates 1191–1921). SIVsmm and SIVmac are shown in black; the eight groups of HIV-2, each of which represents an independent cross-species transmission, are shown in different colors. The phylogenetic tree was estimated using maximum likelihood methods (Guindon and Gascuel 2003). The scale bar represents 0.05 nucleotide substitutions per site.
HOST-SPECIFIC ADAPTATIONS
HIV and SIV must interact with a large number of host proteins to replicate in infected cells (Fu et al. 2009; Ortiz et al. 2009). Because the common ancestor of Old World monkeys and apes existed around 25 million years ago, the divergence of these host proteins may pose an obstacle to cross-species infection. In addition, primates (including humans) encode a number of host restriction factors, which have evolved as part of their innate immune response to protect against infection with a wide variety of viral pathogens (Malim and Emerman 2008; Neil and Bieniasz 2009; Kajaste-Rudnitski et al. 2010). Although viruses have, in turn, found ways to antagonize these restriction factors, these countermeasures are frequently species-specific. Thus, a number of adaptive hurdles have to be overcome before primate lentiviruses can productively infect a new species.
The first evidence of host-specific adaptation of HIV-1 came from an analysis of sites in the viral proteome that were highly conserved in the ape precursors of HIV-1, but changed—in the same way—each time these viruses crossed the species barrier to humans (Wain et al. 2007). This analysis identified one site in the viral matrix protein (Gag-30) that encoded a Met in all known strains of SIVcpzPtt and SIVgor but switched to an Arg in the inferred ancestors of HIV-1 groups M, N, and O, and has subsequently been conserved as a basic amino acid (Arg or Lys) in most strains of HIV-1. The fact that the same nonconservative amino acid substitution occurred on each of the three branches involving cross-species transmission to humans suggested that this matrix residue was under strong host-specific selection pressure. This conclusion was subsequently confirmed by two additional observations. First, it was found that a reciprocal transmission, in which a chimpanzee was experimentally infected with HIV-1, led to the reversion of this host-specific signature; that is, a basic residue at Gag-30 in HIV-1 changed back to a Met on in vivo propagation in a chimpanzee (Mwaengo and Novembre 1998). Second, it was found that in chimpanzee CD4+ T lymphocytes, a virus with a Met at position 30 replicated more efficiently that an otherwise isogenic virus with a Lys at the same position, whereas the opposite was true in human cells (Wain et al. 2007). Interestingly, only one of the two recently discovered HIV-1 group P strains has switched from a Met to a Lys at Gag-30 (Vallari et al. 2011). Although the structure of the HIV-1 matrix protein has been determined (Hill et al. 1996), the function of the amino acid at position 30 is not known, and it remains to be determined why this site is under such strong selection pressure.
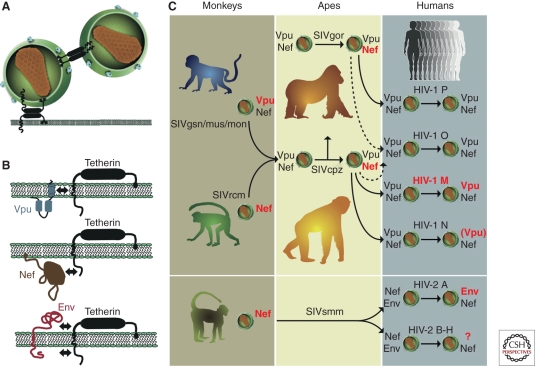
The potential of an SIV to infect a new primate species is also influenced by its ability to counteract different host restriction factors. Three classes of restriction factors have been shown to constitute barriers to SIV cross-species transmission. These include (1) APOBEC3G (apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G), which interferes with reverse transcription (Sheehy et al. 2002); (2) TRIM5α (tripartite motif 5α protein), which interferes with viral uncoating (Stremlau et al. 2004); and (3) tetherin (also termed BST-2 and CD317) which inhibits the budding and release of virions from infected cells (Neil et al. 2008). Of these, tetherin appears to have had the greatest impact on the precursors of HIV-1 and HIV-2 (Fig. 6). Tetherin is comprised of a cytoplasmic amino-terminal region, a trans-membrane domain, a coiled-coiled extracellular domain, and a carboxy-terminal glycosylphosphatidylinositol (GPI) anchor (Fig. 6B). Recent studies have shown that most SIVs use their Nef protein to remove tetherin from the cell surface by targeting its cytoplasmic domain (Jia et al. 2009; Zhang et al. 2009). In contrast, HIV-1 (Neil et al. 2008; Van Damme et al. 2008) as well as SIVs from greater spot-nosed, mona, moustached, and Dent’s monkeys (Sauter et al. 2009; Schmokel et al. 2011) use their Vpu protein to degrade tetherin by binding to its membrane-spanning domain (Iwabu et al. 2009; Rong et al. 2009). Still other viruses use their envelope glycoprotein to interfere with tetherin, by interacting with either its extracellular or its cytoplasmic domain (Bour et al. 1996; Gupta et al. 2009; Le Tortorec and Neil 2009; Serra-Moreno et al. 2011). These various antitetherin responses appear to have emerged as a direct result of host-specific selection pressures following cross-species transmission (Sauter et al. 2009; Evans et al. 2010; Lim et al. 2010).
Figure 6.
Tetherin function and virus specific antagonism in different primate hosts. (A) Mechanism of restricted virion release by tetherin; two alternative models are shown (for details, see Evans et al. 2010); (B) Viral antagonists of tetherin and their sites of interaction (indicated by arrows). Vpu associates with the trans-membrane domain of tetherin, Nef targets the cytoplasmic domain, and Env interacts either with the extracellular or the cytoplasmic domain (Kirchhoff 2010; Serra-Moreno et al. 2011). (C) Antitetherin function in HIV-1 and HIV-2 and their immediate simian precursors. SIVcpz acquired vpu and nef genes from different sources, the SIVgsn/mus/mon and SIVrcm lineages, respectively. During adaptation in chimpanzees, Nef (and not Vpu) evolved to become an effective tetherin antagonist. SIVgor and SIVsmm also use Nef to counteract tetherin. After transmission to humans, SIVcpz, SIVgor, and SIVsmm Nef were unable to antagonize human tetherin because of a deletion in its cytoplasmic domain. HIV-1 group M adapted by regaining Vpu-mediated antitetherin activity. The Nef and Vpu proteins of HIV-1 groups O and P remained poor tetherin antagonists. The Vpu of HIV-1 group N gained modest antitetherin activity, but lost the ability to degrade CD4. HIV-2 group A adapted by gaining Env-mediated antitetherin activity; whether HIV-2 groups B–H gained antitetherin function has not been tested. Proteins that are active against tetherin are highlighted in red, and those that are inactive are shown in gray (adapted from Kirchhoff [2010] and reprinted, with permission, from Elsevier ©2010).
Because of the constant onslaught of viral pathogens, host restriction factors evolve rapidly (Sawyer et al. 2004, 2005; McNatt et al. 2009; Lim et al. 2010). Most notably, the human tetherin gene differs from that of other apes by a five-codon deletion in the region encoding the cytoplasmic domain (Sauter et al. 2009). Because Nef interacts with the cytoplasmic domain of tetherin, this deletion rendered the SIVcpz Nef protein inactive on transmission to humans. Gorilla tetherin does not have this deletion, and thus Nef continued to function as a tetherin antagonist on transmission of SIVcpz to gorillas (Fig. 6C). Thus, to facilitate replication in humans, SIVcpz and SIVgor had to find alternative routes to overcome tetherin. One option was to switch back to using Vpu, as in their monkey ancestors (Fig. 6C). However, this required that the SIVcpz and SIVgor Vpu proteins regained this function because neither has antitetherin activity in chimpanzee or humans cells. Not surprisingly, this was not successful in all instances (Fig. 6C). When representatives of each of the HIV-1 groups were analyzed, only the Vpu proteins of group M viruses showed potent antitetherin activity (Sauter et al. 2009). Group O and P Vpu proteins were completely inactive, whereas the group N Vpu showed only marginal activity (Sauter et al. 2009; Kirchhoff 2010). Moreover, even the latter adaptation came at a cost, because the group N Vpu lost its ability to down-modulate CD4. Thus, of the four transmitted ape viruses, only the precursor of HIV-1 group M succeeded in mounting a full antitetherin defense in human cells. It may thus not be a coincidence that only HIV-1 group M resulted in a global epidemic (Gupta and Towers 2009; Sauter et al. 2009).
Like many other SIVs, SIVsmm does not encode a vpu gene and uses its Nef protein to combat tetherin. Thus, on transmission to humans, SIVsmm had to overcome the same hurdle of a cytoplasmic tail deleted human tetherin. In this case, the envelope glycoprotein (gp41) was recruited as an alternative tetherin antagonist (Le Tortorec and Neil 2009). However, this escape mechanism has thus far only been observed for epidemic HIV-2 group A, and it will be interesting to determine which, if any, of the other HIV-2 groups have acquired a similar antitetherin activity. It seems likely that the cytoplasmic domain deletion in human tetherin has represented a significant barrier to SIV zoonoses, including those viruses that have succeeded in infecting humans.
ORIGIN OF THE AIDS PANDEMIC
HIV-1 evolves around one million times faster than mammalian DNA (Li et al. 1988; Lemey et al. 2006), because the HIV-1 reverse transcriptase is error prone and the viral generation time is short (Ho et al. 1995; Wei et al. 1995). This propensity for rapid genetic change has provided a unique opportunity to gain insight into when and where the AIDS pandemic had its origin. Phylogenetic and statistical analyses have dated the last common ancestor of HIV-1 group M to around 1910 to 1930, with narrow confidence intervals (Korber et al. 2000; Worobey et al. 2008). This indicates that after pandemic HIV-1 first emerged in colonial west central Africa, it spread for some 50 to 70 years before it was recognized. The probable location of the early epidemic has also been identified. Molecular epidemiological studies have indicated that most, if not all, of the early diversification of HIV-1 group M likely occurred in the area around Kinshasa, then called Leopoldville. All of the known HIV-1 group M subtypes were identified there, as well as additional lineages that have remained restricted to this area (Vidal et al. 2000). Leopoldville was also the place where the earliest strains of HIV-1 group M were discovered (Zhu et al. 1998; Worobey et al. 2008). Genetic analysis of infected blood and tissue samples collected from residents of Kinshasa in 1959 and 1960, respectively, revealed that HIV-1 had already diversified into different subtypes by that time (Worobey et al. 2008). Finally, demographic data indicate that pandemic HIV-1 emerged at a time when urban populations in west central Africa were expanding (Worobey et al. 2008). Leopoldville was the largest city in the region at that time and thus a likely destination for a newly emerging infection. Moreover, rivers, which served as major routes of travel and commerce at the time, would have provided a link between the chimpanzee reservoir of HIV-1 group M in southeastern Cameroon and Leopoldville on the banks of the Congo (Sharp and Hahn 2008). Thus, all current evidence points to Leopoldville/Kinshasa as the cradle of the AIDS pandemic.
As HIV-1 group M spread globally, its dissemination involved a number of population bottlenecks—founder events, which led to the predominance of different group M lineages, now called subtypes, in different geographic areas. HIV-1 group M is currently classified into nine subtypes (A–D, F–H, J, K), as well as more than 40 different circulating recombinant forms (CRFs), which were generated when multiple subtypes infected the same population (Taylor et al. 2008). It has been possible to trace the migration pathways of some of these subtypes and CRFs. For example, subtypes A and D originated in central Africa, but ultimately established epidemics in eastern Africa, whereas subtype C was introduced to, and predominates in, southern Africa from where it spread to India and other Asian countries. Subtype B, which accounts for the great majority of HIV-1 infections in Europe and the Americas, arose from a single African strain that appears to have first spread to Haiti in the 1960s and then onward to the US and other western countries (Gilbert et al. 2007). The recombination event that created CRF01 probably occurred in Central Africa, but this viral lineage was first noted in the late 1980s causing a heterosexual epidemic in Thailand, contemporary with subtype B viruses spreading among intravenous drug users (Taylor et al. 2008). CRF01 has gone on to dominate the AIDS epidemic in southeast Asia. Although the initial distribution of these subtypes and CRFs may have been largely caused by chance events, recent studies have suggested that viruses of different subtypes vary in their biological properties, which may influence their epidemiology (Taylor et al. 2008). For example, subtype D has been associated with greater pathogenicity (Kiwanuka et al. 2010) and an increased incidence of cognitive impairment and AIDS dementia (Sacktor et al. 2009). It thus appears that not only the genetic but also the biological diversity of HIV-1 group M subtypes and CRF is increasing.
CONCLUSIONS
Although primate lentiviruses were first identified in the late 1980s, it is only very recently that the complexities of their evolutionary origins, geographic distribution, prevalence, natural history, and pathogenesis in natural and nonnatural hosts have been appreciated. The preceding sections summarize what is known about the origins and evolution of the simian relatives of HIV-1 and HIV-2, their propensity to cross species barriers, and the host factors that govern such transmissions. Given that there are numerous additional primate species that harbor SIV, the question arises what to expect from future zoonoses? Host-specific restriction factors play a major role in preventing cross-species transmission, and they may well be responsible for the fact that only two types of SIV have thus far succeeded in colonizing humans. However, as exemplified by the various HIV-1 and HIV-2 outbreaks, they are certainly not insurmountable. Determining the entire spectrum of host restriction factors and their mechanisms of action will be required to gauge the likelihood of future zoonoses. In this regard, the role of tetherin should be examined further. Because this protein “tethers” virions to the cell surface, a lack of effective antitetherin measures may result in reduced titers of infectious virus in genital secretions. This may explain why the precursors of the rare groups of HIV-1 and HIV-2 were able to infect humans but unable to establish epidemic infections.
From the above, it is also clear that any newly introduced SIV must replicate to some extent to accumulate the necessary mutations that are required to adapt to divergent host proteins and restriction factors. Circumstances that enhance human-to-human passage would thus be expected to increase the chance of such adaptation. It has been suggested that large-scale injection campaigns conducted in west central Africa at the beginning of the twentieth century (Pepin et al. 2006, 2010; Pepin and Labbe 2008), together with the destabilization of social structures (Chitnis et al. 2000), the rapid growth of cities (Worobey et al. 2008), and an increased prevalence in sexually transmitted diseases, including genital ulcers (de Sousa et al. 2010), may have facilitated the early dissemination and adaptation of both HIV-1 and HIV-2. The fact that HIV-1 groups M and O as well as HIV-2 group A all emerged around the same time is consistent with this hypothesis (Korber et al. 2000; Lemey et al. 2003, 2004; Worobey et al. 2008; de Sousa et al. 2010). However, whether these medical interventions and/or social factors really played a role in the emergence of HIV-1 and HIV-2, and more importantly, whether such “jump-starts” were required to spawn the AIDS pandemic, will remain unknown.
Finally, it is important to view the pathogenesis of simian and human AIDS viruses in the context of their evolution (Kirchhoff 2009). One feature of pathogenic HIV-1 and HIV-2 infection that distinguishes them from nonpathogenic SIV infections is a high level of chronic immune activation, which is a strong predictor of disease progression. In HIV-1 infection, this immune activation is fueled, at least in part, by the inability of the Nef protein to down-modulate TCR-CD3, a function that is conserved in most nonpathogenic SIV infections (Schindler et al. 2006; Arhel and Kirchhoff 2009). Lack of this Nef function is associated with increased T-cell activation and apoptosis in vitro and loss of CD4+ T cells in natural SIV infection in vivo (Schindler et al. 2008). A higher state of T- cell activation is associated with enhanced levels of proviral transcription and viral replication, but also with increased expression of interferon-induced restriction factors, such as tetherin. In HIV-1, Vpu compensates for this by providing potent antitetherin activity. It has thus been proposed that to overcome the barriers of cross-species transmission, primate lentiviruses must induce an inflammatory milieu to increase their ability to replicate and accumulate mutations necessary for more adaptation (Kirchhoff 2009). If this were indeed the case, AIDS would be an inevitable consequence of SIV cross-species transmission.
ACKNOWLEDGMENTS
We thank Lilian Pintea for maps of current ape ranges, Frank Kirchhoff for unpublished results, Gerald Learn for phylogenetic tree construction, and Jamie White for artwork and manuscript preparation. This work was supported by grants from the National Institutes of Health (R01 AI50529, R01 AI58715, P30 AI 27767), and the Bristol Myers Freedom to Discover Program.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Aghokeng AF, Bailes E, Loul S, Courgnaud V, Mpoudi-Ngolle E, Sharp PM, Delaporte E, Peeters M 2007. Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages. Virology 360: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghokeng AF, Ayouba A, Mpoudi-Ngole E, Loul S, Liegeois F, Delaporte E, Peeters M 2010. Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect Genet Evol 10: 386–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, Falkenstein S, Bohm R, Koehler J, Traina-Dorge V, et al. 2005. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J Virol 79: 8991–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetrei C, Lerche NW, Pandrea I, Gormus B, Silvestri G, Kaur A, Robertson DL, Hardcastle J, Lackner AA, Marx PA 2006. Kuru experiments triggered the emergence of pathogenic SIVmac. AIDS 20: 317–321 [DOI] [PubMed] [Google Scholar]
- Arhel NJ, Kirchhoff F 2009. Implications of Nef: Host cell interactions in viral persistence and progression to AIDS. Curr Top Microbiol Immunol 339: 147–175 [DOI] [PubMed] [Google Scholar]
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM 2003. Hybrid origin of SIV in chimpanzees. Science 300: 1713. [DOI] [PubMed] [Google Scholar]
- Barouch DH 2008. Challenges in the development of an HIV-1 vaccine. Nature 455: 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, et al. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220: 868–871 [DOI] [PubMed] [Google Scholar]
- Berry N, Jaffar S, Schim van der Loeff M, Ariyoshi K, Harding E, N’Gom PT, Dias F, Wilkins A, Ricard D, Aaby P, et al. 2002. Low level viremia and high CD4% predict normal survival in a cohort of HIV type-2-infected villagers. AIDS Res Hum Retroviruses 18: 1167–1173 [DOI] [PubMed] [Google Scholar]
- Bour S, Schubert U, Peden K, Strebel K 1996. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: A Vpu-like factor? J Virol 70: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC 1981. Kaposi’s sarcoma and Pneumocystis pneumonia among homosexual men—New York City and California. MMWR Morb Mortal Wkly Rep 30: 305–308 [PubMed] [Google Scholar]
- Chakrabarti L, Guyader M, Alizon M, Daniel MD, Desrosiers RC, Tiollais P, Sonigo P 1987. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature 328: 543–547 [DOI] [PubMed] [Google Scholar]
- Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho DD, Marx PA 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: Geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol 70: 3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Luckay A, Sodora DL, Telfer P, Reed P, Gettie A, Kanu JM, Sadek RF, Yee J, Ho DD, et al. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J Virol 71: 3953–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Rawls D, Moore J 2000. Origin of HIV type 1 in colonial French Equatorial Africa? AIDS Res Hum Retroviruses 16: 5–8 [DOI] [PubMed] [Google Scholar]
- Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey MA, Santos-Ferreira MO, Laurent AG, Dauguet C, Katlama C, Rouzioux C, et al. 1986. Isolation of a new human retrovirus from West African patients with AIDS. Science 233: 343–346 [DOI] [PubMed] [Google Scholar]
- Cohen MS, Shaw GM, McMichael AJ, Haynes BF 2011. Acute-HIV-1 Infection: Basic, clinical and public health perspectives. N Engl J Med 364: 1943–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damond F, Descamps D, Farfara I, Telles JN, Puyeo S, Campa P, Lepretre A, Matheron S, Brun-Vezinet F, Simon F 2001. Quantification of proviral load of human immunodeficiency virus type 2 subtypes A and B using real-time PCR. J Clin Microbiol 39: 4264–4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leys R, Vanderborght B, Vanden Haesevelde M, Heyndrickx L, van Geel A, Wauters C, Bernaerts R, Saman E, Nijs P, Willems B, et al. 1990. Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of west-central African origin. J Virol 64: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva TI, Cotten M, Rowland-Jones SL 2008. HIV-2: The forgotten AIDS virus. Trends Microbiol 16: 588–595 [DOI] [PubMed] [Google Scholar]
- de Sousa JD, Muller V, Lemey P, Vandamme AM 2010. High GUD incidence in the early 20th century created a particularly permissive time window for the origin and initial spread of epidemic HIV strains. PLoS One 5: e9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Nerrienet E, Lebreton M, Bibila GT, Foupouapouognigni Y, Rousset D, Nana A, Djoko CF, Tamoufe U, Aghokeng AF, et al. 2011. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DT, Serra-Moreno R, Singh RK, Guatelli JC 2010. BST-2/tetherin: A new component of the innate immune response to enveloped viruses. Trends Microbiol 18: 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PH, Rodrigues A, Douzery EJ 2009. Patterns of macroevolution among primates inferred from a supermatrix of mitochondrial and nuclear DNA. Mol Phylogenet Evol 53: 808–825 [DOI] [PubMed] [Google Scholar]
- Fischer A, Pollack J, Thalmann O, Nickel B, Paabo S 2006. Demographic history and genetic differentiation in apes. Curr Biol 16: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, Ptak RG 2009. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res 37: D417–D422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz PN, Gordon TP, Anderson DC, McClure HM 1990. Prevalence of natural infection with simian immunodeficiency virus and simian T-cell leukemia virus type I in a breeding colony of sooty mangabey monkeys. AIDS 4: 619–625 [DOI] [PubMed] [Google Scholar]
- Gagneux P, Wills C, Gerloff U, Tautz D, Morin PA, Boesch C, Fruth B, Hohmann G, Ryder OA, Woodruff DS 1999. Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc Natl Acad Sci 96: 5077–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224: 500–503 [DOI] [PubMed] [Google Scholar]
- Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, Greene BM, Sharp PM, Shaw GM, Hahn BH 1992. Human infection by genetically diverse SIVsm-related HIV-2 in west Africa. Nature 358: 495–499 [DOI] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397: 436–441 [DOI] [PubMed] [Google Scholar]
- Gifford RJ, Katzourakis A, Tristem M, Pybus OG, Winters M, Shafer RW 2008. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci 105: 20362–20367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M 2007. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci 104: 18566–18570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Maxfield DG, Goodman SM, Feschotte C 2009. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet 5: e1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Locatelli S, Ghobrial L, Mitchell MW, Kujawski JT, Lankester FJ, Stewart CB, Tishkoff SA 2011. Evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proc Natl Acad Sci 108: 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall J 1986. The Chimpanzees of Gombe: Patterns of behavior. Belknap Press, Cambridge, UK [Google Scholar]
- Gray RH, Wawer MJ, Brookmeyer R, Sewankambo NK, Serwadda D, Wabwire-Mangen F, Lutalo T, Li X, vanCott T, Quinn TC 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357: 1149–1153 [DOI] [PubMed] [Google Scholar]
- Greene WC 2007. A history of AIDS: Looking back to see ahead. Eur J Immunol 37 Suppl. 1: S94–S102 [DOI] [PubMed] [Google Scholar]
- Groves C 2001. Primate taxonomy. Smithsonian Institution Press, Washington, DC [Google Scholar]
- Guindon S, Gascuel O 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Gupta RK, Towers GJ 2009. A tail of Tetherin: How pandemic HIV-1 conquered the world. Cell Host Microbe 6: 393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, Towers GJ 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci 106: 20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler LG, Hauser PH, Eberle J, von Brunn A, Knapp S, Zekeng L, Tsague JM, Kaptue L 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol 68: 1581–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyader M, Emerman M, Sonigo P, Clavel F, Montagnier L, Alizon M 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326: 662–669 [DOI] [PubMed] [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM 2000. AIDS as a zoonosis: Scientific and public health implications. Science 287: 607–614 [DOI] [PubMed] [Google Scholar]
- Hamel DJ, Sankale JL, Eisen G, Meloni ST, Mullins C, Gueye-Ndiaye A, Mboup S, Kanki PJ 2007. Twenty years of prospective molecular epidemiology in Senegal: Changes in HIV diversity. AIDS Res Hum Retroviruses 23: 1189–1196 [DOI] [PubMed] [Google Scholar]
- Heeney JL, Rutjens E, Verschoor EJ, Niphuis H, ten Haaft P, Rouse S, McClure H, Balla-Jhagjhoorsingh S, Bogers W, Salas M, et al. 2006. Transmission of simian immunodeficiency virus SIVcpz and the evolution of infection in the presence and absence of concurrent human immunodeficiency virus type 1 infection in chimpanzees. J Virol 80: 7208–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc Natl Acad Sci 93: 3099–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339: 389–392 [DOI] [PubMed] [Google Scholar]
- Hladik F, McElrath MJ 2008. Setting the stage: Host invasion by HIV. Nat Rev Immunol 8: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373: 123–126 [DOI] [PubMed] [Google Scholar]
- Holmes EC 2003. Molecular clocks and the puzzle of RNA virus origins. J Virol 77: 3893–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345: 356–359 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Janssens W, Banor JS, Shinno T, Piedade J, Sata T, Ampofo WK, Brandful JA, Koyanagi Y, Yamamoto N, et al. 2001. Genetic analysis of HIV type 2 from Ghana and Guinea-Bissau, West Africa. AIDS Res Hum Retroviruses 17: 1661–1663 [DOI] [PubMed] [Google Scholar]
- Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, Tanaka Y, Sata T, Tokunaga K 2009. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J Biol Chem 284: 35060–35072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5: e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MJ, Hui H, Robertson DL, Muller MC, Barre-Sinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH 1994a. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J 13: 2935–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MJ, Rogers J, Phillips-Conroy JE, Allan JS, Desrosiers RC, Shaw GM, Sharp PM, Hahn BH 1994b. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: Evidence for cross-species transmission in the wild. J Virol 68: 8454–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A, Pultrone C, Marzetta F, Ghezzi S, Coradin T, Vicenzi E 2010. Restriction factors of retroviral replication: The example of Tripartite Motif (TRIM) protein 5 α and 22. Amino Acids 39: 1–9 [DOI] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ 2007. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci 104: 6261–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313: 523–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J, et al. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460: 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F 2009. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat Rev Microbiol 7: 467–476 [DOI] [PubMed] [Google Scholar]
- Kirchhoff F 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8: 55–67 [DOI] [PubMed] [Google Scholar]
- Kiwanuka N, Robb M, Laeyendecker O, Kigozi G, Wabwire-Mangen F, Makumbi FE, Nalugoda F, Kagaayi J, Eller M, Eller LA, et al. 2010. HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J Acquir Immune Defic Syndr 54: 180–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Klatt NR, Silvestri G, Hirsch V 2011. Nonpathogenic simian immunodeficiency virus infections. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288: 1789–1796 [DOI] [PubMed] [Google Scholar]
- Leendertz SA, Locatelli S, Boesch C, Kucherer C, Formenty P, Liegeois F, Ayouba A, Peeters M, Leendertz FH 2011. No evidence for transmission of SIVwrc from western red colobus monkeys (Piliocolobus badius badius) to wild West African chimpanzees (Pan troglodytes verus) despite high exposure through hunting. BMC Microbiol 11: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Pybus OG, Wang B, Saksena NK, Salemi M, Vandamme AM 2003. Tracing the origin and history of the HIV-2 epidemic. Proc Natl Acad Sci 100: 6588–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Pybus OG, Rambaut A, Drummond AJ, Robertson DL, Roques P, Worobey M, Vandamme AM 2004. The molecular population genetics of HIV-1 group O. Genetics 167: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Pybus OG 2006. HIV evolutionary dynamics within and among hosts. AIDS Rev 8: 125–140 [PubMed] [Google Scholar]
- Le Tortorec A, Neil SJ 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83: 11966–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H, Tanimura M, Sharp PM 1988. Rates and dates of divergence between AIDS virus nucleotide sequences. Mol Biol Evol 5: 313–330 [DOI] [PubMed] [Google Scholar]
- Li Y, Ndjango J-B, Learn G, Robertson J, Takehisa J, Bibollet-Ruche F, Sharp P, Worobey M, Shaw G, Hahn B 2010. Molecular epidemiology of simian immunodeficiency virus in eastern chimpanzees and gorillas. In 17th Conference on Retroviruses and Opportunistic Infections San Francisco, CA [Google Scholar]
- Lim ES, Malik HS, Emerman M 2010. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J Virol 84: 7124–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M 2008. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe 3: 388–398 [DOI] [PubMed] [Google Scholar]
- Mauclere P, Loussert-Ajaka I, Damond F, Fagot P, Souquieres S, Monny Lobe M, Mbopi Keou FX, Barre-Sinoussi F, Saragosti S, Brun-Vezinet F, et al. 1997. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS 11: 445–453 [DOI] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog 5: e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merson MH, O’Malley J, Serwadda D, Apisuk C 2008. The history and challenge of HIV prevention. Lancet 372: 475–488 [DOI] [PubMed] [Google Scholar]
- Mitani JC, Watts DP 1999. Demographic influences on the hunting behavior of chimpanzees. Am J Phys Anthropol 109: 439–454 [DOI] [PubMed] [Google Scholar]
- Mwaengo DM, Novembre FJ 1998. Molecular cloning and characterization of viruses isolated from chimpanzees with pathogenic human immunodeficiency virus type 1 infections. J Virol 72: 8976–8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel C, Etienne L, Li Y, Takehisa J, Rudicell RS, Bass IN, Moudindo J, Mebenga A, Esteban A, Van Heuverswyn F, et al. 2010. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J Virol 84: 1464–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S, Bieniasz P 2009. Human immunodeficiency virus, restriction factors, and interferon. J Interferon Cytokine Res 29: 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451: 425–430 [DOI] [PubMed] [Google Scholar]
- Ortiz M, Guex N, Patin E, Martin O, Xenarios I, Ciuffi A, Quintana-Murci L, Telenti A 2009. Evolutionary trajectories of primate genes involved in HIV pathogenesis. Mol Biol Evol 26: 2865–2875 [DOI] [PubMed] [Google Scholar]
- Paiardini M, Pandrea I, Apetrei C, Silvestri G 2009. Lessons learned from the natural hosts of HIV-related viruses. Annu Rev Med 60: 485–495 [DOI] [PubMed] [Google Scholar]
- Peeters M, Gueye A, Mboup S, Bibollet-Ruche F, Ekaza E, Mulanga C, Ouedrago R, Gandji R, Mpele P, Dibanga G, et al. 1997. Geographical distribution of HIV-1 group O viruses in Africa. AIDS 11: 493–498 [DOI] [PubMed] [Google Scholar]
- Peeters M, Courgnaud V, Abela B, Auzel P, Pourrut X, Bibollet-Ruche F, Loul S, Liegeois F, Butel C, Koulagna D, et al. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg Infect Dis 8: 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Toure-Kane C, Nkengasong JN 2003. Genetic diversity of HIV in Africa: Impact on diagnosis, treatment, vaccine development and trials. AIDS 17: 2547–2560 [DOI] [PubMed] [Google Scholar]
- Pepin J, Labbe AC 2008. Noble goals, unforeseen consequences: Control of tropical diseases in colonial Central Africa and the iatrogenic transmission of blood-borne viruses. Trop Med Int Health 13: 744–753 [DOI] [PubMed] [Google Scholar]
- Pepin J, Plamondon M, Alves AC, Beaudet M, Labbe AC 2006. Parenteral transmission during excision and treatment of tuberculosis and trypanosomiasis may be responsible for the HIV-2 epidemic in Guinea-Bissau. AIDS 20: 1303–1311 [DOI] [PubMed] [Google Scholar]
- Pepin J, Labbe AC, Mamadou-Yaya F, Mbelesso P, Mbadingai S, Deslandes S, Locas MC, Frost E 2010. Iatrogenic transmission of human T cell lymphotropic virus type 1 and hepatitis C virus through parenteral treatment and chemoprophylaxis of sleeping sickness in colonial Equatorial Africa. Clin Infect Dis 51: 777–784 [DOI] [PubMed] [Google Scholar]
- Phillips-Conroy JE, Jolly CJ, Petros B, Allan JS, Desrosiers RC 1994. Sexual transmission of SIVagm in wild grivet monkeys. J Med Primatol 23: 1–7 [DOI] [PubMed] [Google Scholar]
- Pieniazek D, Ellenberger D, Janini LM, Ramos AC, Nkengasong J, Sassan-Morokro M, Hu DJ, Coulibally IM, Ekpini E, Bandea C, et al. 1999. Predominance of human immunodeficiency virus type 2 subtype B in Abidjan, Ivory Coast. AIDS Res Hum Retroviruses 15: 603–608 [DOI] [PubMed] [Google Scholar]
- Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F 2009. A new human immunodeficiency virus derived from gorillas. Nature Med 15: 871–872 [DOI] [PubMed] [Google Scholar]
- Popovic M, Sarngadharan MG, Read E, Gallo RC 1984. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 224: 497–500 [DOI] [PubMed] [Google Scholar]
- Popper SJ, Sarr AD, Gueye-Ndiaye A, Mboup S, Essex ME, Kanki PJ 2000. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: Low virus production in vivo. J Virol 74: 1554–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince AM, Brotman B, Lee DH, Andrus L, Valinsky J, Marx P 2002. Lack of evidence for HIV type 1-related SIVcpz infection in captive and wild chimpanzees (Pan troglodytes verus) in West Africa. AIDS Res Hum Retroviruses 18: 657–660 [DOI] [PubMed] [Google Scholar]
- Pusey AE, Pintea L, Wilson ML, Kamenya S, Goodall J 2007. The contribution of long-term research at Gombe National Park to chimpanzee conservation. Conserv Biol 21: 623–634 [DOI] [PubMed] [Google Scholar]
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ 2009. The challenge of finding a cure for HIV infection. Science 323: 1304–1307 [DOI] [PubMed] [Google Scholar]
- Rong L, Zhang J, Lu J, Pan Q, Lorgeoux RP, Aloysius C, Guo F, Liu SL, Wainberg MA, Liang C 2009. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J Virol 83: 7536–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland-Jones SL, Whittle HC 2007. Out of Africa: What can we learn from HIV-2 about protective immunity to HIV-1? Nat Immunol 8: 329–331 [DOI] [PubMed] [Google Scholar]
- Rudicell RS, Holland Jones J, Wroblewski EE, Learn GH, Li Y, Robertson JD, Greengrass E, Grossmann F, Kamenya S, Pintea L, et al. 2010. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog 6: e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacktor N, Nakasujja N, Skolasky RL, Rezapour M, Robertson K, Musisi S, Katabira E, Ronald A, Clifford DB, Laeyendecker O, et al. 2009. HIV subtype D is associated with dementia, compared with subtype A, in immunosuppressed individuals at risk of cognitive impairment in Kampala, Uganda. Clin Infect Dis 49: 780–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Rodenburg CM, Kamenya S, Bibollet-Ruche F, Gao F, Bailes E, Meleth S, Soong SJ, Kilby JM, Moldoveanu Z, et al. 2002. SIVcpz in wild chimpanzees. Science 295: 465. [DOI] [PubMed] [Google Scholar]
- Santiago ML, Lukasik M, Kamenya S, Li Y, Bibollet-Ruche F, Bailes E, Muller MN, Emery M, Goldenberg DA, Lwanga JS, et al. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J Virol 77: 7545–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Fruteau C, Noe R, Peeters M, Brookfield JF, et al. 2005. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d’Ivoire: Implications for the origin of epidemic human immunodeficiency virus type 2. J Virol 79: 12515–12527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, et al. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6: 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol 2: E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci 102: 2832–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Munch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Muller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, et al. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Schindler M, Schmokel J, Specht A, Li H, Munch J, Khalid M, Sodora DL, Hahn BH, Silvestri G, Kirchhoff F 2008. Inefficient Nef-mediated downmodulation of CD3 and MHC-I correlates with loss of CD4+ T cells in natural SIV infection. PLoS Pathog 4: e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmokel J, Sauter D, Schindler M, Leendertz FH, Bailes E, Dazza MC, Saragosti S, Bibollet-Ruche F, Peeters M, Hahn BH, et al. 2011. The presence of a vpu gene and the lack of Nef-mediated downmodulation of T cell receptor-CD3 are not always linked in primate lentiviruses. J Virol 85: 742–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R, Jia B, Breed M, Alvarez X, Evans DT 2011. Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH 2008. AIDS: Prehistory of HIV-1. Nature 455: 605–606 [DOI] [PubMed] [Google Scholar]
- Sharp PM, Robertson DL, Gao F, Hahn BH 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8: S27–S42 [Google Scholar]
- Sharp PM, Bailes E, Gao F, Beer BE, Hirsch VM, Hahn BH 2000. Origins and evolution of AIDS viruses: Estimating the time-scale. Biochem Soc Trans 28: 275–282 [DOI] [PubMed] [Google Scholar]
- Sharp PM, Shaw GM, Hahn BH 2005. Simian immunodeficiency virus infection of chimpanzees. J Virol 79: 3891–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418: 646–650 [DOI] [PubMed] [Google Scholar]
- Silvestri G 2005. Naturally SIV-infected sooty mangabeys: Are we closer to understanding why they do not develop AIDS? J Med Primatol 34: 243–252 [DOI] [PubMed] [Google Scholar]
- Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin MC, Saragosti S, Georges-Courbot MC, Barre-Sinoussi F, Brun-Vezinet F 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med 4: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Smith SM, Christian D, de Lame V, Shah U, Austin L, Gautam R, Gautam A, Apetrei C, Marx PA 2008. Isolation of a new HIV-2 group in the US. Retrovirology 5: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto PC, Stein LL, Hurtado-Ziola N, Hedrick SM, Varki A 2010. Relative over-reactivity of human versus chimpanzee lymphocytes: Implications for the human diseases associated with immune activation. J Immunol 184: 4185–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquiere S, Bibollet-Ruche F, Robertson DL, Makuwa M, Apetrei C, Onanga R, Kornfeld C, Plantier JC, Gao F, Abernethy K, et al. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J Virol 75: 7086–7096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427: 848–853 [DOI] [PubMed] [Google Scholar]
- Surbeck M, Hohmann G 2008. Primate hunting by bonobos at LuiKotale, Salonga National Park. Curr Biol 18: R906–R907 [DOI] [PubMed] [Google Scholar]
- Switzer WM, Parekh B, Shanmugam V, Bhullar V, Phillips S, Ely JJ, Heneine W 2005. The epidemiology of simian immunodeficiency virus infection in a large number of wild- and captive-born chimpanzees: Evidence for a recent introduction following chimpanzee divergence. AIDS Res Hum Retroviruses 21: 335–342 [DOI] [PubMed] [Google Scholar]
- Takehisa J, Kraus MH, Decker JM, Li Y, Keele BF, Bibollet-Ruche F, Zammit KP, Weng Z, Santiago ML, Kamenya S, et al. 2007. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J Virol 81: 7463–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, Li Y, Rudicell RS, Learn GH, Neel C, et al. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol 83: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM 2008. The challenge of HIV-1 subtype diversity. N Engl J Med 358: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallari A, Bodelle P, Ngansop C, Makamche F, Ndembi N, Mbanya D, Kaptue L, Gurtler LG, McArthur CP, Devare SG, et al. 2010. Four new HIV-1 group N isolates from Cameroon: Prevalence continues to be low. AIDS Res Hum Retroviruses 26: 109–115 [DOI] [PubMed] [Google Scholar]
- Vallari A, Holzmayer V, Harris B, Yamaguchi J, Ngansop C, Makamche F, Mbanya D, Kaptue L, Ndembi N, Gurtler L, et al. 2011. Confirmation of putative HIV-1 group P in Cameroon. J Virol 85: 1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3: 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Haesevelde MM, Peeters M, Jannes G, Janssens W, van der Groen G, Sharp PM, Saman E 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221: 346–350 [DOI] [PubMed] [Google Scholar]
- van der Loeff MF, Awasana AA, Sarge-Njie R, van der Sande M, Jaye A, Sabally S, Corrah T, McConkey SJ, Whittle HC 2006. Sixteen years of HIV surveillance in a West African research clinic reveals divergent epidemic trends of HIV-1 and HIV-2. Int J Epidemiol 35: 1322–1328 [DOI] [PubMed] [Google Scholar]
- van der Loo W, Abrantes J, Esteves PJ 2009. Sharing of endogenous lentiviral gene fragments among leporid lineages separated for more than 12 million years. J Virol 83: 2386–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, et al. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444: 164. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S, et al. 2007. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368: 155–171 [DOI] [PubMed] [Google Scholar]
- van Rensburg EJ, Engelbrecht S, Mwenda J, Laten JD, Robson BA, Stander T, Chege GK 1998. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: Detection of a SIVagm variant from a chacma baboon. J Gen Virol 79: 1809–1814 [DOI] [PubMed] [Google Scholar]
- Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Sema H, Tshimanga K, Bongo B, Delaporte E 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol 74: 10498–10507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain LV, Bailes E, Bibollet-Ruche F, Decker JM, Keele BF, Van Heuverswyn F, Li Y, Takehisa J, Ngole EM, Shaw GM, et al. 2007. Adaptation of HIV-1 to its human host. Mol Biol Evol 24: 1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373: 117–122 [DOI] [PubMed] [Google Scholar]
- Weiss RA, Heeney JL 2009. Infectious diseases: An ill wind for wild chimps? Nature 460: 470–471 [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M 2009. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol 5: e1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Santiago ML, Keele BF, Ndjango JB, Joy JB, Labama BL, Dhed AB, Rambaut A, Sharp PM, Shaw GM, et al. 2004. Origin of AIDS: Contaminated polio vaccine theory refuted. Nature 428: 820. [DOI] [PubMed] [Google Scholar]
- Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, et al. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455: 661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Telfer P, Souquiere S, Hunter M, Coleman CA, Metzger MJ, Reed P, Makuwa M, Hearn G, Honarvar S, et al. 2010. Island biogeography reveals the deep history of SIV. Science 329: 1487. [DOI] [PubMed] [Google Scholar]
- Ylinen LM, Price AJ, Rasaiyaah J, Hue S, Rose NJ, Marzetta F, James LC, Towers GJ 2010. Conformational adaptation of Asian macaque TRIMCyp directs lineage specific antiviral activity. PLoS Pathog 6: e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6: 54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391: 594–597 [DOI] [PubMed] [Google Scholar]