Abstract
Systemic lupus erythematosus (SLE) is a complex autoimmune disease involving multiple organs. The disease is characterized by the production of pathogenic autoantibodies to DNA and certain nuclear antigens, chronic inflammation, and immune dysregulation. Genetic studies involving SLE patients and mouse models have indicated that multiple lupus susceptible genes contribute to the disease phenotype. Notably, the development of SLE in patients and in certain mouse models exhibits a strong sex bias. In addition, several lines of evidence indicates that activation of interferon-α (IFN-α) signaling in immune cells and alterations in the expression of certain immunomodulatory cytokines contribute to lupus pathogenesis. Studies have implicated factors, such as the X chromosomal gene dosage effect and the sex hormones, in gender bias in SLE. However, the molecular mechanisms remain unclear. Additionally, it remains unclear whether these factors influence the “IFN-signature,” which is associated with SLE. In this regard, a mutually positive regulatory feedback loop between IFNs and estrogen receptor-α (ERα) has been identified in immune cells. Moreover, studies indicate that the expression of certain IFN-inducible p200-family proteins that act as innate immune sensors for cytosolic DNA is differentially regulated by sex hormones. In this review, we discuss how the modulation of the expression of the p200-family proteins in immune cells by sex hormones and IFNs contributes to sex bias in SLE. An improved understanding of the regulation and roles of the p200-family proteins in immune cells is critical to understand lupus pathogenesis as well as response (or the lack of it) to various therapies.
Introduction
Systemic lupus erythematosus (SLE) is a complex prototype autoimmune disease (Lahita 1999; Tsokos and Kammer 2000; Crispín and others 2010). The disease is characterized by the development of pathogenic autoantibodies directed against double-stranded DNA (dsDNA) and certain nuclear antigens. The disease involves multiple organs, including the kidneys (Lahita 1999; Tsokos and Kammer 2000). The disease in patients and certain mouse models exhibits a strong sex bias and develops at a female-to-male ratio of 9:1 (Greenstein 2001; Whitacre 2001; Vidaver 2002; Fish 2008; González and others 2010; Rubtsov and others 2010; Weckerle and Niewold 2011). In addition, most SLE patients also exhibit increased serum levels of type I interferon-α (IFN-α), which correlate well with the disease activity (Crow and Wohlgemuth 2003; Baechler and others 2004; Banchereau and others 2004; Banchereau and Pascual 2006). These observations have prompted studies to determine the role of the X chromosomal genes, sex hormones, and the IFN-inducible genes in sex bias in SLE. Consistent with increased serum levels of IFN-α in SLE patients, peripheral blood mononuclear cells (PBMCs) from the patients exhibit increased expression of the IFN-inducible genes referred as the “IFN-signature” (Crow and Wohlgemuth 2003; Baechler and others 2004; Banchereau and Pascual 2006). Notably, genetic studies involving mouse models of SLE have indicated roles for the IFN-inducible p200-family proteins (encoded by the Ifi200-family genes), such as the p202, in the development of pathogenic autoantibodies and the kidney disease [reviewed by Choubey and others (2008)].
Several chromosomal loci and genes are shown to contribute to the lupus phenotype in patients and mouse models (Kono and Theofilopoulos 2006; Graham and others 2009; Moser and others 2009; Morel 2010). Particularly, the NZB autoimmunity 2 (Nba2) locus (∼96–100 cM; the locus is syntenic to the 1q21-23 region in humans), which is derived from the distal region of the chromosome 1 of the New Zealand Black (NZB) mice, has been shown to be a major genetic contribution from the NZB strain of mice to disease susceptibility in the (NZB×NZW)F1 mice (Vyse and others 1997; Rozzo and others 2001). Moreover, these mice spontaneously develop lupus and exhibit a gender bias in the development of the disease (Kono and Theofilopoulos 2006). The B6.Nba2 congenic (congenic for the Nba2 interval on the C57BL/6 genetic background) female mice produced detectable levels of antinuclear pathogenic autoantibodies beginning ∼7 months of age (Rozzo and others 2001). However, these mice do not develop a kidney disease (Rozzo and others 2001).
Studies indicate that the Nba2 interval contains candidate lupus susceptibility genes, including the Fcgr2b (encoding for the inhibitory FcγRIIB receptor) and the IFN-inducible Ifi202 (encoding for the p202 protein) (Wither and others 2000, 2003; Rozzo and others 2001; Xiu and others 2002; Wandstrat and others 2004). Several independent studies involving the generation of the Apcs-null mice, Balb/c.C1 (77–105 cM) mice, B10.Yaa.Bxs2/3 congenic mice, and a comparison of the Ifi202 expression in the MRLlpr/lpr mice revealed that increased expression of the Ifi202 gene in these mice is strongly associated with the development of lupus-like disease (Choubey and others 2008).
Interestingly, generation of the B6.Nba2 subcongenic lines (B6.Nba2-A, -A'B, -B, and -C) and their characterization with respect to autoantibodies and cytokine production at ∼7 months of age has revealed that the B6.Nba2-A'B mice comprising the Fcgr2b and SLAM/CD2 candidate lupus susceptibility genes develop antinuclear antibodies (ANAs) and exhibit higher (higher than the B6.Nba2 and other subcongenic lines) serum levels of IFN-α (Jørgensen and others 2010). In contrast, the B6.Nba2-C subcongenic mice, which harbor the Ifi200-gene cluster, do not develop ANAs and do not produce type I IFN (Jørgensen and others 2010). Moreover, these female mice (age ∼4 months) express detectable levels of the Aim2, but not p202, protein (Panchanathan and others 2010a). However, the age-matched B6.Nba2 female mice exhibit activation of the IFN signaling and increased levels of p202 protein, but undetectable levels of the Aim2 protein (Panchanathan and others 2010a). These observations are consistent with differential roles for the p202 and Aim2 proteins in the development of autoantibodies in the B6.Nba2 mice. Moreover, these observations raise the possibility that epistatic interactions among the Nba2 interval genes contribute to the Nba2 phenotype.
In this review, we discuss how the modulation of the expression of the p200-family proteins by the signaling pathways that are regulated by the X chromosomal genes, sex hormones, and the IFN signaling contributes to sex bias in the development of SLE.
The X Chromosome Gene Dosage Effect
It has been noted that the frequency of Klinefelter's syndrome (47, XXY; males with an extra copy of the X chromosome) is ∼14-fold higher in men with SLE than in men without SLE (Scofield and others 2008; Sawalha and others 2009). Moreover, the risk of SLE in men with the syndrome is comparable with the risk in normal women (46, XX). In contrast to the Klinefelter's syndrome in men, Turner's syndrome [46, X, del(X)(q13)] in women suggests a lower risk of SLE (Cooney and others 2009). These studies suggest that the X chromosome gene dosage effect contributes to increased SLE susceptibility. Moreover, such an effect could be mediated by abnormal activation of genes (see later) or genetic polymorphisms.
Studies involving SLE patients and mouse models of the disease have provided evidence that the X chromosomal genes influence the development of SLE (Fish 2008). In humans, these genes include the TLR7 (Berghöfer and other 2006; Pisitkun and others 2006; Fairhurst and others 2008; Shen and others 2010), MECP2 (Wade 2001; Sawalha and others 2008), and IRAK1 (Gottipati and others 2008; Jacob and others 2009). Notably, TLR7 and IRAK1 proteins are part of an innate immune response, which is initiated after infections and results in type I IFN-production and induction of the IFN-inducible genes. However, it remains a challenge to determine to what extent the gender bias in SLE is due to the X chromosomal gene dosage effect versus the sex hormonal differences. Given that some of the X chromosomal genes (eg, the Tlr7) are also the transcriptional target for the IFN-mediated induction (Green and others 2009), it is likely that a feedforward loop between these gene products and type I IFNs contributes to increased production of IFN-α and the expression of IFN-inducible genes, such as the Ifi200-family genes. In this regard, currently it is not known whether the proteins that are encoded by the X chromosomal genes directly regulate the expression of the p200-family proteins.
Toll-like receptor 7
Toll-like receptor 7 (TLR7) gene (located at Xp22.2) encodes for TLR7 receptor, which plays a critical role in recognition of pathogens and activation of an innate immune response (Berghöfer and other 2006; Pisitkun and others 2006; Fairhurst and others 2008; Shen and others 2010). The receptor recognizes RNA-containing protein complexes and induces the expression of type I IFN (Fairhurst and others 2008; Shen and others 2010). Notably, in lupus-prone BXSB strain of mice, a region of the X chromosome, which is duplicated, translocates to the Y chromosome (Fairhurst and others 2008). The translocation creates the Y-linked autoimmune accelerator (Yaa) locus. The locus, which contains the Tlr7 gene and other genes, in male mice is associated with hyperresponses to RNA-containing antigens by autoreactive B cells and exacerbation of the kidney disease (Fairhurst and others 2008). Studies indicate that in addition to the Tlr7 gene other genes located within the Yaa locus also contribute to the disease phenotype (Santiago-Raber and others 2008). Interestingly, a functional polymorphism in the 3′-untranslated region of the TLR7 gene is associated with SLE in Chinese and Japanese populations (Cheng and others 2007). As expected, the effect is stronger in males than females. Moreover, a TLR7 ligand is shown to induce higher levels of IFN-α production in female SLE patients than healthy individuals (Berghofer and others 2006). These studies are consistent with the idea that increased expression of TLR7 receptor in immune cells contributes to increased production of IFN-α.
Ineterleukin-1 receptor-associated kinase 1
The IRAK1 gene (located in the Xq28 region) encodes for interleukin-1 (IL-1) receptor-associated kinase 1 protein, a serine/threonine protein kinase (Gottipati and others 2008). The protein is involved in the signaling cascade of the Toll/IL-1 receptor (TIR) family of proteins. The family comprises the IL-1 receptor subfamily, which recognizes the endogenous proinflammatory cytokines IL-1 and IL-18 (Gottipati and others 2008). The members of the other subfamily, the Toll-like receptors (TLR), recognize pathogen-associated molecular patterns (PAMPs). The TIR family proteins contain the cytoplasmic TIR domain. The domain serves as a scaffold for protein–protein interactions. These interactions result in activation of a signaling module involving the MyD88 adaptor protein and IRAK family members. The signaling ultimately activates other signaling pathways and transcription factors, including the nuclear factor kappa B (NF-κB). Activation of NF-κB transcription factor results in expression of inflammatory cytokines. Notably, the IRAK1 protein associates with the transcription factor [interferon regulatory factor 5 (IRF5)] (see later) and activates it (Balkhi and others 2008). The expression of the IRF5 protein is dependent on gender in mice: higher in females than the age and strain-matched males (Shen and others 2010).
Genetic studies have identified single-nucleotide polymorphisms (SNPs) in the IRAK1 gene that is associated with increased risk to develop SLE (Jacob and others 2009). Moreover, studies using the Irak1-deficient mice (the B6.IRAK1) suggested that IRAK1 and the Toll/IL-1 pathway play an essential role in T-cell priming (Jacob and others 2009). Given that the activation of IRAK1 is a part of an innate immune response after detection of PAMPs and it can activate the IRF5 (Balkhi and others 2008), it is likely that the IRAK1 and IRF5 axis contributes to sex bias in SLE through increased expression of IFN-α, expression of IFN-inducible genes, and increased expression of Blimp-1, a master regulator of B-cell differentiation (see later).
MECP2
Methyl-CpG-binding protein 2 (MECP2), a chromatin-associated protein, is encoded by the MECP2 gene (located in the region Xq28) (Wade 2001). Given that DNA methylation-sensitive genes are shown to be overexpressed in SLE patients (Sekigawa and others 2003) and that MECP2 protein suppresses the transcription of the methylation-sensitive genes (Wade 2001), the MECP2 gene is a good candidate for a role in the sex bias in SLE. In this regard, a study has reported on the association of the MECP2 gene polymorphisms with susceptibility to develop SLE in 2 independent cohorts of SLE patients and controls (Sawalha and others 2008).
Methylation of the CpG DNA sequence in the 5′-regulatory region (or promoter region) of a gene suppresses gene expression via several mechanisms, including the inability of transcription factors to bind the methylated sequences (Wade 2001). It is known that the expression of certain methylation-sensitive genes, such as TNFSF7 (encoding for the CD70) and CD40LG (encoding for the CD40 ligand), is increased in peripheral CD4+ T cells from female SLE patients when compared with normal female donors (Ballestar and others 2006). Cells that express increased levels of CD40 ligand overstimulate B cells to produce pathogenic autoantibodies. Moreover, in T cells from patients with an active SLE disease, the expression of the enzyme DNA methyltransferase 1, which maintains DNA methylation, is decreased (Zhao and others 2011). Consequently, hypomethylation of certain methylation-sensitive genes results in increased expression in SLE patients, resulting in defects in immune regulation. Although the expression of the human Ifi200-family genes, such as the IFI16 (Choubey and others 2008) and AIM2 (Choubey and others 2010), has been shown to be regulated by methylation, it remains unknown whether the methylation is dependent on gender and whether defects in the methylation contribute to lupus disease.
Sex Hormones and SLE
Studies have indicated that the female and male sex hormones have profound influence on the immune cells and their functions (Wilder 1998; Whitacre 2001; González and others 2010; Rubtsov and others 2010). Both androgens and estrogens regulate the expression of the Th1 and Th2 cytokines. The female sex hormone 17β-estradiol (E2) has immunomodulatory effects [reviewed by Cohen-Solal and others (2008)]: it alters B-cell development to decrease lymphopoiesis, increases the frequency of marginal zone B cells, diminishes B-cell receptor signaling, and allows for the increased survival of high-affinity DNA-reactive B cells. Further, the severity of SLE disease increases in patients during pregnancy when the levels of progesterone and estrogen are higher (Wilder 1998). Notably, the studies of sex hormone levels in female patients with different autoimmune diseases, including SLE, have revealed that the differences in the levels of sex hormones between patients and age-matched healthy individuals are not significant (Verthelyi and others 2001). Therefore, it is likely that the female sex hormone levels in women play an important role in the development of SLE (and possibly in other autoimmune diseases) and other factors, such as the activation of IFN signaling by the female sex hormones, contribute to the overall female bias.
Gender differences in lupus susceptibility are also observed in (NZB×NZW)F1 lupus-prone mice, a spontaneous mouse model of the disease (Walker and others 1994). Studies indicate that the onset of the disease in these mice is significantly delayed in males compared with females and females have shorter lifespan than males. Moreover, castrated (NZB×NZW)F1 males have earlier onset of lupus symptoms and shorter lifespan than their intact littermates (Roubinian and others 1978; Walker and others 1994). Similarly, ovariectomy of female (NZB×NZW)F1 mice significantly delays the onset of the disease, making it similar to that in untreated males. In addition, treatment of mice with the female sex hormone estrogen (E2) exacerbates disease activity and causes early mortality (Roubinian and others 1978). These observations suggest that sex hormones also influence the pathogenesis of the murine lupus.
Autoimmune glomerulonephritis is one of the most serious complications of SLE (Lahita 1999; Tsokos and Kammer 2000). Notably, the female B6.MRLc1(82–100) congenic (congenic for the Mag locus, an MRL-derived glomerulonephritis susceptibility locus) mice develop more severe autoimmune glomerulonephritis than males (Ichii and others 2009). The locus includes the Fcgr2b and Fcgr3 genes that encode for the inhibitory FcγRIIB receptor and activating FcγRIII receptor, respectively. The interval also includes the genomic region harboring the Ifi200-family genes (∼92–94 cM). Interestingly, the expression ratio between the activating FcγRIII receptor and the FcγRIIB inhibitory receptors in the kidneys of the congenic mice was significantly higher in males that developed the severe kidney disease (Ichii and others 2009). Moreover, the study also concluded that the difference in the pathogenesis of autoimmune glomerulonephritis in these congenic mice was due to the inhibitory roles of the male sex hormones, thus suggesting a protective role for the male sex hormones in the development of a kidney disease.
Interestingly, a comparative gene expression analysis identified candidate genes potentially responsible for gender-dependent differences in lupus susceptibility in the (NZB×NZW)F1 mice (Gubbels and others 2008). The study noted gender differences in the expression of a number of genes, including the Csf3r (higher expression in males than females) and Histh1, Serpinb2, Slc6a4, and Cd22 (expression higher in females). Other genes that were found to be regulated by gender included the Tp53 (encoding for the p53) and Tgfb1 (encoding for the TGF-β1; expression higher in males than females; see Table 1). Moreover, the study concluded that the differences in the expression were due to the transcriptional mechanisms. Further, the development of lupus-like disease in certain strains of female mice, including the (NZB×NZW)F1 mice, is associated with the “IFN-signature” (Lu and others 2007). Therefore, these studies support the idea that sex bias in the development of lupus-like disease in mice is also associated with the activation of type I IFN signaling.
Table 1.
Male Sex Hormone Androgen Regulated Immune-Regulatory Genes in Systemic Lupus Erythematosus
| Gene | Cell types | mRNA/protein | Function | References |
|---|---|---|---|---|
| Trp53 | Splenic cells | mRNA (increase) | Cell survival | Gubbels and others (2008) |
| Tgfb1 | Splenic cells | mRNA (increase) | Immune regulation | Gubbels and others (2008) |
| Ifi202 | Splenic cells | mRNA and protein (decrease) | Cell survival IFN signaling |
Panchanathan and others (2009) |
| Aim2 | Splenic cells | mRNA and protein (increase) | IFN signaling Inflammasome |
Panchanathan and others (2010a) |
| Fcgr2b | Splenic cells | mRNA and protein (increase) | Immune regulation | Panchanathan and others (2011) |
IFN, interferon.
Female sex hormone estrogen and its receptors
The female sex hormone estrogen (E2) functions through activation of 2 nuclear receptors, the estrogen receptor (ER)α and ERβ, the expression of which is detectable in immune cells (see later) (Rubtsov and others 2010; Cunningham and Gilkeson 2011). Studies indicate that genetic variations in the ER genes might influence SLE susceptibility through the expression of cytokines, such as IFN-γ, in SLE patients (Lu and others 2009; Kisiel and others 2011). Notably, the ERα and ERβ receptors differentially regulate the maturation and selection of B cells: engagement of either ERα or β can alter B-cell maturation, but only the engagement of ERα is a trigger for the development of autoimmunity (Hill and others 2011). Accordingly, studies have noted that steady-state levels of the ERα mRNA (Inui and others 2007) and protein (Lin and others 2011) are significantly higher in PBMCs from SLE patients when compared with gender- and age-matched normal individuals. Moreover, studies involving mouse models of lupus disease indicate a prominent role of ERα in the development of lupus-like disease (Bynoté and others 2008; Hill and others 2011). Accordingly, ERα-deficient (NZB×NZW)F1 female mice do not develop glomerulonephritis, thus increasing their survival (Bynoté and others 2008). Moreover, the increase in survival of these mice is associated with decreased production of pathogenic anti-dsDNA and anti-chromatin antibodies. Given that E2 promotes the production of IFN-γ by a variety of immune cells, it is not surprising that ERα-deficient (NZB×NZW)F1 female mice exhibit reduced serum levels of the IFN-γ (Bynoté and others 2008). In this regard, it is important to note that the role of IFN-γ in lupus pathogenesis has been demonstrated in SLE patients (Aringer and Smolen 2004) and certain mouse models of the disease (Balomenos and others 1998). Additionally, the (NZB×NZW)F1 mice that are deficient in the IFN-γ receptor or IFN-γ expression do not develop pathogenic autoantibodies and glomerulonephritis (Haas and others 1998). Further, studies have revealed that treatment of immune cells with 17β-estradiol in vitro or in vivo stimulates the expression of genes that are known to regulate levels of the proinflammatory cytokines and cell survival (Table 2).
Table 2.
Female Sex Hormone Estrogen Regulated Immune-Regulatory Genes in Systemic Lupus Erythematosus
| Gene | Cell types | mRNA/protein | Function | Reference |
|---|---|---|---|---|
| PPP3CC | SLE T cells | mRNA | T-cell signaling | Rider and others (2003) |
| Bcl2 | Splenic B cells | Protein | Cell survival | Bynoe and others (2000) |
| Aicda | Splenic B cells | mRNA | SHM and CSR | Pauklin and others (2009) |
| Ifng | Splenic cells | mRNA | Immune regulation | Bynoté and others (2008) |
| II-17 | Splenic cells | Protein | Immune regulation | Khan and others (2010) |
| Cd22 | Splenic B cells | Protein | Negative regulator of BCR signaling | Grimaldi and others (2002) |
| Ptpn6 | Splenic B cells | Protein | BCR signaling | Grimaldi and others (2002) |
| Ifi202 | Splenic B cells | mRNA and protein | B-cell survival IFN signaling | Panchanathan and others (2009) |
| Irf5 | Splenic B cells | mRNA and protein | Transcription of type I IFN genes | Shen and others (2010) |
BCR, B-cell receptor; SLE, systemic lupus erythematosus.
Using an inducible mouse model (the R4A transgenic mice that expresses a transgene encoding the H chain of anti-DNA antibodies) of the lupus disease in which the administration of the female sex hormone estrogen triggers the development of a lupus-like disease, a study (Venkatesh and others 2011) has noted that estrogen treatment of mice alters the normal tolerance mechanisms: high-affinity DNA-reactive B cells mature and acquire a marginal zone phenotype and the mice produce high titers of anti-DNA antibodies. Additionally, the study also noted that 17β-estradiol administration leads to systemic inflammation with increased production of B-cell activating factor (BAFF), increased type I IFN levels, and the induction of the IFN-inducible genes, including the Ifi202 gene (see later).
Estrogen-inducible IRF5
The IRF5 is a key regulator in innate immune responses, including immune complex-induced signaling, host immune signal transduction, and IFN-signaling pathways (Barnes and others 2002; Kozyrev and Alarcon-Riquelme 2007). The human IRF5 gene is a significant genetic risk factor for lupus susceptibility (Kozyrev and Alarcon-Riquelme 2007). It has been reported that the functional SNPs in the human IRF5 gene result in the increased expression of multiple unique isoforms of the IRF5 mRNA (Kozyrev and Alarcon-Riquelme 2007). As a result, the IRF5 SLE risk haplotype is associated with higher serum levels of IFN-α and IFN-induced expression of genes (Niewold and others 2008). Moreover, the risk haplotype-associated disease phenotype is most prominent in patients who develop autoantibodies (Niewold and others 2008).
The murine Irf5 gene is expressed as a full-length transcript with only a single splice variant that is detected in low levels in the bone marrow (BM) of C57BL/6J mice (Paun and others 2008). In vitro, the murine IRF5 is activated by both TBK1 and MyD88 to form homodimers (Barnes and others 2002; Kozyrev and Alarcon-Riquelme 2007). The homodimers can activate transcription of type I IFN and inflammatory cytokine genes. Ubiquitination of IRF5 is important for nuclear translocation. The nuclear IRF5 transcription factor stimulates the transcription of the Ifna genes and the Blimp-1 gene (Lien and others 2010). The Blimp-1 gene encodes for a master regulator of the B-cell differentiation (Martins and Calame 2008), which results in antibody-producing plasma cells.
We found that levels of the Irf5 mRNA and protein were higher in females than in strain- and age-matched male mice (Shen and others 2010). Moreover, splenic cells from the ERα-deficient mice compared with the wild-type female mice expressed relatively lower levels of Irf5 mRNA. Accordingly, treatment of splenic cells with 17β-estradiol increased the Irf5 mRNA levels. Further, splenic B cells from the female mice had relatively more IRF5 protein in the nuclear fraction than the male mice, indicating gender-dependent activation of the IRF5 protein (Shen and others 2010). These observations, which revealed sex bias in the expression and subcellular localization of the murine IRF5 protein, are consistent with increased serum levels of type I IFNs in females. Given that the IRF5 plays a critical role in type I IFN gene expression (Barnes and others 2002; Kozyrev and Alarcon-Riquelme 2007) and the regulation of B-cell differentiation to plasma cells (which produce the pathogenic autoantibodies) (Martins and Calame 2008), it is likely that increased levels as well as activation of the IRF5 in immune cells in female mice contribute to sex bias in the development of lupus.
Male hormone androgen and its receptor
Male hormone androgen signals through the intracellular androgen receptor (AR) (Olsen and Kovacs 2001), which is a member of the nuclear hormone receptors superfamily. Levels of AR are increased by IFN treatment of certain cell types (Basrawala and others 2006). The ligand-dependent activation and nuclear translocation of the AR is followed by its binding to androgen receptor response elements in the promoter region of the target genes to modulate gene expression either positively or negatively (Lamont and Tindall 2010). Interestingly, the expression of AR mRNA has been reported in enriched populations of CD4+ T lymphocytes, CD8+ T lymphocytes, and macrophages (Rubtsov and others 2010). However, the enriched populations of B lymphocytes are reported to express relatively low levels of AR mRNA. Moreover, the expression of AR protein is detectable in splenic B cells and dendritic cells (DCs) (Panchanathan and Choubey, unpublished data). Further, studies have revealed that treatment of immune cells with the male hormone androgen in vitro or in vivo stimulates the expression of genes that regulate signal transduction pathways and cell survival (Table 1). These genes include the absent in melanoma 2 (Aim2) gene, which encodes for a newly identified innate immune sensor for cytosolic DNA (see later) (Choubey and others 2010). Interestingly, the inherited differences in the sensitivity to the male hormone, which are conferred by variations in the length of the CAG repeat in exon 1 of the AR gene, could be correlated with differences in the humoral autoimmunity in men with SLE (Tessnow and others 2011). Therefore, it is likely that levels of the male sex hormone androgen, the genetic polymorphisms in the AR gene, and AR protein levels (increased by the IFNs) influence the lupus phenotype in men.
IFNs and IFN-Inducible Genes in SLE
The IFNs are multifunctional cytokines (Stark and others 1999). The IFN family includes type I, II, and III IFNs among others. The biological activities of the IFNs include activities such as antiviral, cell growth inhibitory, and immunomodulatory. Most IFN-responsive cells respond to type I IFNs through the cell surface receptor. Binding of type I IFN (IFN-α) to the receptor activates the receptor-associated Janus tyrosine kinases, Jak1 and Tyk2. These protein kinases phosphorylate a tyrosine residue in the latent cytoplasmic transcription factors termed signal transducer and activator of transcription (STATs). These activated STATs then form homodimers or heterodimers and translocate into the nucleus to activate transcription of genes that contain a conserved promoter sequences termed interferon-stimulated response element (ISRE), resulting in the transcriptional activation of the IFN-inducible genes (Stark and others 1999). Notably, a transcription factor complex termed IFN-stimulated gene factor 3, which includes the IRF9 factor and a heterodimer of the STATs (STAT1:STAT2), binds to the ISRE sequence to activate the transcription of the target genes. Depending upon the cell type and the strength of the stimulus, the IFNs also activate the transcription of certain IFN-responsive genes through pathway, which do not involve the classical Jak/STAT pathway. Significantly, the IFN-inducible effector proteins mediate the biological activities of the IFNs (Sen and Lengyel 1992).
SLE patients with an active disease show increased serum levels of IFN-α (Crow and Wohlgemuth 2003; Baechler and others 2004; Banchereau and others 2004, 2006). Infections or sterile injury, which often results in cell death, are thought to be a source of autoantigens for the production of type I IFNs [reviewed by Marshak-Rothstein (2006)]. Expectedly, increased serum levels of IFN-α in SLE patients with active disease are associated with increased expression of the IFN-inducible genes in PBMCs (Crow and Wohlgemuth 2003; Baechler and others 2004).
Studies have provided evidence for the role of type I IFN signaling in the mouse models of lupus disease (Theofilopoulos and others 2005). A study involving comparisons of gene expressions between preautoimmune (NZB×NZW)F1 and MRL/lpr mice indicated that mononuclear cells from (NZB×NZW)F1 female mice express higher levels of IFN-α and IFN-α-inducible genes than the MRL/lpr mice (Lu and others 2007). Notably, lupus-prone B6.Nba2 mice that are deficient in the type I receptor signaling do not develop the disease (Jorgensen and others 2007). In addition, our work revealed that the lupus-prone NZB female mice that are deficient in type I IFN receptor develop reduced symptoms of the disease (Santiago-Raber and others 2003). Although these studies provide evidence for a role for IFN signaling in lupus disease, it remains unclear whether the increased levels of IFNs contribute to sex bias in SLE or not. In this regard, our studies have identified the Esr1 gene (encoding for the ERα) as a transcriptional target for activation by both type I and II IFNs (Panchanathan and others 2010b).
A mutual positive feedback loop between the IFNs and ERa
As noted earlier, the murine Esr1 gene is an IFN-inducible gene (Panchanathan and others 2010b). The IFN treatment of splenic cells and B cells increased steady-state levels of ERα mRNA and protein and the increase in the mRNA levels was primarily due to the transcriptional mechanisms. Moreover, the increase was dependent on the expression of STAT1 protein, which is consistent with the presence of 3 potential DNA-binding consensus sites, the ISREs, in the 5′-regulatory region of the Esr1 gene (Panchanathan and others 2010b). The IFN and E2 signaling cooperated to activate the transcription of both IFN and E2-responsive genes, thus indicating a sex bias in the expression of certain IFN and E2-responsive genes. Accordingly, splenic cells from preautoimmune lupus-prone (NZB×NZW)F1 female mice had relatively higher steady-state levels of mRNAs encoded by the IFN and ERα-responsive genes when compared with the age-matched males. Given that serum levels of the IFN-α are higher in most SLE patients (Crow and Wohlgemuth 2003; Baechler and others 2004), it is not surprising that these patients exhibit increased levels of ERα in their PBMCs (Inui and others 2007; Lin and others 2011). The increased levels of ERα in immune cells and its activation by the female sex hormone is likely to contribute to the expression of certain IFN-inducible genes in a gender-dependent manner (Panchanathan and others 2010b).
The p200-Family Proteins
The IFN-inducible p200-family proteins are structurally and functionally related proteins (Lengyel and others 1995; Johnstone and Trapani 1999; Choubey 2000; Asefa and others 2004; Ludlow and others 2005; Gariglio and others 2011). The family includes the murine (eg, p202a, p202b, p203, p204, p205, and Aim2) and human (eg, IFI16, MNDA, IFIX, and AIM2) proteins. All proteins in the family share either 1 or 2 partially conserved repeats of 200-amino acid residues (200-AA repeat; also referred to as the HIN200 domain) toward the C-terminus (Lengyel and others 1995). Additionally, most proteins also share the pyrin domain (PYD; also referred to as the PAAD or DAPIN domain), a homotypic protein–protein interaction domain, in the N-terminus (Choubey and others 2010). The presence of the 200-AA repeat in the p200-family proteins allows them to bind nucleic acids, whereas the PYD allows them to recruit other proteins that contain a PYD domain (Choubey and others 2010).
Complementary approaches, such as detection of the endogenous levels of the p200-family proteins by indirect immunofluorescence and cell fractionation followed by immunoblotting, have revealed that basal constitutive levels of certain proteins (eg, p203 and MNDA proteins) are primarily detected in the nucleus (Choubey 2000; Choubey and others 2010). The nuclear localization of these p200-family proteins is consistent with the presence of a classical nuclear localization signal (NLS). In contrast, depending upon cell type (or when overexpressed in cultured cells), p202, p204, Aim2, AIM2, and IFI16 proteins are detected in the cytoplasm as well as in the nucleus to various extents. Because both p204 (Choubey and Lengyel 1992) and IFI16 (Briggs and others 2001) proteins contain classical NLS and have been detected only in the nucleus, it is likely that their heterodimerization with other p200-family proteins contributes to their localization in the cytoplasm. Similarly, the detection of the Aim2 and AIM2 proteins in the nucleus (these proteins lack the NLS) of certain cell types could be due to a piggyback transport by other p200-family proteins (or other proteins). Further, induction of proteins by IFN treatment of cells potentiates the nuclear localization of the p202a protein (Choubey and Lengyel 1993; Choubey and others 2003). Consistent with the presence of a mitochondrial targeting sequence in the p202a protein in the N-terminus, a significant fraction of p202a protein is associated with the mitochondria (Choubey and others 2003). Considering these observations, it seems likely that subcellular localization of certain p200-family proteins is dependent on cell type and the IFN treatment of cells appears to potentiate the nuclear localization (Choubey and others 2010).
In addition to the IFN signaling, other signaling pathways also regulate the constitutive and inducible expression of the p200-family proteins (Choubey and others 2008). Interestingly, these signaling pathways also regulate the subcellular localization of some of the p200-family proteins (Choubey and others 2008, 2010). Given that the murine (eg, p202, p204, and Aim2) as well as human (eg, AIM2 and IFI16) p200-family proteins can sense dsDNA (the AIM2 protein in the cytoplasm, whereas IFI16 protein in the cytoplasm and nucleus) and can initiate innate immune responses in certain cell types (Choubey and others 2010; Kerur and others 2011), it is likely that the expression levels of these proteins as well as their subcellular localization play an important role in the regulation of the innate immune responses.
After sensing dsDNA in the cytoplasm, the Aim2 and AIM2 proteins through the pyrin domain interact with an adapter protein apoptosis-associated speck-like protein containing a caspase-activating recruitment domain (ASC) to form an inflammasome (Choubey and others 2010). The AIM2/Aim2-ASC inflammasome activates caspase-1, which processes pro-IL-1β and pro-IL-18 for release and induces cell death by pyroptosis (caspase-1-dependent cell death) in macrophages. Presently, it remains unclear whether the Aim2/AIM2 proteins also activate other known inflammasome effector mechanisms, such as unconventional protein secretion, inhibition of glycolysis, cell survival, and caspase-7 activation (Lamkanfi 2011).
The p202 protein can sense cytosolic DNA (Roberts and others 2009). However, it does not contain the PYD to recruit the adaptor protein ASC to form an inflammasome. Interestingly, the knockdown approach has identified the p202 protein as an inhibitor of cytosolic DNA-induced caspase-1 (and caspase-3) activation (Roberts and others 2009). Moreover, caspase-1 activation in macrophages in response to dsDNA correlated inversely with the levels of the p202 protein in 3 strains of mice (Roberts and others 2009). Thus, it has been proposed that the Aim2 protein promotes, whereas the p202 protein represses, the activation of caspase-1 in response to cytoplasmic DNA (Vilaysane and Muruve 2009). In addition, Aim2-deficient cells express higher levels of p202 protein and type I IFN (IFN-β) when compared with wild-type cells (Panchanathan and others 2010a). Further, overexpression of the p202 protein in RAW264.7 cells induced the production of IFN-β and activated the IFN signaling (Panchanathan and others 2011). These observations suggest that the p202 protein stimulates the expression of type I IFN expression and activates the type I IFN signaling. However, the molecular mechanisms remain unknown.
In contrast to the Aim2/AIM2 proteins, upon sensing cytosolic dsDNA, the cytosolic IFI16 protein was reported to recruit the stimulator of interferon genes protein to stimulate the expression of IFN-β through the activation of the transcriptional activity of IRF3 and NF-κB (Unterholzner and others 2010). Accordingly, the knockdown of the expression of IFI16 (or its mouse ortholog p204 protein) by RNA-mediated interference inhibited activation of the transcription factors (the IRF3 and NF-κB) that were induced by cytosolic DNA. However, it remains unclear which immune cells express the IFI16 and p204 proteins (see later). Moreover, a recent study noted that, during Kaposi sarcoma-associated herpesvirus (KSHV) infection of endothelial cells, nuclear IFI16 protein recruited the adaptor ASC and procaspase-1 to form a functional inflammasome (Kerur and others 2011). This protein complex was initially detected in the nucleus and subsequently in the perinuclear area. These observations raise the possibility that IFI16 protein can sense dsDNA in the cytoplasm and nucleus and can initiate an innate immune response.
Studies indicate that the p200-family proteins inhibit cell proliferation, regulate cell cycle progression, modulate cell survival (apoptosis), and promote differentiation (Choubey 2000; Asefa and others 2004; Ludlow and others 2005; Gariglio and others 2011). The p200-family proteins are thought to mediate the biological activities of IFNs primarily by binding and modulating the activities of other proteins, which include various transcription factors, oncoproteins, and tumor suppressor proteins (Choubey 2000; Asefa and others 2004; Ludlow and others 2005; Gariglio and others 2011). However, it remains unclear how the transcriptional modulatory activities of certain p200-family proteins, such as the p202, p204, and IFI16, contribute to the development of autoimmunity.
Regulation and Roles of the p200-Family Proteins in SLE
p202 proteins
Constitutive and induced levels of the p202 (probably both p202a and p202b proteins) protein, a relatively better characterized member of the p200-protein family, depend on the genetic background of the mouse strain (Rozzo and others 2001; Choubey and Kotzin 2002; Choubey and others 2008). We have noted earlier that certain polymorphisms in the 5′-regulatory region of the Ifi202 gene are associated with reduced basal and induced expression of the gene in immune cells of certain nonlupus-prone strains, including the C57BL/6 (B6) strain, of mice (Choubey and others 2008). In contrast, basal and induced levels of the p202 protein are higher in immune cells isolated from certain lupus-prone strains of mice, such as the NZB, MRLlpr, (NZB×NZW)F, and B6.Nba2 (Choubey and Panchanathan 2008). In a variety of cell types, the basal and induced levels of the p202 protein are regulated by the IFN signaling. Accordingly, the B6.Nba2 mice that are deficient in the expression of a subunit of type I IFN receptor express reduced levels (∼2-fold) of the Ifi202 mRNA (Jφrgensen and others 2007). Notably, the reduced expression of the Ifi202 gene in these mice significantly reduced the disease phenotype (production of autoantibodies) (Jφrgensen and others 2007). However, the deficiency of the type I IFN receptor signaling in the NZB mice did not reduce levels of the p202 protein (Santiago-Raber and others 2003). Given that the IFN signaling potentiates the nuclear localization of the p202 protein in the B6.Nba2 cells (Choubey and others 2003), it is likely that defects in the nuclear localization of the p202 protein affect its ability to negatively regulate the transcriptional activities of the factors, such as E2Fs (Panchanathan and others 2008), p53 (Xin and others 2006), and AP-1 (Min and others 1996), which regulate the expression of genes that encode for apoptosis-regulatory proteins. Further, p202 protein modulates the transcriptional activity of NF-κB, a potent regulator of cell survival, in a cell-type-dependent manner (Min and others 1996; Ma and others 2003; Yamauchi and others 2007). Defects in the expression of proapoptotic and antiapoptotic proteins that are targets of these transcription factors result in accumulation of autoreactive immune cells (Peng 2008).
IL-6 through the activation of STAT3 stimulates the transcription of the Ifi202 gene (Pramanik and others 2004). Interestingly, our study revealed that treatment of orchiectomized (NZB×NZW)F1 male mice with the female sex hormone 17β-estradiol significantly increases steady-state levels of the Ifi202 mRNA in splenic cells (Panchanathan and others 2009). However, treatment of mice with the male hormone dihydrotestosterone (DHT) reduces steady-state levels of Ifi202 mRNA. Accordingly, increased levels of the p202 protein in the B6.Nba2 B cells and decreased levels in T cells are associated with increased expression levels of ERα and AR, respectively (Panchanathan and others 2009). Moreover, the steady-state levels of the Ifi202 mRNA are higher in splenic cells from the C57BL/6, B6.Nba2, NZB, and (NZB×NZW)F1 female mice when compared with the age-matched males. We have also investigated the molecular mechanisms by which estrogen through ERα increases the expression of the Ifi202 gene. We found that ERα through the c-Jun/AP-1 DNA-binding site in the promoter region of the Ifi202 gene stimulates the transcription. Accordingly, the ERα preferentially associates with the regulatory region of the Ifi202 gene in the female B6.Nba2 B cells than in males (Panchanathan and others 2009). Further, Ifi202 mRNA levels are detectable in splenic cells of wild-type (Esr1+/+), but not null (Esr1−/−), (NZB×NZW)F1 female mice. These observations demonstrate that the female and male sex hormones differentially regulate the expression of the Ifi202 gene in immune cells. Currently, it is not known how male hormone androgen through the AR represses the expression of the Ifi202 gene.
As noted earlier, increased expression of p202 protein in B6.Nba2 congenic female mice is associated with defects in apoptosis of B cells and production of pathogenic autoantibodies (Rozzo and others 2001; Choubey and Kotzin 2002). The B6.Nba2-C subcongenic mice, which harbor the NZB-derived Ifi200-gene cluster, do not develop autoantibodies (Jørgensen and others 2010). These observations suggest that epistatic interactions of certain Ifi200-family genes with other Nba2 interval genes, such as the Fc receptor genes and SLAM family genes, contribute to the B6.Nba2 phenotype. Consistent with this prediction, we found that the deficiency of the Aim2 gene in the female mice, which resulted in increased levels of p202, production of type I IFN, and activation of IFN signaling, downregulated the expression of the inhibitor Fcγ receptor (FcγRIIB) (Panchanathan and others 2011).
The FcγRIIB receptor plays a critical role in limiting B-cell and DC activation (Nimmerjahn and Ravetch 2008). Moreover, the receptor dampens the signaling strength of the BCR. Interestingly, the deletion of FcγRIIB receptor in R4A BALB/c mice altered the B-cell repertoire, resulting in an expansion and activation of high-affinity DNA-reactive B cells (Venkatesh and others 2009). Accordingly, the R4A×FcγRIIB−/− mice on the BALB/c mice genetic background spontaneously developed anti-DNA antibodies. Notably, these mice also exhibited an induction of IFN-inducible genes, including the Ifi202 gene, and increases in levels of the B-cell survival factor (BAFF). Given that the expression of Ifi202 gene is dependent on gender (Panchanathan and others 2009), it is not surprising that the expression of the FcγRIIB turned out to be dependent on gender (Table 1).
p204 protein
p204 protein expression is detectable in mature monocytes and macrophage cells (Luan and others 2008) and the expression is induced by treatment of cells with the macrophage colony-stimulating factor (M-CSF) or leukemia inhibitory factor (Dauffy and others 2006). Interestingly, steady-state levels of the p204 mRNA are lower in less mature (CD4+ CD8+ double positive) thymocytes than in more mature (CD4+ or CD8+ single positive) thymocytes (Deftos and others 2000), which may be consistent with the potential role of the p204 protein in lymphocytic differentiation. We have noted that steady-state levels of Ifi204 mRNA are higher in splenic cells isolated from females than the age- and strain-matched males (Table 3). Intriguingly, the expression of the Ifi204 gene is not altered in the lupus-prone B6.Nba2 mice when compared with the parental strains of mice (Rozzo and others 2001). Moreover, it remains unclear whether levels of the p204 protein are regulated at the post-transcriptional level. Given that p204 protein can heterodimerize with other members of the p200-family proteins (Choubey 2000) and the protein can sense cytosolic DNA to initiate an innate immune response (Unterholzner and others 2010), which results in increased production of type I IFN, it is likely that increased levels of the p204 protein in immune cells in female mice contribute to increased production of type I IFNs.
Table 3.
Differential Regulation of the Ifi200-Family Genes by Sex Hormones
| Gene | Androgen | Estrogen | Cell types | References |
|---|---|---|---|---|
| Ifi202 | Decrease | Increase | B cells, BMDMs, DCs | Panchanathan and others (2009) |
| Ifi203 | NT | Increase | B cells, BMDMs | UD |
| Ifi204 | NT | Increase | B cells, BMDMs | UD |
| Aim2 | Increase | Decrease | B cells, BMDMs | UD |
| IFI16 | Increase | Decrease | PrECs | Alimirah and others (2006) |
| NT | Increase | THP-1 cells | UD | |
| AIM2 | Increase | NT | THP-1 cells | UD |
BMDMs, bone marrow-derived macrophage; PrECs, prostate epithelial cells; DCs, dendritic cells; NT, not tested; UD, unpublished data (Panchanathan and Choubey).
Aim2 protein
Murine splenic cells, thioglycolate-elicited macrophages (TEMs), bone marrow-derived macrophages (BMDMs), and dendritic cells (DCs) appear to constitutively express the Aim2 protein and the IFN-β treatment of TEMs increases the levels (Rathinam and others 2010; Jones and others 2010). Moreover, we have noted the expression of Aim2 protein in splenic B and T cells (see later). In this regard, it is interesting to note that the NALP1 cytoplasmic sensor, which also forms an inflammasome, is expressed in splenic B and T cells (Kummer and others 2007).
Sequence analysis of the 5′-regulatory region (2430 bp) of the Aim2 gene (from the C57BL/6J strain) has revealed potential DNA-binding sites for AR, NF-κB, and the IRFs. Currently, it is not clear whether sequence polymorphisms in the 5′-regulatory region of the Aim2 gene contribute to its differential expression between nonlupus-prone (C57BL/6 or B6) and certain lupus-prone strains of mice (Choubey and others 2008). Interestingly, we have noted that splenic cells from male mice (B6 and NZB) compared with age- and strain-matched females express higher levels of the Aim2 mRNA and protein (Panchanathan and others 2010a, 2011). These observations indicate that the female and male sex hormones differentially regulate the expression of the Aim2 gene.
Aim2 protein is not needed for IFN-β production after certain bacterial or viral infections (Jones and others 2010; Rathinam and others 2010). Upon infections with certain intracellular pathogens, immune cells (splenic cells and BMDMs) from the Aim2-deficient mice were defective in the activation of caspase-1, secretion of IL-1β and IL-18, and induction of cell death in vitro. Notably, the Aim2 deficiency in mice increases the IFN-β expression, stimulates the expression of the Ifi202 gene, and potentiates the nuclear localization of the p202 protein (Panchanathan and others 2010a). Accordingly, the increased levels of the p202 protein in immune cells were associated with increases in the expression of IFN-β, STAT1, and IFN-inducible genes. Notably, the expression of Aim2 protein in nonlupus-prone (C57BL/6 and B6.Nba2-C) and lupus-prone (B6.Nba2-ABC) mice was inversely correlated with the p202 protein (Panchanathan and others 2010a). Moreover, ectopic expression of p202 protein in RAW264.7 macrophage cells downregulated the expression of Aim2 protein. Further, Aim2 deficiency in mice was associated with reduced expression of the inhibitory receptor FcγRIIB (Panchanathan and others 2011). Given that increased expression of the p202 protein and reduced expression of the Aim2 and FcγRIIB proteins in certain strains of female mice are associated with increased susceptibility to develop lupus disease (Panchanathan and others 2010a, 2011), these observations are consistent with the idea that defects in the expression of the Aim2 gene in female mice contribute to increased susceptibility to lupus disease.
AIM2 protein
The human AIM2 gene is constitutively expressed in the spleen, small intestine, and peripheral leukocytes (DeYoung and others 1997). Moreover, IFN-γ treatment of human HL-60 cell line (DeYoung and others 1997) or IFN-β treatment of human THP-1 cell line (Fernandes-Alnemri and others 2009) increased the steady-state levels of the AIM2 mRNA. Interestingly, the AIM2 gene is silenced by DNA-methylation (Woerner and others 2007) and reduced levels of the AIM2 mRNA have been noted in PBMCs from SLE patients (Flinn 2007). Moreover, we recently reported that the levels of the AIM2 protein increase in the senescent (versus old) human diploid fibroblasts (HDFs) and the knockdown of type I IFN-receptor subunit-α did not result in appreciable decreases in AIM2 protein levels (Duan and others 2011). This observation suggested that the basal levels of the AIM2 protein in HDFs are not maintained by the IFN signaling. However, the IFN-β treatment of the young HDFs, which activated a DNA-damage response, induced the expression of the AIM2 protein. Moreover, IL-1β treatment of HDFs induced the levels of the AIM2 mRNA (Duan and others 2011). However, it remains unclear how the activation of IFN signaling or IL-1β regulates the expression of the AIM2. Although in vitro treatment of cells with DHT increased the levels of AIM2 protein (Table 1), it remains unknown whether the expression of AIM2 protein in human immune cells is regulated in a gender-dependent manner. Further, it remains unknown whether gender-dependent methylation of the AIM2 gene contributes to its reduced expression.
IL-17 regulates the survival and proliferation of human B cells (Nalbandian and others 2009). Moreover, IL-17 promotes differentiation of B cells into immunoglobulin-secreting plasma cells. Accordingly, sera of SLE patients exhibit higher levels of IL-17 than the sera of healthy people. Further, the increased levels of IL-17 are associated with SLE severity and kidney damage (Turner and others 2010). Interestingly, IL-1β plays an important role in promoting IL-17 production by γδ and CD4+ T cells (Lalor and others 2011). Given that the activation of the Aim2/AIM2 inflammasome increases the production of IL-1β in cultured cells in vitro and in mice (Choubey and others 2010), it is likely that increased production of IL-1β by innate immune cells promotes the production of IL-17 by T cells. Therefore, increased production of IL-17 by T cells could account for a poor renal outcome for male lupus patients (Ichii and others 2009).
IFI16 protein
The IFI16 gene encodes for 3 isoforms (A, B, and C) of IFI16 protein through an alternative splicing of mRNA and the B isoform of the IFI16 protein is the predominant form in most cell types (Johnstone and others 1999; Choubey and others 2008). Expression of IFI16 protein is detectable in a variety of normal human tissues and organs and there are indications that the IFI16 is a phosphoprotein. Treatment of a variety of cells with IFNs (α, β, or γ) upregulates the IFI16 expression (Choubey and others 2008). Additionally, AR activates the transcription of the IFI16 gene in PrECs and cancer cell lines (Alimirah and others 2006). Expression of IFI16 gene is silenced by DNA methylation in immortalized HDFs and certain prostate cancer cell lines (Choubey and others 2008). Studies implicate the role of the IFI16 protein in autoimmunity through activation of NF-κB [reviewed recently by Gugliesi and others (2010)]. Moreover, increased levels of IFI16 mRNA have been reported in PBMCs of SLE patients when compared with normal healthy individuals. Interestingly, a recent study noted a role for IFI16 protein in dsDNA-induced activation of human monocyte-derived DCs as well as primary DCs (Kis-Toth and others 2011). Although in vitro treatment of THP-1 monocytic cell line with 17β-estradiol increases levels of the IFI16 protein (Table 3), it remains unknown whether the expression of IFI16 protein in SLE patients is regulated in a gender-dependent manner.
Conclusions
Studies of sex hormone levels in female SLE patients have revealed no significant differences between the patients and age-matched healthy individuals. Therefore, it is likely that other factors, such as the X chromosomal gene dosage effect, ERα receptor polymorphisms, and a recently identified feedforward loop between the ERα receptor and IFNs, which may involve the IRF5 transcription factor, contribute to the overall sex bias in SLE (Fig. 1). Moreover, these studies support a role for the female sex hormone levels in the initiation of SLE. Further, several lines of evidence involving genetic studies indicate that activation of type I IFN signaling in immune cells contributes to the SLE phenotype. In this regard, it is important to note that the IFN-inducible p200-family proteins, such as p202, Aim2/AIM2, and IFI16, have emerged as significant mediators of the immunomodulatory functions of the IFNs. The findings that (i) sex hormones differentially regulate the expression of the Ifi202 lupus susceptibility gene (Panchanathan and others 2009), (ii) the p202 protein stimulates the expression of IFN-β and activates the IFN signaling (Panchanathan and others 2011), (iii) the Aim2 protein negatively regulates type I IFN expression and the expression of IFN-inducible genes (including the Ifi202 gene) (Panchanathan and others 2010a), and (iv) the male sex hormone androgen stimulates the expression of the Aim2 gene (Panchanathan and Choubey, unpublished data) support the idea that the p200-family proteins, such as the p202 and IFI16, contribute to sex bias in the development of SLE through altering the innate immune responses and increasing the survival of autoreactive immune cells (Fig. 2). Consequently, a complete understanding of the roles of the p200-family proteins in immune functions is critical to improve our understanding of the molecular mechanisms that contribute to sex bias in lupus pathogenesis.
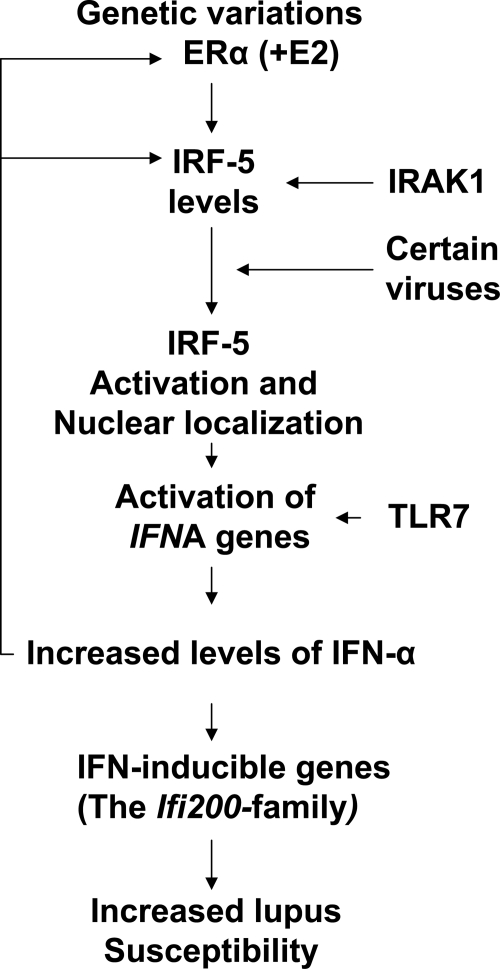
FIG. 1.
A proposed model for sex bias in the development of SLE. The model is based on the demonstrated contribution of the X-chromosomal gene dosage effect, ERα polymorphisms, upregulation of the IRF5 by the female sex hormone estrogen, and a feed-forward loop between the IFNs and ERα, which explains the increased levels of the ERα in peripheral blood mononuclear cells of SLE patients. The model predicts that the female sex hormone levels play a critical role in the initiation of the lupus disease through upregulation of the expression of IFN-α and certain IFN-inducible genes, including the Ifi202 gene. SLE, systemic lupus erythematosus; IFN, interferon; ER, estrogen receptor; IRF5, interferon regulatory factor 5.
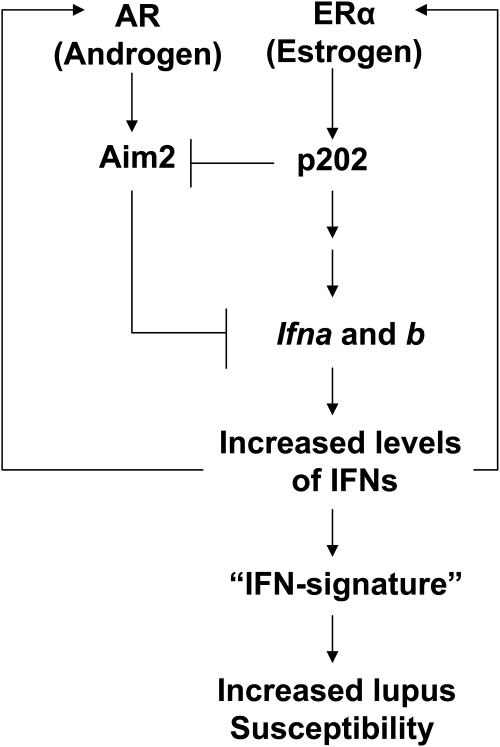
FIG. 2.
A proposed model for the roles of the murine p202 and Aim2 proteins in sex bias in SLE. The model is based on demonstrated (i) upregulation of the p202 expression by the female sex hormone estrogen and downregulation by the male sex hormone androgen (Panchanathan and others 2009), (ii) the ability of the p202 protein to increase the expression of type I IFNs and activate IFN signaling (Panchanathan and others 2011), (iii) increased expression of the Aim2 protein in splenic cells from male mice when compared with age- and strain-matched female mice (Panchanathan and others 2010a), and (iv) induction of IFN-β expression and activation of IFN signaling in Aim2-deficient cells (Panchanathan and others 2010a). The model predicts that the female sex hormone estrogen through the activation of ERα upregulates the expression of the p202 protein. The increased levels of the p202 protein initiate a feedforward loop through upregulation of the expression of IFN-α and certain IFN-inducible genes, resulting in increased survival of autoreactive immune cells in certain strains of female mice.
Acknowledgments
The authors apologize to those investigators in the field whose work was not cited (or was cited through others' review articles) because of space limitations. The relevant research in the authors' laboratory has been supported by the National Institute of Health grants AI066261 and AG025036 and the VA Merit Awards.
Author Disclosure Statement
No competing financial interests exist.
References
- Alimirah F. Chen J. Xin H. Choubey D. Androgen receptor auto-regulates its expression by a negative feedback loop through upregulation of IFI16 protein. FEBS Lett. 2006;580(6):1659–1664. doi: 10.1016/j.febslet.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Aringer M. Smolen JS. Tumour necrosis factor and other proinflammatory cytokines in systemic lupus erythematosus: a rationale for therapeutic intervention. Lupus. 2004;13(5):344–347. doi: 10.1191/0961203303lu1024oa. [DOI] [PubMed] [Google Scholar]
- Asefa B. Klarmann KD. Copeland NG. Gilbert DJ. Jenkins NA. Keller JR. The interferon-inducible p200 family of proteins: a perspective on their role in cell cycle regulation and differentiation. Blood Cells Mol Dis. 2004;32(1):155–167. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Baechler EC. Gregersen PK. Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol. 2004;16(6):801–807. doi: 10.1016/j.coi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Balkhi MY. Fitzgerald KA. Pitha PM. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol Cell Biol. 2008;28(24):7296–7308. doi: 10.1128/MCB.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestar E. Esteller M. Richardson BC. The epigenetic face of systemic lupus erythematosus. J Immunol. 2006;176(12):7143–7147. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- Balomenos D. Rumold R. Theofilopoulos AN. Interferon-gamma is required for lupus-like disease and lymphoaccumulation in MRL-lpr mice. J Clin Invest. 1998;101(2):364–371. doi: 10.1172/JCI750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J. Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25(3):383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Banchereau J. Pascual V. Paluka AK. Autoimmunity through cytokine-induced dendritic cell activation. Immunity. 2004;20(5):539–550. doi: 10.1016/s1074-7613(04)00108-6. [DOI] [PubMed] [Google Scholar]
- Barnes B. Lubyova B. Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res. 2002;22(1):59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- Basrawala Z. Alimirah F. Xin H. Mohideen N. Campbell SC. Flanigan RC. Choubey D. Androgen receptor levels are increased by interferons in human prostate stromal and epithelial cells. Oncogene. 2006;25(19):2812–2817. doi: 10.1038/sj.onc.1209304. [DOI] [PubMed] [Google Scholar]
- Berghöfer B. Frommer T. Haley G. Fink L. Bein G. Hackstein H. TLR7 ligands induce higher IFN-alpha production in females. J Immunol. 2006;177(4):2088–2096. doi: 10.4049/jimmunol.177.4.2088. [DOI] [PubMed] [Google Scholar]
- Briggs LJ. Johnstone RW. Elliot RM. Xiao CY. Dawson M. Trapani JA. Jans DA. Novel properties of the protein kinase CK2-site-regulated nuclear- localization sequence of the interferon-induced nuclear factor IFI 16. Biochem J. 2001;353(Pt 1):69–77. [PMC free article] [PubMed] [Google Scholar]
- Bynoe MS. Grimaldi CM. Diamond B. Estrogen up-regulates Bcl-2 and blocks tolerance induction of naive B cells. Proc Natl Acad Sci USA. 2000;97(6):2703–2708. doi: 10.1073/pnas.040577497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bynoté KK. Hackenberg JM. Korach KS. Lubahn DB. Lane PH. Gould KA. Estrogen receptor-alpha deficiency attenuates autoimmune disease in (NZB x NZW)F1 mice. Genes Immun. 2008;9(3):137–152. doi: 10.1038/sj.gene.6364458. [DOI] [PubMed] [Google Scholar]
- Cheng PL. Eng HL. Chou MH. You HL. Lin TM. Genetic polymorphisms of viral infection-associated Toll-like receptors in Chinese population. Transl Res. 2007;150(5):311–318. doi: 10.1016/j.trsl.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Choubey D. p202: an interferon-inducible negative regulator of cell growth. J Biol Regul Homeost Agents. 2000;14(3):187–192. [PubMed] [Google Scholar]
- Choubey D. Deka R. Ho SM. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci. 2008;13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- Choubey D. Duan X. Dickerson E. Ponomareva L. Panchanathan R. Shen H. Srivastava R. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30(6):371–380. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. Kotzin BL. Interferon-inducible p202 in the susceptibility to systemic lupus. Front Biosci. 2002;7:e252–e262. doi: 10.2741/A921. [DOI] [PubMed] [Google Scholar]
- Choubey D. Lengyel P. Interferon action: nucleolar and nucleoplasmic localization of the interferon-inducible 72-kD protein that is encoded by the Ifi204 gene from the gene 200 cluster. J Cell Biol. 1992;116(6):1333–1341. doi: 10.1083/jcb.116.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. Lengyel P. Interferon action: cytoplasmic and nuclear localization of the interferon-inducible 52-kD protein that is encoded by the Ifi200-gene from the gene 200 cluster. J Interferon Res. 1993;13(1):43–52. doi: 10.1089/jir.1993.13.43. [DOI] [PubMed] [Google Scholar]
- Choubey D. Panchanathan R. Interferon-inducible Ifi200-faily genes in systemic lupus erythematosus. Immunol Lett. 2008;119(1–2):32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. Pramanik R. Xin H. Sub-cellular localization and mechanisms of nucleocytoplasmic distribution of p202, an interferon-inducible candidate for lupus susceptibility. FEBS Lett. 2003;553(3):245–249. doi: 10.1016/s0014-5793(03)01006-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Solal JF. Jeganathan V. Hill L. Kawabata D. Rodriguez-Pinto D. Grimaldi C. Diamond B. Hormonal regulation of B-cell function and systemic lupus erythematosus. Lupus. 2008;17(6):528–532. doi: 10.1177/0961203308089402. [DOI] [PubMed] [Google Scholar]
- Cooney CM. Bruner GR. Aberle T. Namjou-Khales B. Myers LK. Feo L. Li S. D'Souza A. Ramirez A. Harley JB. Scofield RH. 46, X, del(X)(q13) Turner's syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes Immun. 2009;10(5):478–481. doi: 10.1038/gene.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispín JC. Liossis SN. Kis-Toth K. Lieberman LA. Kyttaris VC. Juang YT. Tsokos GC. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends Mol Med. 2010;16(2):47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MK. Wohlgemuth J. Microarray analysis of gene expression in lupus. Arthritis Res Ther. 2003;5(6):279–287. doi: 10.1186/ar1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M. Gilkeson G. Estrogen receptors in immunity and autoimmunity. Clin Rev Allergy Immunol. 2011;40(1):66–73. doi: 10.1007/s12016-010-8203-5. [DOI] [PubMed] [Google Scholar]
- Dauffy J. Mouchiroud G. Bourette RP. The interferon-inducible gene, Ifi204, is transcriptionally activated in response to M-CSF, and its expression favors macrophage differentiation in myeloid progenitor cells. J Leukoc Biol. 2006;79(1):173–183. doi: 10.1189/jlb.0205083. [DOI] [PubMed] [Google Scholar]
- Deftos ML. Huang E. Ojala EW. Forbush KA. Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13(1):73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung KL. Ray ME. Su YA. Anzick SL. Johnstone RW. Trapani JA. Meltzer PS. Trent JM. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15(4):453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- Duan X. Ponomareva L. Veeranki S. Panchanathan R. Dickerson E. Choubey D. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol Cancer Res. 2011;9(5):589–602. doi: 10.1158/1541-7786.MCR-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst AM. Hwang SH. Wang A. Tian XH. Boudreaux C. Zhou XJ. Casco J. Li QZ. Connolly JE. Wakeland EK. Yaa autoimmune phenotypes are conferred by overexpression of TLR7. Eur J Immunol. 2008;38(7):1971–1978. doi: 10.1002/eji.200838138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T. Yu JW. Datta P. Wu J. Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Immunol Rev. 2008;8(9):737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn LJ. Genomic analysis of a human interferon-inducible gene family and systemic lupus erythematosus (a Ph.D. Thesis Abstract) J Interferon Cytokine Res. 2007;27(9):819–834. [Google Scholar]
- Gariglio M. Mondini M. De Andrea M. Landolfo S. The multifaceted interferon-inducible p200 family proteins: from cell biology to human pathology. J Interferon Cytokine Res. 2011;31(1):159–172. doi: 10.1089/jir.2010.0106. [DOI] [PubMed] [Google Scholar]
- González DA. Díaz BB. Rodríguez Pérez Mdel C. Hernández AG. Chico BN. de León AC. Sex hormones and autoimmunity. Immunol Lett. 2010;133(1):6–13. doi: 10.1016/j.imlet.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Gottipati S. Rao NL. Fung-Leung WP. IRAK1: a critical signaling mediator of innate immunity. Cell Signal. 2008;20(2):269–276. doi: 10.1016/j.cellsig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Graham RR. Hom G. Ortmann W. Behrens TW. Review of recent genome-wide association scans in lupus. J Intern Med. 2009;265(6):680–688. doi: 10.1111/j.1365-2796.2009.02096.x. [DOI] [PubMed] [Google Scholar]
- Green NM. Laws A. Kiefer K. Busconi L. Kim YM. Brinkmann MM. Trail EH. Yasuda K. Christensen SR. Shlomchik MJ. Vogel S. Connor JH. Ploegh H. Eilat D. Rifkin IR. van Seventer JM. Marshak-Rothstein A. Murine B cell response to TLR7 ligands depends on an IFN-beta feedback loop. J Immunol. 2009;183(3):1569–1576. doi: 10.4049/jimmunol.0803899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein BD. Lupus: why women? J Womens Health Gend Based Med. 2001;10(3):233–239. doi: 10.1089/152460901300139989. [DOI] [PubMed] [Google Scholar]
- Grimaldi CM. Cleary J. Dagtas AS. Moussai D. Diamond B. Estrogen alters thresholds for B cell apoptosis and activation. J Clin Invest. 2002;109(12):1625–1633. doi: 10.1172/JCI14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MR. Jørgensen TN. Kotzin BL. Identification of candidate genes that influence sex hormone-dependent disease phenotypes in mouse lupus. Genes Immun. 2008;9(1):47–56. doi: 10.1038/sj.gene.6364447. [DOI] [PubMed] [Google Scholar]
- Gugliesi F. De Andrea M. Mondini M. Cappello P. Giovarelli M. Shoenfeld Y. Meroni P. Gariglio M. Landolfo S. The proapoptotic activity of the interferon-inducible gene IFI16 provides new insights into its etiopathogenetic role in autoimmunity. J Autoimmun. 2010;35(2):114–123. doi: 10.1016/j.jaut.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Haas C. Ryffel B. Le Hir M. IFN-gamma receptor deletion prevents autoantibody production and glomerulonephritis in lupus-prone (NZB x NZW)F1 mice. J Immunol. 1998;160(8):3713–3718. [PubMed] [Google Scholar]
- Hill L. Jeganathan V. Chinnasamy P. Grimaldi C. Diamond B. Differential roles of estrogen receptors α and β in control of B-cell maturation and selection. Mol Med. 2011;17(3–4):211–220. doi: 10.2119/molmed.2010.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii O. Konno A. Sasaki N. Endoh D. Hashimoto Y. Kon Y. Onset of autoimmune glomerulo-nephritis derived from the telomeric region of MRL-chromosome 1 is associated with the male sex hormone in mice. Lupus. 2009;18(6):491–500. doi: 10.1177/0961203308098989. [DOI] [PubMed] [Google Scholar]
- Inui A. Ogasawara H. Naito T. Sekigawa I. Takasaki Y. Hayashida Y. Takamori K. Ogawa H. Estrogen receptor expression by peripheral blood mononuclear cells of patients with systemic lupus erythematosus. Clin Rheumatol. 2007;26(10):1675–1678. doi: 10.1007/s10067-007-0568-3. [DOI] [PubMed] [Google Scholar]
- Jacob CO. Zhu J. Armstrong DL. Yan M. Han J. Zhou XJ. Thomas JA. Reiff A. Myones BL. Ojwang JO. Kaufman KM. Klein-Gitelman M. McCurdy D. Wagner-Weiner L. Silverman E. Ziegler J. Kelly JA. Merrill JT. Harley JB. Ramsey-Goldman R. Vila LM. Bae SC. Vyse TJ. Gilkeson GS. Gaffney PM. Moser KL. Langefeld CD. Zidovetzki R. Mohan C. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci USA. 2009;106(15):6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW. Trapani JA. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol Cell Biol. 1999;19(9):5833–5838. doi: 10.1128/mcb.19.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW. Kayagaki N. Broz P. Henry T. Newton K. O'Rourke K. Chan S. Dong J. Qu Y. Roose-Girma M. Dixit VM. Monack DM. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107(21):9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen TN. Alfaro J. Enriquez HL. Jiang C. Loo WM. Atencio S. Bupp MR. Mailloux CM. Metzger T. Flannery S. Rozzo SJ. Kotzin BL. Rosemblatt M. Bono MR. Erickson LD. Development of murine lupus involves the combined genetic contribution of the SLAM and FcgammaR intervals within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;184(2):775–786. doi: 10.4049/jimmunol.0901322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jφrgensen TN. Roper E. Thurman JM. Marrack P. Kotzin BL. Type I interferon signaling is involved in the spontaneous development of lupus-like disease in B6.Nba2 and (B6.Nba2×NZW)F1 mice. Genes Immun. 2007;8(8):653–662. doi: 10.1038/sj.gene.6364430. [DOI] [PubMed] [Google Scholar]
- Kerur N. Veettil MV. Sharma-Walia N. Bottero V. Sadagopan S. Otageri P. Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan D. Dai R. Karpuzoglu E. Ahmed SA. Estrogen increases, whereas IL-27 and IFN-gamma decrease, splenocyte IL-17 production in WT mice. Eur J Immunol. 2010;40(9):2549–2556. doi: 10.1002/eji.201040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiel BM. Kosinska J. Wierzbowska M. Rutkowska-Sak L. Musiej-Nowakowska E. Wudarski M. Olesinska M. Krajewski P. Lacki J. Rell-Bakalarska M. Jagodzinski PP. Tlustochowicz W. Ploski R. Differential association of juvenile and adult systemic lupus erythematosus with genetic variants of oestrogen receptors alpha and beta. Lupus. 2011;20(1):85–89. doi: 10.1177/0961203310381514. [DOI] [PubMed] [Google Scholar]
- Kis-Toth K. Szanto A. Thai TH. Tsokos GC. Cytosolic DNA-activated human dendritic cells are potent activators of the adaptive immune response. J Immunol. 2011;187(3):1222–1234. doi: 10.4049/jimmunol.1100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono DH. Theofilopoulos AN. Genetics of SLE in mice. Springer Semin Immunopathol. 2006;28(2):83–96. doi: 10.1007/s00281-006-0030-7. [DOI] [PubMed] [Google Scholar]
- Kozyrev SV. Alarcon-Riquelme ME. The genetics and biology of Irf5-mediated signaling in lupus. Autoimmunity. 2007;40(8):591–601. doi: 10.1080/08916930701510905. [DOI] [PubMed] [Google Scholar]
- Kozyrev SV. Lewén S. Reddy PM. Pons-Estel B Argentine Collaborative Group; Witte T German Collaborative Group; Junker P. Laustrup H. Gutiérrez C. Suárez A. Francisca González-Escribano M. Martín J Spanish Collaborative Group. Alarcón-Riquelme ME. Structural insertion/deletion variation in IRF5 is associated with a risk haplotype and defines the precise IRF5 isoforms expressed in systemic lupus erythematosus. Arthritis Rheum. 2007;56(4):1234–1241. doi: 10.1002/art.22497. [DOI] [PubMed] [Google Scholar]
- Kummer JA. Broekhuizen R. Everett H. Agostini L. Kuijk L. Martinon F. van Bruggen R. Tschopp J. Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem. 2007;55(5):443–452. doi: 10.1369/jhc.6A7101.2006. [DOI] [PubMed] [Google Scholar]
- Lahita RG. Systemic lupus erythematosus. 3rd. San Diego, CA: Academic Press; 1999. [Google Scholar]
- Lalor SJ. Dungan LS. Sutton CE. Basdeo SA. Fletcher JM. Mills KH. Caspase-1-processed cytokines IL-1{beta} and IL-18 promote IL-17 production by {gamma}{delta} and CD4 T cells that mediate autoimmunity. J Immunol. 2011;186(10):5738–5748. doi: 10.4049/jimmunol.1003597. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M. Emerging inflammasome effector mechanisms. Nat Rev Immunol. 2011;11(3):213–220. doi: 10.1038/nri2936. [DOI] [PubMed] [Google Scholar]
- Lamont KR. Tindall DJ. Androgen regulation of gene expression. Adv Cancer Res. 2010;107:137–162. doi: 10.1016/S0065-230X(10)07005-3. [DOI] [PubMed] [Google Scholar]
- Lengyel P. Choubey D. Li SJ. Datta B. The interferon-activatable gene 200-cluster: From structure toward function. Semin Virol. 1995;6:203–213. [Google Scholar]
- Lien C. Fang CM. Huso D. Livak F. Lu R. Pitha PM. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci USA. 2010;107(10):4664–4668. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HL. Yen JH. Chiou SS. Tsai WC. Ou TT. Wu CC. Liu HW. Estradiol upregulates calcineurin expression via overexpression of estrogen receptor alpha gene in systemic lupus erythematosus. Kaohsiung J Med Sci. 2011;27(4):125–131. doi: 10.1016/j.kjms.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. Shen N. Li ZM. Chen SL. Genomic view of IFN-α response in pre-autoimmune NZB/NZW and MRL/lpr mice. Genes Immun. 2007;8(7):590–603. doi: 10.1038/sj.gene.6364421. [DOI] [PubMed] [Google Scholar]
- Lu ZM. Wang ZE. Liu YQ. Wu CX. Wang CY. Zhang BC. Shao S. Jiao YL. Che ZX. Chen ZJ. Zhao YR. Association of estrogen receptor alpha gene polymorphisms with cytokine genes expression in systemic lupus erythematosus. Croat Med J. 2009;50(2):117–123. doi: 10.3325/cmj.2009.50.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y. Lengyel P. Liu CJ. p204, a p200 family protein, as a multifunctional regulator of cell proliferation and differentiation. Cytokine Growth Factor Rev. 2008;19(5–6):357–369. doi: 10.1016/j.cytogfr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow LE. Johnstone RW. Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308(1):1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Ma XY. Wang H. Ding B. Zhong H. Ghosh S. Lengyel P. The interferon-inducible p202a protein modulates NF-κB activity by inhibiting the binding to DNA of p50/p65 heterodimers and p65 homodimers while enhancing the binding of p50 homodimers. J Biol Chem. 2003;278(25):23008–23019. doi: 10.1074/jbc.M302105200. [DOI] [PubMed] [Google Scholar]
- Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6(11):823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins G. Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- Min W. Ghosh S. Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-kappa B, c-Fos, and c-Jun activities. Mol Cell Biol. 1996;16(1):359–368. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel L. Genetics of SLE: evidence from mouse models. Nat Rev Rheumatol. 2010;6(6):348–357. doi: 10.1038/nrrheum.2010.63. [DOI] [PubMed] [Google Scholar]
- Moser KL. Kelly JA. Lessard CJ. Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10(5):373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian A. Crispín JC. Tsokos GC. Interleukin-17 and systemic lupus erythematosus: current concepts. Clin Exp Immunol. 2009;157(2):209–215. doi: 10.1111/j.1365-2249.2009.03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewold TB. Kelly JA. Flesch MH. Espinoza LR. Harley JB. Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58(8):2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F. Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- Olsen NJ. Kovacs WJ. Effects of androgens on T and B lymphocyte development. Immunol Res. 2001;23(2–3):281–288. doi: 10.1385/IR:23:2-3:281. [DOI] [PubMed] [Google Scholar]
- Panchanathan R. Duan X. Shen H. Rathinam VA. Erickson LD. Fitzgerald KA. Choubey D. Aim2 deficiency stimulates the expression of IFN-inducible Ifi202, a lupus susceptibility murine gene within the Nba2 autoimmune susceptibility locus. J Immunol. 2010a;185(12):7385–7393. doi: 10.4049/jimmunol.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Shen H. Bupp MG. Gould KA. Choubey D. Female and male sex hormones differentially regulate expression of Ifi202, an interferon-inducible lupus susceptibility gene within the Nba2 interval. J Immunol. 2009;183(11):7031–7038. doi: 10.4049/jimmunol.0802665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Shen H. Duan X. Rathinam VA. Erickson LD. Fitzgerald KA. Choubey D. Aim2 deficiency in mice suppresses the expression of the inhibitory Fc{gamma} receptor (Fc{gamma}RIIB) through the induction of the IFN-inducible p202, a lupus susceptibility protein. J Immunol. 2011;186(12):6762–6770. doi: 10.4049/jimmunol.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Shen H. Zhang X. Ho SM. Choubey D. Mutually positive regulatory feedback loop between interferons and estrogen receptor-alpha in mice: implications for sex bias in autoimmunity. PLoS One. 2010b;5:e10868. doi: 10.1371/journal.pone.0010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Xin H. Choubey D. Disruption of mutually negative regulatory feedback loop between interferon-inducible p202 protein and the E2F family of transcription factors in lupus-prone mice. J Immunol. 2008;180(9):5927–5934. doi: 10.4049/jimmunol.180.9.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S. Sernández IV. Bachmann G. Ramiro AR. Petersen-Mahrt SK. Estrogen directly activates AID transcription and function. J Exp Med. 2009;206(1):99–111. doi: 10.1084/jem.20080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun A. Reinert JT. Jiang Z. Medin C. Balkhi MY. Fitzgerald KA. Pitha PM. Functional characterization of murine interferon regulatory factor 5 (IRF-5) and its role in the innate antiviral response. J Biol Chem. 2008;283(21):14295–14308. doi: 10.1074/jbc.M800501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SL. Transcription factors in autoimmune diseases. Front Biosci. 2008;13:4218–40. doi: 10.2741/3001. [DOI] [PubMed] [Google Scholar]
- Pisitkun P. Deane JA. Difilippantonio MJ. Tarasenko T. Satterthwaite AB. Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312(5780):1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Pramanik R. Jorgensen TN. Xin H. Kotzin BL. Choubey D. Interleukin-6 induces expression of Ifi202, an interferon-inducible candidate gene for lupus susceptibility. J Biol Chem. 2004;279(16):16121–16127. doi: 10.1074/jbc.M313140200. [DOI] [PubMed] [Google Scholar]
- Rathinam VA. Jiang Z. Waggoner SN. Sharma S. Cole LE. Waggoner L. Vanaja SK. Monks BG. Ganesan S. Latz E. Hornung V. Vogel SN. Szomolanyi-Tsuda E. Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11(5):395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider V. Keltner S. Abdou NI. Increased estrogen-dependent expression of calcineurin in female SLE T cells is regulated by multiple mechanisms. J Gend Specif Med. 2003;6(2):14–21. [PubMed] [Google Scholar]
- Roberts TL. Idris A. Dunn JA. Kelly GM. Burnton CM. Hodgson S. Hardy LL. Garceau V. Sweet MJ. Ross IL. Hume DA. Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Roubinian JR. Talal N. Greenspan JS. Goodman JR. Siiteri PK. Effect of castration and sex hormone treatment on survival, anti-nucleic acid antibodies, and glomerulonephritis in NZB/NZW F1 mice. J Exp Med. 1978;147(6):1568–1583. doi: 10.1084/jem.147.6.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozzo SJ. Allard JD. Choubey D. Vyse TJ. Izui S. Peltz G. Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15(3):435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- Rubtsov AV. Rubtsova K. Kappler JW. Marrack P. Genetic and hormonal factors in female-biased autoimmunity. Autoimmun Rev. 2010;9(7):494–498. doi: 10.1016/j.autrev.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Raber ML. Baccala R. Haraldsson KM. Choubey D. Stewart TA. Kono DH. Theofilopoulos AN. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197(6):777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Raber ML. Kikuchi S. Borel P. Uematsu S. Akira S. Kotzin BL. Izui S. Evidence for genes in addition to Tlr7 in the Yaa translocation linked with acceleration of systemic lupus erythematosus. J Immunol. 2008;181(2):1556–1562. doi: 10.4049/jimmunol.181.2.1556. [DOI] [PubMed] [Google Scholar]
- Sawalha AH. Harley JB. Scofield RH. Autoimmunity and Klinefelter's syndrome: when men have two X chromosomes. J Autoimmun. 2009;33(1):31–34. doi: 10.1016/j.jaut.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawalha AH. Webb R. Han S. Kelly JA. Kaufman KM. Kimberly RP. Alarcón-Riquelme ME. James JA. Vyse TJ. Gilkeson GS. Choi CB. Scofield RH. Bae SC. Nath SK. Harley JB. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS One. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield RH. Bruner GR. Namjou B. Kimberly RP. Ramsey-Goldman R. Petri M. Reveille JD. Alarcón GS. Vilá LM. Reid J. Harris B. Li S. Kelly JA. Harley JB. Klinefelter's syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58(8):2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekigawa I. Okada M. Ogasawara H. Kaneko H. Hishikawa T. Hashimoto H. DNA methylation in systemic lupus erythematosus. Lupus. 2003;12(2):79–85. doi: 10.1191/0961203303lu321oa. [DOI] [PubMed] [Google Scholar]
- Sen GC. Lengyel P. The interferon system. A bird's eye view of its biochemistry. J Biol Chem. 1992;267(8):5017–5020. [PubMed] [Google Scholar]
- Shen N. Fu Q. Deng Y. Qian X. Zhao J. Kaufman KM. Wu YL. Yu CY. Tang Y. Chen JY. Yang W. Wong M. Kawasaki A. Tsuchiya N. Sumida T. Kawaguchi Y. Howe HS. Mok MY. Bang SY. Liu FL. Chang DM. Takasaki Y. Hashimoto H. Harley JB. Guthridge JM. Grossman JM. Cantor RM. Song YW. Bae SC. Chen S. Hahn BH. Lau YL. Tsao BP. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2010;107(36):15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H. Panchanathan R. Rajavelu P. Duan X. Gould KA. Choubey D. Gender-dependent expression of murine Irf5 gene: implications for sex bias in autoimmunity. J Mol Cell Biol. 2010;2(5):284–290. doi: 10.1093/jmcb/mjq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR. Williams BRG. Silverman RH. Schreiber RD. How cells respond to interferons? Annu Rev Biochem. 1999;67:227–262. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Tessnow AH. Olsen NJ. Kovacs WJ. Expression of humoral autoimmunity is related to androgen receptor CAG repeat length in men with systemic lupus erythematosus. J Clin Immunol. 2011;31(4):567–573. doi: 10.1007/s10875-011-9519-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos AN. Baccala R. Beutler B. Kono DH. Type I interferon (α/β) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Tsokos GC. Kammer GM. Molecular aberrations in human systemic lupus erythematosus. Mol Med Today. 2000;6(11):418–424. doi: 10.1016/s1357-4310(00)01798-6. [DOI] [PubMed] [Google Scholar]
- Turner JE. Paust HJ. Steinmetz OM. Panzer U. The Th17 immune response in renal inflammation. Kidney Int. 2010;77(12):1070–1075. doi: 10.1038/ki.2010.102. [DOI] [PubMed] [Google Scholar]
- Unterholzner L. Keating SE. Baran M. Horan KA. Jensen SB. Sharma S. Sirois CM. Jin T. Latz E. Xiao TS. Fitzgerald KA. Paludan SR. Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh J. Kawabata D. Kim S. Xu X. Chinnasamy P. Paul E. Diamond B. Grimaldi CM. Selective regulation of autoreactive B cells by FcgammaRIIB. J Autoimmun. 2009;32(3–4):149–157. doi: 10.1016/j.jaut.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh J. Yoshifuji H. Kawabata D. Chinnasamy P. Stanevsky A. Grimaldi CM. Cohen-Solal J. Diamond B. Antigen is required for maturation and activation of pathogenic anti-DNA antibodies and systemic inflammation. J Immunol. 2011;186(9):5304–5312. doi: 10.4049/jimmunol.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verthelyi D. Petri M. Ylamus M. Klinman DM. Disassociation of sex hormone levels and cytokine production in SLE patients. Lupus. 2001;10(5):352–358. doi: 10.1191/096120301674365881. [DOI] [PubMed] [Google Scholar]
- Vidaver R. Molecular and clinical evidence of the role of estrogen in lupus. Trends Immunol. 2002;23(5):229–230. doi: 10.1016/s1471-4906(02)02204-4. [DOI] [PubMed] [Google Scholar]
- Vilaysane A. Muruve DA. The innate immune response to DNA. Semin Immunol. 2009;21(4):208–214. doi: 10.1016/j.smim.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Vyse TJ. Rozzo SJ. Drake CG. Izui S. Kotzin BL. Control of multiple autoantibodies linked with a lupus nephritis susceptibility locus in New Zealand black mice. J Immunol. 1997;158(11):5566–5574. [PubMed] [Google Scholar]
- Walker SE. Besch-Williford CL. Keisler DH. Accelerated deaths from systemic lupus erythematosus in NZB x NZW F1 mice treated with the testosterone-blocking drug flutamide. J Lab Clin Med. 1994;124(3):401–407. [PubMed] [Google Scholar]
- Wandstrat AE. Nguyen C. Limaye N. Chan AY. Subramanian S. Tian XH. Yim YS. Pertsemlidis A. Garner HR., Jr Morel L. Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21(6):769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Wade PA. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene. 2001;20(24):3166–3173. doi: 10.1038/sj.onc.1204340. [DOI] [PubMed] [Google Scholar]
- Weckerle CE. Niewold TB. The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin Rev Allergy Immunol. 2011;40(1):42–49. doi: 10.1007/s12016-009-8192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2(9):777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- Wilder RL. Hormones, pregnancy, and autoimmune diseases. Ann NY Acad Sci. 1998;840:45–50. doi: 10.1111/j.1749-6632.1998.tb09547.x. [DOI] [PubMed] [Google Scholar]
- Wither JE. Lajoie G. Heinrichs S. Cai YC. Chang N. Ciofani A. Cheung YH. MacLeod R. Functional dissection of lupus susceptibility loci on the New Zealand black mouse chromosome 1: evidence for independent genetic loci affecting T and B cell activation. J Immunol. 2003;171(4):1697–1706. doi: 10.4049/jimmunol.171.4.1697. [DOI] [PubMed] [Google Scholar]
- Wither JE. Paterson AD. Vukusic B. Genetic dissection of B cell traits in New Zealand black mice. The expanded population of B cells expressing up-regulated co-stimulatory molecules shows linkage to Nba2. Eur J Immunol. 2000;30(2):356–365. doi: 10.1002/1521-4141(200002)30:2<356::AID-IMMU356>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Woerner SM. Kloor M. Schwitalle Y. Youmans H. Doeberitz MK. Gebert J. Dihlmann S. The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Genes Chromosomes Cancer. 2007;46(12):1080–1089. doi: 10.1002/gcc.20493. [DOI] [PubMed] [Google Scholar]
- Xin H. D'Souza S. Jorgensen TN. Vaughan AT. Lengyel P. Kotzin BL. Choubey D. Increased expression of Ifi202, an IFN-activatable gene, in B6.Nba2 lupus susceptible mice inhibits p53-mediated apoptosis. J Immunol. 2006;176(10):5863–5870. doi: 10.4049/jimmunol.176.10.5863. [DOI] [PubMed] [Google Scholar]
- Xiu Y. Nakamura K. Abe M. Li N. Wen XS. Jiang Y. Zhang D. Tsurui H. Matsuoka S. Hamano Y. Fujii H. Ono M. Takai T. Shimokawa T. Ra C. Shirai T. Hirose S. Transcriptional regulation of Fcgr2b gene by polymorphic promoter region and its contribution to humoral immune responses. J Immunol. 2002;169(8):4340–4346. doi: 10.4049/jimmunol.169.8.4340. [DOI] [PubMed] [Google Scholar]
- Yamauchi M. Hashimoto M. Ichiyama K. Yoshida R. Hanada T. Muta T. Komune S. Kobayashi T. Yoshimura A. Ifi202, an IFN-inducible candidate gene for lupus susceptibility in NZB/W F1 mice, is a positive regulator for NF-κB activation in dendritic cells. Int Immunol. 2007;19(8):935–942. doi: 10.1093/intimm/dxm054. [DOI] [PubMed] [Google Scholar]
- Zhao S. Wang Y. Liang Y. Zhao M. Long H. Ding S. Yin H. Lu Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63(5):1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]