Abstract
Several lines of evidence strongly implicate type I interferons (IFN-α and β) and IFN-signaling in the pathogenesis of certain autoimmune inflammatory diseases. Accordingly, genome-wide association studies have identified polymorphisms in the type I IFN-signaling pathways. Other studies also indicate that a feed-forward loop of type I IFN production, which involves sensing of cytoplasmic nucleic acids by sensors, contributes to the development of immunopathology. In addition, a mutually positive regulatory feedback loop between type I IFNs and estrogen receptor-α may contribute to a gender bias, thus resulting in an increased production of type I IFNs and associated immunopathology in women. Increased levels of type I IFNs have numerous immunomodulatory functions for both the innate and adaptive immune responses. Given that the IFN-β also has some anti-inflammatory roles, identifying molecular links among certain genotypes, cytokine profiles, and associated phenotypes in patients with autoimmune inflammatory diseases is likely to improve our understanding of autoimmunity-associated pathogenesis and suboptimal outcomes following standard therapies.
Introduction
Systemic autoimmune diseases, which include systemic sclerosis, rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE), are antigen-driven heterogeneous complex disorders (Lahita 1999; Tsokos and Kammer 2000; Crispín and others 2010). These autoimmune diseases exhibit moderate to strong sex bias in the development: more in women than men (Greenstein 2001; Whitacre 2001; Rubtsov and others 2010; Weckerle and Niewold 2011). Studies indicate that predisposition to the development of systemic autoimmune diseases, in large part, is genetically inherited in humans and in mouse models (Graham and others 2009; Moser and others 2009; Morel 2010). In addition, epigenetic modifications that may arise from exposure of individuals to the environment also contribute to the pathology of autoimmune diseases (Sekigawa and others 2003; Ballestar and others 2006; Zhao and others 2011). Epigenetic modifications, which include CpG-DNA methylation, histone modifications, and microRNAs, influence gene expression and, thus, various cellular functions. In genetically predisposed individuals, the immune system attacks tissues of its own, resulting in inflammation, degeneration, tissue destruction, and organ failure (Lahita 1999). Autoimmune diseases are defined by the tissue that is being targeted by the immune system for destruction. Consequently, autoimmune diseases can be grouped into 2 categories: involving a single organ or multiple organs. For example, type I diabetes is an autoimmune disease that involves a single organ, pancreas: the immune system targets the beta cells. SLE is an example of an autoimmune disease that involves multiple organs: the immune system attacks multiple organs.
Genome-wide association studies involving patients with SLE have identified multiple loci that are associated with the disease susceptibility (Moser and others 2009; Graham and others 2009). Notably, many genetic variations that are linked to SLE (and autoimmunity) may increase the risk of the development of the disease by altering the expression of cytokine and/or cytokine-induced signaling in immune cells (Baechler and others 2004; Banchereau and Pascual 2006; Kariuki and Niewold 2010; Apostolidis and others 2011; Davis and others 2011; Niewold 2011). The altered or deregulated cytokine signaling has potential to decrease the thresholds for both innate and adaptive immune responses in patients (Banchereau and Pascual 2006; Kariuki and Niewold 2010). Given that SLE and certain other autoimmune disease are clinically heterogeneous and the expression of certain cytokines is deregulated, it is likely that a set of cytokine-regulated signaling pathways and genes contribute to differences in disease manifestations among patients.
Patients with autoinflammatory disorder often have relatively higher levels of proinflammatory cytokines [eg, tumor necrosis factor-α, interleukin (IL)-1, and interferon (IFN)-γ], which may result from aberrant activation of innate immune responses (Aringer and Smolen 2004; Apostolidis and others 2011; Astry and others 2011; Davis and others 2011; Niewold 2011). Accordingly, involvement of Toll-like receptors (TLRs) in autoimmune diseases such as SLE has been demonstrated in mouse models (Marshak-Rothstein 2006). In these models, TLR ligands are commonly used as an adjuvant to generate organ-specific autoimmune diseases such as arthritis and encephalitis. Moreover, mice with deficiency of negative regulators for TLR signaling spontaneously develop autoimmune diseases by aberrant production of inflammatory cytokines and type I IFNs (Marshak-Rothstein 2006).
The participation of IFN-γ in autoimmune diseases, such as lupus pathogenesis, has been demonstrated in mouse models (Haas and others 1998; Theofilopoulos and others 2001). Interestingly, the female sex hormone estrogen promotes the IFN-γ production by invariant natural killer (NK) T cells, dendritic cells, and splenocytes (McMurray and others 1997). Consistent with a role for IFN-γ in the development of lupus disease, deletion of the IFN-γ receptor (Haas and others 1998) or depletion of IFN-γ in lupus-prone (NZB×NZW)F1 mice (Lawson and others 2000) prevents autoantibody production and glomerulonephritis. In contrast, studies have shown that IFN-γ can suppress arthritic inflammation in rat models and also contributes to resistance against arthritis (Schurgers and others 2011).
Several lines of evidence involving patients with SLE and certain mouse models strongly suggest that type I IFNs are intimately involved in the pathogenesis of systemic autoimmune diseases (Banchereau and Pascual 2006; Hall and Rosen 2010): (i) IFN immunotherapy in patients is known to induce autoimmunity; (ii) circulating immune complexes, which contain nucleic acids, can initiate type I IFN production and dendritic cell (DC) maturation; (iii) increased IFN-induced gene expression (or “IFN-signature”) in patients with systemic autoimmunity; and (iv) genetic polymorphisms in the IFN pathway genes are associated with an increased risk for the development of systemic autoimmune diseases.
Certain mouse models of lupus also indicate a role for the IFN-signaling in the development of disease (Lu and others 2007). For example, mice that are deficient in type I IFN-signaling do not develop lupus-like disease (Santiago-Raber and others 2003; Jørgensen and others 2007). Additionally, increased levels of IFN-α can induce early lethal lupus in preautoimmune (NZB×NZW)F1 mice. Interestingly, the genetic background of mice appears to play a critical role in lupus susceptibility (Morel 2010). Accordingly, the type I IFN suppresses autoimmunity in the MRL/lpr mice (Hron and Peng 2004). Moreover, blockade of type I IFN activity in the B6.Sle2 congenic mice and C57BL/6 control mice increased serum autoantibody levels (Li and others 2005). Notably, a study using a mouse model of lupus disease has demonstrated that increased levels of IFN-α also render mice relatively resistant to therapeutic intervention (Liu and others 2011). These observations support the genetic background-dependent roles for type I IFN in the development of murine lupus disease and its progression.
Gender bias in the development of autoimmune and inflammatory diseases has been known (Greenstein 2001; Whitacre 2001; Rubtsov and others 2010; Weckerle and Niewold 2011). Studies indicate that, in addition to the X-chromosomal gene dosage effect (Fish 2008; Sawalha and others 2009), sex hormones also contribute to sex bias (Bynoté and others 2008; Cohen-Solal and others 2008; Cunningham and Gilkeson 2011). However, sex hormone levels in female patients with different autoimmune diseases, including lupus, revealed that the differences in the levels of sex hormones between patients and the age-matched healthy individuals were not significant (Verthelyi and others 2001); thus, suggesting that the female sex hormone levels play a role in the initiation of SLE (and possibly in other autoimmune diseases) and other factors, such as the activation of type I IFN-signaling by the female sex hormones, contributes to the overall female bias in the development. Accordingly, studies have indicated that the expression of certain IFN-regulating genes such as the murine Irf5 (Shen and others 2010) and IFN-inducible genes (eg, the Ifi200-family genes; see Choubey and others 2011) is regulated by the sex hormones. Moreover, a mutual positive feedback loop between type I IFN and the estrogen receptor (ER)-α may play a role in gender bias in autoimmune diseases (Panchanathan and others 2010b).
Increased levels of type I IFN, IFN-α, are known to up-regulate expression of TLRs and other cytosolic nucleic acids sensors [eg, the AIM2-like receptor (ALR)-family proteins; see below], extend the activated T-cell response, enhance humoral immunity, and promote antigen presentation by antigen presenting cells (Marshak-Rothstein 2006; Baccala and others 2007). When unchecked, these responses can be pathological. Therefore, systemic autoimmune diseases are likely to result from persistent inflammatory responses that initiate a feedback amplification loop of autoreactive responses (Baccala and others 2007, 2009; Bolland and Garcia-Sastre 2009).
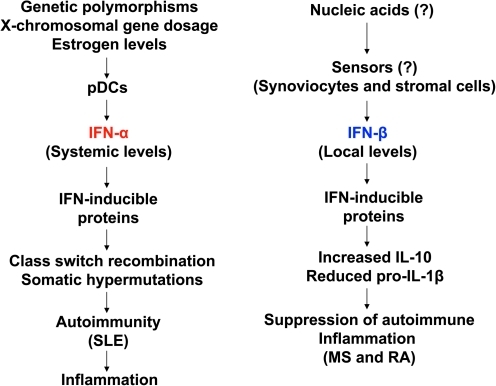
As just noted, a role for type I IFN, IFN-α, in the pathogenesis of SLE is well documented (Fig. 1). However, its role is less clear in certain other autoimmune diseases that are associated with significant inflammation and tissue destruction (Hall and Rosen 2010). Although gene expression analyses of peripheral blood cells from RA and patients with multiple sclerosis (MS) have revealed an “IFN-signature” (van der Pouw Kraan and others 2007, 2008; van Baarsen and others 2006, 2008), which is somewhat similar to the signature seen in patients with lupus (Crow and Wohlgemuth 2003), several lines of evidence support an anti-inflammatory and beneficial role of type I IFN, IFN-β, in patients with RA and in murine arthritis models (van der Pouw Kraan and others 2007, 2008). Similarly, some patients with MS also respond to recombinant IFN-β therapy (van Baarsen and others 2006, 2008). These observations suggest different roles for IFN-α and IFN-β in autoimmune and inflammatory diseases (Fig. 1; see below). In this issue, several articles describe how new insights concerning the regulation and roles of IFNs and other cytokines in autoimmune rheumatic diseases might help identify new approaches to diagnose and treat these diseases.
FIG. 1.
Proposed differential regulation and roles of the IFN-α and IFN-β in autoimmunity and autoimmunity-associated inflammatory diseases. pDCs, plasmacytoid dendritic cells; SLE, systemic lupus erythematosus; MS, multiple sclerosis; RA, rheumatoid arthritis; IFN, interferon.
The IFNs
The IFN family includes 3 distinct classes of IFNs: type I, type II, and type III. These IFNs mediate multiple biological functions (Stark and others 1999; Hall and Rosen 2010). Both Type I IFN (eg, IFN-α, IFN-β, IFN-ω, IFN-ɛ, and IFN-κ) and type III IFN (IFN-λ) are produced by nucleated cells. Both types of IFNs activate essentially same signaling pathways, which result in the transcriptional activation of an overlapping set of genes. Moreover, both types of IFNs are known to mediate the antiviral effects. Interestingly, the receptor for type I IFNs is ubiquitously expressed, whereas the type III IFN receptor appears to have a limited distribution [mainly endothelial cells and plasmacytoid dendritic cells (pDCs)] (Hall and Rosen 2010). Type II IFN (IFN-γ) is produced by NK cells, NK T cells, and T-cell populations. The IFN-γ signals through the IFN-γ receptor, which activates the signaling pathways that are shared with type I and III IFNs (Stark and others 1999; Hall and Rosen 2010). The primary role of the IFN-γ appears to depend on the immune context: regulation of the development and activity of Th17 cells, neutrophil chemotaxis, and enhancing the activity of Treg cells (Saha and others 2010). All 3 classes of IFNs seem to have distinct roles in the development of autoimmune diseases.
Induction of Type I IFN Expression
Most cell types produce low constitutive levels of type I IFNs (Honda and others 2006; Hall and Rosen 2010). However, the production of type I IFNs is greatly induced as a part of an innate immune response that is initiated after infections of cells. Cells that participate in initiating an effective innate immune response after sensing an infection express a number of germ-line pattern recognition receptors (PRRs) (Kawasaki and others 2011). These receptors recognize a wide array of highly conserved pathogen-associated molecular patterns (PAMPs) that are not usually present in the host cell. The PRRs recognize specific viral and bacterial-derived components (eg, nucleic acids), which have specific patterns, and initiate a response that results in the production of type I IFNs (Marshak-Rothstein 2006; Vilaysane and Muruve 2009; Kawasaki and others 2011). Based on the expression pattern and the subcellular localization, 2 types of PRR have been reported: (i) ubiquitously expressed cytoplasmic nucleic acid sensing receptors; and (ii) membrane-bound TLRs. The expression of the TLRs is relatively limited as compared with the cytoplasmic receptors (Kawasaki and others 2011).
Cytoplasmic Nucleic Acid Sensors
Ubiquitously expressed group of cytoplasmic receptors, which recognize and bind unique viral RNA structures, include the retinoic-acid-inducible gene I (RIG-I)-like RNA helicases (Vilaysane and Muruve 2009; Kawasaki and others 2011). The group includes RIG-I and melanoma differentiation-associated gene 5 (MDA5). RIG-I recognizes 5′-triphosphate single-stranded RNA and short double-stranded RNA (dsRNA), whereas MDA5 recognizes long dsRNA structures. Both proteins interact with downstream signaling proteins to induce the production of type I IFNs (Vilaysane and Muruve 2009; Kawasaki and others 2011).
Cytoplasmic sensors of double-stranded DNA (dsDNA), which recognize bacterial and viral DNA, include DNA-dependent activators of IFN-regulatory factors (DAI), NALP3, and members of the p200 protein family (eg, human AIM2 and IFI16 proteins, murine Aim2 and p204 proteins) (Choubey and others 2010; Barber 2011). However, only DAI, IFI16, and p204 are reported to produce type I IFN on sensing cytosolic dsDNA (Barber 2011). Although their expression is cell type-dependent and lower in the steady state, these cytosolic receptors are induced by type I IFNs (Choubey and others 2010; Barber 2011), which enhances their capacity to recognize a pathogen or host-derived nucleic acids as a danger signal.
Sera of patients with SLE contain immune complexes that are bound to nucleic acids (Marshak-Rothstein 2006; Kavai and Szegedi 2007). These immune complexes are bound by immune cells, and the bound nucleic acids are recognized by endosomal TLRs. This sensing of nucleic acids by TLRs activates downstream signaling, which activates the IFN regulatory factors (IRFs), a family of transcription factors (Marshak-Rothstein 2006; Kavai and Szegedi 2007; Hall and Rosen 2010). These factors, when activated, induce the transcription of IFN-α and other immune response genes. Interestingly, genetic variants in the IRF5 and IRF7 genes have been associated with increased lupus susceptibility and increased serum levels of IFN-α in patients with lupus (Barnes and others 2002; Kozyrev and others 2007a; Kozyrev and Alarcon-Riquelme 2007b; Niewold and others 2008; Salloum and Niewold 2011). Therefore, it is likely that polymorphisms in the IRF genes are gain-of-function variants.
In addition to cytosolic nucleic acids sensors that induce expression of type I IFNs, other sensors on sensing cytosolic nucleic acids form inflammasomes (Vilaysane and Muruve 2009; Barber 2011), which results in the production of proinflammatory cytokines such as IL-1β and IL-18 (see below). Interestingly, studies have noted that type I IFN-signaling is necessary for inflammasome activation in response to cytosolic Listeria monocytogenes (Henry and others 2007; Belhocine and Monack 2011). These studies also showed specific connections between type I IFN-signaling and inflammasome activation.
Feedforward Type I IFN Production Loop
Certain systemic autoimmune diseases and type I IFN production share a common aspect: their ability to self-amplify rapidly (Baccala and others 2007, 2009; Hu and others 2008). For example, the IFN receptors, signal transduction molecules, and the transcription factors that drive type I IFN production are auto-stimulated by type I IFNs. Consequently, induction of type I IFN expression in cells further increases the production of type I IFN by neighboring cells, resulting in a “feed-forward” self-amplifying loop, which creates the potential for amplifying immunopathology in systemic autoimmune diseases (Baccala and others 2007, 2009). Additionally, several antigens that are known targets in systemic autoimmune diseases are highly responsive to IFN-mediated induction, thus, further augmenting antigen-driven pathological responses in these diseases. Interestingly, demonstration of a mutually positive feedback loop between type I IFNs and ERα (Panchanathan and others 2010b) and regulation of certain IFN-signaling proteins by the female sex hormone estrogen (Panchanathan and others 2009; Shen and others 2010; Choubey and others 2011) make it likely that increased levels of type I IFNs and the estrogen contribute to observed gender bias in certain systemic autoimmune diseases.
Recent reports (Garcia-Romo and others 2011; Lande and others 2011) suggest that circulating pathogenic autoantibodies in patients with SLE activate neutrophil, which, in turn, releases neutrophil extracellular traps (NETs). These NETs contain complexes of neutrophil-derived DNA and antimicrobial peptide, such as LL37 and human neutrophil peptide. These complexes of DNA and peptides are shown to activate pDCs. The activation of pDCs results in large amounts (200–1000 times more) of IFN-α release (Obermoser and Pascual 2010). The increased release of IFN-α exacerbates disease by potentiating a feed-forward loop of type I IFN production.
Inflammasomes
Protein complexes that are termed inflammasomes sense microbial-derived molecules and endogenous danger signals (Martinon and Tschopp 2007). Inflammasome formation activates the cysteine protease caspase-1. The activated caspase-1 promotes maturation and secretion of the pro-inflammatory cytokines, such as IL-1β and IL-18. Additionally, inflammasomes also induce cell death in macrophages (and possibly other cell types) through pyroptosis to eliminate the infectious agents (Henry and Monack 2007). Interestingly, pathogens antagonize inflammasome pathways by producing virulence factors (Lamkanfi and Dixit 2011). Recent studies suggest that inflammasomes regulate innate and adaptive immune responses (Shaw and others 2011). Moreover, immune complexes that contain danger molecules (either pathogen or cell derived), such as nucleic acids, contribute to auto-inflammatory disorders and autoimmune diseases (eg, MS and type I diabetes) (Shaw and others 2011).
The IL-1 Family
The IL-1 family includes eleven members (Gabay and others 2010; Barber 2011). Of these members, IL-1α and IL-1β are the best-characterized pro-inflammatory cytokines. Although these cytokines signal through a common receptor (IL-1RI), it is evident that these 2 cytokines have nonoverlapping roles. In sterile injuries that result in sterile inflammation, both IL-1α and IL-1β mediate tissue damaging inflammatory response (Gabay and others 2010). Moreover, both IL-1α and IL-1β differ with regard to recruitment of myeloid cells at the site of inflammation (Rider and others 2011): IL-1α recruits neutrophils, whereas IL-1β promotes recruitment of macrophages. The IL-1 production is a 2-stage process. Both IL-1α and IL-1β are produced by cells as precursors in response to sensing of PAMPs. PAMPs are sensed by sensors, such as TLR family members or cytosolic nucleotide-binding oligomerization domain (NOD) proteins. These sensors signal through activation of nuclear factor-κB (NF-κB) and mitogen-activated protein kinase pathways, which regulate the IL-1 gene expression (Gabay and others 2010). Increase in the pro-IL-1α and pro-IL-1β expression is also referred to as “priming”. A second signal or stimulus, which could be a second PAMP, is needed to activate the inactive forms of pro-IL-1β (van de Veerdonk and others 2011). Pro-IL-1β remains inactive and requires a proteolytic cleavage to yield an active released molecule, but pro-IL-1α is active at IL-1RI (Barber 2011; van de Veerdonk and others 2011).
Any damage to the cell membrane or cell death could result in the release of pro-IL-1α, which is followed by a pro-inflammatory response (van de Veerdonk and others 2011). In contrast, as just noted, IL-1β is released after proteolytic cleavage of the pro-IL-1β by activated caspase-1 (Barber 2011). The latter is activated by activation of inflammasomes (Barber 2011). The caspase-1 activating inflammasomes include members of the NOD-like receptor (NLR) family (eg, NLRP1, NLRP3, or NLRC4), RIG-1 receptor, and the DNA-sensing cytosolic human AIM2 and murine Aim2 and related proteins such as the human IFI16 and murine p204 (Choubey and others 2010; Unterholzner and others 2010; Barber 2011; Kerur and others 2011). After sensing the cytosolic dsDNA, a conformational change in the AIM2 protein allows homotypic interaction of its pyrin domain (PYD) with that of the adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) (Fernandes-Alnemri and others 2009; Vilaysane and Muruve 2009). The caspase activation and recruitment domain (CARD) of adaptor protein ASC binds to the CARD domain of pro-caspase-1, thus enabling activation of caspase-1 and secretion of IL-1β from cells (Martinon and Tschopp 2007; Vilaysane and Muruve 2009).
The ALR Family of Proteins
The p200-protein family proteins such as murine Aim2 (Roberts and others 2009), p204 (Unterholzner and others 2010), human AIM2 (Fernandes-Alnemri and others 2009; Vilaysane and Muruve 2009), and IFI16 (Unterholzner and others 2010) can sense cytosolic dsDNA to initiate innate immune responses. These proteins have been termed as the ALRs (the ALR proteins) (Unterholzner and others 2010). Studies indicate that human AIM2 protein is dispensable for production of IFN-β after sensing cytosolic DNA (Fernandes-Alnemri and others 2009; Vilaysane and Muruve 2009). However, it is indispensible for the production of IL-1β after sensing viral and bacterial DNA in the cytoplasm (Vilaysane and Muruve 2009; Jones and others 2010; Rathinam and others 2010). IL-1β is produced after activation of the AIM2 inflammasome, a multi-protein complex that activates caspase-1, with the subsequent cleavage of pro- IL-1β and pro-IL-18 and release of mature IL-1b and IL-18 (Choubey and others 2010). AIM2 protein has the HIN-200 DNA-binding domain, which contains 2 consecutive OB-folds. Additionally, the protein has a PYD that interacts with the PYD of an adapter protein, ASC (Choubey and others 2010). Interestingly, activation of AIM2 protein requires dsDNA of at least 44-base pairs in length, and AIM2 binds to dsDNA in a sequence-independent manner (Roberts and others 2009). Consistent with the role of AIM2 protein in production of IL-1β, the Aim2-deficient mice display reduced survival after infection with Francisella tularensis and mouse cytomegalovirus (MCMV) (Rathinam and others 2010). Notably, the Aim2-deficiency in mice increases the production of type I IFNs and increased expression of IFN-inducible genes, including the lupus susceptible Ifi202 gene (encoding for the p202 protein) (Panchanathan and others 2010a). Moreover, increased expression of p202 protein in certain lupus-prone strains of female mice is associated with inactivation of caspase-1 (Roberts and others 2009; Panchanathan and others 2010a). Accordingly, knockdown of p202 expression in bone marrow-derived macrophages increased the activation of caspase-1 and production of IL-1β (Roberts and others 2009). These observations suggest that reduced levels of AIM2 and Aim2 proteins in immune cells contribute to increased production of type I IFNs.
As just noted, much attention has focused on the ability of the p200-family proteins to sense dsDNA and activate the inflammasome complex, which drives proteolytic processing of inflammatory cytokines; however, these proteins also regulate inflammasome-independent functions in the immune system (Johnstone and Trapani 1999; Choubey 2000; Choubey and Kotzin 2002; Gariglio and others 2011). These functions include the regulation of NF-κB activity, cytokine and chemokine production, and type I IFN production (Choubey and Panchanathan 2008; Choubey and others 2011; Gariglio and others 2011).
Anti-Inflammatory Role of IFN-β
Type I IFN (IFN-β) is being used to treat certain autoimmune and inflammatory diseases, such as relapsing-remitting MS (Billiau 2006), familiar Mediterranean fever (Tweezer-Zaks and others 2008), and Behcet's syndrome (Kotter and others 2004). However, 30%–50% of patients with MS do not respond to IFN-β. Moreover, the molecular mechanisms through which type I IFNs exert the anti-inflammatory effects remain largely unknown. In this regard, a recent study (Guarda and others 2011) noted that the type I IFN inhibits IL-1 production by 2 distinct mechanisms: (i) Type I IFN-signaling, in STAT1 transcription factor-dependent mechanism, repressed the activity of the NLRP1 and NLRP3 inflammasomes, thereby inhibiting caspase-1-dependent IL-1β production; and (ii) type I IFN-signaling induced IL-10 expression in a STAT1-dependent manner; production of autocrine IL-10 then through activation of STAT3 transcription factor reduced the abundance of pro-IL-1α and pro-IL-1β. Interestingly, the study also noted that monocytes from patients with MS who were undergoing IFN-β treatment produced substantially reduced levels of IL-1β than monocytes derived from healthy donors (Guarda and others 2011). Although these observations may provide a molecular basis for the effectiveness of type I IFN in the treatment of inflammatory diseases, the IFN-inducible “effector” proteins that mediate the anti-inflammatory actions of type I IFNs remain to be identified. Additionally, an improved understanding of how IL-10 exerts its anti-inflammatory response in the case of neutrophil-driven inflammatory reactions (which are often seen in patients with RA) (Bazzoni and others 2010) may provide novel clues leading to the therapeutic control.
An improved understanding of the molecular links among certain genotypes, resulting in alterations in cytokine signaling, and associated changes in gene expressions in patients with autoimmune rheumatic diseases are likely to improve our ability to appropriately diagnose and treat these diseases effectively.
As noted in the special Volume I, this volume features articles contributed by the groups of Theofilopoulos (lupus), Niewold (lupus), Choubey (lupus), Lehmann (encephalomyelitis), Matthys (arthritis), and Moudgil (arthritis). A synopsis of each of the articles in the Volume II of the special issue is presented next.
Nucleic Acid Sensors in Autoimmunity
Aberrant activation of innate immune responses by a collection of nucleic acid sensors that can sense nucleic acids derived from pathogens and self may have deleterious consequences for autoimmunity and inflammatory diseases. Theofilopoulos and others (2011; p.867) elegantly review recent advances in our understanding of the role of nucleic acids sensors in the activation of innate immune responses, type I IFN production, and autoimmunity. Additionally, the review presents novel insights for the development of strategies to treat autoimmunity-associated disorders.
IFN-α in Human SLE
Several lines of evidence involving patients and mouse models implicate type I IFN, IFN-α, in autoimmune diseases, including SLE. An article by Niewold (2011; p.887) summarizes experimental evidences to support the idea that increased levels of IFN-α and IFN-induced signaling are the critical mediators of SLE in human patients. Notably, the article provides the genetic basis for an increased production of IFN-α in certain individuals and their increased risk to develop SLE and associated immunopathology.
The p200-Family Proteins in Sex Bias in SLE
Increased levels of type I IFN, IFN-α, and “IFN-signature” are associated with the development of autoimmune diseases, including SLE. Moreover, these autoimmune diseases exhibit a gender bias in the development. Given that the IFN-inducible proteins mediate the immunomodulatory functions of IFNs and sex hormones influence the immune responses, an improved understanding of the role of IFN and sex hormone-regulated proteins is likely to provide new insights into the role of these proteins in sex bias in certain autoimmune diseases. An article by Choubey and others (2011; p.893) details the regulation and roles of the IFN-inducible and sex hormones regulated p200-family proteins in the sex bias in SLE. Notably, the authors suggest that the female sex hormone estrogen may have a role in the initiation of the disease through up-regulation of the expression of certain p200-family proteins.
Multiple Sclerosis
Experimental autoimmune encephalomyelits (EAE), which is induced in experimental naïve mice by injecting a neuroantigen, has been a useful model in defining events that may contribute to autoimmune response and associated central nervous system (CNS) pathology in MS. Kuerten and Lehmann (2011; p.907) have presented a roadmap of different steps involved in the initiation, progression, and regression of autoimmune inflammation during the course of experimental EAE. Specifically, the authors have elaborated the preimmunization neuroantigen-specific T cell repertoire, the dependence of the autoimmune effector response on the type of adjuvant used, the cytokine signature of the effector T cells that mediate EAE, the role of T cell-mediated delayed-type hypersensitivity in the induction of EAE, the entry and activation of T cells in the CNS, the postimmunization peripheral T cell repertoire, the phenomenon of determinant/epitope spreading observed during the course of EAE, and the regression of DTH and other pathogenic effector responses. Also discussed are the reversible and irreversible CNS damage during EAE, the antibody- and CD8+ T cell-mediated pathology, and the multiple facets of MS, particularly the extremely variable clinical features of the disease as well as unpredictable individual patterns.
Rheumatoid Arthritis
Schurgers and others (2011; p.917) have outlined the pathogenesis of human RA and collagen-induced arthritis (CIA), one of the experimental models of RA. The role of the genetic and environmental factors as well as that of different cell types (T cells, B cells, neutrophils, macrophage, osteoclasts, and synovial fibroblasts) and cytokines in the disease process have been reviewed. The authors have highlighted an unexpected disease-attenuating role of endogenous IFN-γ, a prototypic pro-inflammatory cytokine that is generally associated with the initiation and progression of inflammation. This conclusion is drawn from the results of studies involving the administration of IFN-γ, which attenuated CIA, or anti-IFN-γ antibodies, which aggravated CIA. Similarly, mice deficient in the IFN-γ receptor developed more severe CIA than wild-type mice. The likely reasons for the arthritis-protective effects of IFN-γ have been discussed. Also presented is a perspective on the clinical trials of IFN-γ in patients with RA conducted between 1986 and 1998.
Astry and others (2011; p.927) have presented a comprehensive evaluation of the role of cytokines in the pathogenesis of autoimmune arthritis. The authors have described the basic attributes (eg, structure of the cytokine and its receptor, and the cells secreting them) of a large panel of cytokines and critically examined the literature pertaining to the pathogenic versus protective effects of these cytokines in experimental arthritis models as well as RA. For the animal models, most of the information is based on the adjuvant-induced arthritis and CIA models. Among the cytokines, special emphasis has been placed on the IL-17/IL-23 axis and related cytokines such as IL-21 and IL-27. Also outlined is the newly emerging information about IL-32, IL-33, and IL-35. In addition, the authors have provided useful information about clinical trials, either completed or in progress, using neutralizing antibodies directed against specific cytokines, cytokine decoy receptors, or inhibitors of cytokine signaling.
Acknowledgments
The authors apologize to those investigators in the field whose work was not cited (or was cited through others' review articles) because of space limitations. This work was supported by grants R01AT004321 (K.D.M.), R01 AI066261, and R56 AI089775 (D.C.) from the National Institute of Health, Bethesda, MD.
Author Disclosure Statement
No competing financial interests exist.
References
- Apostolidis SA. Lieberman LA. Kis-Toth K. Crispín JC. Tsokos GC. The dysregulation of cytokine networks in systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31(10):769–779. doi: 10.1089/jir.2011.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aringer M. Smolen JS. Tumour necrosis factor and other proinflammatory cytokines in systemic lupus erythematosus: a rationale for therapeutic intervention. Lupus. 2004;13(5):344–347. doi: 10.1191/0961203303lu1024oa. [DOI] [PubMed] [Google Scholar]
- Astry B. Harberts E. Moudgil KD. A cytokine-centric view of the pathogenesis and treatment of autoimmune arthritis. J Interferon Cytokine Res. 2011;31(12):927–940. doi: 10.1089/jir.2011.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccala R. Gonzalez-Quintial R. Lawson BR. Stern ME. Kono DH. Beutler B. Theofilopoulos AN. Sensors of the innate immune system: their mode of action. Nat Rev Rheumatol. 2009;5(8):448–456. doi: 10.1038/nrrheum.2009.136. [DOI] [PubMed] [Google Scholar]
- Baccala R. Hoebe K. Kono DH. Beutler B. Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13(5):543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- Baechler EC. Gregersen PK. Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol. 2004;16(6):801–807. doi: 10.1016/j.coi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Ballestar E. Esteller M. Richardson BC. The epigenetic face of systemic lupus erythematosus. J Immunol. 2006;176(12):7143–7147. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- Banchereau J. Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25(3):383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunol Rev. 2011;243(1):99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- Barnes B. Lubyova B. Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res. 2002;22(1):59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- Bazzoni F. Tamassia N. Rossato M. Cassatella MA. Understanding the molecular mechanisms of the multifaceted IL-10-mediated anti-inflammatory response: lessons from neutrophils. Eur J Immunol. 2010;40(9):2360–2368. doi: 10.1002/eji.200940294. [DOI] [PubMed] [Google Scholar]
- Belhocine K. Monack DM. Francisella infection triggers activation of the AIM2 inflammasome in murine dendritic cells. Cell Microbiol. 2011 doi: 10.1111/j.1462–5822.2011.01700.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiau A. Anti-inflammatory properties of type I interferons. Antiviral Res. 2006;71(2–3):108–116. doi: 10.1016/j.antiviral.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland S. Garcia-Sastre A. Vicious circle: systemic autoreactivity in Ro52/TRIM21-deficient mice. J Exp Med. 2009;206(8):1647–1651. doi: 10.1084/jem.20091507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bynoté KK. Hackenberg JM. Korach KS. Lubahn DB. Lane PH. Gould KA. Estrogen receptor-α deficiency attenuates autoimmune disease in (NZB x NZW)F1 mice. Genes Immun. 2008;9(3):137–152. doi: 10.1038/sj.gene.6364458. [DOI] [PubMed] [Google Scholar]
- Choubey D. p202: an interferon-inducible negative regulator of cell growth. J Biol Regul Homeost Agents. 2000;14(3):187–192. [PubMed] [Google Scholar]
- Choubey D. Duan X. Dickerson E. Ponomareva L. Panchanathan R. Shen H. Srivastava R. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30(6):371–380. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. Kotzin BL. Interferon-inducible p202 in the susceptibility to systemic lupus. Front Biosci. 2002;7:e252–e262. doi: 10.2741/A921. [DOI] [PubMed] [Google Scholar]
- Choubey D. Panchanathan R. Interferon-inducible Ifi200-faily genes in systemic lupus erythematosus. Immunol Lett. 2008;119(1–2):32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. Panchanathan R. Duan X. Liu H. Liu H. Emerging roles for the interferon-inducible p200-family proteins in sex bias in systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31(12):893–906. doi: 10.1089/jir.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Solal JF. Jeganathan V. Hill L. Kawabata D. Rodriguez-Pinto D. Grimaldi C. Diamond B. Hormonal regulation of B-cell function and systemic lupus erythematosus. Lupus. 2008;17(6):528–532. doi: 10.1177/0961203308089402. [DOI] [PubMed] [Google Scholar]
- Crispín JC. Liossis SN. Kis-Toth K. Lieberman LA. Kyttaris VC. Juang YT. Tsokos GC. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends Mol Med. 2010;16(2):47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MK. Wohlgemuth J. Microarray analysis of gene expression in lupus. Arthritis Res Ther. 2003;5(6):279–287. doi: 10.1186/ar1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M. Gilkeson G. Estrogen receptors in immunity and autoimmunity. Clin Rev Allergy Immunol. 2011;40(1):66–73. doi: 10.1007/s12016-010-8203-5. [DOI] [PubMed] [Google Scholar]
- Davis LS. Hutcheson J. Mohan C. The role of cytokines in the pathogenesis and treatment of systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31(10):781–789. doi: 10.1089/jir.2011.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon KB. Stone VV. Type I interferon and systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31(11):803–812. doi: 10.1089/jir.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T. Yu JW. Datta P. Wu J. Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Immunol Rev. 2008;8(9):737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay C. Lamacchia C. Palmer G. IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010;6(4):232–241. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- Garcia-Romo GS. Caielli S. Vega B. Connolly J. Allantaz F. Xu Z. Punaro M. Baisch J. Guiducci C. Coffman RL. Barrat FJ. Banchereau J. Pascual V. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73–20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariglio M. Mondini M. De Andrea M. Landolfo S. The multifaceted interferon-inducible p200 family proteins: from cell biology to human pathology. J Interferon Cytokine Res. 2011;31(1):159–172. doi: 10.1089/jir.2010.0106. [DOI] [PubMed] [Google Scholar]
- Graham RR. Hom G. Ortmann W. Behrens TW. Review of recent genome-wide association scans in lupus. J Intern Med. 2009;265(6):680–688. doi: 10.1111/j.1365-2796.2009.02096.x. [DOI] [PubMed] [Google Scholar]
- Greenstein BD. Lupus: why women? J Women's Health Gender Based Med. 2001;10(3):233–239. doi: 10.1089/152460901300139989. [DOI] [PubMed] [Google Scholar]
- Guarda G. Braun M. Staehli F. Tardivel A. Mattmann C. Förster I. Farlik M. Decker T. Du Pasquier RA. Romero P. Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34(2):213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Haas C. Ryffel B. Le Hir M. IFN-gamma receptor deletion prevents autoantibody production and glomerulonephritis in lupus-prone (NZB x NZW)F1 mice. J Immunol. 1998;160(8):3713–3718. [PubMed] [Google Scholar]
- Hall JC. Rosen A. Type I interferons: crucial participants in disease amplification in autoimmunity. Nat Rev Rheumatol. 2010;6(1):40–49. doi: 10.1038/nrrheum.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T. Brotcke A. Weiss DS. Thompson LJ. Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204(5):987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T. Monack DM. Activation of the inflammasome upon Francisella tularensis infection: interplay of innate immune pathways and virulence factors. Cell Microbiol. 2007;9(11):2543–2551. doi: 10.1111/j.1462-5822.2007.01022.x. [DOI] [PubMed] [Google Scholar]
- Honda K. Takaoka A. Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25(3):349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Hron JD. Peng SL. Type I IFN protects against murine lupus. J Immunol. 2004;173(3):2134–2142. doi: 10.4049/jimmunol.173.3.2134. [DOI] [PubMed] [Google Scholar]
- Hu X. Chakravarty SD. Ivashkiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008;226:41–56. doi: 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW. Trapani JA. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol Cell Biol. 1999;19(9):5833–5838. doi: 10.1128/mcb.19.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW. Kayagaki N. Broz P. Henry T. Newton K. O'Rourke K. Chan S. Dong J. Qu Y. Roose-Girma M. Dixit VM. Monack DM. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107(21):9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen TN. Roper E. Thurman JM. Marrack P. Kotzin BL. Type I interferon signaling is involved in the spontaneous development of lupus-like disease in B6.Nba2 and (B6.Nba2× NZW)F1 mice. Genes Immun. 2007;8(8):653–662. doi: 10.1038/sj.gene.6364430. [DOI] [PubMed] [Google Scholar]
- Kariuki SN. Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2010;155(3):109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavai M. Szegedi G. Immune complex clearance by monocytes and macrophages in systemic lupus erythematosus. Autoimmun Rev. 2007;6(7):497–502. doi: 10.1016/j.autrev.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Kawasaki T. Kawai T. Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243(1):61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N. Veettil MV. Sharma-Walia N. Bottero V. Sadagopan S. Otageri P. Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated Herpesvirus infection. Cell Host Microbe. 2011;9(5):363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotter I. Gunaydin I. Zierhut M. Stubiger N. The use of interferon-alpha in Behcet's disease: review of the literature. Semin Arthritis Rheum. 2004;33(5):320–335. doi: 10.1016/j.semarthrit.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Kozyrev SV. Alarcon-Riquelme ME. The genetics and biology of Irf5-mediated signaling in lupus. Autoimmunity. 2007b;40(8):591–601. doi: 10.1080/08916930701510905. [DOI] [PubMed] [Google Scholar]
- Kozyrev SV. Lewén S. Reddy PM. Pons-Estel B Argentine Collaborative Group; Witte T German Collaborative Group; Junker P. Laustrup H. Gutiérrez C. Suárez A. Francisca González-Escribano M. Martín J Spanish Collaborative Group. Alarcón-Riquelme ME. Structural insertion/deletion variation in IRF5 is associated with a risk haplotype and defines the precise IRF5 isoforms expressed in systemic lupus erythematosus. Arthritis Rheum. 2007a;56(4):1234–1241. doi: 10.1002/art.22497. [DOI] [PubMed] [Google Scholar]
- Kuerten S. Lehmann PV. The immune pathogenesis of experimental autoimmune encephalomyelitis–lessons learned for multiple sclerosis? J Interferon Cytokine Res. 2011;31(12):907–916. doi: 10.1089/jir.2011.0072. [DOI] [PubMed] [Google Scholar]
- Lahita RG. Systemic lupus erythematosus. 3rd. San Diego, CA: Academic Press; 1999. [Google Scholar]
- Lamkanfi M. Dixit VM. Modulation of inflammasome pathways by bacterial and viral pathogens. J Immunol. 2011;187(2):597–602. doi: 10.4049/jimmunol.1100229. [DOI] [PubMed] [Google Scholar]
- Lande R. Ganguly D. Facchinetti V. Frasca L. Conrad C. Gregorio J. Meller S. Chamilos G. Sebasigari R. Riccieri V. Bassett R. Amuro H. Fukuhara S. Ito T. Liu YJ. Gilliet M. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson BR. Prud'homme GJ. Chang Y. Gardner HA. Kuan J. Kono DH. Theofilopoulos AN. Treatment of murine lupus with cDNA encoding IFN-gammaR/Fc. J Clin Invest. 2000;106(2):207–215. doi: 10.1172/JCI10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Liu Y. Xie C. Zhu J. Kreska D. Morel L. Mohan C. Deficiency of type I interferon contributes to Sle2-associated component lupus phenotypes. Arthritis Rheum. 2005;52(10):3063–3072. doi: 10.1002/art.21307. [DOI] [PubMed] [Google Scholar]
- Liu Z. Bethunaickan R. Huang W. Ramanujam M. Madaio MP. Davidson A. IFN-α confers resistance of systemic lupus erythematosus nephritis to therapy in NZB/W F1 mice. J Immunol. 2011;187(3):1506–1513. doi: 10.4049/jimmunol.1004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. Shen N. Li ZM. Chen SL. Genomic view of IFN-α response in pre-autoimmune NZB/NZW and MRL/lpr mice. Genes Immun. 2007;8(7):590–603. doi: 10.1038/sj.gene.6364421. [DOI] [PubMed] [Google Scholar]
- Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6(11):823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F. Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14(1):10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- McMurray RW. Hoffman RW. Nelson W. Walker SE. Cytokine mRNA expression in the B/W mouse model of systemic lupus erythematosus—analyses of strain, gender, and age effects. Clin Immunol Immunopathol. 1997;84(3):260–268. doi: 10.1006/clin.1997.4390. [DOI] [PubMed] [Google Scholar]
- Morel L. Genetics of SLE: evidence from mouse models. Nat Rev Rheumatol. 2010;6(6):348–357. doi: 10.1038/nrrheum.2010.63. [DOI] [PubMed] [Google Scholar]
- Moser KL. Kelly JA. Lessard CJ. Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10(5):373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewold TB. Interferon-alpha as a primary pathogenic factor in human lupus. J Interferon Cytokine Res. 2011;31(12):887–892. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewold TB. Kelly JA. Flesch MH. Espinoza LR. Harley JB. Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58(8):2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G. Pascual V. The interferon-alpha signature of systemic lupus erythematosus. Lupus. 2010;19(9):1012–1019. doi: 10.1177/0961203310371161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Duan X. Shen H. Rathinam VA. Erickson LD. Fitzgerald KA. Choubey D. Aim2 deficiency stimulates the expression of IFN-inducible Ifi202, a lupus susceptibility murine gene within the Nba2 autoimmune susceptibility locus. J Immunol. 2010a;185(12):7385–7393. doi: 10.4049/jimmunol.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Shen H. Bupp MG. Gould KA. Choubey D. Female and male sex hormones differentially regulate expression of Ifi202, an interferon-inducible lupus susceptibility gene within the Nba2 interval. J Immunol. 2009;183(11):7031–7038. doi: 10.4049/jimmunol.0802665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Shen H. Zhang X. Ho SM. Choubey D. Mutually positive regulatory feedback loop between interferons and estrogen receptor-alpha in mice: implications for sex bias in autoimmunity. PLoS One. 2010b;5:e10868. doi: 10.1371/journal.pone.0010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA. Jiang Z. Waggoner SN. Sharma S. Cole LE. Waggoner L. Vanaja SK. Monks BG. Ganesan S. Latz E. Hornung V. Vogel SN. Szomolanyi-Tsuda E. Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11(5):395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider P. Carmi Y. Guttman O. Braiman A. Cohen I. Voronov E. White MR. Dinarello CA. Apte RN. IL-1{alpha} and IL-1{beta} recruit different myeloid cells and promote different stages of sterile inflammation. J Immunol. 2011;187(9):4835–4843. doi: 10.4049/jimmunol.1102048. [DOI] [PubMed] [Google Scholar]
- Roberts TL. Idris A. Dunn JA. Kelly GM. Burnton CM. Hodgson S. Hardy LL. Garceau V. Sweet MJ. Ross IL. Hume DA. Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Rubtsov AV. Rubtsova K. Kappler JW. Marrack P. Genetic and hormonal factors in female-biased autoimmunity. Autoimmun Rev. 2010;9(7):494–498. doi: 10.1016/j.autrev.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B. Jyothi Prasanna S. Chandrasekar B. Nandi D. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine. 2010;50(1):1–14. doi: 10.1016/j.cyto.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Salloum R. Niewold TB. Interferon regulatory factors in human lupus pathogenesis. Transl Res. 2011;157(6):326–331. doi: 10.1016/j.trsl.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Raber ML. Baccala R. Haraldsson KM. Choubey D. Stewart TA. Kono DH. Theofilopoulos AN. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197(6):777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawalha AH. Harley JB. Scofield RH. Autoimmunity and Klinefelter's syndrome: when men have two X chromosomes. J Autoimmun. 2009;33(1):31–34. doi: 10.1016/j.jaut.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurgers E. Billiau A. Matthys P. Collagen-induced arthritis as an animal model for rheumatoid arthritis: focus on interferon-γ. J Interferon Cytokine Res. 2011;31(12):917–926. doi: 10.1089/jir.2011.0056. [DOI] [PubMed] [Google Scholar]
- Sekigawa I. Okada M. Ogasawara H. Kaneko H. Hishikawa T. Hashimoto H. DNA methylation in systemic lupus erythematosus. Lupus. 2003;12(2):79–85. doi: 10.1191/0961203303lu321oa. [DOI] [PubMed] [Google Scholar]
- Shaw PJ. McDermott MF. Kanneganti TD. Inflammasomes and autoimmunity. Trends Mol Med. 2011;17(2):57–64. doi: 10.1016/j.molmed.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H. Panchanathan R. Rajavelu P. Duan X. Gould KA. Choubey D. Gender-dependent expression of murine Irf5 gene: implications for sex bias in autoimmunity. J Mol Cell Biol. 2010;2(5):284–290. doi: 10.1093/jmcb/mjq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR. Williams BRG. Silverman RH. Schreiber RD. How cells respond to interferons? Ann Rev Biochem. 1999;67:227–262. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN. Kono DH. Beutler B. Baccala R. Intracellular nucleic acid sensors and autoimmunity. J Interferon Cytokine Res. 2011;31(12):867–886. doi: 10.1089/jir.2011.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos AN. Koundouris S. Kono DH. Lawson BR. The role of IFN-gamma in systemic lupus erythematosus: a challenge to the Th1/Th2 paradigm in autoimmunity. Arthritis Res. 2001;3(3):136–141. doi: 10.1186/ar290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos GC. Kammer GM. Molecular aberrations in human systemic lupus erythematosus. Mol Med Today. 2000;6(11):418–424. doi: 10.1016/s1357-4310(00)01798-6. [DOI] [PubMed] [Google Scholar]
- Tweezer-Zaks N. Rabinovich E. Lidar M. Livneh A. Interferon-alpha as a treatment modality for colchicine-resistant familial Mediterranean fever. J Rheumatol. 2008;35(7):1362–1365. [PubMed] [Google Scholar]
- Unterholzner L. Keating SE. Baran M. Horan KA. Jensen SB. Sharma S. Sirois CM. Jin T. Latz E. Xiao TS. Fitzgerald KA. Paludan SR. Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarsen LG. van der Pouw Kraan TC. Kragt JJ. Baggen JM. Rustenburg F. Hooper T. Meilof JF. Fero MJ. Dijkstra CD. Polman CH. Verweij CL. A subtype of multiple sclerosis defined by an activated immune defense program. Genes Immun. 2006;7(6):522–531. doi: 10.1038/sj.gene.6364324. [DOI] [PubMed] [Google Scholar]
- van Baarsen LG. Vosslamber S. Tijssen M. Baggen JM. van der Voort LF. Killestein J. van der Pouw Kraan TC. Polman CH. Verweij CL. Pharmacogenomics of interferon-beta therapy in multiple sclerosis: baseline IFN signature determines pharmacological differences between patients. PLoS One. 2008;3(4):e1927. doi: 10.1371/journal.pone.0001927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk FL. Netea MG. Dinarello CA. Joosten LA. Inflammasome activation and IL-1β and IL-18 processing during infection. Trends Immunol. 2011;32(3):110–116. doi: 10.1016/j.it.2011.01.003. [DOI] [PubMed] [Google Scholar]
- van der Pouw Kraan TC. van Baarsen LG. Wijbrandts CA. Voskuyl AE. Rustenburg F. Baggen JM. Dijkmans BA. Tak PP. Verweij CL. Expression of a pathogen-response program in peripheral blood cells defines a subgroup of rheumatoid arthritis patients. Genes Immun. 2008;9(1):16–22. doi: 10.1038/sj.gene.6364438. [DOI] [PubMed] [Google Scholar]
- van der Pouw Kraan TC. Wijbrandts CA. van Baarsen LG. Voskuyl AE. Rustenburg F. Baggen JM. Ibrahim SM. Fero M. Dijkmans BA. Tak PP. Verweij CL. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007;66(8):1008–1014. doi: 10.1136/ard.2006.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verthelyi D. Petri M. Ylamus M. Klinman DM. Disassociation of sex hormone levels and cytokine production in SLE patients. Lupus. 2001;10(5):352–358. doi: 10.1191/096120301674365881. [DOI] [PubMed] [Google Scholar]
- Vilaysane A. Muruve DA. The innate immune response to DNA. Semin Immunol. 2009;21(4):208–214. doi: 10.1016/j.smim.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Weckerle CE. Niewold TB. The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin Rev Allergy Immunol. 2011;40(1):42–49. doi: 10.1007/s12016-009-8192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2(9):777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- Zhao S. Wang Y. Liang Y. Zhao M. Long H. Ding S. Yin H. Lu Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63(5):1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]