Abstract
A collection of molecular sensors has been defined by studies in the last decade that can recognize a diverse array of pathogens and initiate protective immune and inflammatory responses. However, if the molecular signatures recognized are shared by both foreign and self-molecules, as is the case of nucleic acids, then the responses initiated by these sensors may have deleterious consequences. Notably, this adverse occurrence may be of primary importance in autoimmune disease pathogenesis. In this case, microbe-induced damage or mishandled physiologic processes could lead to the generation of microparticles containing self-nucleic acids. These particles may inappropriately gain access to the cytosol or endolysosomes and, hence, engage resident RNA and DNA sensors. Evidence, as reviewed here, strongly indicates that these sensors are primary contributors to autoimmune disease pathogenesis, spearheading efforts toward development of novel therapeutics for these disorders.
Introduction
The pathogenesis of autoimmune diseases has long been addressed from the perspective of abnormalities in the adaptive immune system. Despite impressive advances in defining these abnormalities, a comprehensive picture of how these diseases are initiated has remained relatively difficult to define and integrate into a concise scheme. The recent discovery of an array of cell surface and intracellular germline-encoded innate sensors, which recognize exogenous and endogenous danger signals, has provided a more solid foundation to define the pathogenesis of these disorders. Consequently, it has become clear that, akin to normal adaptive immune responses, pathogenic autoimmune responses almost invariably require a preceding engagement of the innate immune system. Although innate responses are normally beneficial, if excessive or protracted, they can result in pathogenic inflammatory/autoimmune diseases, especially in genetically predisposed individuals. Here, we focus on sensors for nucleic acids, because their ligands are represented in both pathogens and hosts, and many lines of evidence strongly establish them as causative factors for several autoimmune diseases. We describe the diversity of these receptors, their intracellular distribution, trafficking patterns, activation requirements, and the elicited pathogenic mediators.
Complexity of Nucleic Acid Sensors
Cells of the immune system are equipped with a broad range of germline-encoded sensors that recognize primarily microbial substances and, in certain instances, endogenous products released from damaged cells. This ever-expanding list of sensors includes Toll-like receptors (TLRs), retinoid acid-inducible gene (RIG)-like receptors (RLRs), nucleotide oligomerization domain (NOD)-like receptors (NLRs) (Baccala and others 2009; Barbalat and others 2011; Sharma and Fitzgerald 2011), and several other newly identified receptors. These sensors reside on cell surfaces, the cytosol, or within intracellular organelles, thereby safeguarding the integrity of the host. Although all these sensors may directly or indirectly participate in the pathogenesis of autoimmunity, those recognizing DNA or RNA are more likely to play a critical role, as nucleic acids of foreign origin and self-origin are mostly indistinguishable. In particular, nucleic acid sensors are relevant to the pathogenesis of systemic lupus erythematosus (SLE), in which autoantibodies against nucleosomal and spliceosomal antigens typically predominate.
Endolysosomal Nucleic Acid Sensors
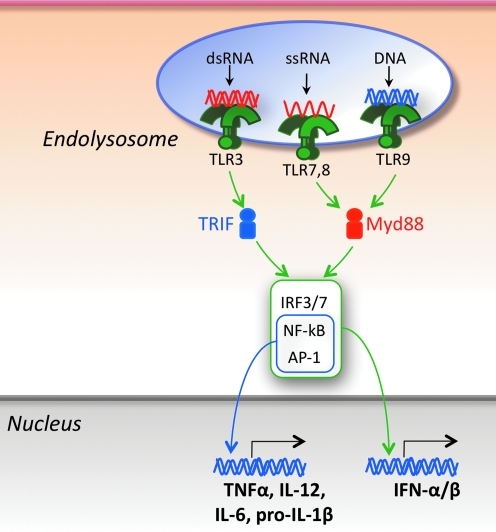
The known innate sensors represented within the endolysosomes are the nucleic acid-sensing TLRs, including TLR3, specific for double-stranded RNA (dsRNA), TLRs 7 and 8 for single-stranded RNA (ssRNA), and TLR9 for DNA (Beutler and others 2006; Uematsu and Akira 2006; Moresco and others 2011) (Fig. 1). These sensors are expressed in several cell types intimately involved in immune responses, including conventional dendritic cells (cDCs), plasmacytoid DCs (pDCs), macrophages, and B cells. The expression profile of nucleic acid-sensing TLRs differs among these cell types, and these differences as well as cell-specific means of transport, intracellular localization, and utilization of TLRs may influence the type of pathogens recognized and the responses elicited. For example, in humans, pDCs selectively express TLRs 7 and 9, whereas cDCs preferentially express TLRs 3 and 8 (in addition to TLRs 1 and 2), monocytes TLR8 (in addition to TLRs 1, 2, 4, and 5), and B cells TLRs 7, 8, and 9 (in addition to TLRs 1, 2, 4, 5, and 6). These distinct TLR expression profiles suggest that efficient innate immune responses require the participation of multiple cell types, rather than a few specialized cells. All TLR family members, including the endolysosomal TLRs, are type I membrane proteins composed of a ligand-binding ectodomain containing several (18 to 25) tandem copies of leucine-rich repeats (LRRs), a single-pass transmembrane domain, and a conserved cytoplasmic Toll/interleukin-1 receptor (TIR) domain for signal transduction. Ligand-induced dimerization and conformational rearrangements of the TIR domains lead to the creation of 2 symmetry-related sites for the binding of the TIR domains of the cognate signaling adaptor molecules (Kawai and Akira 2006; Kenny and O'Neill 2008). Two main adaptors are utilized, ie, myeloid differentiation factor 88 (MyD88) by TLRs 7, 8, and 9, and TRIF (TIR domain-containing adaptor inducing interferon [IFN]-β) by TLR3. These adaptors mediate the recruitment of a series of kinases (IL-1 receptor-associated kinase [IRAK1], TGFβ-activated kinase 1 [TAK1], IkappaB kinase [IKK]αβγ, mitogen-activated protein kinases [MAPKs], TANK-binding kinase 1 [TBK1], IKKɛ) and ubiquitin ligases (TNF receptor-associated factor [TRAF]3 and TRAF6), thus leading to the formation of specific macromolecular signaling platforms. The subunit stochiometry of one of these platforms, termed Myddosome, created by homotypic interactions of the death domains of MyD88 and the kinases IRAK4 and IRAK2 (or the related IRAK1) has been defined (Motshwene and others 2009; Lin and others 2010; Gay and others 2011; Nagpal and others 2011). The TLR signaling cascade results in the activation and nuclear translocation of several transcription factors, including nuclear factor κB (NF-κB), adaptor protein 1 (AP-1), IFN-regulatory factor 3 (IRF3), and IRF7, which together initiate the expression of genes encoding type I IFNs and other cytokines, chemokines, chemokine receptors, and costimulatory molecules. Secreted type I IFNs bind to a specific receptor (IFNAR) present in nearly all cell types and trigger a large number of IFN-stimulated genes through the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway (Schoggins and others 2011).
FIG. 1.
Endolysosomal nucleic acid sensors. The presence of nucleic acids in endolysosomes is detected by TLR3 (dsRNA), TLR7 (ssRNA), TLR8 (ssRNA), and TLR9 (DNA). Ligand binding induces TLR dimerization and recruitment of the main signaling adaptors TRIF (used by TLR3) and MyD88 (used by TLRs 7, 8, and 9). These adaptors provide a nucleating structure for the formation of higher-order oligomeric complexes composed of kinases, ubiquitin ligases, and other signaling molecules that mediate activation of transcription factors (IRF3, IRF7, NF-kB, and AP-1) that, on nuclear translocation, promote expression of type I IFNs and other proinflammatory cytokines.
As is the case for TLR4, which requires the serum lipopolysaccharide (LPS)-binding protein as an accessory molecule for LPS binding, a recent study identified granulin, a cystein-rich serum factor, as a specific contributor to CpG-containing oligodeoxynucleotides (CpG-ODN)-induced TLR9 signaling and production of proinflammatory cytokines by pDCs and cDCs (Park and others 2011). A model proposed to explain this finding is that CpG-ODN binds to granulin or its precursor, progranulin, and the resulting complex is delivered to endosolysosomes on interaction with the lysosomal sorting protein sortilin (Moresco and Beutler 2011).
An unexpected recent finding was that major histocompatibility complex (MHC) class II molecules are required for optimal TLR signaling and induction of type I IFNs and other proinflammatory cytokines (Frei and others 2010). This positive regulation, taking place in endosomes, is mediated by the interaction of MHC class II molecules, via CD40, with the Bruton's tyrosine kinase, which then interacts with the adaptors MyD88 and TRIF (Hassan and Mourad 2011; Liu and others 2011b).
TLR signaling is modulated by multiple endogenous negative regulators that act at the cell membrane or intracellularly by interfering with adaptors, kinases, and transcription factors (Lang and Mansell 2007; Watters and others 2007; Ananieva and others 2008; Lemke and Rothlin 2008; Coll and O'Neill 2010). Inhibitors of TLR signaling may also be encoded by viral and bacterial genes (Stack and others 2005; Cirl and others 2008). Whether alterations in the expression and/or function of these negative regulators play any role in the pathogenesis of autoimmune diseases remains to be fully characterized, but mice deficient in some of these inhibitors, ie, SIGIRR (single Ig interleukin 1 [IL-1] receptor related) or Tyro, Axl and Mer (TAM) receptor tyrosine kinases, have been reported to develop lupus-like manifestations (Rothlin and others 2007; Lech and others 2008).
TLR compartmentalization and trafficking
Spatiotemporal regulation of intracellular trafficking is a determining factor for TLR accessibility to ligands, intensity of signals, and quality of the inflammatory responses, as has been illustrated for the high efficiency of type I IFN production by the TLR7- and TLR9-expressing pDCs (Honda and others 2005). TLRs 3, 7, 8, and 9 are localized in several intracellular compartments, including the endoplasmic reticulum (ER), the endosomes, and lysosomes. Activation of these TLRs, however, occurs only in acidified endolysosomes, as responses are extinguished when acidification is prevented (Hacker and others 1998). These results pose a question of how cells sense nucleic acids before the ER-resident TLRs are mobilized to the acidified endolysosomal compartment. One potential answer is that minimal trafficking of these TLRs takes place at a steady state or in response to low-grade inflammatory stimuli. Indeed, the type of carbohydrates on intracellular TLRs 7 and 9 in nonstimulated cells suggests that these TLRs have trafficked from the ER through the Golgi and gained residence in endolysosomes (Ewald and others 2008; Park and others 2008; Chockalingam and others 2009). Moreover, trafficking of endosomal TLRs can be induced by stimuli other than nucleic acids, such as LPS (Blasius and Beutler 2010).
Several ER-associated proteins have been shown to act as chaperones and to mediate translocation of TLRs to endolysosomes and/or to the cell membrane (Fig. 2). GP96 (an ER paralog of the HSP90 family) affects trafficking of several TLRs, as macrophages deficient in this molecule do not respond to ligands for TLRs 1, 2, 4, 5, 7, or 9 (Randow and Seed 2001; Yang and others 2007a). Trafficking of TLRs 1, 2, and 4 from the ER to the plasma membrane, and TLRs 7 and 9 (but not TLR3) to the endolysosomes, is also affected by PRAT4A (Takahashi and others 2007). Another ER-resident protein central to the trafficking of TLRs 3, 7, and 9 to endolysosomes is Unc93b1. The 3d missense mutation of the Unc93b1 gene, resulting in an H412R substitution in the ninth membrane-spanning region of the encoded protein, causes absence of signaling by TLRs 3, 7, and 9 and susceptibility of the mutant mice to infection by various pathogens (Tabeta and others 2006). Susceptibility to herpes simplex encephalitis was also observed in patients with UNC93B1 gene mutations (Casrouge and others 2006). Further studies showed that the endosomal TLRs bind to the Unc93b1 protein via their transmembrane domains (Brinkmann and others 2007), and that TLRs 7 and 9, but not TLR3, compete for association with this trafficking molecule (Fukui and others 2009). Interestingly, TLR9 binds to Unc93b1 more efficiently than TLR7, thus favoring its translocation to endolysosomes (Fukui and others 2009). Accordingly, TLR9 signaling is normally stronger than that of TLR7, and experimental TLR9 overexpression inhibits TLR7 signaling (Wang and others 2006). The preferential interaction with TLR9 is mediated by the N-terminal domain of Unc93b1, and when this domain is mutated (D34A), then the interaction favors TLR7 (Fukui and others 2009).
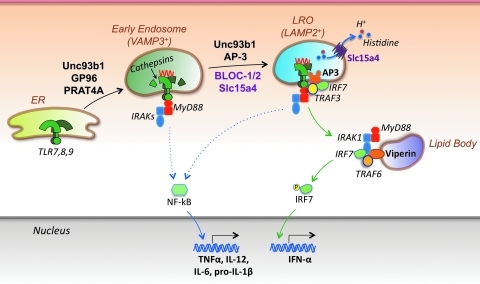
FIG. 2.
TLR compartmentalization and trafficking. Nucleic acid-specific TLRs traffic from the endoplasmic reticulum (ER) to endolysosomes, where they undergo ligand binding and cathepsin-mediated proteolytic cleavage, a process required for efficient signaling. Several proteins have been shown to facilitate TLR translocation to endolysosomes, including the heat-shock protein gp96, PRAT4A, and Unc93b1. Moreover, type I IFN production by pDCs is strictly dependent on TLR localization to specialized compartments termed lysosome-related organelles (LROs). AP-3 (likely together with Unc93b1) mediates TLR translocation to LROs, whereas other molecules in these organelles are required for effective type I IFN induction, including BLOC-1, BLOC-2, and Slc15a4 (a transporter of protons and histidines from the LRO lumen to the cytosol). In addition, the IFN-induced molecule viperin has been shown to localize to lipid bodies and promote assembly of a signaling complex that includes MyD88, IRAK1, and TRAF6, thereby greatly facilitating IRF7 activation and IFN-α production. One study suggests that IRF7 and NF-kB signaling is initiated from LROs and early endosomes, respectively, whereas another study indicates that both pathways are initiated in the LROs.
More recently, an additional set of proteins was shown to play a central role in the trafficking and signaling by nucleic acid-sensing TLRs and production of proinflammatory cytokines. It has long been recognized that signaling through TLR7 and TLR9 can trigger the production of TNFα, IL-12, IL-6, and pro-IL-1β by activating NF-kB and type I IFNs by activating IRF7. A recent study suggested that these two pathways are initiated from distinct cellular compartments (Fig. 2). A key molecule in this trafficking process appears to be AP-3, a 4-subunit (δ, β3A, μ3A, σ3) clathrin-associated adaptor protein complex that recognizes dileucine-based motifs in transmembrane proteins and sorts these proteins to endosomes, lysosomes, or lysosome-related organelles (LRO) (Mattera and others 2011). Thus, pDCs and cDCs from AP-3-deficient mice (Ap3b1-/-, lacking the β3A subunit), as well as pDCs from mice with mutations in the β3A or δ subunits of the AP-3 complex (pearl and mocha mice, respectively), showed defective signaling and production of type I IFNs in response to TLR7 or TLR9 agonists, but NF-kB-mediated induction of IL-12p40 was unaffected (Sasai and others 2010). Further work showed that AP-3 is required for trafficking of TLR9 and Unc93b1 to late endosomes and LROs expressing the lysosomal-associated membrane protein 2 (LAMP2) marker, but not to early endosomes that express vesicle-associated membrane protein 3 (VAMP3). These and additional experiments led to the formulation of a TLR bifurcation signaling model. According to this model, intracellular TLRs, together with Unc93b1, transit first to early endosomes in an AP-3-independent manner, resulting in NF-kB-mediated production of proinflammatory cytokines (NF-kB endosome), and then to late endosomes in an AP-3-dependent manner, thus resulting in TRAF3-mediated attraction and activation of IRF7 and production of type I IFNs (IRF7 endosome or LRO) (Fig. 2). This model suggests that nucleic acid-sensing TLRs, by trafficking into specialized cellular compartments, engage distinct pathways that lead to different cytokine profiles, presumably due to restricted localization of specific signal transduction molecules. It should be noted, however, that in another study with Ap3b1 mutant mice (pearl and bullet gray) and highly purified DC subsets, the defective response to TLR9 engagement was detected in pDCs, but not cDCs, and production of both type I IFNs and TNF-α was reduced (Blasius and others 2010). Thus, rather than a bifurcation model, this latter study suggests that in pDCs, both NF-kB and IRF7 signaling pathways are initiated from a specific LRO present in this cell type.
Of interest, mutations in the AP-3 complex have been linked to the Hermansky–Pudlak syndrome, which defines a group of human autosomal recessive disorders characterized by abnormal biogenesis and function of several types of LROs, such as melanosomes, platelet-dense granules, lamellar bodies of type II alveolar epithelial cells, and lytic granules of cytotoxic T cells and NK cells (Dell'Angelica 2009). The clinical phenotype of this syndrome is characterized by grades of oculocutaneous albinism, a bleeding diathesis due to platelet defects, T lymphocyte dysfunction, and neutropenia. Mutations affecting three ubiquitously expressed protein complexes, named biogenesis of lysosome-related organelles complex (BLOC) -1, -2, and -3, are also associated with Hermansky–Pudlak syndrome (Dell'Angelica 2004). Accordingly, mice carrying the salt and pepper mutation in the Dtnbp1 gene (encoding dysbindin, a component of BLOC-1), or the toffee mutation in the Hps5 gene (encoding a component of BLOC-2), exhibited defective type I IFN responses of pDCs to a TLR9 ligand (Blasius and others 2010). Interestingly, the AP-3 complex interacts via its μ subunit with dysbindin in the BLOC-1 complex (Taneichi-Kuroda and others 2009), and this interaction may be related to the role of both AP-3 and BLOC-1 in endosomal TLR signaling and type I IFN production by pDCs.
Another mutant mouse, feeble, with defective production of type I IFNs as well as TNF-α, IL-6, and IL-12p40 after TLR7 or TLR9 ligation, has been identified (Blasius and others 2010). This defect was confined to pDCs, but the development of these cells was unaffected. The phenotype of these mice was mapped to a mutation in the Slc15a4 gene, which encodes the proton/histidine transporter 1 (PHT1). The solute carrier subfamily 15 (Slc15) contains 4 members (Slc15a1 to Slc15a4) and is a part of the proton-coupled oligopeptide transporter superfamily (Nielsen and Brodin 2003; Daniel and Kottra 2004). Of these, Slc15a1 (also called PEPT1) and Slc15a2 (PEPT2) have broad substrate specificity and transport a large spectrum of di- and tri-peptides, whereas Slc15a3 (PHT2) and Slc15a4 (PHT1) are more specific in that their function is primarily to transport free histidine and certain oligopeptides from inside the endosome to the cytosol (Yamashita and others 1997; Bhardwaj and others 2006). The transport activity of Slc15a4 is pH-dependent, with a higher transport of histidine at pH 5.5 than at pH 7.0 (Yamashita and others 1997), thereby suggesting that the function of this transporter is related to endosomal acidification and lysosomal maturation. Interestingly, Slc15a4 contains a dileucine motif in its N-terminal region that might be recognized by AP-3, thereby suggesting comigration of these two molecules to the endolysosomes. The findings overall suggest that AP-3 primarily affects TLR trafficking, whereas Slc15a4 may be required to create an endolysosomal microenvironment optimal for TLR activation and/or signaling.
The special role of certain organelles in the activation of endosomal TLRs has further been documented by the finding that Viperin, a protein induced by IFNs and even directly by viruses and bacteria (Fitzgerald 2011), is required for TLR7- and TLR9-mediated production of type I IFNs by pDCs (Jiang and Chen 2011; Saitoh and others 2011). Viperin, however, had no role in the production of other inflammatory cytokines (IL-12, TNF-α, and IL-1β) by pDCs and other cell types, nor in the production of type I IFNs by cDCs, macrophages, and fibroblasts, or signaling by other sensors (TLR4 and RLRs). Mechanistically, it was shown that an amphipathic α-helix in the N-terminus of Viperin targets this protein to the cytosolic surface of ER-derived lipid storage organelles, called lipid bodies. These organelles appear to function downstream of the LROs with Viperin serving as a platform for the attraction of signaling effectors, including MyD88, TRAF6, IRAK1, and IRF7. Collectively, the findings with molecules that affect formation and function of specific subcellular compartments, such as AP-3, BLOCs, Slc15a4, and Viperin, have provided novel clues for elucidating the reason that pDCs are such prodigious producers of type I IFNs.
Endosomal TLR activation requires processing by proteases
Activation of nucleic acid-specific TLRs is confined to endolysosomes, thereby precluding their engagement by self-nucleic acids and ensuring that responses are initiated after the breakdown of pathogens in this acidified microenvironment. Recent studies have shown that functionality of these receptors, but not ligand binding, is only acquired in endolysosomes on cleavage of a major portion of the ectodomain by resident acid-dependent proteases, principally cathepsins (Ewald and others 2008; Matsumoto and others 2008; Park and others 2008). Individual cathepsins and/or combinations thereof are required for activation of all endosomal TLRs, and this requirement is conserved across all cell types (Ewald and others 2011). The asparagine endopeptidase (AEP) legumain was also shown to participate in TLR cleavage (Sepulveda and others 2009; Ewald and others 2011). Overall, TLR processing appears to proceed through a two-step mechanism, ie, cleavage of a portion of the ectodomain mediated by multiple cathepsins (perhaps with some participation of legumain) followed by N-terminal trimming (Ewald and others 2011). The reason that ectodomain cleavage is required for MyD88 recruitment and signaling by endosomal TLRs remains to be clarified. However, conformational changes induced by this process may facilitate TLR dimerization, an event critical for efficient signaling (Latz and others 2007). The finding that endosomal TLR activation requires cathepsin-mediated processing explains previous observations in mouse models that pharmacologic inhibition or deletion of cathepsins potently suppressed joint inflammation (Asagiri and others 2008), neuroinflammation (Asagiri and others 2008), autoimmune diabetes (Maehr and others 2005; Hsing and others 2010), and Sjögren's syndrome-like manifestations (Saegusa and others 2002).
Cytosolic Nucleic Acid Sensors
There is also a broad spectrum of receptors that evolved to recognize nucleic acids in the cytosol, and their signaling is critical for the initiation of innate responses against certain microbes. Two main classes of such sensors have been identified, one specific for RNA and the other for DNA. Most of these sensors activate transcription factors leading to expression of type I IFNs and other cytokines, but a few may promote the assembly of an inflammasome and secretion of IL-1β.
RNA sensors
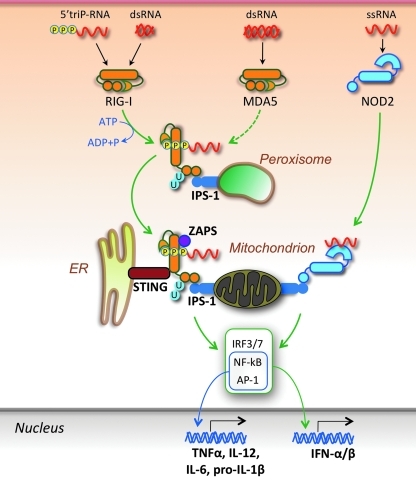
The RNA sensors encompass three helicases, RIG-I, melanoma differentiation-associated gene 5 (MDA5, also known as IFN-induced with helicase C domain 1, IFIH1), and laboratory of genetics and physiology 2 (LGP2) specific for distinct but overlapping sets of viruses (Moore and Ting 2008; Takeuchi and Akira 2008; Matsumiya and Stafforini 2010) (Fig. 3). RIG-I recognizes ssRNA and dsRNA with uncapped 5’-triphosphate ends as well as short dsRNA structures (even without 5’-triphosphates), whereas MDA-5 recognizes long dsRNA species with blunt ends (Pichlmair and others 2006; Schlee and others 2009; Yoneyama and Fujita 2009). In addition to a central DEAD box helicase/ATPase domain, RIG-I and MDA5 also display tandem N-terminal caspase activation and recruitment domains (CARD), and a C-terminal domain (CTD) containing a repressor domain. In resting cells, RIG-I is maintained as a monomer in a latent autoinhibited, “closed” conformation state. After binding of CTD to the 5’-triphosphate moiety of RNA, ATP hydrolysis by the helicase domain results in a conformational change that displaces the CTD and exposes the CARDs, which, through a homotypic interaction, engage the mitochondria-anchored adaptor IFNβ promoter stimulator-1 (IPS-1) (also known as mitochondrial antiviral signaling protein [MAVS], CARD-containing adaptor protein [CARDIF], and virus-induced signaling adaptor [VISA]). This engagement is associated with redistribution of IPS-1 to form speckle-like aggregates, a process mediated by Mitofusin 1 and 2 (Onoguchi and others 2010; Koshiba and others 2011), followed by recruitment of TBK1 and other signaling molecules that drive expression of type I IFNs. IPS-1 was also shown to be present on the surface of peroxisomes, organelles that, in concert with mitochondria, affect metabolism of lipids and reactive oxygen species (Dixit and others 2010). Engagement of peroxisomal IPS-1 precedes that of mitochondrial IPS-1, is short lived, and initiates an early IFN-independent antiviral response that is then potentiated by the mitochondrial IPS-1-induced type I IFN response. Post-translational modifications affect the RIG-I/IPS-1 pathway, with ubiquitination of CARDs by tripartite motif protein 25 (TRIM25) promoting signaling (Yoneyama and Fujita 2009). In this regard, a remarkable finding was that unanchored ubiquitin chains (not conjugated to any target molecule) together with RNA potently activate RIG-I (Zeng and others 2010). The third helicase, LGP2, does not contain CARDs required for signaling, but rather facilitates viral RNA recognition by RIG-I and MDA5, acting either directly by altering RNA conformation or indirectly by dislodging viral ribonucleoproteins (RNPs) in an ATP-dependent manner to expose RNA (Moresco and Beutler 2010; Satoh and others 2010).
FIG. 3.
Cytosolic RNA sensors. RNA in the cytoplasm is primarily sensed by the helicases RIG-I (RNA with uncapped 5′-triphosphates or short dsRNA) and MDA5 (long dsRNA). Ligand binding induces ATP-dependent conformational changes that allow CARD/CARD homotypic interactions between these helicases and the signaling adaptor IPS-1 localized on either peroxisomes or mitochondria. Engagement of peroxisomal IPS-1 promotes a transient IFN-independent response, whereas at the mitochondrial membrane, the engaged IPS-1 relays a signaling cascade that leads to transcription factor activation and expression of type I IFNs and proinflammatory cytokines. RIG-I signaling also requires the participation of the ER-associated STING and is enhanced by the interaction with ZAPS as well as by TRIM25-mediated K63-linked ubiquitination (U) of CARDs, although unanchored ubiquitin chains also promote RIG-I activation. Another helicase, LGP2, lacks CARDs and appears to facilitate RNA binding to RIG-I and MDA5. In addition, NOD2, a CARD-containing member of the NLR family mostly dedicated to the detection of the peptidoglycan component muramyl dipeptide (MDP) found in both Gram-positive and Gram-negative bacteria, can also recognize ssRNA in the cytosol and promote type I IFN and proinflammatory cytokine production through an IPS-1-mediated pathway.
An ER- and mitochondria-associated molecule, stimulator of IFN genes (STING, also known as MITA, MPYS, TMEM173 and ERIS), appears to be essential for efficient type I IFN induction by RIG-I, but not MDA5, and likely acts as an anchor that facilitates the interaction of the sensor with downstream signaling molecules (Ishikawa and Barber 2008; Zhong and others 2008; Ishikawa and others 2009; Barber 2011). In addition, signaling strength and duration are enhanced by the direct interaction of RIG-I with signaling cofactors, including the zinc-finger antiviral protein shorter isoform (ZAPS), a member of the poly(ADP-ribose) polymerase family (Hayakawa and others 2011; Liu and Gale 2011), and the dsRNA-binding protein PACT (Kok and others 2011). A recent study also showed that the protein kinase R regulates the integrity of IFN-β transcripts induced by several MDA5-dependent viruses (Schulz and others 2010).
Interestingly, NOD2, a CARD-containing member of the large NLR family of cytosolic sensors that principally recognizes muramyl dipeptides (MDPs) found in nearly all Gram-positive and Gram-negative bacteria, was also reported to recognize viral ssRNA (Sabbah and others 2009). In fact, NOD2 recognition of MDP leads to NF-kB activation and induction of proinflammatory cytokines, whereas recognition of ssRNA promotes IPS-1-dependent IRF3 activation and type I IFN production. This is an interesting example of a receptor that recognizes two structurally distinct ligands causing induction of alternative signaling pathways, and poses questions about the presumed strict specificity of innate sensors for certain molecular signatures.
Another member of the NLR family, NLRX1, has also been shown to interact with IPS-1, but this interaction leads to inhibition of RIG-I-mediated IFN-β production, thus suggesting that some NLRs might function as modulators of pathogen responses rather than as classical sensors (Moore and others 2008; Allen and others 2011; Xia and others 2011). Other cellular regulators that suppress RLR signaling include the deubiquitinating enzyme A and cylindromatosis tumor suppressor (CYLD), the ubiquitin-ligase RNF125, the autophagy-related Atg5–Atg12 conjugate, and caspase-8 (Gack and others 2008; Moore and Ting 2008; Oshiumi and others 2009; Liu and Gu 2011; Rajput and others 2011). In addition, the intracellular form of the klotho protein has been shown to inhibit the RIG-I-mediated expression of IL-6 and IL-8 induced in senescent cells through the ataxia telangiectasia mutated-IRF1 axis (Liu and others 2011a). As in the case of TLRs, viruses also have acquired several mechanisms to evade and/or suppress production of type I IFNs by the RLR pathway, including expression of proteins that inhibit helicase–ligand interactions or proteases that cleave IPS-1 from the mitochondrion (Takeuchi and Akira 2008).
DNA sensors
Cytosolic dsDNA, particularly of large size, is recognized by a wide spectrum of sensors, thus leading to activation of diverse signaling pathways (Fig. 4). Critically, recognition of cytosolic DNA appears to be sequence independent and largely unaffected by the degree of methylation, thus suggesting that both foreign and self-DNA may initiate such responses. Early studies showed that intracellular administration of the right-handed B-form DNA or a synthetic 45-mer DNA of random sequence lacking CpG motifs triggered type I IFN production in a TLR-independent manner (Ishii and others 2006; Stetson and Medzhitov 2006). A sensor termed DNA-dependent activator of IRFs (DAI, also known as Z-DNA binding protein-1 [ZBP-1] and DLM-1), was subsequently reported to recognize both the B-form and the left-handed Z-form of cytosolic dsDNA and to induce type I IFNs (Takaoka and others 2007). However, this response was not compromised in cells derived from DAI-deficient mice (Ishii and others 2008), thus implying the existence of additional sensors or a cell-specific role for these sensors (Wang and others 2008; DeFilippis and others 2010).
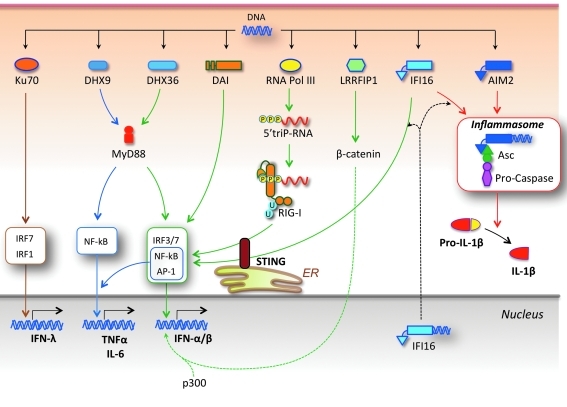
FIG. 4.
Cytosolic DNA sensors. A large panel of sensors is dedicated to the detection of DNA in the cytosol. Among them, Ku70 induces IFN-λ, DHX9 induces proinflammatory cytokines, and DHX36, DAI, RNA polymerase III, and IFI16 induce production of type I IFNs and proinflammatory cytokines. RNA polymerase III acts by transcribing DNA into RNA molecules bearing uncapped 5′-triphosphates, which bind RIG-I and engage the IPS-1/STING pathway (see Fig. 3). Another sensor, LRRFIP1, binds DNA (or dsRNA) and activates β-catenin, which migrates to the nucleus and potentiates IFNB gene transcription by promoting recruitment of the acetyltransferase p300 to the IFN enhanceosome. In addition, IFI16 and AIM2 induce the assembly of an inflammasome by recruiting pro-caspases via the adaptor ASC, thus leading to caspase-mediated activation and secretion of IL-1β and IL-18. IFI16 (and perhaps other sensors) may recognize DNA in the nucleus and then migrate to the cytoplasm to initiate signal activation.
Another means of cytosolic DNA recognition is mediated by DNA-dependent RNA polymerase III. This polymerase transcribes AT-rich dsDNA in a promoter-independent manner to generate dsRNA intermediates bearing uncapped 5′-triphosphates that serve as agonists for RIG-I/IPS-1-dependent type I IFN production, a response that, in part, requires the participation of STING (Ablasser and others 2009; Chiu and others 2009; Barber 2011).
Additional studies showed that interaction with cytosolic DNA is also mediated by human IFI16 and its mouse ortholog p204, which are pyrin domain (PYD)-containing members of the hematopoietic IFN-inducible nuclear (HIN-200) family of proteins (PYHIN) (Unterholzner and others 2010). Although IFI16 is mostly localized in the nucleus, small amounts of this factor might egress into the cytoplasm where recognition of DNA and downstream signaling events lead to type I IFN production. Alternatively, IFI16 recognition of viral DNA may take place in the nucleus followed by migration to the cytoplasm to stimulate signal transduction, as suggested by studies with herpes simplex virus-1 (HSV-1) (Unterholzner and others 2010). As is the case for RIG-I and RNA polymerase III, IFI16 signaling also requires the participation of STING (Unterholzner and others 2010). With regard to the mechanism by which STING contributes to the signaling induced by cytosolic DNA, it has been shown that, on stimulation, STING translocates from the ER to Golgi and finally to cytoplasmic punctate structures to assemble with TBK1, a process that appears to be negatively regulated by the autophagy-related gene 9a (Saitoh and others 2009; Barber 2011).
Four other sensors for cytosolic nucleic acids with unique properties, DHX9, DHX36, LRRFIP1, and Ku70, have been identified. DHX9 and DHX36 are members of the DExD/H box family of helicases that also includes RLRs. They recognize CpG-DNA in pDCs and mediate MyD88-dependent induction of proinflammatory cytokines (via NF-kB) and type I IFNs (via IRF7), respectively (Kim and others 2010). LRRFIP1 notably recognizes both dsRNA and dsDNA and, by activating β-catenin, facilitates recruitment of the acetyltransferase p300 to the IFN enhanceosome, which potentiates IFNB gene transcription (Yang and others 2010). On the other hand, Ku70, a component of the heterodimeric Ku protein required for end-joining DNA repair, VDJ recombination, telomere maintenance, and other nuclear processes, recognizes various types of cytosolic DNA and promotes production of the type III IFN (IFN-λ1) through activation of IRF1 and IRF7 (Zhang and others 2011).
Inflammasome induction by cytosolic nucleic acids
Previous studies noted that intracellular bacterial, viral, and mammalian dsDNA could trigger the production of the potent proinflammatory cytokine IL-1β in a cell-specific manner, thus suggesting the involvement of an inflammasome (Muruve and others 2008). The sensor involved in this response was identified as absent in melanoma 2 (AIM2), a cytoplasmic member of the PYHIN family (Burckstummer and others 2009; Fernandes-Alnemri and others 2009; Hornung and others 2009; Roberts and others 2009). AIM2 binds DNA via its HIN-200 domain, recruits the adaptor apoptosis-associated speck-like protein containing a CARD (ASC) via PYD–PYD interactions, and then ASC attracts pro-caspase-1 through CARD–CARD interactions. On autoactivation, caspase-1 cleaves pro-IL-1β (and pro-IL-18) to a mature secreted form. The AIM2 inflammasome has also been reported to induce a specific form of cell death termed pyroptosis that involves DNA damage but, unlike apoptosis, is associated with a loss of plasma membrane integrity and maintenance of mitochondrial membrane potential (Fernandes-Alnemri and others 2009).
In addition to type I IFN-induction (see above), IFI16 can also mediate the assembly of an ASC-dependent inflammasome in the cytosol after recognition of Kaposi Sarcoma–associated herpes virus (KSHV) in the nucleus (Kerur and others 2011). The findings on nuclear recognition of HSV-1 and KSHV by IFI16 raise the question of how engagement of this sensor by self-nucleic acids is avoided, and masking of cellular DNA by histones and other proteins or recognition restricted for damaged DNA might be potential explanations (Unterholzner and Bowie 2011). Interestingly, in a manner similar to IFI16, the RNA sensor RIG-I has also been shown to induce both type I IFN production and the assembly of an inflammasome, the latter through the recruitment of ASC and pro-caspases 1 and 3 (Kim and Yoo 2008; Rintahaka and others 2008; Poeck and others 2010).
Nucleic Acid Sensors and Autoimmunity
Shortly after or concurrent with the discovery of nucleic acid sensors, an extensive body of evidence has been gathered to suggest that these sensors and the resulting induction of proinflammatory cytokines play an important effector role in the pathogenesis of autoimmune diseases, with the bulk of the evidence, at present, related to endosomal TLRs and systemic autoimmunity.
Endosomal TLRs and systemic autoimmunity
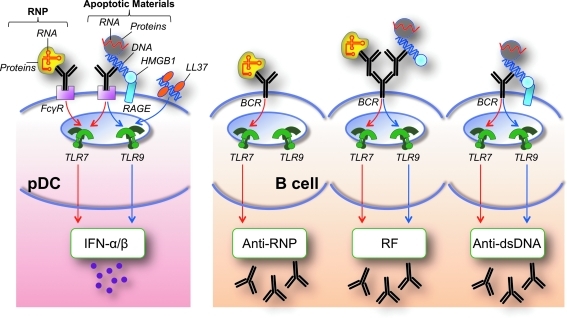
As discussed, specific mechanisms have evolved to prevent the engagement of intracellular innate sensors by self-nucleic acids, including compartmentalization of functional TLRs to endolysosomes, rarity and/or chemical modification of stimulatory motifs in mammalian nucleic acids (eg, methylation, capping of 5′-triphosphates of RNA molecules), and clearance of self-nucleic acids and apoptotic/necrotic materials containing such ligands by nucleases and other processes. However, several pathways have been identified by which these barriers can be overcome, thus leading to pathological consequences, particularly in lupus (Fig. 5). Specifically with regard to nucleic acid-sensing TLRs, these pathways involve autoantibodies bound to subcellular particles containing self-nucleic acids, which are taken up by pDCs via FcR, or by B cells via BCRs specific for either antigenic determinants in the complex or the Fc portion of the autoantibodies (Ronnblom and others 2006; Marshak-Rothstein and Rifkin 2007; Theofilopoulos and others 2010). The synergistic engagement of BCR and TLR induces enhanced B cell proliferation, whereas production of type I IFN by pDCs leads to B cell differentiation and immunoglobulin isotype switching. Free nucleic acids or particles containing such molecules (ie, nucleosomes, RNPs) can also interact with specific BCRs (Viglianti and others 2003), and recent evidence suggests that BCR signaling induces fusion of TLR9-containing endosomes with the internalized BCR into autophagosomes, thereby facilitating B cell activation (Chaturvedi and others 2008; Monroe and Keir 2008).
FIG. 5.
Mechanisms by which barriers for TLR recognition of self-nucleic acids are breached in systemic autoimmunity. Various mechanisms usually prevent TLR engagement by self-nucleic acids, including nuclease-mediated degradation, exclusion from endosomes, and methylation. Autoantibodies and BCRs specific for nucleic acid-containing particles (eg, anti-RNP, anti-DNA) or exhibiting rheumatoid factor activity (RF) can overcome these mechanisms, mediating nucleic acid delivery to endosomes in pDCs (via FcγR) and B cells (via BCR), and promoting type I IFN (IFN-α/β) and autoantibody production. Nucleic acid-binding accessory proteins such as HMGB1 (in part via RAGE) and LL37 can also facilitate DNA uptake and TLR engagement. Production of anti-RNP autoantibodies requires TLR7, whereas production of anti-dsDNA and RF autoantibodies requires either TLR7 or TLR9. Thus, the major antigen for autoantibodies to RNP contains RNA (red) and proteins (particle in yellow), whereas the antigenic target of anti-DNA autoantibodies contains DNA (blue), RNA, and other accessory molecules (particle in gray).
The uptake of nucleic acids and related complexes by pDCs and B cells is enhanced by certain accessory molecules, such as the high-mobility group box (HMGB) proteins and the antimicrobial peptide LL37. HMGB proteins are highly expressed in the nucleus, where they regulate chromatin structure and transcription, but they are also present in the cytosol and are released from necrotic cells and cells stimulated with TLR ligands or cytokines, thus potentially inducing inflammatory responses (Scaffidi and others 2002; Bianchi and Manfredi 2007). Initial studies showed that HMGB1 enhanced TLR9-mediated signaling in response to CpG-DNA or nucleic acid-containing complexes, likely by facilitating ligand uptake either directly or through the receptor for advanced glycation end-products (RAGE) (Ivanov and others 2007; Tian and others 2007). Interestingly, DNA-C3a complexes were also reported to stimulate type IFN production in a RAGE-dependent manner (Ruan and others 2010). Moreover, increased levels of HMGB1 in serum of patients with lupus have been shown to correlate with disease activity (Andersson and Rauvala 2011; Urbonaviciute and Voll 2011). Strikingly, a more recent study expanded the role of HMGB proteins by demonstrating that HMGB1, HMGB2, and HMGB3 act as universal sentinels for both DNA and RNA and that their presence is required for efficient signaling by cytosolic and endolysosomal nucleic acid sensors (Yanai and others 2009; Yanai and others 2011). The exact mechanism by which HMGB proteins deliver nucleic acids to these sensors and the circumstances under which this function may exert beneficial or detrimental effects to the organism remain to be elucidated.
DNA and RNA delivery to endosomes leading to TLR engagement in pDCs, myeloid DCs, and B cells are also promoted by the nucleic acid-binding LL37 cathelicidin, a mammalian antimicrobial amphypathic peptide (Lande and others 2007; Ganguly and others 2009; Hurtado and Peh 2010). LL37 is produced by keratinocytes and released after skin injury, and this response may be involved in the pathogenesis of psoriasis (Lande and others 2007). However, other studies have presented evidence that the Aim2/DNA inflammasome may be involved in psoriasis, and that LL37 induced by vitamin D may inhibit this inflammasome (Dombrowski and others 2011). This finding suggests a potential beneficial role of LL37 and is consonant with the known therapeutic effects of vitamin D in this condition. Whether LL37 or Aim2 have any relevance to dermatologic manifestations of lupus, particularly after UV exposure, remains to be addressed.
Several studies in humans and mice have demonstrated the primary role of endosomal TLR engagement and type I IFN production in the pathogenesis of lupus (Theofilopoulos and others 2010). In SLE, there is an “IFN signature” (a dominance of genes affected by type I IFNs) that appears to correlate with clinical severity (Baechler and others 2004; Kirou and others 2005; Banchereau and Pascual 2006; Nikpour and others 2008; Petri and others 2009; Obermoser and Pascual 2010). Polymorphic variants of TLR7 that may associate with increased TLR7 transcripts and more pronounced IFN signatures were reported to constitute a predisposing factor in Eastern Asian patients with lupus (Shen and others 2010; Kawasaki and others 2011), and greater TLR7 gene copy number increased risk of SLE in the Mexican population (Garcia-Ortiz and others 2010). However, other studies found no detectable association between lupus and TLR7 gene copy number or polymorphic variants (Demirci and others 2007; Kelley and others 2007; Sanchez and others 2009), thus suggesting contributions by additional genetic factors. Genome-wide and other studies have also revealed an association of SLE susceptibility with genetic variants of IRF5, IRF7, IRAK1, STAT4, and Tyk2, molecules involved in type I IFN signaling pathways (Harley and others 2009; Fu and others 2011). Interestingly, underexpression of the microRNA miR-146a, a negative regulator of the IFN pathway, correlated with disease activity in patients with SLE (Tang and others 2009), and targeted deletion of this microRNA in mice was associated with some lupus-like manifestations (Boldin and others 2011). Expression and/or function of several other microRNAs have been implicated in SLE and mouse models, but the exact mechanisms of action and whether they affect innate responses and cytokine production remain to be clarified (Pauley and others 2009; Vinuesa and others 2009; Xiao and Rajewsky 2009; Dai and others 2010).
The pathogenic role of endosomal TLRs and type I IFNs has been directly documented in the classical spontaneous lupus strains of mice. Previous studies showed that NZB mice deleted of the common receptor for type I IFNs had significantly reduced disease (Santiago-Raber and others 2003), whereas, conversely, administration of recombinant IFN-α, plasmids encoding this cytokine or synthetic endosomal TLR ligands exacerbated disease in NZBxW, NZBxBXSB, NZM2328, and B6.Sle123 mice (Mathian and others 2005; Fairhurst and others 2008; Jacob and others 2011; Ramanujam and others 2009; Triantafyllopoulou and others 2010). Moreover, MRL-Faslpr deficient in IRF5 showed decreased serologic, cellular, and histologic disease characteristics and increased survival (Tada and others 2011). A recent study showed that treatment of NZBxW mice with IFN-α led to the induction of short-lived, but not long-lived, plasma cells, presumably due to changes in the niches that sustain plasma cell survival (Mathian and others 2011), whereas a proteasome inhibitor (bortezomib) that depletes both short- and long-lived plasma cells protected lupus-predisposed mice (Neubert and others 2008). Interestingly, bortezomib was also shown to suppress function and survival of pDCs by disrupting the coordinated translocation of TLRs and Unc93b1 and disturbing ER homeostasis (Hirai and others 2011). IFN-α-driven autoantibody production and nephritis in NZBxW mice was dependent on CD4 T cell help (Liu and others 2011c), as expected based on the fact that contributions by both innate and adaptive systems are required for full disease expression (Baccala and others 2007).
The strongest evidence for the role of endosomal TLRs in systemic autoimmunity was obtained in male BXSB mice bearing the Y-linked autoimmune accelerating locus (Yaa), where a translocation from the X to Y chromosome leads to the duplication of several genes, including Tlr7 (Pisitkun and others 2006; Subramanian and others 2006). The principal role of the Tlr7 gene duplication was shown by significant disease reduction in male BXSB mice carrying half of the Tlr7 gene dosage (due to Tlr7 null mutation in the X chromosome) and, conversely, the appearance of autoimmune manifestations in transgenic normal background mice with increased copy numbers of the Tlr7 gene (Deane and others 2007). However, Tlr7 duplication in itself is insufficient but rather, as expected in this polygenic disorder, requires additional genetic contributions, as demonstrated by absence of disease in normal background mice consomic for the Yaa chromosome (Theofilopoulos and Dixon 1985). Moreover, genes other than Tlr7 in the duplicated segment also appear to contribute, as introduction of a Tlr7 null mutation on the X chromosome of C57BL/6.Nba2 mice congenic for the Yaa chromosome led to incomplete disease resolution (Santiago-Raber and others 2008). MRL-Faslpr mice lacking TLR7 also showed considerable decreases in the levels of anti-Sm autoantibodies and a modest reduction of renal disease (Christensen and others 2006). Further documentation for the central role of nucleic acid-specific TLRs in murine lupus was obtained with BXSB and C57BL/6-Faslpr mice congenic for the Unc93b1 3d mutation, which extinguishes signaling through TLRs 3, 7, and 9 (Kono and others 2009). Interestingly, patients deficient for UNC93B1, despite showing defective central and peripheral B cell tolerance and accumulation of large numbers of autoreactive mature B cells in their blood, did not develop autoantibodies or histologic autoimmune manifestations, thus further documenting the central role of UNC93B1 and endosomal TLRs in promoting systemic autoimmunity (Isnardi and others 2008).
However, there is considerable controversy with regard to the role of TLR9 in lupus. This is because, paradoxically, deficiency of TLR9 caused an overall disease enhancement in the apoptosis-defective MRL-Faslpr mice, despite reductions in anti-dsDNA autoantibody titers (Christensen and others 2005; Christensen and others 2006; Wu and Peng 2006). This finding parallels that in TLR8-deficient C57BL/6 mice, which developed lupus-like serologic and histopathologic characteristics (Demaria and others 2010). Interestingly, in both TLR9- and TLR8-deleted mice, DCs overexpressed TLR7 and showed stronger activation on stimulation with a TLR7 ligand; whereas combined deletion of TLR9 or TLR8 with TLR7 abolished autoimmunity (Demaria and others 2010; Santiago-Raber and others 2010). Moreover, the mutation D34A in the N-terminus of Unc93b1, which abolishes interaction with TLR9, rendered DC hyperesponsive to TLR7 (Fukui and others 2009), with a concomitant induction of systemic inflammation associated with expansion of Th1 and Th17 cells and various autoantibodies, including ANA (Fukui and others 2011).
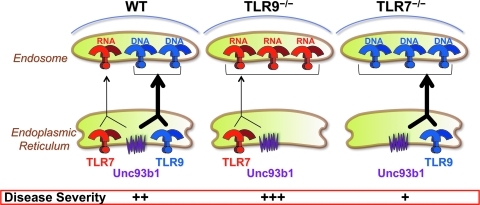
The presumed protective effect of TLR9 contrasts with the following: (a) TLR9 engagement by DNA-containing immune complexes promoted B cell proliferation (Leadbetter and others 2002), (b) TLR7-deleted MRL-Faslpr mice showed considerably more residual disease than mice lacking both TLR7 and TLR9 (Nickerson and others 2010), and (c) disease was prevented in 3d mutant C57BL/6-Faslpr mice (Kono and others 2009). We, therefore, hypothesized that both TLRs exert an adverse effect, but the pathogenic contribution of TLR7 is stronger than that of TLR9 (Fig. 6). This hypothesis is supported by the finding that Unc93b1 mediates the trafficking of all nucleic acid-sensing TLRs to endolysosomes. Since TLR9 exhibits higher affinity for this molecule, it is reasonable to suggest that the absence of TLR9 will result in increased trafficking of TLR7 and enhanced disease, as, in fact, occurred in the examples just given. Although it is still unclear why TLR7 is more pathogenic than TLR9, likely possibilities include differences in signaling intensity and/or higher availability of particles that contain TLR7-engaging ligands such as snRNPs (Theofilopoulos and others 2010).
FIG. 6.
Differential Unc93b1-mediated trafficking of TLR7 versus TLR9 may affect lupus pathogenesis in predisposed mice. Increased disease severity in TLR9-deleted lupus-predisposed mice may be due to increased availability of the common Unc93b1 trafficking partner, thus leading to enhanced translocation of the more pathogenic TLR7 to endolysosomes.
Thus, apart from minor incongruities, the published data strongly indicate that engagement of nucleic acid-specific TLRs is critical for the pathogenesis of lupus. The emerging scenario is that signaling by these TLRs leads to the induction of proinflammatory cytokines, of which type I IFNs predominate. These pleiotrophic cytokines induce maturation of DCs, upregulation of MHC and costimulatory molecules, production of B cell-trophic factors (B cell activating factor [BAFF], and a proliferation-inducing ligand [APRIL]), and activation of previously quiescent autoreactive T and B cells. The outcome is production of autoantibodies and creation of immune complexes with particles containing nucleic acids, which, by engaging endosomal TLRs on several cellular targets (pDCs, DCs, and B cells), perpetuate and amplify the pathogenic autoimmune process. Type II IFN-γ, produced by innate and adaptive immune cells, also contributes to lupus, particularly at later stages of the disease, as suggested by increased IFN-γ levels in serum and IFN-γ-induced transcripts in PBMC of patients with SLE; disease inhibition in lupus-predisposed mice after deletion of IFN-γ or its receptor; and blockade of IFN-γ signaling by using antibodies, recombinant soluble receptors, or a plasmid encoding this receptor (Baccala and others 2005).
Although we focus here on the role of endosomal TLRs and type I IFNs in lupus, considerable evidence suggests that similar inflammatory pathways may also be involved in other autoimmune diseases, including rheumatoid arthritis, Sjögren's syndrome, type I diabetes, myasthenia gravis, hemolytic anemia, neuromyelitis optica, polymyositis/dermatomyositis, and psoriasis (Baccala and others 2005; Theofilopoulos and others 2005).
Cytosolic innate sensors and autoimmunity
Cytosolic nucleic acid sensors may also play a role in the pathogenesis of autoimmune diseases, but the evidence is mostly indirect. Nonetheless, incompletely digested DNA can provoke inflammatory responses, as inferred by a few patients with SLE with mutations in DNase I (Yasutomo and others 2001), lupus-like manifestations in mice deficient in DNase I (Napirei and others 2000), and anemia or arthritis in mice deficient in DNase II (Nagata 2008). DNase I is the major serum endonuclease that degrades extracellular dsDNA, whereas DNase II is localized in lysosomes and degrades chromosomal DNA from apoptotic cells and nuclei expelled from erythroid precursors. Moreover, mutations in the ER-localized 3’ repair exonuclease 1 (Trex1, also known as DNase III) were found in a few patients with SLE (Lee-Kirsch and others 2007). In addition, mutations in Trex1 or RNaseH2 have been associated with the Aicardi–Goutières syndrome and chilblain lupus, two conditions with some clinical and pathophysiological similarities to SLE (Crow and others 2006a; Crow and others 2006b; Lee-Kirsch and others 2007; Kavanagh and others 2008; Perrino and others 2009). Interestingly, autoimmunity was also observed in mice with Trex1-deficiency and attributed to accumulation of ssDNA in the ER (Yang and others 2007b), possibly derived from reverse transcription of endogenous retroelements (Stetson and others 2008). In all these conditions, disease is presumably mediated by type I IFN-inducing cytosolic sensors, as autoimmunity in Trex1-deficient mice was prevented by genetic ablation of IFNAR or IRF3 (Stetson and others 2008), and Aicardi-Goutières syndrome is also associated with mutations in SAMHD1, a negative regulator of antiviral responses (Rice and others 2009).
Inflammasome assembly after Aim2/DNA interaction and production of IL-1β and IL-18 may also contribute to systemic autoimmunity. In support of this possibility, IL-1β levels are increased in most mouse lupus models, recombinant IL-1β aggravated nephritis in NZB/W mice, and treatment with a soluble IL-1 receptor reduced disease in MRL-Faslpr mice (Schorlemmer and others 1993). Levels of IL-18 are also increased in sera of MRL-Faslpr mice, and injections of this cytokine accelerated kidney pathology, whereas induction of anti-IL-18 antibodies conferred protection (Bossu and others 2003). Interestingly, an allelic variant of another DNA-binding member of the HIN-200 family, p202 (encoded by ifi202 in mice), was reported to be hyperexpressed in lymphoid cells and contribute to disease in NZB mice (Rozzo and others 2001). However, the mode by which p202 might promote disease in mice is unclear, as this molecule lacks a PYD domain and is, therefore, unlikely to induce an inflammasome and, in fact, p202 inhibited the formation of the Aim2-DNA inflammasome (Roberts and others 2009). Nonetheless, studies in mice have implied an interplay between p202 and Aim2, with deficiency in Aim2 causing induction of type I IFNs and p202, and p202 suppressing expression of the inhibitory FcγRIIB (Panchanathan and others 2010; Panchanathan and others 2011). Perhaps of relevance, upregulation of all 4 human HIN-200 homologs (MNDA, IFIX, IFI16, and AIM2) and a significant association with certain allelic variants of IFIX and IFI16 have been reported in one ethnic group of patients with SLE (Kimkong and others 2009, 2010).
Indirect evidence also suggests the potential involvement of the RLR sensing system in the pathogenesis of autoimmune diseases. Thus, viral 5’-triphosphate RNA aggravated lupus nephritis in MRL-Faslpr mice (Allam and others 2008). Moreover, increased levels of RIG-I were detected in the epidermis of patients with psoriasis (Kitamura and others 2007; Prens and others 2008), synovial tissues from patients with rheumatoid arthritis (Imaizumi and others 2008), and epithelial cells lining the gut mucosa (Kawaguchi and others 2009). In addition, loss-of-function mutations in RIG-I, and more frequently in MDA5 (encoded by IFIH1), were associated with resistance to type I diabetes (Nejentsev and others 2009; Shigemoto and others 2009; Downes and others 2010), and mice lacking one copy of MDA5 developed transient hyperglycemia after infection with a β cell-tropic virus (McCartney and others 2011). These findings support a viral pathogenesis of type I diabetes, but self-nucleic acids derived from phagocytosed apoptotic materials may also contribute.
Apoptotic materials as triggers in lupus
Despite the evidence that self-nucleic acids, either free or complexed with IgG autoantibodies, promote systemic autoimmunity, there is a relative paucity of information regarding the exact composition of nuclear materials presented in vivo to the immune system. In fact, considering the broad specificity of autoantibodies in lupus, it has been proposed that nucleic acids and other associated self-antigens may be presented in the form of “particles” generated during apoptotic or necrotic cell death (Hardin 1986; Tan 1989; Casciola-Rosen and others 1994; Riemekasten and Hahn 2005; Munoz and others 2010). This possibility is supported by the finding that materials from apoptotic cells bind antinuclear and antinucleosomal antibodies in vitro, and such complexes are isolated from the plasma of patients with lupus (Ullal and others 2011). Other evidence of the inflammatory properties of apoptotic materials is the finding that dying cells taken up by cDCs trigger TLR-independent type I IFN production and enhance adaptive antigen-specific responses (Janssen and others 2006). Moreover, SLE is characterized by excessive production of apoptotic cells and/or defective clearance of apoptotic materials (Viorritto and others 2007). The potential contribution of defects in clearance of apoptotic materials is further supported by the development of lupus in humans and mice with C1q deficiency (Botto and others 1998; Lewis and Botto 2006), and in mice with genetic deletion of the peroxisome proliferator-activated receptor-δ, which decreases expression of C1q (Mukundan and others 2009). In addition, lupus-like autoimmunity develops in mice lacking serum amyloid P (Bickerstaff and others 1999; Paul and Carroll 1999), TAM receptor protein tyrosine kinases (Tyro3, Axl, and Mer) (Lu and Lemke 2001; Cohen and others 2002; Lemke and Rothlin 2008), or milk fat globule-epidermal growth factor 8 (MFG-E8), all molecules that contribute to the removal of apoptotic materials (Hanayama and others 2004). Further, injection of apoptotic cells or apoptosis-promoting agents enhanced autoantibody production and kidney disease, especially in lupus-predisposed mice (Mevorach and others 1998; Denny and others 2006), whereas disease was reduced when clearance of apoptotic cells was enhanced by opsonization with adiponectin (Takemura and others 2007). Finally, nuclear fragmentation of apoptotic cells by the endonuclease caspase-activated DNase (CAD) was required for induction of antichromatin and anti-snRNP autoantibodies in pristane-treated mice (Frisoni and others 2007).
Although the role of apoptotic materials as potential triggers of innate responses has primarily been addressed by using splenocytes or lymphocytes, other types of dying hematopoietic cells, such as neutrophils, may also release materials that can engage endolysosomal or cytosolic sensors for nucleic acids. Thus, the number of circulating apoptotic neutrophils in SLE positively correlated with disease activity (Courtney and others 1999), and a granulopoiesis gene expression signature was detected in PBMC of some patients with SLE (Bennett and others 2003). Recent studies have provided mechanistic evidence of how neutrophil-derived apoptotic materials are generated and may contribute to lupus pathogenesis (Garcia-Romo and others 2011; Lande and others 2011; Mantovani and others 2011). It was determined that mature neutrophils primed in vivo by type I IFNs die when exposed to anti-RNP autoantibodies, releasing web-like structures known as neutrophil-extracellular traps (NETs), which contain DNA as well as the accessory factors LL37 and HMGB1. These NETs were detected in SLE sera and were shown to activate pDCs to produce high levels of type I IFN in a DNA- and TLR9-dependent manner. LL37-containing NETs were also reported to be associated with autoimmune small-vessel vasculitis (Kessenbrock and others 2009).
Microbial Triggers and Systemic Autoimmunity
Pathogens themselves or in combination with materials released from damaged tissues may constitute the trigger that initiates chronic inflammatory processes when responses are excessive or unchecked. Specifically, microbial infections are frequently associated with lupus disease flares, and Epstein-Barr virus (EBV) has been considered a major environmental risk factor for this disease (Poole and others 2006). Moreover, disease is enhanced in lupus-predisposed mice injected with bacterial or viral TLR ligands or mimics (Theofilopoulos and others 2005). Type I IFNs are the ultimate mediators of pathogenesis, as disease enhancement is not observed in IFNAR-deficient mice that are similarly injected (Braun and others 2003).
A clear example of requirement for microbial stimuli in the induction of systemic inflammatory diseases is that of mice carrying a hypomorphic mutation of the Ptpn6 gene (Croker and others 2008). Mice homozygous for this mutation (termed spin for spontaneous inflammation) displayed inflammation and mononuclear cell infiltration in feet, salivary glands, and lungs, as well as antichromatin antibodies. The Ptpn6 gene encodes the Src-homology-2-domain-containing protein tyrosine phosphatase SHP-1, which downregulates signaling from TCRs, BCRs, TLRs, integrins, and receptors for several cytokines and chemokines. The spin phenotype resembled, in an attenuated form, that of the viable motheaten (mev) mice, which also carry hypomorphic Ptpn6 mutations. Likewise, B cell-specific deficiency of SHP-1 promoted B-1a cell development and caused serologic and histologic manifestations of systemic autoimmunity (Pao and others 2007). With regard to spin mice, compound mutations of Ptpn6 with MyD88, IRAK4, or IL-1R inhibited disease development, whereas those with STAT1 (affecting type I and II IFN signaling) or TNF-α had no effect, thus indicating that disease in this model is primarily driven by IL-1β. The most striking finding was that disease was absent when spin mice were derived and bred in a germ-free environment, but reemerged when these mice were reconventionalized in a pathogen-free environment. This indicates that superimposition of a microbial trigger, likely derived from the enteric flora, is required for disease expression (Croker and others 2008). The overall scenario for disease in this model is that an initial TLR stimulus induces expression of pro-IL-1β, which, in a second step likely mediated by an inflammasome, is converted to mature, secreted IL-1β. On binding its receptor, IL-1β then creates an autoamplification loop that sustains the autoimmune/inflammatory process (Beutler 2009).
Conclusion
The interplay of genetic and environmental forces in the pathogenesis of diseases is a familiar concept, but the precise definition of these forces is often extremely difficult. This has certainly been the case for autoimmune diseases. The detailed characterization of the central pathways involved in the initiation of innate immune responses has now provided us with some clearer hypotheses. A wide spectrum of sensors has been identified that are solely dedicated to the detection and containment of infection and cellular damage. However, the associated beneficial inflammatory responses may sometimes be excessive or uncontrolled, thereby contributing to the initiation of pathogenic autoimmunity. Microbes may be involved in this process and act as triggers or long-term drivers of pathologic responses in predisposed individuals. However, in some instances, microbes may be entirely excluded, and autoimmune responses may develop under “sterile” conditions as a true dysfunction of “self.” However, the combined participation of both microbes and self-tissue-derived stimuli may also occur. In all these cases, the primary stimulus may be nucleic acids recognized by endosomal or cytosolic sensors, thereby leading to induction of type I IFNs and other proinflammatory cytokines. This concept may be applicable to a wide spectrum of autoimmune diseases, although it is most clearly documented for lupus. This mechanistic reformulation for the pathogenesis of autoimmune diseases will likely lead to new therapeutic avenues directed toward the inhibition of nucleic acid recognition, trafficking of sensors to specific cellular organelles where recognition occurs, downstream signaling events, and/or inhibition of end-stage mediators. It can be envisaged that interventions in these processes will soon find a place in modern medicine for the treatment of these frequently intractable disorders.
Acknowledgments
This is article number 21377 from The Scripps Research Institute, Department of Immunology and Microbial Science. The work of the authors cited here was made possible by Grant Numbers AR39555, AR30103, AR053228, and AR053731 from NIAMS/NIH, and by a Grant from the Guthy–Jackson Charitable Foundation.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Ablasser A. Bauernfeind F. Hartmann G. Latz E. Fitzgerald KA. Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10(10):1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allam R. Pawar RD. Kulkarni OP. Hornung V. Hartmann G. Segerer S. Akira S. Endres S. Anders HJ. Viral 5'-triphosphate RNA and non-CpG DNA aggravate autoimmunity and lupus nephritis via distinct TLR-independent immune responses. Eur J Immunol. 2008;38(12):3487–3498. doi: 10.1002/eji.200838604. [DOI] [PubMed] [Google Scholar]
- Allen IC. Moore CB. Schneider M. Lei Y. Davis BK. Scull MA. Gris D. Roney KE. Zimmermann AG. Bowzard JB. Ranjan P. Monroe KM. Pickles RJ. Sambhara S. Ting JP. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34(6):854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananieva O. Darragh J. Johansen C. Carr JM. McIlrath J. Park JM. Wingate A. Monk CE. Toth R. Santos SG. Iversen L. Arthur JS. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat Immunol. 2008;9(9):1028–1036. doi: 10.1038/ni.1644. [DOI] [PubMed] [Google Scholar]
- Andersson U. Rauvala H. Introduction: HMGB1 in inflammation and innate immunity. J Intern Med. 2011;270(4):296–300. doi: 10.1111/j.1365-2796.2011.02430.x. [DOI] [PubMed] [Google Scholar]
- Asagiri M. Hirai T. Kunigami T. Kamano S. Gober HJ. Okamoto K. Nishikawa K. Latz E. Golenbock DT. Aoki K. Ohya K. Imai Y. Morishita Y. Miyazono K. Kato S. Saftig P. Takayanagi H. Cathepsin K-dependent Toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319(5863):624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- Baccala R. Gonzalez-Quintial R. Lawson BR. Stern ME. Kono DH. Beutler B. Theofilopoulos AN. Sensors of the innate immune system: their mode of action. Nat Rev Rheumatol. 2009;5(8):448–456. doi: 10.1038/nrrheum.2009.136. [DOI] [PubMed] [Google Scholar]
- Baccala R. Hoebe K. Kono DH. Beutler B. Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13(5):543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- Baccala R. Kono DH. Theofilopoulos AN. Interferons as pathogenic effectors in autoimmunity. Immunol Rev. 2005;204:9–26. doi: 10.1111/j.0105-2896.2005.00252.x. [DOI] [PubMed] [Google Scholar]
- Baechler EC. Gregersen PK. Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol. 2004;16(6):801–807. doi: 10.1016/j.coi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Banchereau J. Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25(3):383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Barbalat R. Ewald SE. Mouchess ML. Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011;23(1):10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L. Palucka AK. Arce E. Cantrell V. Borvak J. Banchereau J. Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197(6):711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol Rev. 2009;227(1):248–263. doi: 10.1111/j.1600-065X.2008.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B. Jiang Z. Georgel P. Crozat K. Croker B. Rutschmann S. Du X. Hoebe K. GENETIC analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- Bhardwaj RK. Herrera-Ruiz D. Eltoukhy N. Saad M. Knipp GT. The functional evaluation of human peptide/histidine transporter 1 (hPHT1) in transiently transfected COS-7 cells. Eur J Pharm Sci. 2006;27(5):533–542. doi: 10.1016/j.ejps.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Bianchi ME. Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol Rev. 2007;220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- Bickerstaff MC. Botto M. Hutchinson WL. Herbert J. Tennent GA. Bybee A. Mitchell DA. Cook HT. Butler PJ. Walport MJ. Pepys MB. Serum amyloid P component controls chromatin degradation and prevents antinuclear autoimmunity. Nat Med. 1999;5(6):694–697. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]
- Blasius AL. Arnold CN. Georgel P. Rutschmann S. Xia Y. Lin P. Ross C. Li X. Smart NG. Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107(46):19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL. Beutler B. Intracellular Toll-like receptors. Immunity. 2010;32(3):305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Boldin MP. Taganov KD. Rao DS. Yang L. Zhao JL. Kalwani M. Garcia-Flores Y. Luong M. Devrekanli A. Xu J. Sun G. Tay J. Linsley PS. Baltimore D. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossu P. Neumann D. Del Giudice E. Ciaramella A. Gloaguen I. Fantuzzi G. Dinarello CA. Di Carlo E. Musiani P. Meroni PL. Caselli G. Ruggiero P. Boraschi D. IL-18 cDNA vaccination protects mice from spontaneous lupus-like autoimmune disease. Proc Natl Acad Sci U S A. 2003;100(24):14181–14186. doi: 10.1073/pnas.2336094100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto M. Dell'Agnola C. Bygrave AE. Thompson EM. Cook HT. Petry F. Loos M. Pandolfi PP. Walport MJ. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19(1):56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- Braun D. Geraldes P. Demengeot J. Type I Interferon controls the onset and severity of autoimmune manifestations in lpr mice. J Autoimmun. 2003;20(1):15–25. doi: 10.1016/s0896-8411(02)00109-9. [DOI] [PubMed] [Google Scholar]
- Brinkmann MM. Spooner E. Hoebe K. Beutler B. Ploegh HL. Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177(2):265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T. Baumann C. Bluml S. Dixit E. Durnberger G. Jahn H. Planyavsky M. Bilban M. Colinge J. Bennett KL. Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10(3):266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Casciola-Rosen LA. Anhalt G. Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179(4):1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A. Zhang SY. Eidenschenk C. Jouanguy E. Puel A. Yang K. Alcais A. Picard C. Mahfoufi N. Nicolas N. Lorenzo L. Plancoulaine S. Senechal B. Geissmann F. Tabeta K. Hoebe K. Du X. Miller RL. Heron B. Mignot C. de Villemeur TB. Lebon P. Dulac O. Rozenberg F. Beutler B. Tardieu M. Abel L. Casanova JL. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- Chaturvedi A. Dorward D. Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28(6):799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH. Macmillan JB. Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chockalingam A. Brooks JC. Cameron JL. Blum LK. Leifer CA. TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol Cell Biol. 2009;87(3):209–217. doi: 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SR. Kashgarian M. Alexopoulou L. Flavell RA. Akira S. Shlomchik MJ. Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J Exp Med. 2005;202(2):321–331. doi: 10.1084/jem.20050338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SR. Shupe J. Nickerson K. Kashgarian M. Flavell RA. Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25(3):417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Cirl C. Wieser A. Yadav M. Duerr S. Schubert S. Fischer H. Stappert D. Wantia N. Rodriguez N. Wagner H. Svanborg C. Miethke T. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14(4):399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- Cohen PL. Caricchio R. Abraham V. Camenisch TD. Jennette JC. Roubey RA. Earp HS. Matsushima G. Reap EA. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J Exp Med. 2002;196(1):135–140. doi: 10.1084/jem.20012094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll RC. O'Neill LA. New insights into the regulation of signalling by Toll-like receptors and nod-like receptors. J Innate Immun. 2010;2(5):406–421. doi: 10.1159/000315469. [DOI] [PubMed] [Google Scholar]
- Courtney PA. Crockard AD. Williamson K. Irvine AE. Kennedy RJ. Bell AL. Increased apoptotic peripheral blood neutrophils in systemic lupus erythematosus: relations with disease activity, antibodies to double stranded DNA, and neutropenia. Ann Rheum Dis. 1999;58(5):309–314. doi: 10.1136/ard.58.5.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker BA. Lawson BR. Berger M. Eidenschenk C. Blasius AL. Moresco EM. Sovath S. Cengia L. Shultz LD. Theofilopoulos AN. Pettersson S. Beutler BA. Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc Natl Acad Sci U S A. 2008;105(39):15028–15033. doi: 10.1073/pnas.0806619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ. Hayward BE. Parmar R. Robins P. Leitch A. Ali M. Black DN. van Bokhoven H. Brunner HG. Hamel BC. Corry PC. Cowan FM. Frints SG. Klepper J. Livingston JH. Lynch SA. Massey RF. Meritet JF. Michaud JL. Ponsot G. Voit T. Lebon P. Bonthron DT. Jackson AP. Barnes DE. Lindahl T. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006a;38(8):917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow YJ. Leitch A. Hayward BE. Garner A. Parmar R. Griffith E. Ali M. Semple C. Aicardi J. Babul-Hirji R. Baumann C. Baxter P. Bertini E. Chandler KE. Chitayat D. Cau D. Dery C. Fazzi E. Goizet C. King MD. Klepper J. Lacombe D. Lanzi G. Lyall H. Martinez-Frias ML. Mathieu M. McKeown C. Monier A. Oade Y. Quarrell OW. Rittey CD. Rogers RC. Sanchis A. Stephenson JB. Tacke U. Till M. Tolmie JL. Tomlin P. Voit T. Weschke B. Woods CG. Lebon P. Bonthron DT. Ponting CP. Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006b;38(8):910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Dai R. Zhang Y. Khan D. Heid B. Caudell D. Crasta O. Ahmed SA. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One. 2010;5(12):e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel H. Kottra G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch. 2004;447(5):610–618. doi: 10.1007/s00424-003-1101-4. [DOI] [PubMed] [Google Scholar]
- Deane JA. Pisitkun P. Barrett RS. Feigenbaum L. Town T. Ward JM. Flavell RA. Bolland S. Control of Toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27(5):801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis VR. Alvarado D. Sali T. Rothenburg S. Fruh K. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol. 2010;84(1):585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16(4):458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009;21(4):552–559. doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Demaria O. Pagni PP. Traub S. de Gassart A. Branzk N. Murphy AJ. Valenzuela DM. Yancopoulos GD. Flavell RA. Alexopoulou L. TLR8 deficiency leads to autoimmunity in mice. J Clin Invest. 2010;120(10):3651–3662. doi: 10.1172/JCI42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci FY. Manzi S. Ramsey-Goldman R. Kenney M. Shaw PS. Dunlop-Thomas CM. Kao AH. Rhew EY. Bontempo F. Kammerer C. Kamboh MI. Association study of Toll-like receptor 5 (TLR5) and Toll-like receptor 9 (TLR9) polymorphisms in systemic lupus erythematosus. J Rheumatol. 2007;34(8):1708–1711. [PubMed] [Google Scholar]
- Denny MF. Chandaroy P. Killen PD. Caricchio R. Lewis EE. Richardson BC. Lee KD. Gavalchin J. Kaplan MJ. Accelerated macrophage apoptosis induces autoantibody formation and organ damage in systemic lupus erythematosus. J Immunol. 2006;176(4):2095–2104. doi: 10.4049/jimmunol.176.4.2095. [DOI] [PubMed] [Google Scholar]
- Dixit E. Boulant S. Zhang Y. Lee AS. Odendall C. Shum B. Hacohen N. Chen ZJ. Whelan SP. Fransen M. Nibert ML. Superti-Furga G. Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141(4):668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski Y. Peric M. Koglin S. Kammerbauer C. Goss C. Anz D. Simanski M. Glaser R. Harder J. Hornung V. Gallo RL. Ruzicka T. Besch R. Schauber J. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3(82):82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes K. Pekalski M. Angus KL. Hardy M. Nutland S. Smyth DJ. Walker NM. Wallace C. Todd JA. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS One. 2010;5(9):ii–e12646. doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE. Engel A. Lee J. Wang M. Bogyo M. Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208(4):643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE. Lee BL. Lau L. Wickliffe KE. Shi GP. Chapman HA. Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456(7222):658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst AM. Mathian A. Connolly JE. Wang A. Gray HF. George TA. Boudreaux CD. Zhou XJ. Li QZ. Koutouzov S. Banchereau J. Wakeland EK. Systemic IFN-alpha drives kidney nephritis in B6.Sle123 mice. Eur J Immunol. 2008;1960;38(7):1948. doi: 10.1002/eji.200837925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T. Yu JW. Datta P. Wu J. Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA. The interferon inducible gene: Viperin. J Interferon Cytokine Res. 2011;31(1):131–135. doi: 10.1089/jir.2010.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei R. Steinle J. Birchler T. Loeliger S. Roduit C. Steinhoff D. Seibl R. Buchner K. Seger R. Reith W. Lauener RP. MHC class II molecules enhance Toll-like receptor mediated innate immune responses. PLoS One. 2010;5(1):e8808. doi: 10.1371/journal.pone.0008808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni L. McPhie L. Kang SA. Monestier M. Madaio M. Satoh M. Caricchio R. Lack of chromatin and nuclear fragmentation in vivo impairs the production of lupus anti-nuclear antibodies. J Immunol. 2007;179(11):7959–7966. doi: 10.4049/jimmunol.179.11.7959. [DOI] [PubMed] [Google Scholar]
- Fu Q. Zhao J. Qian X. Wong JL. Kaufman KM. Yu CY. Mok MY. Harley JB. Guthridge JM. Song YW. Cho SK. Bae SC. Grossman JM. Hahn BH. Arnett FC. Shen N. Tsao BP. Association of a functional IRF7 variant with systemic lupus erythematosus. Arthritis Rheum. 2011;63(3):749–754. doi: 10.1002/art.30193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui R. Saitoh S. Matsumoto F. Kozuka-Hata H. Oyama M. Tabeta K. Beutler B. Miyake K. Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J Exp Med. 2009;206(6):1339–1350. doi: 10.1084/jem.20082316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui R. Saitoh SI. Kanno A. Onji M. Shibata T. Ito A. Matsumoto M. Akira S. Yoshida N. Miyake K. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity. 2011;35(1):69–81. doi: 10.1016/j.immuni.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Gack MU. Kirchhofer A. Shin YC. Inn KS. Liang C. Cui S. Myong S. Ha T. Hopfner KP. Jung JU. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci U S A. 2008;105(43):16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D. Chamilos G. Lande R. Gregorio J. Meller S. Facchinetti V. Homey B. Barrat FJ. Zal T. Gilliet M. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. 2009;206(9):1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ortiz H. Velazquez-Cruz R. Espinosa-Rosales F. Jimenez-Morales S. Baca V. Orozco L. Association of TLR7 copy number variation with susceptibility to childhood-onset systemic lupus erythematosus in Mexican population. Ann Rheum Dis. 2010;69(10):1861–1865. doi: 10.1136/ard.2009.124313. [DOI] [PubMed] [Google Scholar]
- Garcia-Romo GS. Caielli S. Vega B. Connolly J. Allantaz F. Xu Z. Punaro M. Baisch J. Guiducci C. Coffman RL. Barrat FJ. Banchereau J. Pascual V. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay NJ. Gangloff M. O'Neill LA. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011;32(3):104–109. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Hacker H. Mischak H. Miethke T. Liptay S. Schmid R. Sparwasser T. Heeg K. Lipford GB. Wagner H. CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 1998;17(21):6230–6240. doi: 10.1093/emboj/17.21.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R. Tanaka M. Miyasaka K. Aozasa K. Koike M. Uchiyama Y. Nagata S. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304(5674):1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- Hardin JA. The lupus autoantigens and the pathogenesis of systemic lupus erythematosus. Arthritis Rheum. 1986;29(4):457–460. doi: 10.1002/art.1780290401. [DOI] [PubMed] [Google Scholar]
- Harley IT. Kaufman KM. Langefeld CD. Harley JB. Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10(5):285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan GS. Mourad W. An unexpected role for MHC class II. Nat Immunol. 2011;12(5):375–376. doi: 10.1038/ni.2023. [DOI] [PubMed] [Google Scholar]
- Hayakawa S. Shiratori S. Yamato H. Kameyama T. Kitatsuji C. Kashigi F. Goto S. Kameoka S. Fujikura D. Yamada T. Mizutani T. Kazumata M. Sato M. Tanaka J. Asaka M. Ohba Y. Miyazaki T. Imamura M. Takaoka A. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol. 2011;12(1):37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- Hirai M. Kadowaki N. Kitawaki T. Fujita H. Takaori-Kondo A. Fukui R. Miyake K. Maeda T. Kamihira S. Miyachi Y. Uchiyama T. Bortezomib suppresses function and survival of plasmacytoid dendritic cells by targeting intracellular trafficking of Toll-like receptors and endoplasmic reticulum homeostasis. Blood. 2011;117(2):500–509. doi: 10.1182/blood-2010-05-284737. [DOI] [PubMed] [Google Scholar]
- Honda K. Ohba Y. Yanai H. Negishi H. Mizutani T. Takaoka A. Taya C. Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434(7036):1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- Hornung V. Ablasser A. Charrel-Dennis M. Bauernfeind F. Horvath G. Caffrey DR. Latz E. Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing LC. Kirk EA. McMillen TS. Hsiao SH. Caldwell M. Houston B. Rudensky AY. LeBoeuf RC. Roles for cathepsins S, L, and B in insulitis and diabetes in the NOD mouse. J Autoimmun. 2010;34(2):96–104. doi: 10.1016/j.jaut.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado P. Peh CA. LL-37 promotes rapid sensing of CpG oligodeoxynucleotides by B lymphocytes and plasmacytoid dendritic cells. J Immunol. 2010;184(3):1425–1435. doi: 10.4049/jimmunol.0902305. [DOI] [PubMed] [Google Scholar]
- Imaizumi T. Arikawa T. Sato T. Uesato R. Matsumiya T. Yoshida H. Ueno M. Yamasaki S. Nakajima T. Hirashima M. Sakata K. Ishibashi Y. Toh S. Ohyama C. Satoh K. Involvement of retinoic acid-inducible gene-I in inflammation of rheumatoid fibroblast-like synoviocytes. Clin Exp Immunol. 2008;153(2):240–244. doi: 10.1111/j.1365-2249.2008.03685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ. Coban C. Kato H. Takahashi K. Torii Y. Takeshita F. Ludwig H. Sutter G. Suzuki K. Hemmi H. Sato S. Yamamoto M. Uematsu S. Kawai T. Takeuchi O. Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7(1):40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishii KJ. Kawagoe T. Koyama S. Matsui K. Kumar H. Kawai T. Uematsu S. Takeuchi O. Takeshita F. Coban C. Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451(7179):725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H. Ma Z. Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnardi I. Ng YS. Srdanovic I. Motaghedi R. Rudchenko S. von Bernuth H. Zhang SY. Puel A. Jouanguy E. Picard C. Garty BZ. Camcioglu Y. Doffinger R. Kumararatne D. Davies G. Gallin JI. Haraguchi S. Day NK. Casanova JL. Meffre E. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29(5):746–757. doi: 10.1016/j.immuni.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S. Dragoi AM. Wang X. Dallacosta C. Louten J. Musco G. Sitia G. Yap GS. Wan Y. Biron CA. Bianchi ME. Wang H. Chu WM. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007;110(6):1970–1981. doi: 10.1182/blood-2006-09-044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob N. Guo S. Mathian A. Koss MN. Gindea S. Putterman C. Jacob CO. Stohl W. B Cell and BAFF dependence of IFN-{alpha}-exaggerated disease in systemic lupus erythematosus-prone NZM 2328 mice. J Immunol. 2011;186(8):4984–4993. doi: 10.4049/jimmunol.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen E. Tabeta K. Barnes MJ. Rutschmann S. McBride S. Bahjat KS. Schoenberger SP. Theofilopoulos AN. Beutler B. Hoebe K. Efficient T cell activation via a Toll-Interleukin 1 Receptor-independent pathway. Immunity. 2006;24(6):787–799. doi: 10.1016/j.immuni.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Jiang X. Chen ZJ. Viperin links lipid bodies to immune defense. Immunity. 2011;34(3):285–287. doi: 10.1016/j.immuni.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh D. Spitzer D. Kothari PH. Shaikh A. Liszewski MK. Richards A. Atkinson JP. New roles for the major human 3'-5' exonuclease TREX1 in human disease. Cell Cycle. 2008;7(12):1718–1725. doi: 10.4161/cc.7.12.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S. Ishiguro Y. Imaizumi T. Mori F. Matsumiya T. Yoshida H. Ota K. Sakuraba H. Yamagata K. Sato Y. Tanji K. Haga T. Wakabayashi K. Fukuda S. Satoh K. Retinoic acid-inducible gene-I is constitutively expressed and involved in IFN-gamma-stimulated CXCL9-11 production in intestinal epithelial cells. Immunol Lett. 2009;123(1):9–13. doi: 10.1016/j.imlet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Kawai T. Akira S. TLR signaling. Cell Death Differ. 2006;13(5):816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- Kawasaki A. Furukawa H. Kondo Y. Ito S. Hayashi T. Kusaoi M. Matsumoto I. Tohma S. Takasaki Y. Hashimoto H. Sumida T. Tsuchiya N. TLR7 single-nucleotide polymorphisms in the 3' untranslated region and intron 2 independently contribute to systemic lupus erythematosus in Japanese women: a case-control association study. Arthritis Res Ther. 2011;13(2):R41. doi: 10.1186/ar3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J. Johnson MR. Alarcon GS. Kimberly RP. Edberg JC. Variation in the relative copy number of the TLR7 gene in patients with systemic lupus erythematosus and healthy control subjects. Arthritis Rheum. 2007;56(10):3375–3378. doi: 10.1002/art.22916. [DOI] [PubMed] [Google Scholar]
- Kenny EF. O'Neill LA. Signalling adaptors used by Toll-like receptors: an update. Cytokine. 2008;43(3):342–349. doi: 10.1016/j.cyto.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Kerur N. Veettil MV. Sharma-Walia N. Bottero V. Sadagopan S. Otageri P. Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K. Krumbholz M. Schonermarck U. Back W. Gross WL. Werb Z. Grone HJ. Brinkmann V. Jenne DE. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med. 2009;15(6):623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ. Yoo JY. Active caspase-1-mediated secretion of retinoic acid inducible gene-I. J Immunol. 2008;181(10):7324–7331. doi: 10.4049/jimmunol.181.10.7324. [DOI] [PubMed] [Google Scholar]
- Kim T. Pazhoor S. Bao M. Zhang Z. Hanabuchi S. Facchinetti V. Bover L. Plumas J. Chaperot L. Qin J. Liu YJ. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase a helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107(34):15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimkong I. Avihingsanon Y. Hirankarn N. Expression profile of HIN200 in leukocytes and renal biopsy of SLE patients by real-time RT-PCR. Lupus. 2009;18(12):1066–1072. doi: 10.1177/0961203309106699. [DOI] [PubMed] [Google Scholar]
- Kimkong I. Avihingsanon Y. Hirankarn N. Association of IFI200 gene polymorphisms with susceptibility to systemic lupus erythematosus. J Rheumatol. 2010;37(7):1544–1547. doi: 10.3899/jrheum.091255. [DOI] [PubMed] [Google Scholar]
- Kirou KA. Lee C. George S. Louca K. Peterson MG. Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52(5):1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- Kitamura H. Matsuzaki Y. Kimura K. Nakano H. Imaizumi T. Satoh K. Hanada K. Cytokine modulation of retinoic acid-inducible gene-I (RIG-I) expression in human epidermal keratinocytes. J Dermatol Sci. 2007;45(2):127–134. doi: 10.1016/j.jdermsci.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Kok KH. Lui PY. Ng MH. Siu KL. Au SW. Jin DY. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. 2011;9(4):299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Kono DH. Haraldsson MK. Lawson BR. Pollard KM. Koh YT. Du X. Arnold CN. Baccala R. Silverman GJ. Beutler BA. Theofilopoulos AN. Endosomal TLR signaling is required for anti-nucleic acid and rheumatoid factor autoantibodies in lupus. Proc Natl Acad Sci U S A. 2009;106(29):12061–12066. doi: 10.1073/pnas.0905441106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T. Yasukawa K. Yanagi Y. Kawabata S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci Signal. 2011;4(158):ra7. doi: 10.1126/scisignal.2001147. [DOI] [PubMed] [Google Scholar]
- Lande R. Ganguly D. Facchinetti V. Frasca L. Conrad C. Gregorio J. Meller S. Chamilos G. Sebasigari R. Riccieri V. Bassett R. Amuro H. Fukuhara S. Ito T. Liu YJ. Gilliet M. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Gregorio J. Facchinetti V. Chatterjee B. Wang YH. Homey B. Cao W. Wang YH. Su B. Nestle FO. Zal T. Mellman I. Schroder JM. Liu YJ. Gilliet M. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449(7162):564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- Lang T. Mansell A. The negative regulation of Toll-like receptor and associated pathways. Immunol Cell Biol. 2007;85(6):425–434. doi: 10.1038/sj.icb.7100094. [DOI] [PubMed] [Google Scholar]
- Latz E. Verma A. Visintin A. Gong M. Sirois CM. Klein DC. Monks BG. McKnight CJ. Lamphier MS. Duprex WP. Espevik T. Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8(7):772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- Leadbetter EA. Rifkin IR. Hohlbaum AM. Beaudette BC. Shlomchik MJ. Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416(6881):603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- Lech M. Kulkarni OP. Pfeiffer S. Savarese E. Krug A. Garlanda C. Mantovani A. Anders HJ. Tir8/Sigirr prevents murine lupus by suppressing the immunostimulatory effects of lupus autoantigens. J Exp Med. 2008;205(8):1879–1888. doi: 10.1084/jem.20072646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Kirsch MA. Gong M. Chowdhury D. Senenko L. Engel K. Lee YA. de Silva U. Bailey SL. Witte T. Vyse TJ. Kere J. Pfeiffer C. Harvey S. Wong A. Koskenmies S. Hummel O. Rohde K. Schmidt RE. Dominiczak AF. Gahr M. Hollis T. Perrino FW. Lieberman J. Hubner N. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39(9):1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- Lemke G. Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8(5):327–336. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ. Botto M. Complement deficiencies in humans and animals: links to autoimmunity. Autoimmunity. 2006;39(5):367–378. doi: 10.1080/08916930600739233. [DOI] [PubMed] [Google Scholar]
- Lin SC. Lo YC. Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. Gu J. Retinoic acid inducible gene-I, more than a virus sensor. Protein Cell. 2011;2(5):351–357. doi: 10.1007/s13238-011-1045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. Wu S. Ren H. Gu J. Klotho suppresses RIG-I-mediated senescence-associated inflammation. Nat Cell Biol. 2011a;13(3):254–262. doi: 10.1038/ncb2167. [DOI] [PubMed] [Google Scholar]
- Liu HM. Gale M., Jr ZAPS electrifies RIG-I signaling. Nat Immunol. 2011;12(1):11–12. doi: 10.1038/ni0111-11. [DOI] [PubMed] [Google Scholar]
- Liu X. Zhan Z. Li D. Xu L. Ma F. Zhang P. Yao H. Cao X. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat Immunol. 2011b;12(5):416–424. doi: 10.1038/ni.2015. [DOI] [PubMed] [Google Scholar]
- Liu Z. Bethunaickan R. Huang W. Lodhi U. Solano I. Madaio MP. Davidson A. Interferon-alpha accelerates murine systemic lupus erythematosus in a T cell-dependent manner. Arthritis Rheum. 2011c;63(1):219–229. doi: 10.1002/art.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM. Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. Lemke G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science. 2001;293(5528):306–311. doi: 10.1126/science.1061663. [DOI] [PubMed] [Google Scholar]
- Maehr R. Mintern JD. Herman AE. Lennon-Dumenil AM. Mathis D. Benoist C. Ploegh HL. Cathepsin L is essential for onset of autoimmune diabetes in NOD mice. J Clin Invest. 2005;115(10):2934–2943. doi: 10.1172/JCI25485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A. Cassatella MA. Costantini C. Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11(8):519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- Marshak-Rothstein A. Rifkin IR. Immunologically active autoantigens: the role of Toll-like receptors in the development of chronic inflammatory disease. Annu Rev Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- Mathian A. Gallegos M. Pascual V. Banchereau J. Koutouzov S. Interferon-alpha induces unabated production of short-lived plasma cells in pre-autoimmune lupus-prone (NZBxNZW)F1 mice but not in BALB/c mice. Eur J Immunol. 2011;41(3):863–872. doi: 10.1002/eji.201040649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathian A. Weinberg A. Gallegos M. Banchereau J. Koutouzov S. IFN-alpha induces early lethal lupus in preautoimmune (New Zealand Black x New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005;174(5):2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- Matsumiya T. Stafforini DM. Function and regulation of retinoic acid-inducible gene-I. Crit Rev Immunol. 2010;30(6):489–513. doi: 10.1615/critrevimmunol.v30.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto F. Saitoh S. Fukui R. Kobayashi T. Tanimura N. Konno K. Kusumoto Y. Akashi-Takamura S. Miyake K. Cathepsins are required for Toll-like receptor 9 responses. Biochem Biophys Res Commun. 2008;367(3):693–699. doi: 10.1016/j.bbrc.2007.12.130. [DOI] [PubMed] [Google Scholar]
- Mattera R. Boehm M. Chaudhuri R. Prabhu Y. Bonifacino JS. Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J Biol Chem. 2011;286(3):2022–2030. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney SA. Vermi W. Lonardi S. Rossini C. Otero K. Calderon B. Gilfillan S. Diamond MS. Unanue ER. Colonna M. RNA sensor-induced type I IFN prevents diabetes caused by a beta cell-tropic virus in mice. J Clin Invest. 2011;121(4):1497–1507. doi: 10.1172/JCI44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevorach D. Zhou JL. Song X. Elkon KB. Systemic exposure to irradiated apoptotic cells induces autoantibody production. J Exp Med. 1998;188(2):387–392. doi: 10.1084/jem.188.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JG. Keir ME. Bridging Toll-like- and B cell-receptor signaling: meet me at the autophagosome. Immunity. 2008;28(6):729–731. doi: 10.1016/j.immuni.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Moore CB. Bergstralh DT. Duncan JA. Lei Y. Morrison TE. Zimmermann AG. Accavitti-Loper MA. Madden VJ. Sun L. Ye Z. Lich JD. Heise MT. Chen Z. Ting JP. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451(7178):573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Moore CB. Ting JP. Regulation of mitochondrial antiviral signaling pathways. Immunity. 2008;28(6):735–739. doi: 10.1016/j.immuni.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Moresco EM. Beutler B. LGP2: positive about viral sensing. Proc Natl Acad Sci U S A. 2010;107(4):1261–1262. doi: 10.1073/pnas.0914011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco EM. Beutler B. Special delivery: granulin brings CpG DNA to Toll-like receptor 9. Immunity. 2011;34(4):453–455. doi: 10.1016/j.immuni.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco EM. Lavine D. Beutler B. Toll-like receptors. Curr Biol. 2011;21(13):R488–493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Motshwene PG. Moncrieffe MC. Grossmann JG. Kao C. Ayaluru M. Sandercock AM. Robinson CV. Latz E. Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284(37):25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan L. Odegaard JI. Morel CR. Heredia JE. Mwangi JW. Ricardo-Gonzalez RR. Goh YP. Eagle AR. Dunn SE. Awakuni JU. Nguyen KD. Steinman L. Michie SA. Chawla A. PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat Med. 2009;15(11):1266–1272. doi: 10.1038/nm.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz LE. Janko C. Schulze C. Schorn C. Sarter K. Schett G. Herrmann M. Autoimmunity and chronic inflammation—two clearance-related steps in the etiopathogenesis of SLE. Autoimmun Rev. 2010;10(1):38–42. doi: 10.1016/j.autrev.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Muruve DA. Petrilli V. Zaiss AK. White LR. Clark SA. Ross PJ. Parks RJ. Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452(7183):103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Nagata S. Rheumatoid polyarthritis caused by a defect in DNA degradation. Cytokine Growth Factor Rev. 2008;19(3–4):295–302. doi: 10.1016/j.cytogfr.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Nagpal K. Plantinga TS. Sirois CM. Monks BG. Latz E. Netea MG. Golenbock DT. Natural loss-of-function mutation of myeloid differentiation protein 88 disrupts its ability to form Myddosomes. J Biol Chem. 2011;286(13):11875–11882. doi: 10.1074/jbc.M110.199653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napirei M. Karsunky H. Zevnik B. Stephan H. Mannherz HG. Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25(2):177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- Nejentsev S. Walker N. Riches D. Egholm M. Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert K. Meister S. Moser K. Weisel F. Maseda D. Amann K. Wiethe C. Winkler TH. Kalden JR. Manz RA. Voll RE. The proteasome inhibitor bortezomib depletes plasma cells and protects mice with lupus-like disease from nephritis. Nat Med. 2008;14(7):748–755. doi: 10.1038/nm1763. [DOI] [PubMed] [Google Scholar]
- Nickerson KM. Christensen SR. Shupe J. Kashgarian M. Kim D. Elkon K. Shlomchik MJ. TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J Immunol. 2010;184(4):1840–1848. doi: 10.4049/jimmunol.0902592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CU. Brodin B. Di/tri-peptide transporters as drug delivery targets: regulation of transport under physiological and patho-physiological conditions. Curr Drug Targets. 2003;4(5):373–388. doi: 10.2174/1389450033491028. [DOI] [PubMed] [Google Scholar]
- Nikpour M. Dempsey AA. Urowitz MB. Gladman DD. Barnes DA. Association of a gene expression profile from whole blood with disease activity in systemic lupus erythaematosus. Ann Rheum Dis. 2008;67(8):1069–1075. doi: 10.1136/ard.2007.074765. [DOI] [PubMed] [Google Scholar]
- Obermoser G. Pascual V. The interferon-alpha signature of systemic lupus erythematosus. Lupus. 2010;19(9):1012–1019. doi: 10.1177/0961203310371161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoguchi K. Onomoto K. Takamatsu S. Jogi M. Takemura A. Morimoto S. Julkunen I. Namiki H. Yoneyama M. Fujita T. Virus-infection or 5'ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 2010;6(7):e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H. Matsumoto M. Hatakeyama S. Seya T. Riplet/RNF135, a RING finger protein, Ubiquitinates RIG-I to promote interferon-{beta} induction during the early phase of viral infection. J Biol Chem. 2009;284(2):807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- Panchanathan R. Duan X. Shen H. Rathinam VA. Erickson LD. Fitzgerald KA. Choubey D. Aim2 deficiency stimulates the expression of IFN-inducible Ifi202, a lupus susceptibility murine gene within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;185(12):7385–7393. doi: 10.4049/jimmunol.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R. Shen H. Duan X. Rathinam VA. Erickson LD. Fitzgerald KA. Choubey D. Aim2 deficiency in mice suppresses the expression of the inhibitory Fc{gamma} receptor (Fc{gamma}RIIB) through the induction of the IFN-Inducible p202, a lupus susceptibility protein. J Immunol. 2011;186(12):6762–6770. doi: 10.4049/jimmunol.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao LI. Lam KP. Henderson JM. Kutok JL. Alimzhanov M. Nitschke L. Thomas ML. Neel BG. Rajewsky K. B cell-specific deletion of protein-tyrosine phosphatase Shp1 promotes B-1a cell development and causes systemic autoimmunity. Immunity. 2007;27(1):35–48. doi: 10.1016/j.immuni.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Park B. Brinkmann MM. Spooner E. Lee CC. Kim YM. Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9(12):1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. Buti L. Lee S. Matsuwaki T. Spooner E. Brinkmann MM. Nishihara M. Ploegh HL. Granulin is a soluble cofactor for Toll-like receptor 9 signaling. Immunity. 2011;34(4):505–513. doi: 10.1016/j.immuni.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Paul E. Carroll MC. SAP-less chromatin triggers systemic lupus erythematosus. Nat Med. 1999;5(6):607–608. doi: 10.1038/9450. [DOI] [PubMed] [Google Scholar]
- Pauley KM. Cha S. Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32(3–4):189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino FW. Harvey S. Shaban NM. Hollis T. RNaseH2 mutants that cause Aicardi-Goutieres syndrome are active nucleases. J Mol Med. 2009;87(1):25–30. doi: 10.1007/s00109-008-0422-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri M. Singh S. Tesfasyone H. Dedrick R. Fry K. Lal P. Williams G. Bauer J. Gregersen P. Behrens T. Baechler E. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 2009;18(11):980–989. doi: 10.1177/0961203309105529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A. Schulz O. Tan CP. Naslund TI. Liljestrom P. Weber F. Reis ESC. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5' phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pisitkun P. Deane JA. Difilippantonio MJ. Tarasenko T. Satterthwaite AB. Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312(5780):1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Poeck H. Bscheider M. Gross O. Finger K. Roth S. Rebsamen M. Hannesschlager N. Schlee M. Rothenfusser S. Barchet W. Kato H. Akira S. Inoue S. Endres S. Peschel C. Hartmann G. Hornung V. Ruland J. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11(1):63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Poole BD. Scofield RH. Harley JB. James JA. Epstein-Barr virus and molecular mimicry in systemic lupus erythematosus. Autoimmunity. 2006;39(1):63–70. doi: 10.1080/08916930500484849. [DOI] [PubMed] [Google Scholar]
- Prens EP. Kant M. van Dijk G. van der Wel LI. Mourits S. van der Fits L. IFN-alpha enhances poly-IC responses in human keratinocytes by inducing expression of cytosolic innate RNA receptors: relevance for psoriasis. J Invest Dermatol. 2008;128(4):932–938. doi: 10.1038/sj.jid.5701087. [DOI] [PubMed] [Google Scholar]
- Rajput A. Kovalenko A. Bogdanov K. Yang SH. Kang TB. Kim JC. Du J. Wallach D. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity. 2011;34(3):340–351. doi: 10.1016/j.immuni.2010.12.018. [DOI] [PubMed] [Google Scholar]
- Ramanujam M. Kahn P. Huang W. Tao H. Madaio MP. Factor SM. Davidson A. Interferon-alpha treatment of female (NZW x BXSB)F(1) mice mimics some but not all features associated with the Yaa mutation. Arthritis Rheum. 2009;60(4):1096–1101. doi: 10.1002/art.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F. Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3(10):891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- Rice GI. Bond J. Asipu A. Brunette RL. Manfield IW. Carr IM. Fuller JC. Jackson RM. Lamb T. Briggs TA. Ali M. Gornall H. Couthard LR. Aeby A. Attard-Montalto SP. Bertini E. Bodemer C. Brockmann K. Brueton LA. Corry PC. Desguerre I. Fazzi E. Cazorla AG. Gener B. Hamel BC. Heiberg A. Hunter M. van der Knaap MS. Kumar R. Lagae L. Landrieu PG. Lourenco CM. Marom D. McDermott MF. van der Merwe W. Orcesi S. Prendiville JS. Rasmussen M. Shalev SA. Soler DM. Shinawi M. Spiegel R. Tan TY. Vanderver A. Wakeling EL. Wassmer E. Whittaker E. Lebon P. Stetson DB. Bonthron DT. Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemekasten G. Hahn BH. Key autoantigens in SLE. Rheumatology. 2005;44(8):975–982. doi: 10.1093/rheumatology/keh688. [DOI] [PubMed] [Google Scholar]
- Rintahaka J. Wiik D. Kovanen PE. Alenius H. Matikainen S. Cytosolic antiviral RNA recognition pathway activates caspases 1 and 3. J Immunol. 2008;180(3):1749–1757. doi: 10.4049/jimmunol.180.3.1749. [DOI] [PubMed] [Google Scholar]
- Roberts TL. Idris A. Dunn JA. Kelly GM. Burnton CM. Hodgson S. Hardy LL. Garceau V. Sweet MJ. Ross IL. Hume DA. Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Ronnblom L. Eloranta ML. Alm GV. The type I interferon system in systemic lupus erythematosus. Arthritis Rheum. 2006;54(2):408–420. doi: 10.1002/art.21571. [DOI] [PubMed] [Google Scholar]
- Rothlin CV. Ghosh S. Zuniga EI. Oldstone MB. Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131(6):1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Rozzo SJ. Allard JD. Choubey D. Vyse TJ. Izui S. Peltz G. Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15(3):435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- Ruan BH. Li X. Winkler AR. Cunningham KM. Kuai J. Greco RM. Nocka KH. Fitz LJ. Wright JF. Pittman DD. Tan XY. Paulsen JE. Lin LL. Winkler DG. Complement C3a, CpG oligos, and DNA/C3a complex stimulate IFN-alpha production in a receptor for advanced glycation end product-dependent manner. J Immunol. 2010;185(7):4213–4222. doi: 10.4049/jimmunol.1000863. [DOI] [PubMed] [Google Scholar]
- Sabbah A. Chang TH. Harnack R. Frohlich V. Tominaga K. Dube PH. Xiang Y. Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10(10):1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa K. Ishimaru N. Yanagi K. Arakaki R. Ogawa K. Saito I. Katunuma N. Hayashi Y. Cathepsin S inhibitor prevents autoantigen presentation and autoimmunity. J Clin Invest. 2002;110(3):361–369. doi: 10.1172/JCI14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T. Fujita N. Hayashi T. Takahara K. Satoh T. Lee H. Matsunaga K. Kageyama S. Omori H. Noda T. Yamamoto N. Kawai T. Ishii K. Takeuchi O. Yoshimori T. Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106(49):20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T. Satoh T. Yamamoto N. Uematsu S. Takeuchi O. Kawai T. Akira S. Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity. 2011;34(3):352–363. doi: 10.1016/j.immuni.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Sanchez E. Callejas-Rubio JL. Sabio JM. Gonzalez-Gay MA. Jimenez-Alonso J. Mico L. de Ramon E. Camps M. Suarez A. Gutierrez C. Garcia-Portales R. Tolosa C. Ortego-Centeno N. Sanchez-Roman J. Garcia-Hernandez FJ. Gonzalez-Escribano MF. Martin J. Investigation of TLR5 and TLR7 as candidate genes for susceptibility to systemic lupus erythematosus. Clin Exp Rheumatol. 2009;27(2):267–271. [PubMed] [Google Scholar]
- Santiago-Raber ML. Baccala R. Haraldsson KM. Choubey D. Stewart TA. Kono DH. Theofilopoulos AN. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197(6):777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Raber ML. Dunand-Sauthier I. Wu T. Li QZ. Uematsu S. Akira S. Reith W. Mohan C. Kotzin BL. Izui S. Critical role of TLR7 in the acceleration of systemic lupus erythematosus in TLR9-deficient mice. J Autoimmun. 2010;34(4):339–348. doi: 10.1016/j.jaut.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Santiago-Raber ML. Kikuchi S. Borel P. Uematsu S. Akira S. Kotzin BL. Izui S. Evidence for genes in addition to Tlr7 in the Yaa translocation linked with acceleration of systemic lupus erythematosus. J Immunol. 2008;181(2):1556–1562. doi: 10.4049/jimmunol.181.2.1556. [DOI] [PubMed] [Google Scholar]
- Sasai M. Linehan MM. Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329(5998):1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T. Kato H. Kumagai Y. Yoneyama M. Sato S. Matsushita K. Tsujimura T. Fujita T. Akira S. Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107(4):1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P. Misteli T. Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418(6894):191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Schlee M. Hartmann E. Coch C. Wimmenauer V. Janke M. Barchet W. Hartmann G. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009;227(1):66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Schoggins JW. Wilson SJ. Panis M. Murphy MY. Jones CT. Bieniasz P. Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorlemmer HU. Kanzy EJ. Langner KD. Kurrle R. Immunoregulation of SLE-like disease by the IL-1 receptor: disease modifying activity on BDF1 hybrid mice and MRL autoimmune mice. Agents Actions. 1993;39:C117–120. doi: 10.1007/BF01972740. Spec No. [DOI] [PubMed] [Google Scholar]
- Schulz O. Pichlmair A. Rehwinkel J. Rogers NC. Scheuner D. Kato H. Takeuchi O. Akira S. Kaufman RJ. Reis e Sousa C. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7(5):354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda FE. Maschalidi S. Colisson R. Heslop L. Ghirelli C. Sakka E. Lennon-Dumenil AM. Amigorena S. Cabanie L. Manoury B. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31(5):737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Sharma S. Fitzgerald KA. Innate immune sensing of DNA. PLoS Pathog. 2011;7(4):e1001310. doi: 10.1371/journal.ppat.1001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N. Fu Q. Deng Y. Qian X. Zhao J. Kaufman KM. Wu YL. Yu CY. Tang Y. Chen JY. Yang W. Wong M. Kawasaki A. Tsuchiya N. Sumida T. Kawaguchi Y. Howe HS. Mok MY. Bang SY. Liu FL. Chang DM. Takasaki Y. Hashimoto H. Harley JB. Guthridge JM. Grossman JM. Cantor RM. Song YW. Bae SC. Chen S. Hahn BH. Lau YL. Tsao BP. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2010;107(36):15838–15843. doi: 10.1073/pnas.1001337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto T. Kageyama M. Hirai R. Zheng J. Yoneyama M. Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284(20):13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack J. Haga IR. Schroder M. Bartlett NW. Maloney G. Reading PC. Fitzgerald KA. Smith GL. Bowie AG. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J Exp Med. 2005;201(6):1007–1018. doi: 10.1084/jem.20041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB. Ko JS. Heidmann T. Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB. Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24(1):93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Subramanian S. Tus K. Li QZ. Wang A. Tian XH. Zhou J. Liang C. Bartov G. McDaniel LD. Zhou XJ. Schultz RA. Wakeland EK. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103(26):9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K. Hoebe K. Janssen EM. Du X. Georgel P. Crozat K. Mudd S. Mann N. Sovath S. Goode J. Shamel L. Herskovits AA. Portnoy DA. Cooke M. Tarantino LM. Wiltshire T. Steinberg BE. Grinstein S. Beutler B. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7(2):156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Tada Y. Kondo S. Aoki S. Koarada S. Inoue H. Suematsu R. Ohta A. Mak TW. Nagasawa K. Interferon regulatory factor 5 is critical for the development of lupus in MRL/lpr mice. Arthritis Rheum. 2011;63(3):738–748. doi: 10.1002/art.30183. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Shibata T. Akashi-Takamura S. Kiyokawa T. Wakabayashi Y. Tanimura N. Kobayashi T. Matsumoto F. Fukui R. Kouro T. Nagai Y. Takatsu K. Saitoh S. Miyake K. A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J Exp Med. 2007;204(12):2963–2976. doi: 10.1084/jem.20071132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A. Wang Z. Choi MK. Yanai H. Negishi H. Ban T. Lu Y. Miyagishi M. Kodama T. Honda K. Ohba Y. Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takemura Y. Ouchi N. Shibata R. Aprahamian T. Kirber MT. Summer RS. Kihara S. Walsh K. Adiponectin modulates inflammatory reactions via calreticulin receptor-dependent clearance of early apoptotic bodies. J Clin Invest. 2007;117(2):375–386. doi: 10.1172/JCI29709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O. Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20(1):17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Tan EM. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- Taneichi-Kuroda S. Taya S. Hikita T. Fujino Y. Kaibuchi K. Direct interaction of Dysbindin with the AP-3 complex via its mu subunit. Neurochem Int. 2009;54(7):431–438. doi: 10.1016/j.neuint.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Tang Y. Luo X. Cui H. Ni X. Yuan M. Guo Y. Huang X. Zhou H. de Vries N. Tak PP. Chen S. Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN. Baccala R. Beutler B. Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–335. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN. Dixon FJ. Murine models of systemic lupus erythematosus. Adv Immunol. 1985;37:269–390. doi: 10.1016/s0065-2776(08)60342-9. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN. Gonzalez-Quintial R. Lawson BR. Koh YT. Stern ME. Kono DH. Beutler B. Baccala R. Sensors of the innate immune system: their link to rheumatic diseases. Nat Rev Rheumatol. 2010;6(3):146–156. doi: 10.1038/nrrheum.2009.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J. Avalos AM. Mao SY. Chen B. Senthil K. Wu H. Parroche P. Drabic S. Golenbock D. Sirois C. Hua J. An LL. Audoly L. La Rosa G. Bierhaus A. Naworth P. Marshak-Rothstein A. Crow MK. Fitzgerald KA. Latz E. Kiener PA. Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8(5):487–196. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- Triantafyllopoulou A. Franzke CW. Seshan SV. Perino G. Kalliolias GD. Ramanujam M. van Rooijen N. Davidson A. Ivashkiv LB. Proliferative lesions and metalloproteinase activity in murine lupus nephritis mediated by type I interferons and macrophages. Proc Natl Acad Sci U S A. 2010;107(7):3012–3017. doi: 10.1073/pnas.0914902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S. Akira S. Toll-like receptors and innate immunity. J Mol Med. 2006;84(9):712–725. doi: 10.1007/s00109-006-0084-y. [DOI] [PubMed] [Google Scholar]
- Ullal AJ. Reich CF., 3rd Clowse M. Criscione-Schreiber LG. Tochacek M. Monestier M. Pisetsky DS. Microparticles as antigenic targets of antibodies to DNA and nucleosomes in systemic lupus erythematosus. J Autoimmun. 2011;36(3–4):173–180. doi: 10.1016/j.jaut.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Unterholzner L. Bowie AG. Innate DNA sensing moves to the nucleus. Cell Host Microbe. 2011;9(5):351–353. doi: 10.1016/j.chom.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Unterholzner L. Keating SE. Baran M. Horan KA. Jensen SB. Sharma S. Sirois CM. Jin T. Latz E. Xiao TS. Fitzgerald KA. Paludan SR. Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonaviciute V. Voll RE. HMGB1 represents a potential marker of disease activity and novel therapeutic target in SLE. J Intern Med. 2011;270(4):309–318. doi: 10.1111/j.1365-2796.2011.02432.x. [DOI] [PubMed] [Google Scholar]
- Viglianti GA. Lau CM. Hanley TM. Miko BA. Shlomchik MJ. Marshak-Rothstein A. Activation of autoreactive B cells by CpG dsDNA. Immunity. 2003;19(6):837–847. doi: 10.1016/s1074-7613(03)00323-6. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG. Rigby RJ. Yu D. Logic and extent of miRNA-mediated control of autoimmune gene expression. Int Rev Immunol. 2009;28(3–4):112–138. doi: 10.1080/08830180902934909. [DOI] [PubMed] [Google Scholar]
- Viorritto IC. Nikolov NP. Siegel RM. Autoimmunity versus tolerance: can dying cells tip the balance? Clin Immunol. 2007;122(2):125–134. doi: 10.1016/j.clim.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Shao Y. Bennett TA. Shankar RA. Wightman PD. Reddy LG. The functional effects of physical interactions among Toll-like receptors 7, 8, and 9. J Biol Chem. 2006;281(49):37427–37434. doi: 10.1074/jbc.M605311200. [DOI] [PubMed] [Google Scholar]
- Wang Z. Choi MK. Ban T. Yanai H. Negishi H. Lu Y. Tamura T. Takaoka A. Nishikura K. Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105(14):5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters TM. Kenny EF. O'Neill LA. Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol Cell Biol. 2007;85(6):411–419. doi: 10.1038/sj.icb.7100095. [DOI] [PubMed] [Google Scholar]
- Wu X. Peng SL. Toll-like receptor 9 signaling protects against murine lupus. Arthritis Rheum. 2006;54(1):336–342. doi: 10.1002/art.21553. [DOI] [PubMed] [Google Scholar]
- Xia X. Cui J. Wang HY. Zhu L. Matsueda S. Wang Q. Yang X. Hong J. Songyang Z. Chen ZJ. Wang RF. NLRX1 negatively regulates TLR-Induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity. 2011;34(6):843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C. Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Yamashita T. Shimada S. Guo W. Sato K. Kohmura E. Hayakawa T. Takagi T. Tohyama M. Cloning and functional expression of a brain peptide/histidine transporter. J Biol Chem. 1997;272(15):10205–10211. doi: 10.1074/jbc.272.15.10205. [DOI] [PubMed] [Google Scholar]
- Yanai H. Ban T. Taniguchi T. Essential role of HMGB proteins in nucleic acid-mediated innate immune responses. J Intern Med. 2011;270(4):301–308. doi: 10.1111/j.1365-2796.2011.02433.x. [DOI] [PubMed] [Google Scholar]
- Yanai H. Ban T. Wang Z. Choi MK. Kawamura T. Negishi H. Nakasato M. Lu Y. Hangai S. Koshiba R. Savitsky D. Ronfani L. Akira S. Bianchi ME. Honda K. Tamura T. Kodama T. Taniguchi T. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462(7269):99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- Yang P. An H. Liu X. Wen M. Zheng Y. Rui Y. Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11(6):487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Yang Y. Liu B. Dai J. Srivastava PK. Zammit DJ. Lefrancois L. Li Z. Heat shock protein gp96 is a master chaperone for Toll-like receptors and is important in the innate function of macrophages. Immunity. 2007a;26(2):215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG. Lindahl T. Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007b;131(5):873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Yasutomo K. Horiuchi T. Kagami S. Tsukamoto H. Hashimura C. Urushihara M. Kuroda Y. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28(4):313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- Yoneyama M. Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227(1):54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Zeng W. Sun L. Jiang X. Chen X. Hou F. Adhikari A. Xu M. Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141(2):315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Brann TW. Zhou M. Yang J. Oguariri RM. Lidie KB. Imamichi H. Huang DW. Lempicki RA. Baseler MW. Veenstra TD. Young HA. Lane HC. Imamichi T. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011;186(8):4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B. Yang Y. Li S. Wang YY. Li Y. Diao F. Lei C. He X. Zhang L. Tien P. Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]