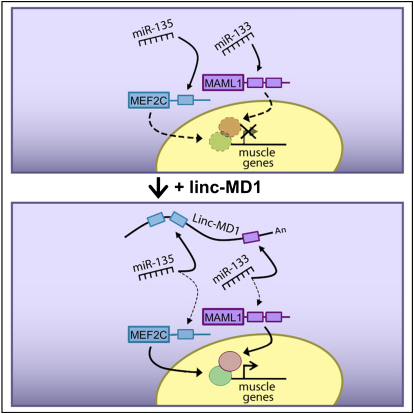
Summary
Recently, a new regulatory circuitry has been identified in which RNAs can crosstalk with each other by competing for shared microRNAs. Such competing endogenous RNAs (ceRNAs) regulate the distribution of miRNA molecules on their targets and thereby impose an additional level of post-transcriptional regulation. Here we identify a muscle-specific long noncoding RNA, linc-MD1, which governs the time of muscle differentiation by acting as a ceRNA in mouse and human myoblasts. Downregulation or overexpression of linc-MD1 correlate with retardation or anticipation of the muscle differentiation program, respectively. We show that linc-MD1 “sponges” miR-133 and miR-135 to regulate the expression of MAML1 and MEF2C, transcription factors that activate muscle-specific gene expression. Finally, we demonstrate that linc-MD1 exerts the same control over differentiation timing in human myoblasts, and that its levels are strongly reduced in Duchenne muscle cells. We conclude that the ceRNA network plays an important role in muscle differentiation.
Graphical Abstract
Highlights
► linc-MD1 is a long noncoding cytoplasmic RNA expressed during myoblast differentiation ► linc-MD1 acts as a competitive endogenous RNA (ceRNA) for miR-133 and miR-135 targets ► Through these miRNAs, linc-MD1 controls MEF2C and MAML1 and myoblast differentiation ► linc-MD1 is conserved in humans and levels are reduced in Duchenne Muscular Dystrophy
A long noncoding RNA, linc-MD1, governs muscle cell differentiation by competitively binding to microRNAs that target muscle-specific transcription factors. linc-MD1 functions not only as a decoy RNA but also as a precursor pri-microRNA.
Introduction
One of the greatest surprises of high throughput transcriptome analysis of the last years has been the discovery that the mammalian genome is pervasively transcribed into many different complex families of RNA. In addition to a large number of alternative transcriptional start sites, termination and splicing patterns, a complex collection of new antisense, intronic and intergenic transcripts was found. Moreover, almost half of the polyadenylated species resulted to be non-protein-coding RNAs. Although many studies have helped unveiling the function of many small noncoding RNAs, very little is known about the long noncoding (lncRNA) counterpart of the transcriptome. In spite of their very low levels of expression in specific body compartments and thanks to the availability of sensitive detection techniques, specific patterns of lncRNA expression in specific cell types, tissues and developmental conditions (Amaral and Mattick, 2008; Qureshi et al., 2010) have been defined.
So far, a large range of functions has been attributed to lncRNAs (Mattick, 2011; Nagano and Fraser, 2011), such as modulation of apoptosis and invasion (Khaitan et al., 2011), reprogramming of induced pluripotent stem cells (Loewer et al., 2010), marker of cell fate (Ginger et al., 2006) and parental imprinting (Sleutels et al., 2002), indicating that they may represent a major regulatory component of the eukaryotic genome.
A specific mode of action in mediating epigenetic changes through recruitment of the Polycomb Repressive Complex (PRC) was described for the Xist and HOTAIR transcripts (Chaumeil et al., 2006; Rinn et al., 2007). lncRNAs were also found to act in the nucleus as antisense transcripts or as decoy for splicing factors leading to splicing malfunctioning (Beltran et al., 2008; Tripathi et al., 2010). In the cytoplasm, lncRNAs were described to transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements (Gong and Maquat, 2011) or, in the case of pseudogenes, to compete for miRNA binding, thereby modulating the derepression of miRNA targets (Poliseno et al., 2010; Salmena et al., 2011).
These findings have prompted studies directed toward the identification of the circuitries that are regulated by these molecules.
Muscle differentiation is a powerful system for these investigations, because it can be both recapitulated in vitro and because the networks of transcription factors coordinating the expression of genes involved in muscle growth, morphogenesis, and differentiation are well known and evolutionarily conserved (Buckingham and Vincent, 2009). Moreover, recent studies have shown that these myogenic transcription factors not only control protein-coding genes but also regulate the expression of specific miRNAs (Zhao et al., 2005; Rao et al., 2006). These miRNAs act at different levels in the modulation of muscle differentiation and homeostasis and their expression was found to be altered in several muscular disorders such as myocardial infarction, Duchenne Muscular Dystrophy, and other myopathies (Eisenberg et al., 2007; Cacchiarelli et al., 2010).
Among miRNAs specifically expressed in muscle tissue, the most widely studied are members of the miR-1/206 and miR-133a/133b families, which originate from three separate chromosomes (Chen et al., 2006). miR-206 differs from other members of its family because it is exclusive of skeletal muscles (McCarthy, 2008). Moreover, at variance with other myomiRs mainly expressed in mature muscle fibers, miR-206 expression is enriched in differentiating satellite cells, where it represses the stemness factor Pax7, a crucial player in the regeneration process, as we recently demonstrated (Cacchiarelli et al., 2010).
In this study, through a detailed analysis of the genomic region of miR-206/133b, we discovered the existence of a muscle specific lncRNA and defined its expression profile and function. We demonstrated that this lncRNA is involved in the timing of muscle differentiation and acts as a natural decoy for miRNAs, playing a crucial role in the control of factors involved in the myogenic program.
Results
linc-MD1 Is Expressed during Myoblasts Differentiation
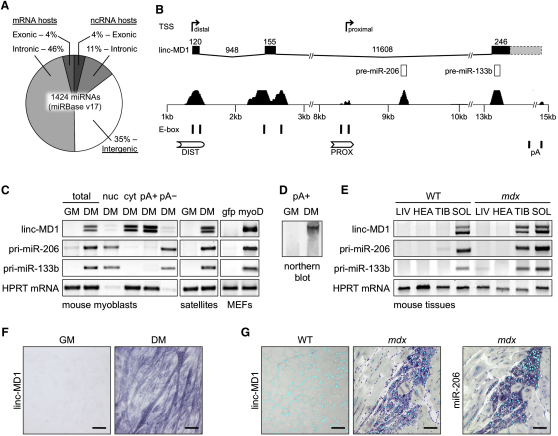
miRNA coding regions display different genomic organizations: while 50% are encoded in introns or exons of protein coding genes, the other half map in ncRNA host genes or, when no host transcript can be identified, in intergenic regions (Figure 1A). According to this classification, muscle specific pre-miR-206 and pre-miR-133b were annotated as overlapping with a noncoding RNA (Williams et al., 2009). With the aim of better understanding the transcriptional regulation of these two miRNAs, we carried out a detailed analysis in order to identify their transcriptional start sites (TSS) and promoter elements. 5′ RACE analysis, performed in differentiating myoblasts with reverse primers surrounding the pre-miR-206 sequence, demonstrated the existence of a proximal TSS mapping about 600 bp upstream of the pre-miR-206 sequence (proximal, Figure 1B). This region contains E-box sequences (CANNTG) previously shown to be functional for MyoD association (Rao et al., 2006) and mir-206 expression (Williams et al., 2009). The same analysis was also performed with reverse primers surrounding pre-miR-133b. A strong TSS, mapping approximately 13 Kb upstream of pre-miR-133b sequence, was identified (distal, Figure 1B). Analysis of the genomic region revealed the existence of a transcript composed of three exons and two introns; with respect to this structure, pre-miR-206 maps in the second intron, while pre-miR-133b in the third exon (Figure 1B and Figure S1A available online). Even if short reading frames can be detected in the mature transcript, neither of their AUGs shows the Kozak consensus, nor their sequences are more or less conserved than the surrounding regions (Figure S1B), making it very unlikely for them to be coding. Therefore, the identified transcript was classified as a bona fide long intergenic noncoding (linc) RNA, hereafter termed linc-MD1. Phylogenetic analysis of linc-MD1 revealed high conservation in exon 1 and 2, while homology is limited to the pre-mir-133b sequence in exon 3 (Figure 1B). All splice junctions are conserved as well. In silico analysis highlighted the presence of conserved E-boxes both in the distal (DIST) and proximal (PROX) regions (Figure 1B) as well as in the regions surrounding the second exon where minor alternative TSSs were mapped (data not shown).
Figure 1.
Muscle-Specific lincRNA Profiling
(A) Human miRNA genomic location relative to their host genes.
(B) Schematic representation of the murine miR-206/133b genomic locus. Upper panel shows transcriptional start sites (TSS, indicated by arrows) mapped through 5′ RACE analysis in differentiated myoblasts. The genomic structure of the identified linc-MD1 and the exon-intron lengths are shown (conserved exons in black, nonconserved 3′-portions in dash) as well as pre-miRNA sequences. Lower panel shows conserved regions among vertebrates together with E-boxes and regulatory elements (DIST, PROX and pA signals).
(C) RT-PCR for linc-MD1, pri-miR-206, and pri-miR-133b expression performed in mouse myoblasts in growth (GM) or differentiation (DM) conditions. The total, nuclear (nuc), cytoplasmic (cyt), polyadenylated (pA+) and nonpolyadenylated (pA-) RNA fractions are shown. The same analysis was also performed in primary satellite cells in GM and DM conditions and in mouse embryonic fibroblasts (MEFs) infected with a MyoD expressing lentivirus (MyoD) or control (gfp). HPRT mRNA was used as endogenous control.
(D) Northern blot analysis for linc-MD1 in the poly-A+ fraction from mouse myoblasts in GM and DM conditions.
(E) RT-PCR for linc-MD1, pri-miR-206, pri-miR-133b performed on RNA from liver (LIV), heart (HEA), tibialis anterior (TIB) and soleus (SOL) of wild-type (WT) and mdx mice.
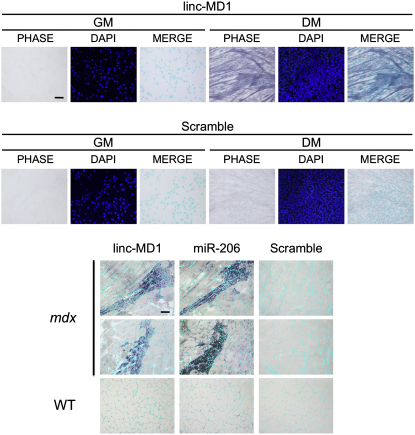
(F) In situ analysis with a DIG-labeled linc-MD1 probe in C2 myoblasts in GM and DM. (G) In situ analysis with DIG-labeled linc-MD1 and miR-206 probes in wild-type (WT) and mdx gastrocnemius cryosections; DAPI staining (light blue) is also shown.
Original magnification is 20×. The scale bar represents 100 μm. Additional fields and controls are show in Figure S2.
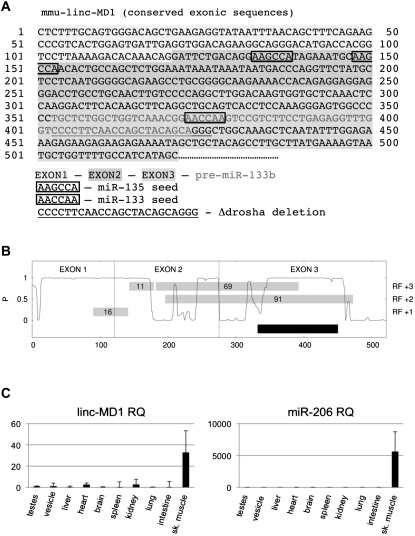
Figure S1.
Related to Figure 1
(A) Murine linc-MD1 with exons identification, miRNA binding sites and sequence deleted in Δdrosha constructs. The nucleotide sequence refers to the portion of the transcript which is conserved between human and mouse. Two 3′-end isoforms mapping at two pA-sites have been mapped in a non conserved region at 1060 and 1450 nt downstream the indicated sequence (Figure 1B).
(B) Conservation pattern of the linc-MD1 transcript expressed as probability of negative selection taken from the Mammal Cons track. RF+1, +2 and +3 refer to the reading frames, gray boxes indicate the position of ORFs in number of triplets. The mature transcript contains four open reading frames (defined as any sequence starting with AUG and containing at least 10 non terminator codons) ranging in size from 11 to 91 triplets. None of the AUG shows a Kozak consensus (Kozak 1989), nor are the sequences more or less conserved than the surrounding regions.
(C) linc-MD1 and miR-206 relative quantifications in the indicated tisses. Values are normalized for HPRT or U6 snRNA respectively and shown with respect to testes sample set to a value of 1.
RT-PCR analysis (Figure 1C) indicates that linc-MD1 is localized in the cytoplasm and is polyadenylated. Moreover, while absent in growth conditions (GM), linc-MD1 is activated upon shift to differentiation (DM) of mouse myoblasts, satellite cells and MyoD-transdifferentiated fibroblasts. The expression level of linc-MD1 parallels that of miR-133b upon induction of differentiation, while it is uncoupled from miR-206, which is already present in proliferating C2 myoblasts. The two bands detected by RT-PCR reveal the presence of a 70 nucleotide splice variant in exon 2. Northern blot analysis of poly-A+ RNA from differentiating myoblasts indicates that linc-MD1 is indeed the major pA+ product originating from this region, even though the two alternative splice forms are not distinguishable on this gel (Figure 1D). In situ analysis confirms that linc-MD1 is not expressed in proliferating conditions while it is induced upon myoblast differentiation (Figure 1F).
RT-PCR analysis of linc-MD1 in mouse tissues (Figure 1E) indicates that it is highly expressed in skeletal muscles of dystrophic mdx animals (TIB and SOL), paralleling miR-206 and miR-133b synthesis. Notably, in wild-type animals linc-MD1 is expressed at low levels only in the soleus, while it is absent in tibialis and other skeletal muscles (data not shown). No linc-MD1 expression is observed in nonmuscle tissues (LIV and Figure S1C) nor in heart (HEA), thus indicating that also linc-MD1, similarly to miR-206, is restricted to skeletal muscles. In situ analysis (Figure 1G) on WT and mdx muscles indicates that linc-MD1 expression occurs exclusively in newly regenerating fibers (characterized by centronucleated fibers), abundant in dystrophic conditions, similarly to what previously shown for miR-206 (Cacchiarelli et al., 2010, Yuasa et al., 2008). No expression is instead detected in mature terminally differentiated fibers, as shown in wild-type animals devoid of regenerating fibers. The low level of linc-MD1 found in the soleus would therefore suggest that some degree of regeneration occurs in this district known to have a high content of satellite cells (Chargè and Rudniki, 2004).
These data indicate that linc-MD1 is muscle-specific and is activated upon myoblast differentiation.
Identification of Regulatory Elements Directing linc-MD1 and miR-206/133b Expression
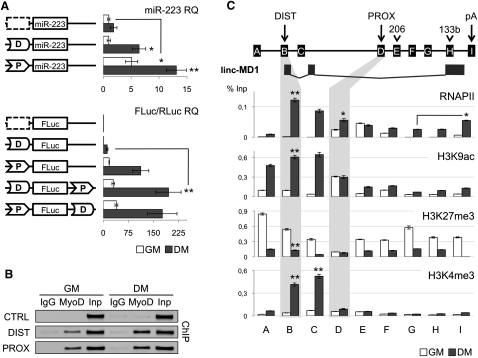
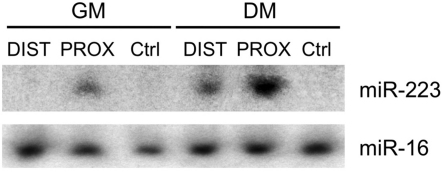
Promoter fusion experiments with the Distal (DIST) and Proximal (PROX) regions were performed in order to test their role in transcription. 810 and 310 nucleotides of DIST and PROX regions respectively, were cloned upstream of either the murine pre-miR-223 (Ballarino et al., 2009) sequence (D-miR-223 and P-miR-223) or the firefly luciferase coding region (D-FLuc and P-FLuc). Their promoter activity was tested in mouse C2 myoblasts in proliferation (GM, white bars) versus differentiation (DM, black bars) conditions. Figure 2A shows that the PROX element is already active in GM, in agreement with basal miR-206 expression (see Figure 1C). Upon induction of differentiation, the proximal region is able to further induce the expression of both reporter genes (miR-223 and FLuc). On the contrary, the DIST element is inactive in GM while, upon shift to differentiation, is able to activate transcription. Notably, when the PROX and DIST elements are present on the same construct, they act synergistically providing the strongest activation (D-Fluc-P and P-Fluc-D).
Figure 2.
Transcriptional and Epigenetic Regulation of miR-206/133b Genomic Locus
(A) Promoter activity assay. Upper panel shows that distal (box D) and proximal (box P) regions were cloned upstream of murine pre-miR-223 and tested in C2 myoblasts in GM and DM conditions. A region between miR-206 and miR-133b was used as negative control (dashed box). miR-223 expression was measured by northern blot (Figure S3) and qRT-PCR. U6 snRNA was used as endogenous control. miR-223 relative quantification (RQ) is shown with respect to control vector in GM set to a value of 1. Lower panel shows that the same regions were cloned, alone or in combinations, in a firefly luciferase reporter construct (FLuc) and tested in GM and DM conditions. A luciferase construct (RLuc) was transfected as control. Luciferase activity, from three independent experiments, was measured as FLuc/RLuc RQ shown with respect to the negative control vector in GM set to a value of 1.
(B) ChIP analysis for MyoD enrichment on distal (DIST), proximal (PROX) and negative control (CTRL) regions performed on chromatin extracted from C2 myoblasts in GM and DM conditions. Amplifications of IgG control immunoprecipitations (IgG) and 10% of input chromatin (Inp) are shown.
(C) Upper panel is a schematic representation of the miR-206/133b genomic locus. Capital letters indicate regions analyzed in ChIP experiments. Location of regulatory regions (DIST, PROX and pA), pre-miRNAs (206, 133b) and linc-MD1 are shown along the locus as in Figure 1B. Lower panel displays ChIP analyses for RNA polymerase II (RNAP II), H3K9ac, H3K27me3, and H3K4me3 performed on chromatin extracted from myoblasts in GM and DM. Values derived from three independent experiments were normalized for background signals (IgG) and expressed as percentage of Input chromatin (% Inp). Unless specifically indicated, statistical significance was calculated with respect to GM conditions. Data are shown as mean ± SD. One asterisk indicates p < 0.05; two asterisks: p < 0.01.
As indicated in Figure 1B, both regions contain E-box elements and indeed both of them are able to bind MyoD in vivo, as demonstrated by chromatin immunoprecipitation analysis (Figure 2B). MyoD binding to DIST in GM conditions is in line with the notion that MyoD binds promoters prior to transcriptional activation, which occurs upon its acetylation.
Nine regions spanning the entire locus (A-I, Figure 2C) were tested for the major histone modifications in both GM and DM conditions. Consistently with the promoter fusion analysis, RNA polymerase II (RNAPII) enrichment is observed on the PROX promoter already in GM. Interestingly, in these conditions, no polymerase is found on miR-133b indicating that the PROX promoter does not direct transcriptional read-through into this region. These data are in agreement with the observation that miR-133b expression is uncoupled from that of miR-206. Upon induction of differentiation, RNAPII immunoprecipitates on the DIST promoter and on the entire region: RNAPII enrichment decreases gradually along the cluster and increases at the 3′ end in a fashion similar to that of many transcriptional units (Moore and Proudfoot, 2009). Histone-H3-lysine-9 acetylation (H3K9ac) and Histone-H3-lysine-27 tri-methylation (H3K27me3) patterns are in agreement with the differential transcriptional activity of the two promoters: low H3K27me3 and high H3K9ac immunoprecipitation levels are found on the PROX element already in GM and are maintained in DM (highlighted in gray). Conversely, DIST displays low H3K9ac and high H3K27me3 signature in GM, while the pattern is reverted upon differentiation, in line with transcriptional activation (highlighted in gray). Notably, the H3K4me3 marker, enriched around TSS of active RNAPII promoters (Okitsu et al., 2010), confirmed the presence of TSS on the distal region in DM conditions (Figure 2C, lower panel). Interestingly, H3K4me3 was detected also in region C where minor TSS were mapped (data not shown), suggesting the presence of additional transcripts in this region.
Altogether, our data indicate that the PROX promoter is responsible for miR-206 expression in growth conditions, whereas upon differentiation, both PROX and DIST cooperate to drive transcription of the locus.
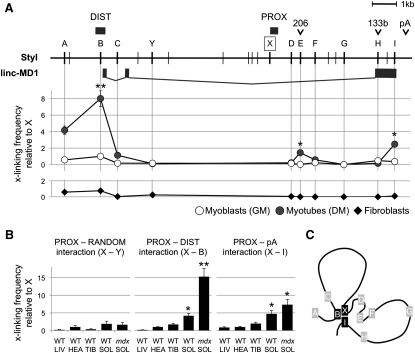
Distal and Proximal Promoters Are Involved in Long-Distance Interactions
Since promoter fusion assays demonstrated the cooperation between the DIST and PROX elements, we investigated whether these two regions could physically interact in vivo. Gene loops have been shown to be transcriptionally dependent, as they are absent in nontranscribing conditions (West and Fraser, 2005). Chromosome conformation capture (3C) analysis (Tan-Wong et al., 2008) was utilized to determine relative crosslinking frequencies among regions of interest. The conformation of the miR-206/133b genomic locus was initially tested in myoblasts, both in GM and DM conditions, as well as in fibroblasts where the two miRNAs are not expressed. A common reverse primer (indicated by X in Figure 3A) mapping in the PROX region was used in combination with a set of primers along the genomic locus and interactions were analyzed by qPCR (Figure 3A; note that A-I sites correspond to the same regions analyzed in ChIP experiments). A specific interaction between PROX and DIST region (X-B) is observed upon induction of differentiation. A less prominent but reproducible interaction is also detected between X and the I, which identifies the polyadenylation region (pA). No specific long-range interactions were detected in fibroblasts where the locus is silent.
Figure 3.
Tridimensional Architecture of miR-206/133b Genomic Locus
(A) Upper panel shows StyI restriction sites (vertical lines), regulatory regions (DIST, PROX, pA), pre-miRNAs (206, 133b), and linc-MD1. Capital letters (A–I, X, and Y) indicate the position of StyI sites analyzed in 3C experiments. A–I sites identify the same regions analyzed in ChIP experiments in Figure 2C. A common reverse primer identifying the proximal region (X) was used in combination with different reverse primers. Lower panel shows 3C analysis performed on chromatin extracted from C2 myoblasts in GM and DM conditions and from fibroblast cell cultures. Crosslinking frequencies relative to X region were measured both by semiquantitative PCR (data not shown) and qRT-PCR. Data from multiple experiments were normalized for primer amplification efficiency and reported with respect to X-B interaction in GM set to a value of 1. Statistical significance was calculated with respect to GM conditions.
(B) The same 3C analysis was performed in liver (LIV), heart (HEA), tibialis anterior (TIB), soleus (SOL) of wild-type (WT), and mdx animals. For each set of interactions analyzed, crosslinking frequencies relative to X region derived from multiple experiments were reported with respect to WT HEA interaction set to a value of 1. Statistical significance was calculated with respect to WT HEA tissue. Data are shown as mean ± SD. One asterisk indicates p < 0.05; two asterisks: p < 0.01.
(C) Schematic representation of the interactions detected upon induction of differentiation.
An interaction, clearly distinguishable from the background, is also found with the A region. This can be due to its proximity to the DIST element, or it can point to the existence of an additional enhancer region.
3C analysis was also performed in different types of mouse tissues from WT and mdx animals. Figure 3B shows that the interaction between the PROX and DIST regions only occurs in skeletal muscles and it is characteristic of muscles with high regeneration rate, such as the soleus (Chargé and Rudnicki, 2004). Notably, PROX-DIST interaction is particularly enhanced in mdx muscles, known to undergo intense regeneration (mdx SOL). The same specificity was also detected for the PROX-pA interaction (Figure 3B, X-I); on the contrary, no relevant interaction was detected between PROX and a negative control region (X-Y).
From these data, we conclude that the long-distance interaction between the DIST and PROX is functional to both linc-MD1 and miRNAs expression. Figure 3C schematically shows the looping structure correlated with the activation state of the locus.
Modulation of linc-MD1 Expression Affects Myogenic Differentiation
Figure 4A shows the expression profiles of myogenic proteins (Myogenin - MYOG and Myosin Heavy Chain - MHC), linc-MD1 and muscle miRNAs (miR-206, miR-1 and miR-133) during in vitro C2 myoblast differentiation. The analysis reveals that: (1) miR-206 is already expressed in GM (in line with the observed basal activity of the PROX promoter and its active chromatin signature, Figures 2A–2C); (2) miR-1 and miR-133 expression is delayed with respect to miR-206 (note that the probes used do not distinguish between miR-133a and miR-133b); (3) linc-MD1 expression starts from the third day of differentiation.
Figure 4.
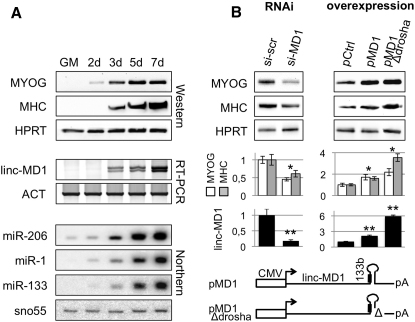
Modulation of Linc-MD1 Expression Affects Myogenesis
(A) RNA and protein samples were extracted from myoblasts in proliferation (GM) and after shift to differentiation medium (DM) for the indicated times. Upper panels show western blot analysis for myogenin (MYOG), myosin heavy chain (MHC), and HPRT. Middle panels show RT-PCR analysis for linc-MD1 and Actin mRNA (ACT) expression. Lower panels show northern blots for miR-206, miR-1, miR-133, and snoRNA55.
(B) Left panel: siRNAs for linc-MD1 (si-MD1) or scramble control (si-scr) transfected in C2 myoblasts and maintained in DM for 5 days. Right panel: linc-MD1 overexpression was obtained by transfection of linc-MD1 (pMD1) and linc-MD1-Δdrosha (pMD1-Δdrosha) constructs (see schematic representation below) together with a control GFP cDNA (pCtrl). Samples were collected 4 days after induction of differentiation. Densitometric analysis, normalized for HPRT, is shown below. Lower panels display the levels of linc-MD1, normalized for HPRT, measured by qRT-PCR.
Data are shown with respect to control experiments set to a value of 1. Data are shown as mean ± SD. One asterisk indicates: p < 0.05; two asterisks: p < 0.01.
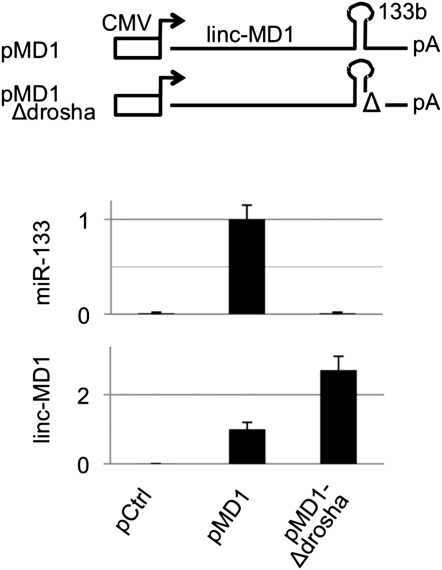
In order to understand the role of linc-MD1 in skeletal muscle differentiation, we modulated its expression through RNA interference and overexpression experiments. The left panel in Figure 4B shows that, in C2 myoblasts, MYOG and MHC protein levels decrease after 5 days of linc-MD1 interference (si-MD1) with respect to control siRNA (si-scr). Two different constructs were used for ectopic expression of linc-MD1 (see scheme in Figure 4B): pMD1, carrying the conserved portion of linc-MD1 (Figure S1A), and pMD1-Δdrosha, containing a mutation in the miR-133b flanking region that prevents Drosha cleavage and miR-133b release. The use of both constructs should permit to distinguish the effect of linc-MD1 from that of miR-133b that can be produced in the nucleus from Drosha cleavage of the linc-MD1 precursor. Figure S4 demonstrates that pMD1 is indeed able to express high levels of miR-133b, while pMD1-Δdrosha is not. Figure 4B (right panel) shows that both types of constructs give rise to an increase of myogenic markers, MYOG and MHC, with respect to control treatment (pCtrl). Interestingly, pMD1-Δdrosha displayed a slightly stronger activity (more evident for MHC), indicating that the observed effects are not due to miR-133b production but rather to linc-MD1 overdosage. Lower panels of Figure 4B indicate the relative quantification of linc-MD1 with respect to controls. Considering the disproportion between linc-MD1 abundance and the effects on myogenic target synthesis, it is reasonable to postulate the existence of a threshold level above which the system cannot be further influenced.
Figure S4.
Related to Figure 4
Schematic representation of pMD-1 and pMD-1-Δdrosha constructs used in linc-MD1 overexpression experiments. Histograms show miR-133 and linc-MD1 RNA relative expression measured by qRT-PCR normalized for U6 snRNA or HPRT respectively and shown with respect to pCtrl set to a value of 1.
linc-MD1 Is a Target of miR-133 and miR-135
Bioinformatics analysis (see Extended Experimental Procedures) for miRNA recognition sequences on linc-MD1 revealed the presence of thirty-six highly conserved putative miRNA sites listed in Table S1. We discarded miRNAs not expressed in muscle as well as miRNAs whose targets are not expressed or do not have a known function in muscle physiology. The two remaining miRNA were miR-135, with two predicted sites on linc-MD1 and miR-133, with one site (see Figure 5A and Table S1; note that both members, a and b, of the miR-135 and miR-133 families can associate with those sites on linc-MD1). Interestingly the 70 nucleotide shorter isoform of linc-MD1 (see Figure 1C) lacks the two miR-135 sites. In all subsequent experiments we concentrated on the longest isoform containing the miR-135 sites (linc-MD1 cDNA).
Extended Experimental Procedures.
Bionformatic Analyses
The mature miR sequences were taken from the miRBase database (release 17) (Griffiths-Jones et al., 2008). Linc-MD1 was displayed using the UCSC genome browser (Fujita et al., 2010); genomic locus conservation was evaluated using the Mammal Cons phastCons30wayPlacental (Siepel et al., 2005).
The likelihood of binding of a mature miRNA to linc-MD1 was evaluated using the miRanda package (Enright et al., 2003). After filtering for conservation, 36 putative target sites were identified and are listed in Table S1.
We identified miRNAs not expressed in muscle (Cacchiarelli et al., 2010; Cardinali et al., 2009) and discarded them. Next, we searched for putative targets of the remaining miRNAs using TargetScan (Friedman et al., 2009). The expression profile of the putative targets in myoblasts (as reported in the GEO database (Barrett et al., 2011) in datasets gds2412 and gds586 (Chen et al., 2006, Tomczak et al., 2004)) were analyzed to discard miRNAs whose targets are not expressed in muscle or do not represent valuable targets in muscle physiology according to Ashburner et al. (2000) (see Table S1).
Transcription factor binding sites were predicted using RVista Algorithm (Loots et al., 2004).
RNA and Protein Analyses
Total RNA was prepared from liquid nitrogen-powdered tissues or cell cultures using miRNeasy (QIAGEN). miRNA and mRNA analyses were performed using miScript System (QIAGEN). Relative quantification was performed using, as endogenous controls, U6 snRNA for miRNAs and HPRT1 for mRNAs. PolyA+ RNA fraction was obtained using oligodT affinity purification (QIAGEN). Northern blot for miRNAs was performed according to Cacchiarelli et al. (2010) while Northern blot for linc-MD1 was performed on purified polyA+ RNA, using a radioactive probe obtained by nick-translation. RNA in situ hybridization was performed in formaldehyde and carbodiimide (EDC)-fixed gastrocnemius cryosections or cell cultures, according to Cacchiarelli et al. (2010). Primers sequences for ncRNA detection are listed in supplementary experimental procedures ; Western blot on total extracts were performed as described in Denti et al. (2006).
RACE Analyses
5′ RACE analyses were performed choosing reverse primers surrounding pre-miRNA sequences while 3′RACE forward primers were designed to validate putative polyadenylation sites indicated in Figure 1B. cDNA synthesis, PCR and nested-PCR were performed according to manufacturer's specifications (Invitrogen).
Reporter Constructs and Luciferase Assays
Distal (DIST) and proximal (PROX) elements were tested using two types of reporter constructs. DIST and PROX were cloned in a Pgl3basic (Promega) modified plasmid in which firefly luciferase gene was substituted with murine pre-miR-223 sequence. The same regions were also cloned in a Pgl4.10 FLuc reporter plasmid (Promega) individually (D-FLuc or P-FLuc) or in combination as enhancer assay (D-FLuc-P or P-FLuc-D). Transfection efficiency of these constructs were assessed by cotransfection of pRLTK plasmid (Promega) encoding for renilla luciferase gene.
Exon 1, exon 2 and exon 3 sequence of linc-MD1 cDNA (RLuc-MD1-WT) were amplified by PCR and cloned in Ψcheck2 plasmid (Promega), downstream renilla luciferase gene (RLuc). The same plasmid also contains the firefly luciferase gene (FLuc) to normalize for transfection efficiency. Mutant derivatives (RLuc-MD1-Δ133 and RLuc-MD1-Δ135) were obtained by deletion of miR-133 and miR-135 binding sites indicated in Figure S1 by inverse PCR. The same procedure was followed for the production of maml1 and mef2c 3′UTR reporter constructs (RLuc-maml1-WT and RLuc-mef2c-WT) and their mutant derivatives (RLuc-maml1-mut and RLuc-mef2c-mut).
RLuc and FLuc activities were measured by Dual Glo Luciferase assay (Promega).
Overexpression Constructs
Constructs for the overexpression of linc-MD1 were obtained by cloning linc-MD1 cDNA (Figure S1) in pCDNA3.1- plasmid (Invitrogen) and all mutants were obtained by inverse PCR. Lentiviral constructs were obtained by subcloning the CMV-lincMD1-BGH cassette into HpaI site of PCCL-gfp plasmid (Incitti et al., 2010).
miRNA overexpression constructs were obtained by cloning 100 nucleotides upstream and downstream from the pre-miRNA of interest into the U1snRNA expression cassette (Cacchiarelli et al., 2010).
Oligonucleotides
murine linc-MD1 detection by RT-PCR
mmu_LINC_MD1_FW – tggagtgattgaggtggaca
mmu_LINC_MD1_RV – tgatggcaaaaccagcatta
murine and human linc-MD1 detection by qRT-PCR
mmu_LINC_MD1_FW – gcaagaaaaccacagaggagg
mmu_LINC_MD1_RV – gtgaagtccttggagtttgag
hsa_LINC_MD1_FW – cactgccagctctggaaaat
hsa_LINC_MD1_RV – acttggttccgtttgaccag
murine linc-MD1 cloning in ψcheck2 and pCDNA3.1-
mmu_LINC_MD1_cdna_FW – ctctttgcagtgggacagct
mmu_LINC_MD1_cdna_RV – tgatggcaaaaccagcatta
5′ RACE reverse oligos for RT and subsequent nested PCR
MIR206_RT - atgtagccaaggaacgaaga
MIR206_PCR_OUTER – tcacgcagaaaggaaaagc
MIR206_PCR_INNER – acttcatccattctacactccc
MIR133B_RT – cttcttgggaacataaggcta
MIR133B_PCR_OUTER – tgaagtccttggagtttgagc
MIR133B_PCR_INNER – ggagtttgagcaccacttgtc
3′ RACE forward oligos for nested PCR
3′RACE_outer – catctaaattacaagaaaacaaga
3′RACE_inner – ctataactgtattccattttcgtg
PROMOTER CLONING
Prox_FW – ggacccttcttctcctctta
Prox_RV – caggcgctattgtacttc
Dist_FW – atggctaccttgtcagcacttcc
Dist_RV – gcctcttcccttttgtactttcc
Oligonucleotide sequences for Chip, 3C as well as plasmids and other material are available upon request.
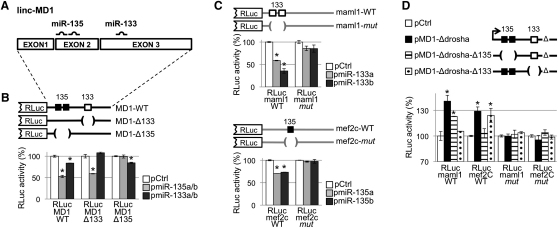
Figure 5.
linc-MD1 Acts as a Natural Decoy for miR-135 and miR-133
(A) Positions of miR-135 and miR-133 binding sites on linc-MD1.
(B) linc-MD1 (RLuc-MD1-WT) and mutant derivatives devoid of miR-133 or miR-135 binding sites (RLuc-MD1-Δ133 and Rluc-MD1-Δ135) were cloned downstream the Renilla luciferase coding region (RLuc). These constructs were cotransfected in C2 myoblasts together with plasmids expressing miR-135 (pmiR-135a/b) or miR-133 (pmiR-133a/b) or with a control vector (pCtrl).
(C) The 3′UTR of MAML1 and of MEF2C were cloned downstream RLuc (RLuc-maml1-WT, RLuc-mef2c-WT) together with mutant derivatives lacking miRNA recognition sequences (RLuc-maml1-mut and RLuc-mef2c-mut). These constructs were cotransfected in C2 myoblasts with plasmids expressing miR-133 (pmiR-133a/b) or miR-135 (pmiR-135a/b) or with a control vector (pCtrl). (D) RLuc-maml1-WT, RLuc-mef2c-WT and their corresponding mutant derivatives (-mut) were transfected in C2 myoblasts with pMD1-Δdrosha or its mutant derivatives depleted in either miR-135 (pMD1-Δdrosha-Δ135) or miR-133 (pMD1-Δdrosha-Δ133) recognition elements. A GFP construct was used as control (pCtrl). For miRNA overexpression experiments, unless specifically stated, a mix of a and b pre-miRNA expression plasmids was used. For all the experiments, histograms indicate the values of luciferase measured 24 hr after transfection. Data, derived from three independent experiments, are shown with respect to RLuc control vector set to a value of 100%. Data are shown as mean ± SD. One asterisk indicates p < 0.05.
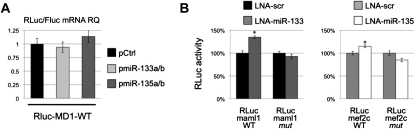
The linc-MD1 cDNA (RLuc-MD1-WT) and mutant derivatives lacking the putative miR-133 and miR-135 recognition sequences (RLuc-MD1-Δ133 and RLuc-MD1-Δ135) were cloned downstream of the luciferase gene (Figure 5B) and transfected in C2 myoblasts together with either miR-135 (pmiR-135a/b) or miR-133 (pmiR-133a/b) coding plasmids. Figure 5B shows that luciferase expression is reduced by 50% and 20% with respect to the control plasmid (pCtrl) when miR-135 and mir-133 were respectively expressed. These effects are abolished when mutant substrates for either miRNA were utilized. qRT-PCR for RLuc mRNA revealed that overexpression of both miRNAs do not affect luciferase mRNA stability (Figure S5A). These data demonstrate that linc-MD1 can bind both miR-135 and miR-133.
Figure S5.
Related to Figure 5
(A) RLuc/FLuc mRNA ratio measured by qRT-PCR in C2 myoblasts transfected with RLuc-MD1-WT construct together with plasmids expressing miR-133a/b, miR-135a/b or control. Values are normalized for HPRT and shown with respect to pCtrl set to a value of 1.
(B) The constructs described in Figure 5 were cotransfected in C2 myoblasts with scamble LNA (LNA-scr) or LNA for the indicated miRNA. Histograms indicate the values of renilla luciferase measured 48 hr after transfection in DM. Data are shown with respect to the values of RLuc control vector set to a value of 100%. One asterisk: p < 0.05.
The different levels of repression exerted by the two miRNAs could be due to the fact that linc-MD1 contains two miR-135 recognition elements and only one for miR-133. However, it cannot be excluded that the presence of a pre-miR-133b hairpin structure in the linc-MD1 sequence could limit miR-133 association.
linc-MD1 Controls miR-133 and miR-135 Targets
Among the many predicted targets of miR-135 and miR-133, we concentrated on MEF2C (with one miR-135 site) and MAML1 (with two miR-133b sites) mRNAs since they encode for transcription factors known to play a relevant role in myogenic differentiation (Shen et al., 2006). Interestingly, comparative analysis revealed that miRNA putative target sites in MEF2C and MAML1 3′UTR are highly conserved in mammals. The 3′UTRs of MAML1 and MEF2C were fused to the Luciferase coding region (RLuc-maml1-WT and RLuc-mef2c-WT, Figure 5C) and transfected in C2 myoblasts with plasmids encoding miR-133 (pmiR-133a/b) or miR-135 (pmiR-135a/b) in parallel to a control plasmid (pCtrl). Luciferase assays show that MAML1 and MEF2C are targets of miR-133 and miR-135, respectively (Figure 5C). The use of mutant derivatives (-mut) in the miRNA recognition sites confirms the specificity of the repressing activity. Moreover, LNA against miR-133 or miR-135 were able to prevent the repression by endogenous miRNAs on RLuc-maml1-WT and RLuc-mef2c-WT, respectively (Figure S5B).
RLuc-maml1-WT and RLuc-mef2c-WT constructs were subsequently transfected in C2 myoblasts together with pMD1-ΔDrosha or mutant derivatives (pMD1-Δ135 and pMD1-Δ133; see Figure 5D). Luciferase assays indicate that, in the presence of the pMD1-ΔDrosha, both 3′UTR reporter constructs are upregulated (Figure 5D, black bars). This indicates that linc-MD1, by binding miR-133 and miR-135, acts as a decoy abolishing miRNA repressing activity on both MAML1 and Mef2C 3′UTR. On the contrary, when the pMD1-Δdrosha -Δ133 was used, RLuc-maml1-WT repression is restored, as is also the case for pMD1-Δdrosha−Δ135 on RLuc-mef2c-WT (dotted and dashed bars, respectively). These effects were lost when both RLuc-maml1-mut and RLuc-mef2C-mut were utilized.
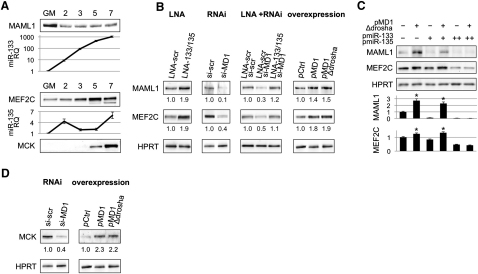
Figure 6A shows MAML1 and MEF2C expression in parallel with that of miR-133 and miR-135 during C2 myoblast differentiation. The effect of linc-MD1 on MAML1 and MEF2C endogenous proteins in combination with a modulation of miRNA levels was monitored by different approaches shown in Figure 6B: (1) LNA against miR-133 and miR-135; (2) RNAi against linc-MD1; (3) RNAi against linc-MD1 in combination with LNA against miR-133 and miR-135; and (4) overexpression of linc-MD1 either in its wild-type form or in its Δdrosha mutant derivative. The results indicate that the levels of MAML1 and MEF2C increase in the presence of LNA against miR-133 and miR-135, while they decrease in the absence of linc-MD1. Notably, LNA are able to resume synthesis of both proteins when linc-MD1 was downregulated by RNAi. Finally, the overexpression of linc-MD1 either in its wild-type form or in its Δdrosha mutant derivative produced an increase of MAML1 and MEF2C expression. These data indicate the existence of a specific crosstalk between the linc-MD1 RNA and MAML1 and MEF2C mRNAs through competition for miR-133 and miR-135 binding.
Figure 6.
linc-MD1 Modulates MAML1 and MEF2C Expression in Muscle
(A) Western blot analysis for MAML1, MEF2C, and MCK, in growth medium (GM) and at different days upon shift to differentiation conditions (DM). The relative expression of miR-133 and miR-135 is also reported.
(B) MAML1 and MEF2C levels upon modulation of miRNA and linc-MD1 levels in C2 myoblasts maintained in DM for 5 days. Modulation was obtained with: anti-miR-133 and anti-miR-135 LNAs, RNAi against linc-MD1, and linc-MD1 overexpression (pMD1 and pMD1-Δdrosha). Scrambled LNA (LNA-scr) and siRNA (si-scr) were used as control. HPRT was used as a loading control. Below each panel, relative quantifications with respect to control samples set to a value of 1 are displayed.
(C) Western blot for the same proteins from C2 myoblasts transfected with different combinations of pMD1-Δdrosha and pmiR-133/pmiR-135 expression plasmids. (+) corresponds to 1.5 μg pMD1-Δdrosha and to 50 ng of pmiR-133/miR-135, while (++) corresponds to 300 ng of pmiR-133/pmiR-135. Control myoblasts were transfected with a GFP plasmid (−). The values, derived from densitometric analysis, are reported with respect to mock samples set to a value of 1. Data are shown as mean ± SD. One asterisk indicates p < 0.05.
(D) MCK measured in cells treated with the indicated LNA or siRNAs as in panel (B).
If linc-MD1 effectively acts as a decoy, one would expect that the relative concentration of the decoy and the miRNAs affects the expression of the target mRNAs. We gradually increased the amount of miRNAs in the presence of increasing amount of linc-MD1-Δdrosha. Figure 6C indicates that the levels of the endogenous MAML1 and MEF2C are higher in excess of linc-MD1 and are gradually reduced when miRNA levels are increased. This further proves that there is an interplay among the three components.
Since muscle creatine kinase (MCK, which increases during muscle differentiation as shown in Figure 6A) was previously shown to be controlled by MEF2C in concert with MAML1 (Shen et al., 2006), we tested the effect of linc-MD1 knockdown and overexpression on this downstream target. Figure 6D shows that the amount of MCK directly correlates with that of its transcriptional activators, demonstrating that the linc-MD1 and miR-133/135 circuitry indeed impinges on muscle gene expression.
Altogether these data indicate that linc-MD1, by binding miR-133 and miR-135, acts as a competing endogenous RNA (ceRNA) for their mRNA targets, including MAML1 and MEF2C, which encode crucial myogenic factors required for the activation of muscle-specific genes. In line with a decoy mechanism, the predicted ΔG of binding (Enright et al., 2003) of the miRNAs with linc-MD1 is lower than that with the respective targets (Figure S6).
Figure S6.
Related to Figure 5
Base pairing of the miRNAs with their predicted binding sites on linc-MD1 and on their mRNA targets. ΔG values were obtained from miRanda (Enright et al., 2003)
The linc-MD1 Function Is Conserved in Human Myoblasts
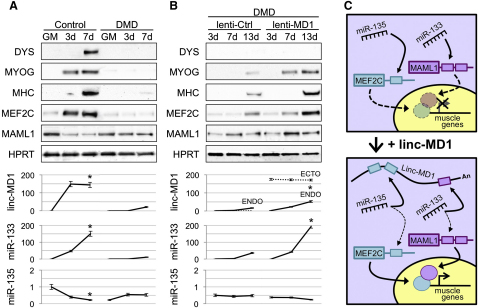
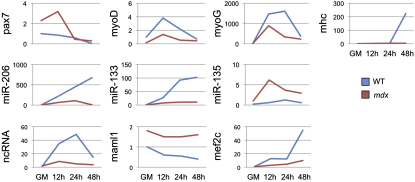
Taking advantage of the presence of conserved regions in linc-MD1, we amplified a linc-MD1 human homolog from differentiated primary myoblasts. We confirmed the exon/intron organization and, in particular, the conservation around the recognition motifs for miR-135 and miR-133. Human primary myoblasts were analyzed in parallel with Duchenne myoblasts (DMD), characterized by mutations in the dystrophin gene and known to have a reduced ability of undergoing terminal differentiation (Cacchiarelli et al., 2011). Figure 7A shows that, compared to control cells, DMD myoblasts display a reduced and delayed accumulation of the muscle-specific markers MYOG and MHC. Notably, in DMD cells the linc-MD1 levels are strongly reduced. This, together with the unaffected accumulation of miR-135, likely determines low levels of MEF2C; vice versa, the strong downregulation of miR-133 correlates with the upregulation of MAML1. The same results were also obtained during differentiation of satellite cells derived from wild-type and mdx mice (Figure S7).
Figure 7.
linc-MD1 Is Conserved in Humans, and It Improves Differentiation of Duchenne Myoblasts
(A) Myoblasts derived from healthy (Control) and Duchenne Muscular Dystrophy (DMD) individuals were induced to differentiate for the indicated times and RNA and protein samples were collected. Western blot analysis for the indicated proteins was performed in parallel with qRT-PCR for the expression of linc-MD1, miR-133 and miR-135.
(B) DMD myoblasts were infected with lentiviral vectors carrying a control GFP-expressing cassette (lenti-Ctrl) or the linc-MD1-Δdrosha construct under the control of the CMV promoter (lenti-MD1). Cells were induced to differentiate for the indicated times and RNA and protein samples were collected. Western blot analysis for the indicated proteins was performed in parallel with qRT-PCR for the expression of linc-MD1, miR-133 and miR-135. Data are shown as mean ± SD. One asterisk indicates p < 0.05.
(C) Schematic representation of the circuitry linking linc-MD1, miR-135, miR-133, and muscle differentiation.
Figure S7.
Related to Figure 7
qRT-PCR for the indicated mRNAs and miRNAs in satellite cells purified from WT or mdx mice differentiated for the indicated hours. Values were normalized for HPRT or U6 snRNA respectively and shown with respect to GM set to a value of 1.
Interestingly, when DMD myoblasts were infected with a lentiviral construct expressing the murine pMD1-Δdrosha, the expression levels of MYOG and MHC as well as those of MEF2C are restored toward control levels (Figure 7B). Despite the upregulation of miR-133, which parallels linc-MD1 overexpression, MAML1 levels increase indicating that the amount of linc-MD1 is sufficient to overcome miR-133 repression activity.
In conclusion, these data indicated that linc-MD1 RNA is expressed also in human muscle cells where it modulates miR-133 and miR-135 targets, playing an important role in the timing control of myoblast differentiation.
Discussion
It is becoming largely accepted that the noncoding portion of the genome rather than its coding counterpart is likely to account for the greater complexity of higher eukaryotes. Many new functions have been assigned to noncoding RNAs both in the nucleus and in the cytoplasm (Mattick, 2011; Nagano and Fraser, 2011). Likewise, similar to what happened for the well-known small noncoding RNAs, long noncoding RNAs are now attracting much interest. Recent data suggest that coding and noncoding RNAs can regulate one another through their ability to compete for miRNA binding; these molecules have been termed competing endogenous RNA (ceRNA, Salmena et al., 2011). ceRNAs can sequester miRNAs, thereby protecting their target RNAs from repression (Karreth et al., 2011 [this issue of Cell]; Sumazin et al., 2011 [this issue of Cell]; Tay et al., 2011 [this issue of Cell]).
In this paper, we identify a muscle-specific long noncoding RNA (linc-MD1) that displays decoy activity for two specific miRNAs and, in doing so, regulates their targets in a molecular circuitry affecting the differentiation program.
We show that linc-MD1 is encoded by a genomic locus containing the miR-206 and miR-133b coding regions and demonstrate that there is a complex architecture in terms of transcriptional control in this locus: while miR-206 is expressed autonomously from its own proximal promoter, miR-133b is cotranscribed with linc-MD1 RNA which derives from a 13 Kb distal promoter.
Here, we provide evidence of the existence of two distinct promoters: (1) miR-206 is already expressed in growing myoblasts, whereas miR-133b and linc-MD1 are activated only upon differentiation; (2) 5′ RACE and promoter fusion experiments indicate the existence of two transcriptional regulatory elements (DIST and PROX); (3) ChIP experiments for RNA Polymerase II and different markers of chromatin activity indicate a well-defined chromatin organization of the two transcriptional units. In particular, in growth conditions, only the PROX promoter displayed markers of transcriptional activation, and no RNAPII loading was detected on the miR-133b region. These data suggest that miR-133b mainly originates from the DIST promoter by processing of linc-MD1.
linc-MD1 accumulates as a cytoplasmic poly-A+ RNA, supporting the conclusion that this species is the remaining portion of the transcript that escaped Drosha cleavage inside the nucleus. We prove that indeed miR-133b is produced with ectopic expression of linc-MD1. In order to avoid a possible confusion between the effect of linc-MD1 and that of miR-133b release, a mutant linc-MD1 derivative lacking the ability to release miR-133b was utilized in most of the overexpression experiments. Future work will address the mechanism regulating the relative ratio between miR-133b processing and the export of the unprocessed precursor.
Notably, we show that transcriptional activation of the linc-MD1 promoter correlates with the formation of a DNA loop in which the distal and proximal promoters (and the polyadenylation region) are connected in a functional/structural interaction. So far, gene loops have been shown to be transcription-dependent, because they are absent in nontranscribing conditions and have been suggested to represent specific structural domains of active chromatin (Tan-Wong et al., 2008; West and Fraser, 2005). Therefore, a drastic structural change occurs in the miR-206/miR-133b locus; in growth conditions, only the proximal promoter is active and no long-distance interactions occur, while upon differentiation, a DNA loop is observed between distantly located regions, and this correlates with activation of the distal promoter and consolidation of the overall transcription of the locus.
As far as the function of linc-MD1 is concerned, we show that its modulation impinged on myogenesis. linc-MD1 RNAi-dependent downregulation in mouse myoblasts produced a decrease in the accumulation of myogenic markers, while its overexpression led to increased synthesis. linc-MD1 was found to be conserved in human cells: high levels were observed upon induction of differentiation in wild-type cells, whereas strongly reduced levels were found in Duchenne myoblasts. This observation is in line with the well-known delay observed in the differentiation program of DMD myoblasts (Cacchiarelli et al., 2011). Notably, when linc-MD1 expression was restored to wild-type levels in DMD myoblasts, the timing and expression level of the myogenic factors were partially rescued toward wild-type levels.
According to the ceRNA hypothesis, lncRNAs may elicit their biological activity through their ability to act as endogenous decoys for miRNAs; such activity would in turn affect the distribution of miRNAs on their targets (Salmena et al., 2011). We searched for miRNA recognition motifs in the linc-MD1 sequence and found that the presence of recognition sites for miR-133 and miR-135 could be reliably predicted. linc-MD1 was validated as target for both these miRNAs since they induced translational repression of a reporter gene.
Among the many different putative targets for these miRNAs, we discovered two mRNAs encoding for proteins with a relevant function in myogenesis: the Myocyte-specific enhancer factor 2C (MEF2C), targeted by miR-135 and Mastermind-like-1 (MAML1) controlled by miR-133.
Consistent with linc-MD1 being a decoy for miR-133 and miR-135, we proved that its depletion reduced the levels of both MAML1 and MEF2C while its overexpression produced an increase in protein accumulation. These data are consistent with the idea that decoy lincRNAs are transmodulators of gene expression through miRNA binding.
The identification of the targets indirectly controlled by linc-MD1 can be instrumental to explain the myogenic alterations observed upon its deregulation. MEF2C belongs to a family of transcription factors that bind the control regions of numerous muscle-specific genes activating their expression (Lin et al., 1997). Moreover, it was shown to play a key role in differentiation of muscle cells (Lilly et al., 1995) and in the maintenance of sarcomere integrity (Potthoff et al., 2007).
On the other side, the Mastermind-like genes encode critical transcriptional coactivators for Notch signaling. Additionally, the MAML proteins were described as transcriptional coactivators in other signal transduction pathways including muscle differentiation: mice with a targeted disruption of the MAML1 gene had severe muscular dystrophy and MAML1-null embryonic fibroblasts failed to undergo MyoD-induced myogenic differentiation (Shen et al., 2006). Moreover, ectopic MAML1 expression in mouse myoblasts dramatically enhanced myotube formation and increased the expression of muscle-specific genes, while MAML1 knockdown inhibited differentiation.
Even more interesting is the finding that MAML1 and MEF2C specifically interact and act synergistically to activate several genes required for muscle development and function, including muscle creatine kinase (MCK). The MAML1 promyogenic effects were completely blocked upon activation of Notch signaling, which was associated with recruitment of MAML1 away from MEF2C to the Notch transcriptional complex (Wilson-Rawls et al., 1999). Therefore, a crosstalk between MAML1 and Notch was postulated to influence myogenic differentiation.
In light of these notions, we proved that depletion of linc-MD1 led to repression of both MAML1 and MEF2C, while its overexpression restored their synthesis at high levels. Notably, in conditions of linc-MD1 excess, titrated repression of both MAML1 and MEF2C could be obtained by increasing miR-133 and miR-135 levels. This indicated a direct competition for miRNA binding between linc-MD1 and mRNAs, allowing us to conclude that the three components crosstalk with one another at the post-trascriptional level. Notably, MCK, a known target of MEF2C, coherently behaved as part of the circuitry: it increased upon linc-MD1 overexpression and decreased upon linc-MD1 RNAi.
In Duchenne muscle cells the rescue of linc-MD1 through lentiviral-mediated expression produced the recovery of both MAML1 and MEF2C synthesis and partial rescue of the correct timing of the differentiation program. These data allowed us to conclude, that also long noncoding RNAs play a relevant role in the complex network of regulatory interactions governing muscle terminal differentiation. Moreover, the discovery of the decoy role of lncRNA opens the road to the prediction and identification of new regulatory networks acting through miRNA competition.
Experimental Procedures
Cell Cultures and Treatments
C2 myoblasts (C2.7 clone) were transfected with plasmid DNA using lipofectamine-2000. (Invitrogen). siRNA molecules designed against linc-MD1 exon2 and exon3 sequences (see supplementary experimental procedures) where transfected using HiPerfect (QIAGEN). LNA oligos against miR-133a/b and miR-135a/b (EXIQON) were transfected using XtremeGene (Roche). All transfections were performed according to manufacturer's specifications.
Control and Duchenne primary myoblasts carrying exon 44 deletion (obtained from Telethon Biobank), were grown and infected with lenti-Ctrl and lenti-MD1 constructs according to Incitti et al. (2010). Muscle satellite cells were cultured and differentiated as described in Cacchiarelli et al. (2010).
Chromatin Immunoprecipitation Assays
ChIP analyses were performed on chromatin extracts from myoblasts (GM) and myotubes (DM) according to manufacturer's specifications (MAGnify ChIP - Invitrogen) with the following antibodies: RNA Polymerase II, MyoD (Santa Cruz), anti-acetyl-HistoneH3 (Lys9), anti-acetyl-HistoneH3 (Lys4), and anti-trimethyl Histone H3 (Lys27) (Millipore).
A standard curve was generated for each primer pair testing 5-point dilutions of input sample. Fold enrichment was quantified using qRT-PCR (QuantiTect SYBR Green - QIAGEN) and calculated as a percentage of Input chromatin (% Inp). Data were normalized to an unrelated genomic region and are representative of three independent experiments. Primer sequences are available upon request.
3C Analysis
The 3C assay was performed as described by Tan-Wong et al., 2008. Briefly, chromatin was crosslinked with 1% formaldehyde and nuclei were isolated by using Nonidet P-40. DNA was digested with 800 units of StyI restriction enzyme and ligated in 1X ligation buffer (NEB). Ligation products were purified using QIAquick PCR purification kit (QIAGEN). Two types of controls were included in the analysis. First, to ensure that primer efficiency does not introduce bias, a control template was generated by digesting and ligating equimolar amounts of all possible PCR products and used to calculate amplification efficiency of each primer pairs. Second, a loading control was generated by amplifying part of HPRT promoter to evaluate total amount of DNA used in the 3C analysis. We confirmed that all 3C primers amplified an artificial control template but not undigested and ligated, or digested but not ligated chromatin. Therefore we verified that the sequence of all 3C products was correct (data not shown).
3C products detection was done in triplicate by GoTaq qPCR (Promega) according to manufacturer's instructions. Data were analyzed according Abou El Hassan and Bremner, (2009). Primer sequences are available upon request.
Statistical Analyses
The data shown in the histograms are the result of at least three independent experiments performed on at least three samples or animals. Unless stated otherwise, data are shown as mean ± standard deviation (SD) and statistical significance of differences between means was assessed by two-tailed t test and p < 0.05 was considered significant.
Figure S2.
Related to Figure 1
(A) In situ analysis for linc-MD1, in C2 myoblasts in GM and DM. DAPI staining is also shown alone (DAPI) or together with DIG staining (MERGE).
(B) In situ analysis for linc-MD1, miR-206 in tibialis cryosections from WT and mdx animals. DAPI staining is also shown in light blue. Original magnification X20, scale bar 100 μm. In both analyses scramble oligo is used as control.
Figure S3.
Related to Figure 2
Northern blot analysis for murine mir-223 expression in C2 myoblasts transfected with D-miR-223 and P-miR-223 constructs and maintained in GM or DM conditions. Ctrl plasmid contains a region between miR-206 and miR-133b, used as negative control. miR-16 was used as loading control.
Acknowledgments
We thank N. Proudfoot and K. Perkins for introducing M.C. to the 3C analysis; M. Mora and the Telethon Neuromuscular Biobank for providing material; J. Martone, V. Cazzella for useful discussion; and M. Marchioni for technical support. D.C. is a recipient of a Microsoft research PhD fellowship. This work was partially supported by grants from: Telethon (GGP07049), Parent Project Italia, EU project SIROCCO (LSHG-CT-2006-037900), KAUST KUK-I1-012-43, AIRC, IIT “SEED,” FIRB, PRIN, and BEMM.
Supplemental Information
Conservation was evaluated using the Mammal Cons phastCons30wayPlacental. (1) miRNAs not expressed in muscle (see Extended Experimental Procedures). (2) miRNA whose targets are not expressed in muscle or do not have a known function in muscle physiology (see Extended Experimental Procedures).
References
- Abou El Hassan M., Bremner R. A rapid simple approach to quantify chromosome conformation capture. Nucleic Acids Res. 2009;37:e35. doi: 10.1093/nar/gkp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral P.P., Mattick J.S. Noncoding RNA in development. Mamm. Genome 19, 454–492. Genome Res. 2008;17:556–565. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Ballarino M., Pagano F., Girardi E., Morlando M., Cacchiarelli D., Marchioni M., Proudfoot N.J., Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M., Puig I., Peña C., García J.M., Alvarez A.B., Peña R., Bonilla F., de Herreros A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M., Vincent S.D. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 2009;19:444–453. doi: 10.1016/j.gde.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D., Martone J., Girardi E., Cesana M., Incitti T., Morlando M., Nicoletti C., Santini T., Sthandier O., Barberi L. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12:341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D., Incitti T., Martone J., Cesana M., Cazzella V., Santini T., Sthandier O., Bozzoni I. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011;12:136–141. doi: 10.1038/embor.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J., Le Baccon P., Wutz A., Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chargé S.B., Rudnicki M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., Amato A.A., Lidov H.G., Kang P.B., North K.N., Mitrani-Rosenbaum S. Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger M.R., Shore A.N., Contreras A., Rijnkels M., Miller J., Gonzalez- Rimbau M.F., Rosen J.M. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl. Acad. Sci. USA. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incitti T., De Angelis F.G., Cazzella V., Sthandier O., Pinnarò C., Legnini I., Bozzoni I. Exon skipping and Duchenne Muscular Dystrophy therapy: Selection of the most active U1 snRNA-antisense able to induce dystrophin exon 51skipping. Mol. Ther. 2010;18:1675–1682. doi: 10.1038/mt.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth F.A., Tay Y., Perna D., Ala U., Tan S.M., Rust A.G., DeNicola G., Webster K.A., Weiss D., Perez-Mancera P.A. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitan D., Dinger M.E., Mazar J., Crawford J., Smith M.A., Mattick J.S., Perera R.J. The melanoma-upregulated long noncoding RNA SPRY4-IN1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- Lilly B., Zhao B., Ranganayakulu G., Paterson B.M., Schulz R.A., Olson E.N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin Q., Schwarz J., Bucana C., Olson E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.J. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J.S. The central role of RNA in human development and cognition. FEBS Lett. 2011;585:1600–1616. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Proudfoot N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Okitsu C.Y., Hsieh J.C., Hsieh C.L. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol. Cell. Biol. 2010;30:2933–2946. doi: 10.1128/MCB.01478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff M.J., Arnold M.A., McAnally J., Richardson J.A., Bassel-Duby R., Olson E.N. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2C. Mol. Cell. Biol. 2007;27:8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi I.A., Mattick J.S., Mehler M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;18:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. The ceRNA hypothesis: the Rosetta stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., McElhinny A.S., Cao Y., Gao P., Liu J., Bronson R., Griffin J.D., Wu L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F., Zwart R., Barlow D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sumazin P., Yang X., Chiu H.-S., Chung W.-J., Iyer A., Llobet-Navas D., Rajbhandari P., Bansal M., Guarnieri P., Silva J. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong S.M., French J.D., Proudfoot N.J., Brown M.A. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc. Natl. Acad. Sci. USA. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y., Kats L., Salmena L., Weiss D., Tan S.M., Ala U., Karreth F., Poliseno L., Provero P., Di Cunto F. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14(Spec No 1):R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- Williams A.H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J.L., Bassel-Duby R., Sanes J.R., Olson E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Rawls J., Molkentin J.D., Black B.L., Olson E.N. Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C. Mol. Cell. Biol. 1999;19:2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K., Hagiwara Y., Ando M., Nakamura A., Takeda S., Hijikata T. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct. Funct. 2008;33:163–169. doi: 10.1247/csf.08022. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed]
- Barrett, T., Troup, D.B., Wilhite, S.E., Ledoux, P., Evangelista, C., Kim, I.F., Tomashevsky, M., Marshall, K.A., Phillippy, K.H., Sherman, P.M., et al. (2011). NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 39 (Database issue), D1005–D1010. [DOI] [PMC free article] [PubMed]
- Cardinali, B., Castellani, L., Fasanaro, P., Basso, A., Alemà, S., Martelli, F., and Falcone, G. (2009). Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE 4, e7607. [DOI] [PMC free article] [PubMed]
- Denti, M.A., Rosa, A., D'Antona, G., Sthandier, O., De Angelis, F.G., Nicoletti, C., Allocca, M., Pansarasa, O., Parente, V., Musarò, A., et al. (2006). Body-wide gene therapy of Duchenne Muscular Dystrophy in the mdx mouse model. Proc. Natl. Acad. Sci. USA 103, 3758–3763. [DOI] [PMC free article] [PubMed]
- Friedman, R.C., Farh, K.K., Burge, C.B., and Bartel, D.P. (2009). Most Mammalian mRNAs Are Conserved Targets of MicroRNAs. Genome Res. 19, 92–105. [DOI] [PMC free article] [PubMed]
- Fujita, P.A., et al. (2010). The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39, D876–D882. [DOI] [PMC free article] [PubMed]
- Griffiths-Jones, S., Saini, H.K., van Dongen, S., and Enright, A.J. (2008). miRBase: tools for microRNAgenomics. Nucleic Acids Res. 36, D154–D158. [DOI] [PMC free article] [PubMed]
- Kozak, M. (1989). The scanning model for translation: an update. J. Cell Biol. 108, 229–241. [DOI] [PMC free article] [PubMed]
- Loots, G., and Ovcharenko, I. (2004). rVista 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32, W217–W221. [DOI] [PMC free article] [PubMed]
- Siepel, A., Bejerano, G., Pedersen, J.S., Hinrichs, A.S., Hou, M., Rosenbloom, K., Clawson, H., Spieth, J., Hillier, L.W., Richards, S., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. [DOI] [PMC free article] [PubMed]
- Tomczak, K.K., Marinescu, V.D., Ramoni, M.F., Sanoudou, D., Montanaro, F., Han, M., Kunkel, L.M., Kohane, I.S., and Beggs, A.H. (2004). Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 18, 403–405. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conservation was evaluated using the Mammal Cons phastCons30wayPlacental. (1) miRNAs not expressed in muscle (see Extended Experimental Procedures). (2) miRNA whose targets are not expressed in muscle or do not have a known function in muscle physiology (see Extended Experimental Procedures).