Abstract
Wolf–Hirschhorn syndrome is caused by anomalies of the short arm of chromosome 4. About 55% of cases are due to de novo terminal deletions, 40% from unbalanced translocations and 5% from other abnormalities. The facial phenotype is characterized by hypertelorism, protruding eyes, prominent glabella, broad nasal bridge and short philtrum. We used dense surface modelling and pattern recognition techniques to delineate the milder facial phenotype of individuals with a small terminal deletion (breakpoint within 4p16.3) compared to those with a large deletion (breakpoint more proximal than 4p16.3). Further, fine-grained facial analysis of several individuals with an atypical genotype and/or phenotype suggests that multiple genes contiguously contribute to the characteristic Wolf–Hirschhorn syndrome facial phenotype.
Keywords: Wolf–Hirschhorn syndrome, facial dysmorphism, 3D shape analysis
Introduction
Wolf–Hirschhorn syndrome (WHS; OMIM 194190) is a contiguous gene syndrome caused by deletion of the short arm of chromosome 4. The first report of the condition1 was re-published 4 years later alongside another by Wolf et al.2, 3 Since then, more than 150 patients have been reported.4 Previously, approximately 75% of cases were considered to be due to de novo terminal deletions,5, 6 13% from unbalanced translocations and the remainder from a more unusual cytogenetic abnormality. More recent studies have suggested the relative frequencies of deletions, unbalanced translocations and other causes to be 55%, 40% and 5%, respectively.7 Translocations between chromosomes 4 and 8 occur more frequently than expected.7, 8, 9, 10, 11
WHS is characterized by pre- and postnatal growth delay, microcephaly, seizures, hypotonia, developmental delay, congenital anomalies and a recognizable facial appearance, which includes: hypertelorism, protruding eyes, epicanthus, arched eyebrows, prominent nasal bridge, downturned corners of the mouth, micrognathia and short philtrum, and, with increasing age, a more prominent nose. Dental anomalies, some occurring in more than half of WHS patients, have been described: delayed eruption and hypodontia, retained primary teeth, peg-shaped teeth and taurodontism.4, 12 Midline defects occur in approximately 50% of cases and include cleft lip and palate and cardiac defects.13
There is considerable variation in the phenotypic spectrum, and, more recently, a correlation with the size of the deletion has been recognized; children with submicroscopic deletions within 4p16.3 tend to have a milder phenotype.14, 15 Some patients with microdeletions within 4p16.3 were initially reported as having Pitt–Rogers–Danks syndrome, now considered to be within, and at the milder end of, the WHS spectrum.16, 17 We use small and large, respectively, to refer to a terminal deletion with a break point within 4p16.3, a deletion size of about 3.1 Mb or less, and one with a break point more proximal than 4p16.3.
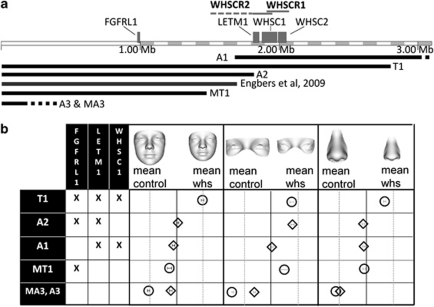
The loss of genetic material on chromosome 4 is variable, with some deletions being visible using standard cytogenetic techniques, while others, being submicroscopic, require the use of fluorescence in situ hybridization probes for their detection. Two Wolf–Hirschhorn critical regions, WHSCR1 and WHSCR2, have been identified within 4p16.3.18, 19 WHSCR2 is distal to, and overlaps with, WHSCR1, and three genes LETM1, WHSC1 and WHSC2 have been identified within these regions. The gene LETM1 is thought to be associated with seizures. It has been suggested that the facial phenotype arises from loss of WHSC1,20 but a later communication21 reported one patient without the facial phenotype with WHSC1 deleted and another patient with the facial phenotype, but with WHSC1 retained. Two recent studies22, 23 have suggested that the fibroblast growth factor receptor FGFRL1 is involved in the aetiology of WHS and in particular influences craniofacial development. Figure 1a shows the relative positions of FGFRL1, LETM1, WHSC1 and WHSC2 in 4p16.3, corresponding approximately to the terminal 3.1 Mb region. In WHS patients with oligodontia, the gene MSX1 has been found to be deleted.24 MSX1 lies proximally to 4p16.3.
Figure 1.
(a) The terminal 3.1 Mb region of chromosome 4p corresponding approximately to 4p16.3 is shown annotated by the two critical regions WHSCR1 and WHSCR2, and four genes (FGFRL1, LETM1, WHSC1 and WHSC2). The five black bands depict the deletions of patients T1, MT1, A1–A3 and MA3 selected for fine-grained facial analysis and the grey band that of a previously cited patient linked with FGFRL1. (b) Mean face patch classification positions for the six selected patients with reference to the particular genes affected by their deletion (X means gene is deleted or curtailed).
Wieczorek and co-workers14 previously reported a correlation in WHS between postnatal head circumference and deletion size. The original aim of this study was to analyse the effects of deletion size on facial morphology in WHS. To test this hypothesis in an objective manner, we undertook a quantitative facial analysis using three-dimensional (3D) photographs and dense surface modelling (DSM) techniques.25, 26 Previously, studies using 3D DSMs of face shape have delineated common facial features in a range of neurodevelopmental conditions, often, in addition, establishing accurate discriminating characteristics or assisting the determination of phenotype–genotype correlations.26, 27, 28, 29, 30, 31, 32, 33, 34 We have shown that DSM-based analysis provides a very accurate instrument for classifying faces or facial regions along the control–WHS spectrum. In addition, for the first time, we have used DSMs to construct mean face surfaces matched for age and size for fine-grained analysis of six individuals, so that subtle face shape differences can be related to the underlying genotype. Our findings support the hypothesis that multiple genes contribute to the facial phenotype of Wolf–Hirschhorn syndrome in a contiguous manner.
Subjects and methods
3D face images of individuals with WHS were collected using a commercial photogrammetric device at family support group meetings in the United Kingdom and United States, and during a scanning trip to Italy. The accuracy of such 3D imaging devices has been shown to be highly reliable.35 Some images were unusable because the coverage of the face was incomplete or the subject was not sufficiently co-operative to capture a usable image. In addition, several hundred images of unrelated, unaffected individuals of a wide age range (3 months to over 70 years) were drawn from an existing collection with a view to making comparisons between the WHS group and matched controls. All images were collected in association with informed written consent and research ethics approval (UCLH: JREC00/E042; Sheffield: MREC/03/4/022). Details of the specific 4p abnormality were obtained from clinical records.
In the analysis of younger subjects, the WHS data set comprised 100 individuals of white, European descent (1.0–19.5 years; mean 7.9 years) with 81 confirmed deletions of 4p with known breakpoints (2 interstitial; 23 small (3.1 Mb or less); 28 large (more than 3.1 Mb); 28 translocations) and 19 with breakpoints reported as 4p16, which could not be further subclassified. These were matched ethnically with a control group of 200 individuals between 0.2 and 20.7 years (mean 8.6 years). A small group of five children under 5 years with an ethnic background other than white European was also included in the study. For the analysis of older subjects, we used an overlapping set of 43 individuals with WHS between 9 and 32.3 years (mean 15.6 years) and 141 controls between 9 and 29.5 years (mean 16.1 years). Finally, six individual cases were subjected to more refined facial analysis: one with a small deletion and a typical WHS phenotype; his mother with an even smaller deletion; and four others without the classical WHS facial features. Approximately 400 control faces were used for the fine-grained analysis.
Cases T1 and MT1
T1 and MT1 were first reported by Faravelli et al.,36 where full details can be found. At the time of this study, they were 18.1 and 57.5 years. T1 had a 2.8 Mb 4p terminal deletion including both critical regions and a typical phenotype for his diagnosis of WHS. In contrast, his mother, MT1, had a 1.35–1.47 Mb terminal deletion distal to both critical regions and removing only FGFRL1. MT1's face was considered WHS-like with hypertelorism, high nasal bridge, large protruding eyes and down-slanting palpebral fissures. Her cognitive abilities were within lower limits of normal. Her motor development was normal and she experienced a few febrile seizures before the age of 5 years. When analyzing younger subjects, we included T1 in training sets as someone with a small deletion and typical phenotype. When analysing older subjects, we excluded T1 and MT1 from model generation, so that we could compare their faces blinded and detect differences due to distinct genotypes.
Case A1
Case A1 was a 1.8-year-old male patient with an interstitial deletion of approximately 4.3 Mb removing LETM1, WHSC1 and WHSC2, but retaining FGFRL1. Intrauterine growth retardation was noted during pregnancy. Delivery was at 39 weeks and birth weight was 2.5 kg. Parents were concerned about his weight gain at age 4 months, and at 6 months an atrial septal defect not requiring treatment was detected. Feeding continued to be difficult and a gastrostomy tube was used from 10 months to 3 years. He sat at 1 year and at 3.8 years was walking if held by both hands. Following removal of tonsils and adenoids at 1.9 years, he spoke single words and now uses short phrases. He has duplex ureters, no reflux and no seizures to date.
Case A2
Case A2 was a 4.6-year-old female patient published as one of two cases of 4p16.3 deletions excluding both WHS critical regions. FGFRL1 and a portion of the LETM1 gene were absent in a terminal deletion of approximately 1.78 Mb distal to WHSCR1 with a breakpoint near or within the distal boundary of WHSCR2. She had growth retardation, development delay, lacked typical WHS facial features, but was described as having a prominent forehead, mild telecanthus and normal philtral length.37 At 6 months of age, myoclonic jerking was investigated and clinical episodes of atypical absence seizures have recurred.
Cases A3 and MA3
Case A3 was a 6.3-year-old female patient with a maternally inherited terminal deletion of 160–400 kb. Following concern over foetal growth, she was delivered by Caesarean section at 38 weeks. Her head circumference was around the 0.4th centile, but then fell away from the centile, whereas her weight was between the 2nd and 9th centiles. She did not have a history of seizures, but required surgical repair of an atrial septal defect and a right inguinal hernia. Her development was delayed; she attended mainstream education with 1:1 support. Her parents were healthy and had normal head circumferences. The deleted region had not been reported within the Database of Genomic Variation and was not observed in more than 400 control cases. Therefore, her deletion may be a rare benign copy number variation, a conclusion consistent with the absence of a clinical phenotype in the mother (MA3). However, a contribution to A3's phenotype cannot be excluded.
Figure 1a illustrates the 3.1 Mb terminal region of chromosome 4p corresponding approximately to 4p16.3, with annotations showing the WHS critical regions, important genes and the deletions of the selected cases T1, MT1, A1–A3 and MA3. Also represented in the figure is the case where a role for FGFRL1 in the WHS facial phenotype was first proposed.22 This patient has a terminal deletion with a breakpoint in between those of MT1 and A2. Supplementary Table ST1 gives more detailed descriptions of the breakpoints for these six individuals.
Methods
Comparison of linear regressions
Linear regressions were undertaken for various facial measurements and DSM-based markers against age. The approach of Wuensch et al.38 was used to determine significant differences in slope and/or intercept in comparisons of separate regressions for control and WHS subgroups.
Image preparation
Each captured face surface was annotated manually by one individual (PH) at 22 anatomical locations: left and right endocanthion, exocanthion, palpebrale superius, palpebrale inferius, otobasion inferius, crista philtrum, cheilion and alare; nasion, pronasale, subnasale, labiale superius, labiale inferius and gnathion (Supplementary Figure S1). These landmarks have previously been studied for their reproducibility and accuracy.39 Generally speaking, landmarks on the lips and eyes are the most reproducible. All landmarking and dense surface model analyses were undertaken using the software developed in-house.30, 40
DSM building and closest mean classification
DSMs were constructed using techniques described elsewhere.25, 26 A DSM refers to the set of principal components (PCs) or principal component analysis (PCA) models accounting for the shape variation in the surfaces included. For a set of faces with wide age variation, the first mode of variation (PC1) follows the shape of a typical growth curve and is highly correlated with age. With closest mean (CM) classification, average faces are computed for the control and affected subgroups and each face is classified according to which average is closest using the DSM representation. For ‘blinded' or unseen testing, we randomly generated twenty 90–10% training-test set pairs sampled in a stratified manner from the control and WHS groups. We used the training sets to build DSMs and the test sets for unseen or blinded classification. The accuracy of classification of the control or WHS group was estimated as the mean of the areas under the twenty receiver operating characteristic curves arising from the unseen testing. Alongside CM classification, a useful measure defining outlier status is the distance of a face from the line joining unaffected and affected means in the DSM representation.
PCA models are used generally to reduce dimensionality of complex multi-variate data sets. In such models, the modes are listed in terms of largest to smallest variation coverage. In pattern recognition, a typical approach is to retain modes covering 95–99% of shape variation on the assumption that omitted modes represent noise. To optimize discrimination, we performed classification testing for all possible leading subsequences of modes in the models and for each of three pattern recognition classification algorithms –CM; linear discriminant analysis (LDA); and support vector machines (SVMs).
Results
Facial growth in WHS is significantly delayed, but does not correlate with deletion size
PC1 in a DSM for a reduced face patch (no ears) computed for the control and WHS subjects under 20 years is highly correlated with age (Supplementary Figure S3). Separate linear regressions of PC1 against age for control and WHS subgroups highlight both delay and diminished rate of facial growth in WHS. The difference in slope was statistically significant (t=2.5724, d.f.=296, P=0.0107) and the difference in intercept highly significant (t=8.6024, d.f.=296, P<0.0001). These results are predictably consistent with the universal clinical observation of reduced birth size and increasing size difference throughout the life of a child with WHS. Restricting the facial growth analysis to individuals with small and large deletions or translocations does not suggest any correlation between facial growth and deletion size (Figure 3). The inclusion of A1–A3 in the figure demonstrated that each had delay in facial growth.
Facial dysmorphology in WHS is visualized effectively using static and dynamic comparisons
Comparisons of the mean faces of the WHS and control subgroups confirmed many of the well-documented facial characteristics. In Figure 2a, the two mean faces are shown to the same scale using surface shape alone. A combined shape and facial appearance DSM provided similar, but more photorealistic mean surfaces (Figure 2b). The third comparison is a heat map of the mean WHS face reflecting location differences between corresponding points on the mean control and WHS faces along a normal to the mean WHS face surface. The red–green–blue spectrum depicts inward–null–outward displacement along the surface normal (Figure 2c). The abundance of red clearly demonstrates how the comparison is overwhelmed by the gross size difference in the two means, despite their corresponding mean ages differing by only months. By adjusting for size using the ratio of nasion–gnathion length for each mean surface, a shape-only comparison was possible, and by focusing on position differences parallel to three orthogonal axes, more subtle shape differences were revealed. For example, a comparison of left–right horizontal differences in corresponding points in Figure 2d indicates relative hypertelorism and the stronger colouring at the outer canthi indicates relatively wider palpebral fissures in WHS. Relative to face length (used in the resizing), the WHS mean face is also shown to have a wider zygomatic arch, to be broader at the tragion level, but to have a narrower gonial width reflecting a much smaller mandible. The vertical axial comparison of Figure 2e emphasizes the upward sweep of the supra-orbit and associated eyebrow arching; the shortened philtrum (blue generally on upper lip) and concave curling of the upper lip (dark blue at the centre of upper lip); reduced anterior mandible height (blue on chin); and low-set ear position (red on ears). The prominent glabella, proptosis and reduced mandible size are all confirmed in the depth-wise comparison of Figure 2f.
Figure 2.
(a) Comparisons of the mean faces of the control and WHS subgroups computed from a shape-only DSM; (b) comparisons of the mean faces of the control and WHS subgroups computed from a combined shape and appearance DSM; (c) a heat-map comparison of the raw mean faces reflecting displacement normal to the surface of the WHS mean; (d–f) size-adjusted heat-map comparisons parallel to three orthogonal axes; (g–i) heat-map comparisons of mean WHS nose surface to mean control nose surface with size adjustment and reflecting displacement along three orthogonal axes; and (j and k) cartoons showing nasal cartilage substructures.
Similar visualizations are achievable by building DSMs for patches of face surface. More subtle shape differences, swamped by larger ones in a full-face model, are revealed by focusing on smaller regions of the face. For example, from a DSM for a nasal patch, the size (nasion–subnasale)-adjusted shape comparison emphasizes a broader nasal root and bridge (Figure 2g); longer nose and vertically diminished alae (Figure 2h); and depth-wise flattening of the nose tip (Figure 2i). A comparison with the cartoon in Figures 2j–k further supports the hypothesis that these nose shape differences are very much due to altered positioning and shape of the underlying nasal cartilages (especially Figure 2g).
Finally, dynamic morphs between the control and WHS mean faces very clearly demonstrate the important differences in face shape either with size retained or more emphatically when it is adjusted for (see Supplementary Movie S1 and S2).
Facial dysmorphism in WHS is milder for individuals with small deletions
In a single DSM for a reduced face patch (without ears) for 200 controls and 100 individuals with WHS and under 20 years, we calculated the normalized position of each constituent face between the mean control and WHS faces as described earlier. Figure 3b shows the classification position of the faces of individuals with small and large deletions relative to the overall control and WHS average faces normalized to the ends of the x axis interval (−1, +1). The y axis represents generalized Euclidean distance between a face and the hyperline joining the two means in the DSM, also defined earlier. Thus, position on the x axis reflects similarity to one of the means, whereas position on the y axis reflects difference from both means with higher values reflecting outlier status for one or both subgroups. The shaded convex hulls show that 50% of the small deletion subgroup are closer to the control mean than the large deletion subgroup; that 50% or so of the large deletion subgroup overlap the small deletion subgroup; and that the remainder are either more extreme than the mean WHS (further out on the x axis) or more outlying from both means (further out on the y axis).
Figure 3.
(a) A scatter of age against PC1 is shown for a single DSM for a reduced face patch (no ears) for both the control and WHS subgroups. The scatter is restricted to those individuals known to have small or large deletions or unbalanced translocations. The younger selected patients A1–A3 are also represented. (b) Closest mean classification positions of all controls (n=200) and those individuals with WHS who have a small or large deletion (n=51).
DSM analysis supports high levels of discrimination between controls and individuals with WHS
We used 20 training-unseen test set pairs for the multi-folded face classification of the younger study group of Caucasian individuals. The graphs in Supplementary Figure S2 show how the estimated discrimination accuracy varies according to the number of modes retained in the DSMs and to the pattern recognition algorithms employed –CM; LDA; and SVMs. Supplementary Table 1 summarizes the best discrimination rates for the face (without ears) and patches around the eyes, nose and mouth and for each of the three pattern recognition algorithms. Almost perfect classification was obtained in each case. An analogous multi-folded classification analysis of the older subjects produced similar almost perfect rates of discrimination for each algorithm.
We also classified the face shape of five affected children under 5.2 years with an ethnic origin other than white European, four had large deletions and one a small deletion. There were insufficient numbers of ethnically matched subjects for a separate classificatory cross-validation, so instead we used the models based on European subjects. The difference in ethnic background had no effect on the success of the face shape classification.
Fine-grained facial phenotype–genotype analysis of six selected individuals
To prepare for visual analysis of the faces of the selected cases (T1, MT1, A1–A3, MA3), we computed running means of contiguously aged same-sex subgroups of size 40 for 400 or so controls. We then selected same-sex running mean subgroups whose mean ages matched those of the selected cases to compute age/sex-matched mean faces. Using nasion–gnathion length, we then made size-adjusted comparisons analogous to the size-adjusted ‘control mean–WHS mean' comparisons described above. The heat-map comparisons of these patients to age–sex-size-matched means in Figure 4 need careful comparison with Figures 2d–f.
Figure 4.
(a–i) Heat maps comparing the younger subjects with an age-matched average face from the running mean sequence of 40 age-contiguous control faces. Three axial comparisons are given. (j–r) Heat maps comparing the older selected subjects with an age-matched average face from the running mean sequence of 40 age-contiguous control faces. Three axial comparisons are given.
Figures 4d–f for subject A2 show similarities to Figures 2d–f: some hypertelorism (red/blue patches on eyes in Figure 4d); wider palpebral fissures (stronger colouring at exocanthi in Figure 4d); some shortening of the philtrum (blue on upper lip in Figure 4e); a prominent glabella (blue on forehead in Figure 4f); and a little (asymmetric) proptosis (blue on eyes in Figure 4f). For A1, there is no hypertelorism (Figure 4a), a slight vertical displacement in the supra-orbital region (Figure 4b), no reduction in philtrum length (Figure 4b), no proptosis (Figure 4c) and a little asymmetric prominence of the glabella (Figure 4c). A3 shows no signs of the WHS facial phenotype (Figure 4g–i) – the shortening of the philtrum (Figure 4h) was likely due to facial expression stretching the upper lip. The mother of A3, MA3, shows no WHS-like features (Figure 4p–r). As expected from his deletion and WHS diagnosis, T1 shows hypertelorism (Figure 4j); a prominent nasal bridge (Figure 4l); shortened/curled philtrum (Figure 4k); an upsweep of the supra-orbital ridge (Figure 4k); and proptosis and prominence of the nasal bridge (Figure 4l). In contrast, his mother, MT1, showed no hypertelorism (Figure 4m); an upward sweep of the supra-orbital ridge (Figure 4n); and a prominent nasal bridge (Figure 4o). The proptosis and prominence of the glabella that are detectable by eyes were disguised by mild mid-facial hypoplasia and fullness of the cheeks of the face (Figure 4o). This resulted in the flattened areas being painted red and the periorbit green – but still reflected relative proptosis. Specific size-adjusted patch comparisons of the periorbital and perinasal regions also highlight the proptosis and the glabella prominence (not shown).
To complement the visual analysis, the DSMs constructed for the two cross-validations conducted for younger and older subjects were re-used to classify the face/face patches of the six selected cases in a blinded or unseen manner. Their average CM classification positions for face, periorbital and perinasal patches with 95% confidence intervals are shown in Figure 1 along with a summary of the genes affected by their respective deletions. MA3 is classified close to the mean control face and even further into the control groups for the periorbital and perinasal patches. A3 is classified close to the control–WHS borderline, suggesting some facial dysmorphism overall, but with much more control-like than WHS-like features for the smaller face patches. MT1, whose deletion removes only FGFRL1, is also classified facially close to the control–WHS border. However, consistent with the size-adjusted surface comparisons above, her periorbital patch is classified as more WHS like. The interstitial deletion of A1, affecting LETM1 and WHSC1, results in classification positions on or just control side of the control–WHS borderline. This is consistent with the size-adjusted surface comparisons, suggesting very mild dysmorphism with more control-like than WHS-like features. A2's loss/disruption of FGFRL1 and LETM1 results in classification positions consistent with the mild hypertelorism and glabella prominence identified in the size-adjusted surface patch comparisons. Finally, the loss of FGRL1, LETM1 and WHSC1 in T1's deletion results in classifications on or close to the mean WHS position.
Discussion
Mouse models of the two WHS critical regions have failed to recapitulate the classical phenotype of WHS. The human gene FGFRL1, more distal than both critical regions, was implicated recently in the craniofacial phenotype of a single patient22 whose terminal deletion has a breakpoint between those of A2 and MT1. A targeted deletion of the mouse gene Fgfrl1 recapitulated much of the WHS phenotype.23 Fgfrl1 is expressed in cartilaginous structures such as primordia of bones and permanent cartilage of the trachea, ribs and nose.41 Our DSM analysis of a nasal patch of the face surface (Figure 2) detected altered shape that appears to coincide with nasal cartilage structures, especially those of the lower nasal cartilage. These findings support the hypothesis that FGFRL1 is influential in the WHS facial phenotype.
Subjects MA3 and A3 retain all the highlighted genes and have no significant features that would be given a diagnosis of WHS. The interstitial deletion of patient A1 removes LETM1, WHSC1 and WHSC2, but retains FGFRL1. Our analysis suggests that this patient does not have WHS facial characteristics, but does have some mildly dysmorphic features. The terminal deletion of patient A2 spares WHSC1 and WHSC2, interferes with the distal end of LETM1 and removes FGFRL1. The fine-grained facial analysis suggests that patient A2 has some isolated facial features that are also part of the WHS facial phenotype. A1 and A2 are in a grey area, but could not be given a diagnosis of WHS. MT1's deletion involves FGFRL1 alone, but does appear to produce some WHS facial characteristics, as was suggested for the patient in Engbers et al,22 whom we have not analysed. Thus, the loss or disruption of FGFRL1 or WHSC1 alone does affect facial morphology, with FGFRL1 possibly having a slightly greater effect. However, when the deletion is a contiguous sequence including both FGFRL1 and WHSC1, as with patient T1, the full complement of the WHS facial characteristics results. T1 would certainly be given a diagnosis of WHS, but MT1 would not.
In this study, we first demonstrated that small terminal deletions with a breakpoint within 4p16.3 are associated with milder facial dysmorphology than large deletions. This was achieved using dense surface models to develop sensitive and accurate visualization and quantitative tools for discerning control–WHS face shape difference. Armed with these instruments, we undertook fine-grained facial analysis of six selected patients, suggesting that WHS facial features are due to the combined deletion of multiple genes adding further weight to the hypothesis that WHS is a true contiguous gene disorder as suggested previously.20
Although facial dysmorphism in WHS can be clinically recognizable, an objective, quantitative evaluation is particularly valuable when assessing phenotypically or genotypically unusual cases. Such individuals are likely to be particularly informative in genotype–phenotype correlation analysis and the clarification of the influence of individual genes. In the future, DSM-based analysis will enable the recognition of WHS facial characteristics in patients without 4p deletions. Genetic/genomic analysis of such patients may identify mutations and in combination with detailed facial analysis enable unequivocal identification of causative genes.
Acknowledgments
We offer of sincere thanks to all of the families who volunteered for the study and allowed their faces to be scanned. The WHS family support groups of the United Kingdom, and United States of America deserve special thanks for their continued co-operation and hospitality during this long study. NewLife and the National Institutes of Health generously provided financial support that enabled several generations of 3D cameras to be purchased and many field trips to be undertaken. We also thank Dr Helen Cox for comments on the manuscript and the Sheffield Children's appeal for travel grants to OQ.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Cooper H, Hirschhorn K. Apparent deletion of short arms of one chromosome (4 or 5) in a child with defects of midline fusion. Mamm Chrom Nwsl. 1961;4:14. doi: 10.1007/BF00279124. [DOI] [PubMed] [Google Scholar]
- Wolf U, Reinwein H, Porsch R, Schröter R, Baitsch H. Deficiency on the short arms of a chromosome no. 4. Humangenetik. 1965;1:397–413. [PubMed] [Google Scholar]
- Hirschhorn K, Cooper HL, Firschein IL. Deletion of short arms of chromosome 4–5 in a child with defects of midline fusion. Humangenetik. 1965;1:479–482. doi: 10.1007/BF00279124. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Carey JC, South ST, Wright TJ.Wolf-Hirschhorn Syndromein: Pagon RA, Bird TD, Dolan CR, Stephens K, (eds). GeneReviews 2010 [Internet] Seattle, WA: University of Washington; 1993–2002. updated 17 June 2010). [Google Scholar]
- Lurie IW, Lazjuk GI, Ussova YI, Presman EB, Gurevich DB. The Wolf–Hirschhorn syndrome. I. Genetics. Clin Genet. 1980;17:375–384. doi: 10.1111/j.1399-0004.1980.tb00167.x. [DOI] [PubMed] [Google Scholar]
- Shannon NL, Maltby EL, Rigby AS, Quarrell OWJ. An epidemiological study of Wolf–Hirschhorn syndrome: life expectancy and cause of mortality. J Med Genet. 2001;38:674–679. doi: 10.1136/jmg.38.10.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Whitby H, Battaglia A, Carey JC, Brothman AR. Comprehensive analysis of Wolf–Hirschhorn syndrome using array CGH indicates a high prevalence of translocations. Eur J Hum Genet. 2008;16:45–52. doi: 10.1038/sj.ejhg.5201915. [DOI] [PubMed] [Google Scholar]
- Giglio S, Calvari V, Gregato G, et al. Heterozygous submicroscopic inversions involving olfactory receptor–gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollino M, Lecce R, Selicorni A, et al. A double cryptic chromosome imbalance is an important factor to explain phenotypic variability in Wolf–Hirschhorn syndrome. Eur J Hum Genet. 2004;12:797–804. doi: 10.1038/sj.ejhg.5201203. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, et al. Unexpected high frequency of de novo unbalanced translocations in patients with Wolf–Hirschhorn syndrome (WHS) J Med Genet. 2000;37:798–804. doi: 10.1136/jmg.37.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tönnies H, Stumm S, Neumann L, et al. Two further cases of WHS with unbalanced de novo translocation t(4;8) characterised by CGH and FISH. J Med Genet. 2001;38:e21. doi: 10.1136/jmg.38.6.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia A, Carey JC, Wright TJ. Wolf–Hirschhorn (4p−) syndrome. Adv Pediatr. 2001;48:75–113. [PubMed] [Google Scholar]
- Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf–Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, et al. Effect of the size of the deletion and clinical manifestation in Wolf–Hirschhorn syndrome: analysis of 13 patients with a de novo deletion. Eur J Hum Genet. 2000;8:519–526. doi: 10.1038/sj.ejhg.5200498. [DOI] [PubMed] [Google Scholar]
- Zollino M, Di Stefano C, Zampino G, et al. Genotype–phenotype correlations and clinical diagnostic criteria in Wolf–Hirschhorn syndrome. Am J Med Genet. 2000;94:254–261. doi: 10.1002/1096-8628(20000918)94:3<254::aid-ajmg13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Clemens M, Martsolf JT, Rogers JG, Mowery-Rushton P, Surti U, McPherson E. Pitt–Rogers–Danks syndrome: the result of a 4p microdeletion. Am J Med Genet. 1996;66:95–100. doi: 10.1002/(SICI)1096-8628(19961202)66:1<95::AID-AJMG26>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Lindeman-Kusse MC, Van Haeringen A, Hoorweg-Nijman JJ, Brunner HG. Cytogenetic abnormalities in two new patients with Pitt–Rogers–Danks phenotype. Am J Med Genet. 1996;66:104–112. doi: 10.1002/(SICI)1096-8628(19961202)66:1<104::AID-AJMG28>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Rauch A, Schellmoser S, Kraus C, et al. First known microdeletion within the Wolf–Hirschhorn syndrome critical region refines genotype–phenotype correlation. Am J Med Genet. 2001;99:338–342. doi: 10.1002/ajmg.1203. [DOI] [PubMed] [Google Scholar]
- Zollino M, Lecce R, Fischetto R, et al. Mapping the Wolf–Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region WHSCR-2. Am J Hum Genet. 2003;72:590–597. doi: 10.1086/367925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buggenhout G, Melotte C, Dutta B, et al. Mild Wolf–Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype–phenotype map. J Med Genet. 2004;41:691–698. doi: 10.1136/jmg.2003.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas NM, Van Buggenhout G, Hannes F, et al. Genotype–phenotype correlation in 21 patients with Wolf–Hirschhorn syndrome using high resolution array comparative genome hybridisation (CGH) J Med Genet. 2008;45:71–80. doi: 10.1136/jmg.2007.052910. [DOI] [PubMed] [Google Scholar]
- Engbers H, van der Smagt JJ, van't Slot R, Vermeesch JR, Hochstenbach R, Poot M. Wolf–Hirschhorn syndrome facial dysmorphic features in a patient with a terminal 4p16.3 deletion telomeric to the WHSCR and WHSCR 2 regions. Eur J Hum Genet. 2009;17:129–132. doi: 10.1038/ejhg.2008.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te Welscher P. Multiple congenital malformations of Wolf–Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Model Mech. 2009;2:283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen P, Kotilainen J, Aalto Y, Knuutila S, Pirinen S, Thesleff I. MSX1 gene is deleted in Wolf–Hirschhorn syndrome patients with oligodontia. J Dent Res. 2003;82:1013–1017. doi: 10.1177/154405910308201215. [DOI] [PubMed] [Google Scholar]
- Hutton TJ.Dense surface models of the human facePhD thesis,University College London, London; 2004 [Google Scholar]
- Hammond P, Hutton TJ, Allanson J, et al. Discriminating power of localised 3D facial morphology. Am J Hum Genet. 2005;77:999–1010. doi: 10.1086/498396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan Z, Klein M, Hammond P, et al. Genotype–phenotype correlations of 39 patients with Cornelia de Lange syndrome: the Dutch experience. J Med Genet. 2006;43:568–575. doi: 10.1136/jmg.2005.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Brinkman J, Vedder A, Hollak C, et al. 3D face shape in Fabry disease. Eur J Hum Gen. 2007;15:535–542. doi: 10.1038/sj.ejhg.5201798. [DOI] [PubMed] [Google Scholar]
- Hammond P, Hutton TJ, Allanson JE, et al. 3D analysis of facial morphology. Am J Med Genet. 2004;A 126:339–348. doi: 10.1002/ajmg.a.20665. [DOI] [PubMed] [Google Scholar]
- Hammond P. The use of 3D face shape modelling in dysmorphology. Arch Dis Child. 2007;92:1120–1126. doi: 10.1136/adc.2006.103507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P, Forster-Gibson C, Chudley AE, et al. Face–brain asymmetry in autism spectrum disorders. Mol Psychol. 2008;13:614–623. doi: 10.1038/mp.2008.18. [DOI] [PubMed] [Google Scholar]
- Hennessy RJ, McLearie S, Kinsella A, Waddington JL. Facial surface analysis by 3D laser scanning and geometric morphometrics in relation to sexual dimorphism in cerebral–craniofacial morphogenesis and cognitive function. J Anat. 2005;207:283–295. doi: 10.1111/j.1469-7580.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Hammond P, Karmiloff-Smith A, et al. GTF2IRD1 in craniofacial development of humans and mice. Science. 2006;310:1184–1187. doi: 10.1126/science.1116142. [DOI] [PubMed] [Google Scholar]
- Tobin JL, DiFranco M, Eichers E, et al. Defects of Shh transduction and neural crest cell migration underlie craniofacial dysmorphology in Bardet–Biedl syndrome. Proc Natl Acad Sci. 2008;105:6714–6719. doi: 10.1073/pnas.0707057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge K, Boyadjiev SA, Capone GT, DeLeon VB, Richtsmeier JT. Precision and error of three-dimensional phenotypic measures acquired from 3dMD photogrammetric images. Am J Med Genet. 2005;A 138:247–253. doi: 10.1002/ajmg.a.30959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faravelli F, Murdolo M, Marangi G, Bricarelli FD, Di Rocco M, Zollino M. Mother to son amplification of a small subtelomeric deletion: a new mechanism of familial recurrence in microdeletion syndromes. Am J Med Genet. 2007;A 143A:1169–1173. doi: 10.1002/ajmg.a.31723. [DOI] [PubMed] [Google Scholar]
- South ST, Bleyl SB, Carey JC. Two unique patients with novel microdeletions in 4p16.3 that exclude the WHS critical regions: implications for critical region designation. Am J Med Genet. 2007;143A:2137–2142. doi: 10.1002/ajmg.a.31900. [DOI] [PubMed] [Google Scholar]
- Wuensch KL, Jenkins KW, Poteat G. Misanthropy, idealism and attitudes towards animals. Anthrozoos. 2002;15:139–149. [Google Scholar]
- Gwilliam JR, Cunningham SJ, Hutton TJ. Reproducibility of soft tissue landmarks on three-dimensional facial scans. Eur J Ortho. 2006;28:408–415. doi: 10.1093/ejo/cjl024. [DOI] [PubMed] [Google Scholar]
- Hutton TJ, Buxton BF, Hammond P, Potts HWW. Estimating average growth trajectories in shape–space using kernel smoothing. IEEE Trans Med Imag. 2003;22:747–753. doi: 10.1109/TMI.2003.814784. [DOI] [PubMed] [Google Scholar]
- Trueb B, Taeschler S. Expression of FGFRL1, a novel fibroblast growth factor receptor, during embryonic development. Int J Mol Med. 2006;17:617–620. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.